Abstract
IMPORTANCE
Clostridium difficile is a major cause of health care–associated infection, but disagreement between diagnostic tests is an ongoing barrier to clinical decision making and public health reporting. Molecular tests are increasingly used to diagnose C difficile infection (CDI), but many molecular test-positive patients lack toxins that historically defined disease, making it unclear if they need treatment.
OBJECTIVE
To determine the natural history and need for treatment of patients who are toxin immunoassay negative and polymerase chain reaction (PCR) positive (Tox−/PCR+) for CDI.
DESIGN, SETTING, AND PARTICIPANTS
Prospective observational cohort study at a single academic medical center among 1416 hospitalized adults tested for C difficile toxins 72 hours or longer after admission between December 1, 2010, and October 20, 2012. The analysis was conducted in stages with revisions from April 27, 2013, to January 13, 2015.
MAIN OUTCOMES AND MEASURES
Patients undergoing C difficile testing were grouped by US Food and Drug Administration–approved toxin and PCR tests as Tox+/PCR+, Tox−/PCR+, or Tox−/PCR−. Toxin results were reported clinically. Polymerase chain reaction results were not reported. The main study outcomes were duration of diarrhea during up to 14 days of treatment, rate of CDI-related complications (ie, colectomy, megacolon, or intensive care unit care) and CDI-related death within 30 days.
RESULTS
Twenty-one percent (293 of 1416) of hospitalized adults tested for C difficile were positive by PCR, but 44.7% (131 of 293) had toxins detected by the clinical toxin test. At baseline, Tox−/PCR+ patients had lower C difficile bacterial load and less antibiotic exposure, fecal inflammation, and diarrhea than Tox+/PCR+ patients (P < .001 for all). The median duration of diarrhea was shorter in Tox−/PCR+ patients (2 days; interquartile range, 1-4 days) than in Tox+/PCR+ patients (3 days; interquartile range, 1-6 days) (P = .003) and was similar to that in Tox−/PCR− patients (2 days; interquartile range, 1-3 days), despite minimal empirical treatment of Tox−/PCR+ patients. No CDI-related complications occurred in Tox−/PCR+ patients vs 10 complications in Tox+/PCR+ patients (0% vs 7.6%, P < .001). One Tox−/PCR+ patient had recurrent CDI as a contributing factor to death within 30 days vs 11 CDI-related deaths in Tox+/PCR+ patients (0.6% vs 8.4%, P = .001).
CONCLUSIONS AND RELEVANCE
Among hospitalized adults with suspected CDI, virtually all CDI-related complications and deaths occurred in patients with positive toxin immunoassay test results. Patients with a positive molecular test result and a negative toxin immunoassay test result had outcomes that were comparable to patients without C difficile by either method. Exclusive reliance on molecular tests for CDI diagnosis without tests for toxins or host response is likely to result in overdiagnosis, overtreatment, and increased health care costs.
Clostridium difficile is one of the most common causes of health care–associated infection in US hospitals, affecting almost 1% of hospitalized patients each year.1-3 Since 2000, the incidence of C difficile infection (CDI) has increased more than 200% while the rates of other health care–associated infections have decreased.1,2,4-6 More than 300 000 hospitalizations involve a CDI each year, at an annual cost of $1.0 to $4.9 billion to the US health care system.2,7
Initial increases in the rate of CDI were attributed to the emergence of a novel, hypervirulent strain during a period when at least 95% of hospitals used toxin immunoassays for diagnosis (2000-2008).3,5,8-10 More recent increases have been linked to greater C difficile detection after the introduction of molecular tests, which are more sensitive and detect microbial DNA instead of toxin.10-15 Individual hospitals have reported a 50% to 100% increase in the rate of CDI after switching from toxin tests to molecular tests.11,12,14 Similar increases have been observed in the rate of publicly reported CDI as reporting facilities adopted molecular tests.15
For decades, toxin tests were favored over culture for diagnosis of CDI because toxins mediate disease and toxin detection was faster and provided evidence of toxin production in vivo that typically correlated better with clinical disease.3,10,16-18 Molecular tests such as polymerase chain reaction (PCR) target toxin genes but are similar to culture in detecting C difficile bacteria regardless of toxin production, making it unclear whether positive PCR results reflect clinical disease.3,10,19-21 The uncertain clinical significance of positive PCR results is problematic in inpatient health care facilities, where C difficile colonization is 5 to 10 times more common than CDI and noninfectious causes of diarrhea are also common.22-26 Nonetheless, concern that patients with CDI were being missed by toxin tests prompted many laboratories to switch to molecular tests in 2009, when they became available.10,19,27 As of the first quarter of 2014, a total of 44% of acute care hospitals participating in the National Healthcare Safety Network (NHSN) reported using molecular tests alone or in combination with other tests for diagnosis of CDI (NHSN, written communication, September 15, 2014). Therefore, there is an urgent need to determine whether patients with negative toxin test results and positive molecular test results have CDI or are simply colonized with another cause of symptoms.
To address this need, we prospectively tested hospitalized adults with suspected CDI at the University of California Davis Medical Center with molecular tests while maintaining our existing toxin test for clinical diagnosis. We then collected clinical outcome and treatment data to enable us to ask 3 related questions. First, what is the natural history of PCR-positive patients with negative toxin immunoassay results? Second, how do outcomes in these patients compare with outcomes in patients with positive toxin and PCR results or completely negative C difficile test results? Third, do PCR-positive patients with negative toxin results require treatment for CDI?
Methods
Study Design and Population
Hospitalized adults with a diarrheal stool sample submitted for C difficile testing 72 hours or longer after admission to the University of California Davis Medical Center between December 1, 2010, and October 20, 2012, were included in the study. Only the first sample was analyzed for each patient. Samples received after discharge were excluded. Patients with C difficile detected by culture and no other test were excluded from the study. The study protocol was approved by the University of California Davis Institutional Review Board. Informed consent was waived for the initial screening and symptom verification and overall outcome and safety analysis. A subset of patients had written informed consent obtained for additional in-person follow-up.
Laboratory Testing
All stool samples had a US Food and Drug Administration (FDA)–approved C difficile toxin immunoassay (C difficile Premier toxins A and B; Meridian Biosciences) performed and reported clinically. Formed stools were rejected. Eligible samples also had 1 or more FDA-approved molecular C difficile tests (Xpert C. difficile/Epi; Cepheid; and illumigene C. difficile; Meridian Biosciences) performed but not reported, allowing patients to be grouped by C difficile toxin immunoassay and PCR results as toxin immunoassay positive and PCR positive (Tox+/PCR+), Tox−/PCR+, or Tox−/PCR−. Additional tests were performed to characterize the nature of the C difficile colonization and host inflammatory response. The PCR-positive samples had toxin quantitated (xCELLigence System for Real-Time Cellular Analysis, version 2; ACEA Biosciences) and the concentration of C difficile DNA determined as a measure of bacterial load (Xpert C. difficile/Epi; Cepheid).28-30 The Tox−/PCR+ samples were tested by a cell cytotoxin assay (C. difficile Tox-B; TechLab), the more sensitive historical standard for C difficile toxin detection and diagnosis, to determine the number of samples that would have been positive if this test had been used instead of the toxin immunoassay. Culture was performed to recover C difficile isolates for ribotyping and verification of capacity to produce toxins. Lactoferrin was measured in PCR+ samples and random PCR− samples as a marker of inflammation (Leuko EZ Vue; TechLab; and IBD-Scan; TechLab). Lactoferrin results were classified as high if they exceeded the 95th percentile of results in PCR− patients. See the eMethods in the Supplement for additional details.
