Summary
Regular exercise is effective in the prevention of chronic diseases and confers a lower risk of death in individuals displaying risk factors such as hypertension and dyslipidaemia. Thus, knowledge of the molecular responses to exercise provides a valuable contrast for interpreting investigations of disease and can highlight novel therapeutic targets. While exercise is an everyday experience and can be conceptualized in simple terms, exercise is a complex physiological phenomena and investigation of exercise responses requires sophisticated analytical techniques and careful standardization of the exercise stimulus. Proteomic investigation of exercise is in its infancy but the ability to link changes in function with comprehensive changes in protein expression and post-translational modification holds great promise for advancing physiology. This review highlights recent pioneering work investigating the effects of exercise in skeletal and cardiac muscle that has uncovered novel mechanisms underling the benefits of physical activity.
Keywords: Skeletal muscle proteomics, cardiac muscle proteomics, exercise training, cardioprotection, 2-D gel electrophoresis, mass spectrometry, post-translational modification, heat shock proteins, ATP synthase beta.
Background
Regular exercise is fundamental to maintaining health throughout the lifespan and an individual’s maximum aerobic capacity (i.e. VO2max) is an independent indicator of their health status [1]. Low aerobic capacity is a risk factor for diseases, including type 2 diabetes mellitus and cardiovascular disease [2], and a stronger predictor of early mortality than other established risk factors such as cigarette smoking or hypertension [3]. Accordingly, regular exercise prevents chronic diseases and confers a lower risk of death in individuals displaying the established risk factors of cigarette smoking, hypertension and dyslipidaemia [4]. Whole-body aerobic capacity is determined by central (i.e. cardiovascular) and peripheral (i.e. skeletal muscle) components and regular physical activity produces specific molecular adaptations in both cardiac and skeletal muscles that confer protection against chronic disease.
A large body of literature exists regarding adaptations in striated muscle in response to exercise. In the majority this information arises from investigations testing specific hypotheses using reductionist approaches, and these works have illuminated many fundamental features of exercise-induced adaptation. More latterly, post-genomic techniques, such as gene expression profiling using microarrays, have been applied in exercise physiology. In particular, transcriptome analyses have illustrated extensive differences in mRNA expression between muscles of trained and untrained individuals [5, 6] and brought forth new insight of the complexity of molecular responses to exercise (e.g. [7–9]). In light of this knowledge it is clear physiological phenomena cannot be explained at the level of individual genes or signaling pathways, instead, networks of gene circuits interact to bring about changes in function. Thus, systems biology approaches embracing comprehensive analysis hold the potential to extend the boundaries of knowledge.
While transcriptome analysis is a powerful discovery tool the quantitative link between the abundance of a protein and the expression of its gene can be tenuous [10] and protein analysis is necessary to verify mRNA findings. Such validation may be especially important when investigating exercise because muscle activity affects the rate of protein turnover [11] and induces changes in post-transcriptional processing [12], translational regulation [13], and protein degradation [14] that may each influence the quantitative relationship between the abundance of a protein and the transcription of its gene. Further to validation of mRNA findings, proteomic analysis is necessary to discover post-translational modifications that can alter the functional characteristics of the cell in the absence of changes in gene transcription.
Technological developments including robust two-dimensional (2-D) gel electrophoresis and mass spectrometry (MS) now enable large-scale investigations and inductive ‘discovery science’ approaches to be applied at the protein level [15]. However, because proteins exhibit broad differences in physical-chemical properties and copy number, comprehensive analysis is difficult particularly in skeletal muscle, which expresses a relatively small number of myofibrillar proteins in high abundance. At present, proteome analysis cannot resolve the large numbers of features detected by microarray technology; in particular, low abundance proteins and hydrophobic membrane proteins may be absent in 2-D gel investigations. Nevertheless, the proteome is the interface between the genome and the environment, and is what defines a cell and dictates its functional characteristics. Thus, purposeful exploration of muscle proteome responses to exercise has the potential to advance physiology and our understanding of the health benefits of a physically active lifestyle. Excellent reviews of proteomic analyses of cardiac [16] and skeletal [17] muscle have recently been published in this journal and only a brief contextual introduction is included here prior to considering recent findings relating to exercise.
Proteomic Analysis of Striated Muscle
Skeletal muscle comprises a heterogeneous mixture of myofibres, in human muscle 3 fibre subtypes are recognized based on their contractile and metabolic properties [18]. Fast-twitch fatigable fibres rely predominantly on glycolytic metabolism and are designated FG (fast glycolytic), whereas, fast-twitch fatigue-resistant and slow-twitch fibres have relatively greater mitochondrial content and are designated FOG (fast oxidative glycolytic) and SO (slow oxidative), respectively. The fibre type-specific differences in contractile function are due to differential expression of a diverse range of isoforms of each myofibrillar protein [19]. Myosin heavy chain (MyHC) isoforms are intimately associated with myofibre contractile and energetic properties and are commonly used molecular markers: FG fibres express MyHC IIx/d, FOG fibres express MyHC IIa and SO fibres express MyHC I, which is also the predominant isoform in adult human myocardium. Phenotyping of muscle based on its profile of FG, FOG and SO fibres or relative expression of MyHC isoforms has provided important insights regarding muscle function and plasticity, but such characterization is relatively simplistic. For example, changes in isoform expression, splice variation or post-translational modification of other myofibrillar proteins, such as troponins and myosin light chains, can alter contractile characteristics in the absence of changes in MyHC. Moreover, each myofibre type is metabolically heterogeneous and can exhibit a broad spectrum of different metabolic enzyme concentrations that determines substrate utilisation and resistance to fatigue.
Proteomics affords the capability to analyse large numbers of contractile and metabolic proteins simultaneously and, therefore, develop a more sophisticated understanding of muscle diversity and plasticity. Early investigations produced 2-D gel maps of rodent heart [20] and mixed-fibre gastrocnemius muscle [21], and compared [22–25] archetypal muscles such as rat extensor digitorum longus (EDL) and soleus that comprise predominantly fast- or slow-twitch fibres, respectively. These and other proteome mining works provide important catalogues of accessible muscle proteins and extend earlier biochemical and histochemical studies describing muscle diversity. Indeed, proteome mining of human muscle [26] reveals the relative expression of many myofibrillar and metabolic proteins lies within the extremes observed in archetypal rodent fast- and slow-twitch muscle, which is consistent with the differences in myofibre profile between