Clinical Data Collection
Diarrheal symptoms were verified at the time of C difficile testing. Patients were considered to have diarrhea if they had at least 3 unformed bowel movements or at least 600 mL of rectal or colostomy output recorded in the electronic health record (EHR) within 24 hours on the day of or before sample collection. Patients not meeting the threshold for diarrhea in the EHR had their nurse called to verify diarrheal status. Other data were obtained from laboratory, EHR, and administrative databases. See the eMethods in the Supplement for additional details.
Outcomes and Clinical Case Attribution
The primary outcome was duration of diarrhea for the 15-day period encompassing the day of sample collection (day 1) and up to 14 days of treatment. Secondary outcomes included rate of CDI-related complications (ie, megacolon, colectomy for fulminant colitis, and intensive care unit [ICU] care related to CDI) and CDI-related deaths within 30 days. The CDI-related complications and deaths were analyzed separately to distinguish patients with complicated CDI disease of the colon from patients with CDI as a contributing cause of death but not necessarily complicated CDI of the colon. Repeat C difficile tests and treatment were analyzed within 14 days of day 1 as an indication of ongoing clinical suspicion or empirical treatment for CDI in Tox−/PCR+ patients and to determine how many became positive with repeat testing. Clostridium difficile tests and treatment 15 to 30 days after day 1 were analyzed as a proxy for recurrent or prolonged CDI occurring after the initial treatment period. Ten or more days of metronidazole or oral vancomycin therapy was considered full treatment. Duration of diarrhea was determined from nurse-recorded stool counts and rectal or colostomy outputs in the EHR, excluding formed stools. Each day was categorized as a diarrhea day if at least 3 unformed stools or at least 600 mL of fecal output was recorded. Days with less stool output were categorized as a no-diarrhea day. Duration of diarrhea was the sum of days from day 1 to the last day with diarrhea, followed by 2 or more days without diarrhea. Cases of CDI-related megacolon and colectomies were identified by searching for patients with a procedure or billing code for abdominal radiology, colonoscopy, colectomy, or diagnosis of megacolon or pseudomembranous colitis within 30 days (eTable 1 in the Supplement). Clinical and surgical notes and radiology, endoscopy, and pathology reports were reviewed to confirm or exclude CDI-related megacolon or colectomy. Partially treated complications diagnosed before day 1 were excluded. Intensive care unit care related to CDI was determined as follows. First, patients located in or transferred into the ICU on day 1 (±1 day) were identified. The ICU care was then determined to be CDI related (ie, attributable to or contributed to by CDI) or unrelated by blinded EHR review by 2 board-certified infectious diseases physicians (H.H.N., L.W.L., J.V.S., or S.H.C.). The physician adjudicators were blinded to PCR results but otherwise were provided with all relevant clinical, procedural, diagnostic, and outcome information available in the EHR. Disagreements were resolved by a third infectious diseases physician (H.H.N., L.W.L., J.V.S., or S.H.C.). Deaths were identified by discharge disposition codes and EHR review of PCR-positive patients with unknown mortality status at 30 days. Attribution of deaths as CDI related or unrelated was determined by blinded infectious diseases physician EHR review (L.W.L., J.V.S., or S.H.C.) in the same manner as for ICU care.
Statistical Analysis
Baseline data were summarized and tested for differences. The Kruskal-Wallis test was used for continuous variables except for age, which was compared with an analysis of variance. For categorical variables, including outcomes, a χ2 test or Fisher exact test was used. Kaplan-Meier estimates were used to show time to resolution of diarrhea for each group, with censoring of patients who were discharged or died during the follow-up, and compared with the log-rank test. A Cox proportional hazards model was used to estimate the effect of Tox+/PCR+ or Tox−/PCR+ status compared with Tox−/PCR− status on the duration of diarrhea, adjusting for age, comorbidities, ICU status on day 1 (±1 day), prior antibiotic days, prior metronidazole or oral vancomycin exposure, maximum white blood cell count on day 1 (±1 day), C difficile ribotype, and fecal lactoferrin level. See the eMethods in the Supplement for additional details.
Results
Patient Cohort and Baseline Characteristics
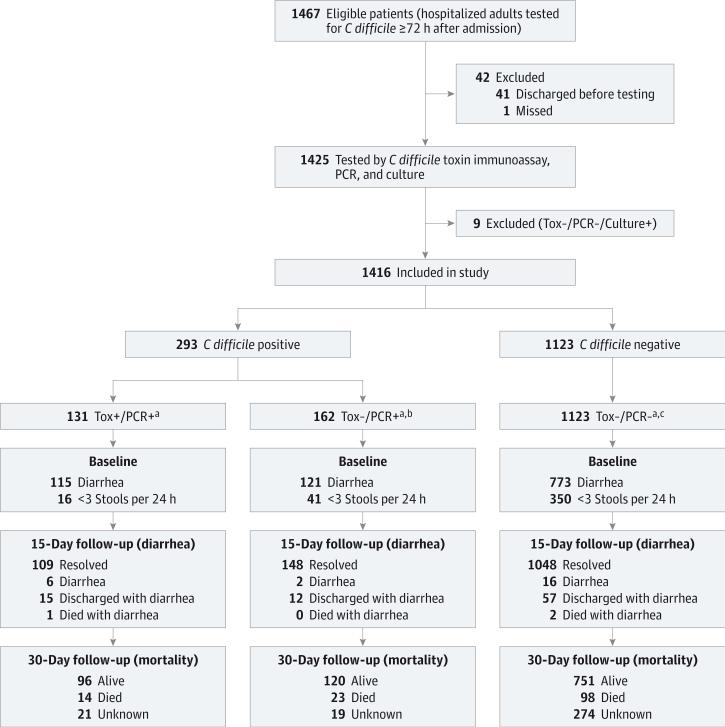
An overview of the study design, patient cohort, and follow-up is shown in Figure 1. In total, 1416 hospitalized adults were analyzed, including 131 Tox+/PCR+ patients (9.3%), 162 Tox−/PCR+ patients (11.4%), and 1123 Tox−/PCR− patients (79.3%).
Figure 1. Flow of Patients Through Testing and Follow-up.
Tox+/PCR+ indicates Clostridium difficile toxin immunoassay positive and polymerase chain reaction positive; Tox−/PCR+, C difficile toxin immunoassay negative and polymerase chain reaction positive; and Tox−/PCR−, C difficile toxin immunoassay negative and polymerase chain reaction negative.
a Clostridium difficile test group based on US Food and Drug Administration–approved toxin immunoassay and polymerase chain reaction results.
b Includes one patient with false-positive immunoassay.
c Includes 20 patients with false-positive immunoassay.
The groups were similar in age, sex, number of comorbidities, nonantibiotic medication exposures, and proportions with leukopenia, renal insufficiency, and hypoalbuminemia except for fewer comorbidities in Tox−/PCR− patients (Table 1 and eTable 2 in the Supplement). However, the Tox+/PCR+ group had more prior antibiotic exposure, more patients with leukocytosis, and more diarrhea on day 1. In feces, Tox+/PCR+ patients had an increased C difficile bacterial load, higher toxin concentration, and greater frequency of hypervirulent C difficile strain than Tox−/PCR+ patients. Correspondingly, Tox+/PCR+ patients had significantly more fecal lactoferrin than Tox−/PCR+ patients, and 36.8% (43 of 117) had a lactoferrin level greater than the 95th percentile of Tox−/PCR− patients. In contrast, few Tox−/PCR+ patients (13.4% [19 of 142]) had a lactoferrin level above the 95th percentile of Tox−/PCR− patients, and 79.0% (15 of 19) of these patients had an alternative explanation for fecal inflammation, a previous diagnosis of CDI, or anti–C difficile treatment before testing (eTable 3 in the Supplement).
Table 1.
Baseline Characteristics of the Study Population by Clostridium difficile Test Group
|
C difficile Positive |
C difficile Negative |
|||
|---|---|---|---|---|
| Characteristic | Tox+/PCR+b (n = 131) | Tox–/PCR+b,c (n = 162) | Tox–/PCR–b,d (n = 1123) | P Valuea |
| Age, median (IQR), y | 64 (52-71) | 58 (48-68) | 59 (47-71) | .12 |
| Female sex, No. (%) | 64 (48.9) | 83 (51.2) | 530 (47.2) | .61 |
| Comorbidities, median (IQR) | 4 (2-6) | 4 (2-5) | 3 (2-5) | .01 |
| APR-DRG risk of mortality subclass 3 or 4, No. (%) | 104 (79.4) | 128 (79.0) | 787 (70.1) | .008 |
| Intensive care unit care on day 1 ±1 d, No. (%)e | 30 (22.9) | 57 (35.2) | 435 (38.7) | .002 |
| Hospital days before day 1, median (IQR)e | 10 (6-24) | 8 (5-12) | 8 (5-12) | <.001 |
| Admitted from health care facility, No. (%) | 40 (30.5) | 34 (21.0) | 160 (14.2) | <.001 |
| C difficile positive within 90 d before day 1e | 5 (3.8) | 10 (6.2) | 13 (1.2) | <.001 |
| Antibiotic days within 90 d before day 1, median (IQR)e | 16 (7-32) | 10 (4-27) | 8 (4-18) | <.001 |
| Other diarrheal or gastrointestinal inflammatory process, No. (%)f | 8 (6.1) | 27 (16.7) | 161 (14.3) | .02 |
| Metronidazole or oral vancomycin within 48 h before day 1, No. (%)e | 3 (2.3) | 32 (19.8) | 184 (16.4) | <.001 |
| WBC count ≥15 000 cells/μL on day 1 ±1 d, No./total No. tested (%)e | 54/129 (41.9) | 50/154 (32.5) | 323/1101 (29.3) | .01 |
| WBC count <4000 cells/μL on day 1 ±1 d, No./total No. tested (%)e | 20/129 (15.5) | 32/154 (20.8) | 200/1101 (18.2) | .52 |
| Creatinine level >1.5 mg/dL on day 1 ±1 d, No./total No. tested (%)e | 36/127 (28.3) | 45/156 (28.8) | 297/1102 (27.0) | .85 |
| Albumin level <2.5 g/dL on day 1 ±1 d, No./total No. tested (%)e | 29/48 (60.4) | 50/70 (71.4) | 318/475 (66.9) | .46 |
| Diarrhea present on day 1 ±1 d, No. (%)e | 121 (92.4) | 143 (88.3) | 927 (82.5) | .004 |
| Stool count on day 1, median (IQR)e | 5 (3-6) | 3 (2-5) | 3 (2-5) | <.001 |
| C difficile toxin B, median (IQR), ng/mL | 640.8 (172.5-1194.0) | 1.1 (0.3-2.5) | NA | <.001 |
| Hypervirulent C difficile ribotype RT027/078, No. (%) | 68 (51.9) | 39 (24.1) | NA | <.001 |
| C difficile binary toxin positive, No. (%) | 71 (54.2) | 45 (27.8) | NA | <.001 |
| Log10 C difficile DNA copies/mL, median (IQR) | 7.3 (6.6-7.7) | 4.9 (4.4-6.2) | NA | <.001 |
| Fecal lactoferrin level, median (IQR), μg/mL, | 37.7 (8.8-261.5) | 20.1 (5.0-50.3) | 7.8 (0.5-32.6) | <.001 |
| Normal lactoferrin level, No./total No. testedg | 25/117 (21.4) | 44/142 (31.0) | 89/188 (47.3) | <.001 |
| High lactoferrin level, No./total No. testedh | 43/117 (36.8) | 19/142 (13.4) | 9/188 (4.8) | <.001 |
Abbreviations: APR-DRG, all-patient refined diagnosis-related group; IQR, interquartile range; NA, not applicable; Tox+/PCR+, C difficile toxin immunoassay positive and polymerase chain reaction positive; Tox–/PCR+, C difficile toxin immunoassay negative and polymerase chain reaction positive; Tox–/PCR–, C difficile toxin immunoassay negative and polymerase chain reaction negative; WBC, white blood cell.
SI conversion factors: To convert WBC count to ×109/L, multiply by 0.001; to convert creatinine level to micromoles per liter, multiply by 88.4; to convert albumin level to grams per liter, multiply by 10.
P value for significance across 3 groups except for characteristics not applicable to Tox–/PCR– group.
Clostridium difficile test group based on US Food and Drug Administration–approved toxin immunoassay and PCR results.
Includes one patient with false-positive toxin immunoassay.
Includes 20 patients with false-positive toxin immunoassay.
Day 1 is the day of sample collection for the C difficile toxin test.
Includes inflammatory bowel diseases, functional diarrheal disorders, diverticulitis, appendicitis, ischemic colitis, other infectious or noninfectious enterocolitis, graft-vs-host disease, and peritoneal, mesenteric, or retroperitoneal infections.
Normal fecal lactoferrin level defined as within the upper limit of a healthy person's reference range per the manufacturer's package insert.
High fecal lactoferrin level defined as exceeding the 95th percentile fecal lactoferrin level in Tox–/PCR– patients (>89.05 μg/mL).
Duration of Diarrhea
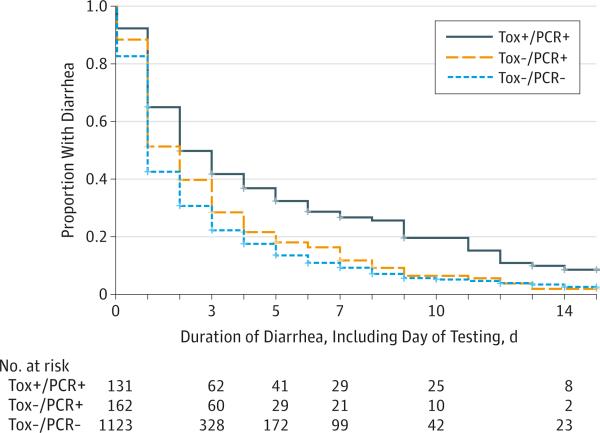
The Tox+/PCR+ patients had a longer duration of diarrhea than Tox−/PCR+ patients and Tox−/PCR− patients (P < .001) and had a greater risk of diarrhea during the follow-up (Figure 2 and Table 2). In contrast, Tox−/PCR+ patients and Tox−/PCR− patients had a similar risk of diarrhea on most days.
Figure 2. Kaplan-Meier Curves of Time to Resolution of Diarrhea by Clostridium difficile Test Group.
The median duration of diarrhea for patients with at least 1 day was 3 days (interquartile range, 1-6 days) for Tox+/PCR+ (121 of 131), 2 days (interquartile range, 1-4 days) for Tox−/PCR+, and 2 days (interquartile range, 1-3 days) for Tox−/PCR− (927 of 1123) (P < .001). Log-rank P values are P < .001 for all groups, P = .003 for Tox+/PCR+ vs Tox−/PCR+, (143 of 162) P < .001 for Tox+/PCR+ vs Tox−/PCR−, and P < .001 for Tox−/PCR+ vs Tox−/PCR−. Tox+/PCR+ indicates C difficile toxin immunoassay positive and polymerase chain reaction positive; Tox−/PCR+, C difficile toxin immunoassay negative and polymerase chain reaction positive; Tox−/PCR−, C difficile toxin immunoassay negative and polymerase chain reaction negative.
Table 2.
Relative Risk (95% CI) of Diarrhea Each Day
| Day | Comparison | ||
|---|---|---|---|
| Tox+/PCR+ vs Tox–/PCR+ | Tox+/PCR+ vs Tox–/PCR– | Tox–/PCR+ vs Tox–/PCR– | |
| 1 | 1.05 (0.97-1.13) | 1.12 (1.06-1.18) | 1.07 (1.01-1.14) |
| 2 | 1.27 (1.03-1.56) | 1.46 (1.29-1.73) | 1.18 (0.99-1.40) |
| 3 | 1.28 (0.98-1.67) | 1.62 (1.32-1.98) | 1.27 (1.02-1.58) |
| 4 | 1.51 (1.07-02.13) | 1.87 (1.46-2.40) | 1.24 (0.93-1.66) |
| 5 | 1.75 (1.15-2.65) | 2.04 (1.53-2.73) | 1.17 (0.82-1.67) |
| 6 | 1.88 (1.17-3.02) | 2.31 (1.67-3.20) | 1.23 (0.81-1.85) |
| 7 | 1.71 (1.02-2.85) | 2.51 (1.73-3.64) | 1.47 (0.95-2.29) |
| 8 | 2.30 (1.25-4.22) | 2.72 (1.82-4.06) | 1.18 (0.69-2.04) |
| 9 | 3.09 (1.54-6.20) | 3.90 (2.52-6.03) | 1.26 (0.66-2.42) |
| 10 | 3.18 (1.37-7.38) | 3.67 (2.18-6.19) | 1.16 (0.53-2.53) |
| 11 | 3.18 (1.37-7.38) | 4.06 (2.39-6.90) | 1.28 (0.58-2.81) |
| 12 | 2.89 (1.14-7.30) | 3.64 (2.00-6.62) | 1.26 (0.54-2.96) |
| 13 | 3.09 (0.99-9.63) | 3.30 (1.63-6.68) | 1.07 (0.38-3.02) |
| 14 | 4.95 (1.07-22.90) | 2.98 (1.36-6.53) | 0.60 (0.14-2.53) |
| 15 | 3.71 (0.76-18.08) | 3.22 (1.28-8.07) | 0.87 (0.20-3.73) |
Abbreviations: Tox+/PCR+, Clostridium difficile toxin immunoassay positive and polymerase chain reaction positive; Tox–/PCR+, C difficile toxin immunoassay negative and polymerase chain reaction positive; Tox–/PCR–, C difficile toxin immunoassay negative and polymerase chain reaction negative.
In the multivariable model, Tox+/PCR+ status had the strongest effect on duration of diarrhea, decreasing the probability of diarrhea being resolved by 37% each day relative to the Tox−/PCR− reference group (hazard ratio, 0.63; 95% CI, 0.48-0.83). Age, white blood cell count, and lactoferrin level were also significant predictors of duration of diarrhea, but their relative contribution was small (≤2% each) (eTable 4 in the Supplement). The Tox−/PCR+ status and pretest exposure to metronidazole or oral vancomycin were not significant predictors in the multivariable model.
CDI-Related Complications and Mortality Within 30 Days
The frequency of CDI-related complications (ie, megacolon, colectomy for fulminant colitis, and ICU care related to CDI) and deaths is summarized in Table 3. The Tox+/PCR+ patients had more CDI-related complications than Tox−/PCR+ patients and Tox−/PCR− patients (10 [7.6%] of 131 vs 0 [0%] of 162 vs 3 [0.3%] of 1123, P < .001). In contrast, the rate of CDI-related complications was similar between Tox−/PCR+ patients and Tox−/PCR− patients (0% vs 0.3%, P > .99). The Tox+/PCR+ patients also had more CDI-related deaths than Tox−/PCR+ patients and Tox−/PCR− patients (11 [8.4%] of 131 vs 1 [0.6%] of 162 vs 0 [0%] of 1123, P < .001) while the rate was similar between Tox−/PCR+ patientsandTox−/PCR− patients (0.6% vs 0%, P = .13). Two deaths in the Tox+/PCR+ group were directly attributable to CDI, and 9 had CDI as a contributing factor. One Tox−/PCR+ patient (patient 1641 in eTable 3 in the Supplement) had an uncomplicated, recurrent CDI that resolved before care was withdrawn for severe underlying illness, but CDI was considered a contributing factor to death.
Table 3.
Nondiarrheal Outcomes and Treatment by Clostridium difficile Test Group
|
C difficile Positive |
C difficile Negative |
|||
|---|---|---|---|---|
| Outcome | Tox+/PCR+ (n =131) | Tox–/PCR+ (n = 162) | Tox–/PCR– (n = 1123) | P Valuea |
| C difficile-Related Complication or Death Within 30 d, No. (%) | ||||
| Complicationb | 10 (7.6) | 0 | 3 (0.3) | <.001 |
| Deathc | 11 (8.4) | 1 (0.6) | 0 | <.001 |
| Complication or death | 18 (13.7) | 1 (0.6) | 3 (0.3) | <.001 |
| Repeat C difficile Testing Within 14 d, No. (%) | ||||
| Retested | 14 (10.7) | 61 (37.7) | 374 (33.3) | <.001 |
| Positive toxin test result | 3 (2.3) | 13 (8.0) | 17 (1.5) | <.001 |
| Repeat C difficile Testing at 15-30 d, No. (%) | ||||
| Tested | 26 (19.8) | 18 (11.1) | 106 (9.4) | .001 |
| Positive toxin test result | 14 (10.7) | 5 (3.1) | 10 (0.9) | <.001 |
| Treatment Within 14 d | ||||
| Metronidazole or oral vancomycin, No. (%)d | 131 (100) | 66 (40.7) | 361 (32.1) | <.001 |
| Duration of metronidazole or oral vancomycin, if treated, median (IQR), d | 14 (11-14) | 6 (3-11) | 5 (2-9) | <.001 |
| Non-C difficile antibiotic, No. (%) | 98 (74.8) | 141 (87.0) | 912 (81.2) | .03 |
| Duration of non-C difficile antibiotic, if treated, median (IQR), d | 11 (3-14) | 10 (4-14) | 10 (4-14) | .13 |
| Treatment at 15-30 d | ||||
| Metronidazole or oral vancomycin, No. (%) | 75 (57.3) | 35 (21.6) | 137 (12.2) | <.001 |
| Duration of metronidazole or oral vancomycin, if treated, median (IQR), d | 9 (3-14) | 4 (3-15) | 6 (3-9) | <.001 |
Abbreviations: IQR, interquartile range; Tox+/PCR+, C difficile toxin immunoassay positive and polymerase chain reaction positive; Tox–/PCR+, C difficile toxin immunoassay negative and polymerase chain reaction positive; Tox–/PCR–, C difficile toxin immunoassay negative and polymerase chain reaction negative.
P value for significance across 3 groups.
Intensive care unit care, colectomy, or megacolon related to C difficile infection. The Tox+/PCR+ complications included 3 fulminant colitis or megacolon and 7 intensive care unit care related to C difficile infection. Two Tox–/PCR+ patients with partially treated complications diagnosed as having a positive toxin test result before day 1 were excluded. The Tox–/PCR– complications included 3 intensive care unit care related to C difficile infection. P < .001 for Tox+/PCR+ vs Tox–/PCR+ and P > .99 for Tox–/PCR+ vs Tox–/PCR–.
All-cause mortality within 30 days was 14 (10.7%), 23 (14.2%), and 98 (8.7%), respectively, for the 3 groups. P = .08 for all groups and P = .21 for Tox+/PCR+ vs Tox–/PCR+. For C difficile infection-related death, P < .001 for Tox+/PCR+ vs Tox–/PCR+ and P = .13 Tox–/PCR+ vs Tox–/PCR–.
Full treatment (≥10 days) and partial treatment (1-9 days) values were 119 (90.8%) and 12 (9.2%), respectively; 21 (13.0%) and 45 (27.8%), respectively; and 82 (7.3%) and 279 (24.8%), respectively, for the 3 groups.
Repeat C difficile Testing and Treatment Within 14 Days
Repeat C difficile testing and treatment within 14 days of day 1 was analyzed as an indication of ongoing clinical suspicion or empirical treatment for CDI in Tox−/PCR+ patients (Table 3). During this period, 61 Tox−/PCR+ patients (37.7%) were retested, and 13 (8.0%) had toxins detected (mean time to positive result, 5.7 days; 95% CI, 3.2-8.2 days). None of these patients developed a C difficile–related complication. However, one patient (patient 1641 in eTable 3 in the Supplement) had CDI that was considered a contributing factor to death, although symptoms had resolved before care was withdrawn for other reasons. During the same period, most Tox−/PCR+ patients (59.3% [96 of 162]) received no treatment, 45 patients (27.8% [45 of 162]) received partial treatment (1-9 days), and 21 patients (13.0% [21 of 162]) received the equivalent of full treatment (≥10 days).
Clostridium difficile Testing and Treatment Between 15 and 30 Days
Clostridium difficile tests and treatment 15 to 30 days after day 1 were analyzed as a proxy for recurrent or prolonged CDI (Table 3). During this period, Tox+/PCR+ patients were retested almost twice as often as Tox−/PCR+ patients (19.8% vs 11.1%, P = .04) and were positive 3 times more often (10.7% vs 3.1%, P < .001). During the same period, most Tox−/PCR+ patients (78.4% [127 of 162]) received no treatment, while 13 patients (8.0% [13 of 162]) received treatment for at least 10 days.
Additional Analyses to Evaluate the Robustness of the Study Findings
Outcome differences between the Tox−/PCR+ and Tox+/PCR+ groups remained significant when comparisons were limited to the subgroup of Tox−/PCR+ patients who received full or partial treatment within 14 days (P = .04 for time to resolution of diarrhea and P = .004 for CDI-related complication or death) or no treatment (P = .003 for time to resolution of diarrhea and P < .001 for CDI-related complication or death). No significant outcome differences were observed between the Tox−/PCR− group and individual Tox−/PCR+ subgroups with or without treatment.
If the historical cell cytotoxin assay had been used for diagnosis instead of a toxin immunoassay, 48 additional Tox−/PCR+ patients (29.6%) would have been reported positive. However, this subgroup had a low toxin concentration (median, 10 ng/mL; interquartile range, 2-81 ng/mL) and outcomes that were similar to cell cytotoxin–negative Tox−/PCR+ patients (P = .47 for time to resolution of diarrhea and P = .30 for CDI-related complication or death), with no difference in treatment (P = .61), and better than Tox+/PCR+ patients (P < .001 for time to resolution of diarrhea and P = .03 for CDI-related complication or death).
Discussion
This study addresses an important question for physicians, hospitals, and policy makers: do toxin-negative patients with a positive C difficile PCR test result require treatment? To answer this question, we prospectively tested 1416 hospitalized patients with FDA-approved PCR tests while maintaining our existing toxin test for clinical diagnosis to determine the natural history of toxin-negative patients with positive PCR results. We found that 55.3% (162 of 293) of patients with a positive C difficile PCR test result lacked toxin by the clinical toxin immunoassay test and had outcomes that were comparable to patients with no C difficile detected. These Tox−/PCR+ patients had milder symptoms at the time of testing and a shorter duration of diarrhea than toxin-positive patients. In total, 58.7% (95 of 162) were never retested, and only 13.0% (21 of 162) received the equivalent of a full course of treatment. Repeat analyses with the treated Tox−/PCR+ patients removed did not change our conclusions. Overall, 18 of 19 C difficile–related complications and deaths (94.7%) occurred in toxin-positive patients. Only one of 162 toxin-negative patients (0.6%) was considered to have CDI as a contributing factor to death.
Our findings are consistent with the conventional view that CDI is a toxin-mediated inflammatory disease preceded by antibiotic exposure and C difficile overgrowth.3 Toxin-negative patients had less antibiotic exposure, C difficile DNA, and inflammation and manifested milder symptoms and no complications, despite minimal or no treatment. These findings strongly suggest that most patients with negative toxin test results and C difficile detected by PCR do not need treatment for CDI. We suspect that most of these patients were colonized with C difficile and had another cause of diarrhea. This hypothesis is supported by studies22-26,31 showing that C difficile colonization and immunity are common in hospitalized patients and most nosocomial diarrhea is noninfectious. It is possible that some toxin-negative patients have mild or early infection because clinical toxin tests can miss toxin at low concentrations, and occasional toxin-negative patients become positive on repeat testing.3,10,18,32-35 Correspondingly, we detected toxin in 29.6% (48 of 162) of Tox−/PCR+ patients by the historical cell cytotoxin assay, and 8.0% (13 of 162) of Tox−/PCR+ patients retested positive by the clinical toxin immunoassay in a subsequent sample. However, the relative lack of adverse events in this subgroup suggests that these patients are also at lower risk of complications than clinical toxin immunoassay–positive patients and routine treatment is unnecessary.
These results are consistent with a large retrospective study36 that found no C difficile–related complications and lower mortality among hospitalized patients with negative toxin results. Our findings also agree with several smaller studies11,14,37-41 and one large, multicenter study21 that reported milder symptoms or a lower mortality rate in toxin-negative patients with positive PCR results. Other studies42-45 that have investigated clinical characteristics of Tox−/PCR+ patients were generally underpowered or not designed to compare outcomes. Finally, there are reports of patients with severe or complicated CDI missed by toxin tests,43,46 but our data suggest that such patients are rare.
Strengths of our study include the prospective study design, large sample size, nonreporting of PCR results, measurement of duration of diarrhea, inclusion of patients without C difficile for comparison, and rigorous evaluation of C difficile–related complications and deaths. We quantified fecal C difficile DNA, toxins, and inflammation to provide mechanistic insight into the reasons for the different test results and outcomes. The primary weakness of the study was the inability to achieve equivalent risk allocation between groups. In addition, we cannot exclude the possibility that empirical treatment affected outcomes in some Tox−/PCR+ patients, but the outcome differences we observed remained when these patients were removed. It is also possible that our outcome adjudicators were influenced by positive toxin results, but 26 of 42 Tox+/PCR+ patients with ICU care or death (61.9%) were judged not to have a CDI-related outcome, indicating that the adjudication was a highly discriminatory process overall. Finally, we cannot exclude the possibility that systematic underrecording of stools in patients with negative toxin results could account for the shorter duration of diarrhea in these patients. However, our requirement of 2 or more days without diarrhea to end an episode would make it unlikely that underrecording by individual nurses would have a significant effect on our diarrhea measure.
Molecular tests have the potential benefits of decreasing the need for repeat testing and empirical treatment because of their high negative predictive value and may have a role in infection prevention if Tox−/PCR+ patients contribute to the spread of C difficile in health care facilities.34,43,47 However, our results offer compelling evidence that as many as half of the patients with positive C difficile PCR test results are likely to be overdiagnosed and exposed to unnecessary treatment at institutions using molecular tests. The number of patients potentially affected by this issue is massive. Most institutions experience a 50% to 100% increase in reported CDI after switching to molecular tests, and the proportion of institutions using molecular C difficile tests has increased dramatically since initiation in 2009 of the first FDA-approved molecular test.11-15 In 2014, almost 44% of NHSN acute care facilities reported using molecular tests for CDI diagnosis (NHSN, written communication, September 15, 2014).
Therefore, there is an urgent need to educate physicians that molecular tests are not specific for CDI, even in the presence of symptoms, and patients with positive PCR results do not necessarily need treatment. Similarly, while underdiagnosis may occur with lack of testing,48 policy makers should be aware that molecular C difficile tests are a major cause of overdiagnosis and consider the potential costs of overtreatment in recommendations and analyses. Laboratories need to be aware that rejection of formed stool samples is not sufficient to ensure that all positive molecular C difficile results represent disease.
We concur with authors in the United Kingdom that molecular tests should not be used as a stand-alone diagnostic test for CDI and diagnostic recommendations should move back in the direction of defining clinical disease as a positive toxin result in patients with diarrhea.21,49 Most toxin-negative patients with C difficile do not need specific treatment, although there may be a role for identifying carriers to prevent transmission.21,43 Future studies should focus on developing diagnostic approaches to accurately distinguish patients with active infection vs colonization, which may include quantitation of C difficile DNA, toxins, or host response. In the meantime, 2-step testing with a screening test, such as PCR or glutamate dehydrogenase antigen detection, followed by a toxin test to confirm active infection is a reasonable diagnostic strategy.21,49
Conclusions
Up to half of the patients with positive molecular test results for C difficile do not experience adverse events without treatment and do not need treatment for CDI. Exclusive reliance on molecular tests for C difficile diagnosis is likely to result in over diagnosis, unnecessary treatment, and increased health care costs.
Supplementary Material
Acknowledgments
Funding/Support: This study was supported by grants UL1 RR024146 and TL1 RR024145-06 from the National Center for Research Resources (Dr Polage), grant UL1TR000002 and linked award TL1TR000133 from the National Center for Advancing Translational Sciences (Dr Chin), and grant T32HS022236 from the Agency for Healthcare Research and Quality through the Quality Safety Comparative Effectiveness Research Training Program (Dr Chin). Clostridium difficile diagnostic test kits and reagents were received from Meridian Biosciences, Cepheid, TechLab, and Alere.
Role of the Funder/Sponsor: The industry contributors to this study had no role in the design and conduct of the study; the collection, management, analysis, and interpretation of the data; or the preparation, review, or approval of the manuscript.
Footnotes
Author Contributions: Dr Polage had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the analysis.
Study concept and design: Polage, Chin, Romano, Cohen.
Acquisition, analysis, or interpretation of data: Polage, Gyorke, Kennedy, Leslie, Chin, Wang, Nguyen, Huang, Tang, Lee, Panacek, Goodell, Solnick, Cohen.
Drafting of the manuscript: Polage.
Critical revision of the manuscript for important intellectual content: All authors.
Statistical analysis: Polage, Kim, Taylor.
Obtained funding: Polage, Romano.
Administrative, technical, or material support: Polage, Panacek.
Study supervision: Polage.
Conflict of Interest Disclosures: All authors have completed and submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Dr Polage reported receiving research materials (ie, Clostridium difficile diagnostic test kits and reagents) from Meridian Biosciences, Cepheid, TechLab, and Alere; and honoraria for a webinar about diagnostic testing for C difficile and nosocomial diarrhea from Alere. Dr Nguyen reported receiving research funding for pharmaceutical clinical trials of treatment for patients with C difficile from Merck, Cubist, and Viropharma. Dr Tang reported receiving research funding and materials from ACEA Biosciences. Dr Cohen reported serving on an advisory board for Merck and receiving research funding for pharmaceutical clinical trials of treatment for patients with C difficile from Merck, Viropharma, Seres Health, and Actelion. No other disclosures were reported.
Additional Contributions: We thank the following for their contributions without compensation: staff of the University of California Davis Medical Center Clinical Microbiology Laboratory for assistance with testing and processing samples; Jennifer Brown, MD (University of California Davis School of Medicine) for blinded electronic health record reviews; and all the volunteers who participated as undergraduates at the University of California Davis or California State University, Sacramento, in patient enrollment, clinical data collection, or sample processing, including Sameer Aggarwal, BS, Robert Araiza, BS, Adeel Ashfaq, BS, Mercy Bechtold, BS, Joel Breck, BA, MPH, Jenna DeYoung, BS, Tu Dinh, BS, Timothy Fong, BS, MPH, Kaitlin Gee, BS, Megan Gilbert, BS, Breana Hill, BS, Chae Ji Kim, BS, Bryan Lee, BS, Dan O'Brien, BS, Thomas Osterberg-Diess, BS, Erik Pennell, BS, Roberto Ramos, BS, Daniel Sadoma, BS, Natalie Telis, BS, Joanna Tripet-Diel, BS, MPH, Andrew Tubbs, BS, William Valcheck, BS, and Alvin Au Yeung, BS.
Supplemental content at jamainternalmedicine.com
REFERENCES
- 1.Magill SS, Edwards JR, Bamberg W, et al. Emerging Infections Program Healthcare-Associated Infections and Antimicrobial Use Prevalence Survey Team. Multistate point-prevalence survey of health care–associated infections. N Engl J Med. 2014;370(13):1198–1208. doi: 10.1056/NEJMoa1306801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lucado J, Gould C, Elixhauser A. Clostridium difficile Infections (CDI) in Hospital Stays, 2009: Statistical Brief #124. Rockville, MD: Agency for Health Care Policy and Research; 2006-2012. Healthcare Cost and Utilization Project (HCUP) Statistical Briefs. [PubMed] [Google Scholar]
- 3.Rupnik M, Wilcox MH, Gerding DN. Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nat Rev Microbiol. 2009;7(7):526–536. doi: 10.1038/nrmicro2164. [DOI] [PubMed] [Google Scholar]
- 4.McDonald LC, Lessa F, Sievert D, et al. Centers for Disease Control and Prevention (CDC). Vital signs: preventing Clostridium difficile infections. MMWR Morb Mortal Wkly Rep. 2012;61(9):157–162. [PubMed] [Google Scholar]
- 5.Zilberberg MD, Shorr AF, Kollef MH. Increase in adult Clostridium difficile–related hospitalizations and case-fatality rate, United States, 2000-2005. Emerg Infect Dis. 2008;14(6):929–931. doi: 10.3201/eid1406.071447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kang J, Sickbert-Bennett EE, Brown VM, Weber DJ, Rutala WA. Changes in the incidence of health care–associated pathogens at a university hospital from 2005 to 2011. Am J Infect Control. 2014;42(7):770–775. doi: 10.1016/j.ajic.2014.03.019. [DOI] [PubMed] [Google Scholar]
- 7.Dubberke ER, Olsen MA. Burden of Clostridium difficile on the healthcare system. Clin Infect Dis. 2012;55(suppl 2):S88–S92. doi: 10.1093/cid/cis335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McDonald LC, Killgore GE, Thompson A, et al. An epidemic, toxin gene–variant strain of Clostridium difficile. N Engl J Med. 2005;353(23):2433–2441. doi: 10.1056/NEJMoa051590. [DOI] [PubMed] [Google Scholar]
- 9.Pépin J, Valiquette L, Alary ME, et al. Clostridium difficile–associated diarrhea in a region of Quebec from 1991 to 2003: a changing pattern of disease severity. CMAJ. 2004;171(5):466–472. doi: 10.1503/cmaj.1041104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burnham CA, Carroll KC. Diagnosis of Clostridium difficile infection: an ongoing conundrum for clinicians and for clinical laboratories. Clin Microbiol Rev. 2013;26(3):604–630. doi: 10.1128/CMR.00016-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Longtin Y, Trottier S, Brochu G, et al. Impact of the type of diagnostic assay on Clostridium difficile infection and complication rates in a mandatory reporting program. Clin Infect Dis. 2013;56(1):67–73. doi: 10.1093/cid/cis840. [DOI] [PubMed] [Google Scholar]
- 12.Moehring RW, Lofgren ET, Anderson DJ. Impact of change to molecular testing for Clostridium difficile infection on healthcare facility–associated incidence rates. Infect Control Hosp Epidemiol. 2013;34(10):1055–1061. doi: 10.1086/673144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fong KS, Fatica C, Hall G, et al. Impact of PCR testing for Clostridium difficile on incident rates and potential on public reporting: is the playing field level? Infect Control Hosp Epidemiol. 2011;32(9):932–933. doi: 10.1086/661789. [DOI] [PubMed] [Google Scholar]
- 14.Koo HL, Van JN, Zhao M, et al. Real-time polymerase chain reaction detection of asymptomatic Clostridium difficile colonization and rising C. difficile–associated disease rates. Infect Control Hosp Epidemiol. 2014;35(6):667–673. doi: 10.1086/676433. [DOI] [PubMed] [Google Scholar]
- 15.Gould CV, Edwards JR, Cohen J, et al. Clostridium difficile Infection Surveillance Investigators, Centers for Disease Control and Prevention. Effect of nucleic acid amplification testing on population-based incidence rates of Clostridium difficile infection. Clin Infect Dis. 2013;57(9):1304–1307. doi: 10.1093/cid/cit492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bartlett JG, Taylor NS, Chang T, Dzink J. Clinical and laboratory observations in Clostridium difficile colitis. Am J Clin Nutr. 1980;33(11)(suppl):2521–2526. doi: 10.1093/ajcn/33.11.2521. [DOI] [PubMed] [Google Scholar]
- 17.Gerding DN, Johnson S, Peterson LR, Mulligan ME, Silva J., Jr Clostridium difficile–associated diarrhea and colitis. Infect Control Hosp Epidemiol. 1995;16(8):459–477. doi: 10.1086/648363. [DOI] [PubMed] [Google Scholar]
- 18.Bartlett JG, Gerding DN. Clinical recognition and diagnosis of Clostridium difficile infection. Clin Infect Dis. 2008;46(suppl 1):S12–S18. doi: 10.1086/521863. [DOI] [PubMed] [Google Scholar]
- 19.Wilcox MH, Planche T, Fang FC, Gilligan P. What is the current role of algorithmic approaches for diagnosis of Clostridium difficile infection? J Clin Microbiol. 2010;48(12):4347–4353. doi: 10.1128/JCM.02028-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dubberke ER, Han Z, Bobo L, et al. Impact of clinical symptoms on interpretation of diagnostic assays for Clostridium difficile infections. J Clin Microbiol. 2011;49(8):2887–2893. doi: 10.1128/JCM.00891-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Planche TD, Davies KA, Coen PG, et al. Differences in outcome according to Clostridium difficile testing method: a prospective multicentre diagnostic validation study of C difficile infection. Lancet Infect Dis. 2013;13(11):936–945. doi: 10.1016/S1473-3099(13)70200-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kyne L, Warny M, Qamar A, Kelly CP. Asymptomatic carriage of Clostridium difficile and serum levels of IgG antibody against toxin A. N Engl J Med. 2000;342(6):390–397. doi: 10.1056/NEJM200002103420604. [DOI] [PubMed] [Google Scholar]
- 23.Samore MH, DeGirolami PC, Tlucko A, Lichtenberg DA, Melvin ZA, Karchmer AW. Clostridium difficile colonization and diarrhea at a tertiary care hospital. Clin Infect Dis. 1994;18(2):181–187. doi: 10.1093/clinids/18.2.181. [DOI] [PubMed] [Google Scholar]
- 24.Leekha S, Aronhalt KC, Sloan LM, Patel R, Orenstein R. Asymptomatic Clostridium difficile colonization in a tertiary care hospital: admission prevalence and risk factors. Am J Infect Control. 2013;41(5):390–393. doi: 10.1016/j.ajic.2012.09.023. [DOI] [PubMed] [Google Scholar]
- 25.Alasmari F, Seiler SM, Hink T, Burnham CA, Dubberke ER. Prevalence and risk factors for asymptomatic Clostridium difficile carriage. Clin Infect Dis. 2014;59(2):216–222. doi: 10.1093/cid/ciu258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Polage CR, Solnick JV, Cohen SH. Nosocomial diarrhea: evaluation and treatment of causes other than Clostridium difficile. Clin Infect Dis. 2012;55(7):982–989. doi: 10.1093/cid/cis551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brecher SM, Novak-Weekley SM, Nagy E. Laboratory diagnosis of Clostridium difficile infections: there is light at the end of the colon. Clin Infect Dis. 2013;57(8):1175–1181. doi: 10.1093/cid/cit424. [DOI] [PubMed] [Google Scholar]
- 28.Ryder AB, Huang Y, Li H, et al. Assessment of Clostridium difficile infections by quantitative detection of tcdB toxin by use of a real-time cell analysissystem. JClinMicrobiol. 2010;48(11):4129–4134. doi: 10.1128/JCM.01104-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leslie JL, Cohen SH, Solnick JV, Polage CR. Role of fecal Clostridium difficile load in discrepancies between toxin tests and PCR: is quantitation the next step in C. difficile testing? Eur J Clin Microbiol Infect Dis. 2012;31(12):3295–3299. doi: 10.1007/s10096-012-1695-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang B, Jin D, Zhang J, et al. Real-time cellular analysis coupled with a specimen enrichment accurately detects and quantifies Clostridium difficile toxins in stool. J Clin Microbiol. 2014;52(4):1105–1111. doi: 10.1128/JCM.02601-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Viscidi R, Laughon BE, Yolken R, et al. Serum antibody response to toxins A and B of Clostridium difficile. J Infect Dis. 1983;148(1):93–100. doi: 10.1093/infdis/148.1.93. [DOI] [PubMed] [Google Scholar]
- 32.Cardona DM, Rand KH. Evaluation of repeat Clostridium difficile enzyme immunoassay testing. J Clin Microbiol. 2008;46(11):3686–3689. doi: 10.1128/JCM.00931-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Borek AP, Aird DZ, Carroll KC. Frequency of sample submission for optimal utilization of the cell culture cytotoxicity assay for detection of Clostridium difficile toxin. J Clin Microbiol. 2005;43(6):2994–2995. doi: 10.1128/JCM.43.6.2994-2995.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khanna S, Pardi DS, Rosenblatt JE, Patel R, Kammer PP, Baddour LM. An evaluation of repeat stool testing for Clostridium difficile infection by polymerase chain reaction. J Clin Gastroenterol. 2012;46(10):846–849. doi: 10.1097/MCG.0b013e3182432273. [DOI] [PubMed] [Google Scholar]
- 35.Deshpande A, Pasupuleti V, Patel P, et al. Repeat stool testing to diagnose Clostridium difficile infection using enzyme immunoassay does not increase diagnostic yield. Clin Gastroenterol Hepatol. 2011;9(8):665–669. e1. doi: 10.1016/j.cgh.2011.04.030. doi:10.1016/j.cgh.2011 .04.030. [DOI] [PubMed] [Google Scholar]
- 36.Polage CR, Chin DL, Leslie JL, Tang J, Cohen SH, Solnick JV. Outcomes in patients tested for Clostridium difficile toxins. Diagn Microbiol Infect Dis. 2012;74(4):369–373. doi: 10.1016/j.diagmicrobio.2012.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gerding DN, Olson MM, Peterson LR, et al. Clostridium difficile–associated diarrhea and colitis in adults: a prospective case-controlled epidemiologic study. Arch Intern Med. 1986;146(1):95–100. [PubMed] [Google Scholar]
- 38.Walker AS, Eyre DW, Wyllie DH, et al. Infections in Oxfordshire Research Database. Relationship between bacterial strain type, host biomarkers, and mortality in Clostridium difficile infection. Clin Infect Dis. 2013;56(11):1589–1600. doi: 10.1093/cid/cit127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de Jong E, de Jong AS, Bartels CJ, van der Rijt-van den Biggelaar C, Melchers WJ, Sturm PD. Clinical and laboratory evaluation of a real-time PCR for Clostridium difficile toxin A and B genes. Eur J Clin Microbiol Infect Dis. 2012;31(9):2219–2225. doi: 10.1007/s10096-012-1558-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beaulieu C, Dionne LL, Julien AS, Longtin Y. Clinical characteristics and outcome of patients with Clostridium difficile infection diagnosed by PCR versus a three-step algorithm. Clin Microbiol Infect. 2014;20(10):1067–1073. doi: 10.1111/1469-0691.12676. [DOI] [PubMed] [Google Scholar]
- 41.Baker I, Leeming JP, Reynolds R, Ibrahim I, Darley E. Clinical relevance of a positive molecular test in the diagnosis of Clostridium difficile infection. J Hosp Infect. 2013;84(4):311–315. doi: 10.1016/j.jhin.2013.05.006. [DOI] [PubMed] [Google Scholar]
- 42.Kaltsas A, Simon M, Unruh LH, et al. Clinical and laboratory characteristics of Clostridium difficile infection in patients with discordant diagnostic test results. J Clin Microbiol. 2012;50(4):1303–1307. doi: 10.1128/JCM.05711-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guerrero DM, Chou C, Jury LA, Nerandzic MM, Cadnum JC, Donskey CJ. Clinical and infection control implications of Clostridium difficile infection with negative enzyme immunoassay for toxin. Clin Infect Dis. 2011;53(3):287–290. doi: 10.1093/cid/cir361. [DOI] [PubMed] [Google Scholar]
- 44.Humphries RM, Uslan DZ, Rubin Z. Performance of Clostridium difficile toxin enzyme immunoassay and nucleic acid amplification tests stratified by patient disease severity. J Clin Microbiol. 2013;51(3):869–873. doi: 10.1128/JCM.02970-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Berry N, Sewell B, Jafri S, et al. Real-time polymerase chain reaction correlates well with clinical diagnosis of Clostridium difficile infection. J Hosp Infect. 2014;87(2):109–114. doi: 10.1016/j.jhin.2014.03.005. [DOI] [PubMed] [Google Scholar]
- 46.Lashner BA, Todorczuk J, Sahm DF, Hanauer SB. Clostridium difficile culture–positive toxin-negative diarrhea. Am J Gastroenterol. 1986;81(10):940–943. [PubMed] [Google Scholar]
- 47.Sydnor ER, Lenhart A, Trollinger B, et al. Antimicrobial prescribing practices in response to different Clostridium difficile diagnostic methodologies. Infect Control Hosp Epidemiol. 2011;32(11):1133–1136. doi: 10.1086/662381. [DOI] [PubMed] [Google Scholar]
- 48.Davies KA, Longshaw CM, Davis GL, et al. Underdiagnosis of Clostridium difficile across Europe: the European, Multicentre, Prospective, Biannual, Point-Prevalence Study of Clostridium difficile Infection in Hospitalised Patients With Diarrhoea (EUCLID). Lancet Infect Dis. 2014;14(12):1208–1219. doi: 10.1016/S1473-3099(14)70991-0. [DOI] [PubMed] [Google Scholar]
- 49.Updated guidance on the diagnosis and reporting of Clostridium difficile. Department of Health; United Kingdom: [July 8, 2015]. https://www.gov.uk/government/publications/updated-guidance-on-the-diagnosis-and-reporting-of-clostridium-difficile. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.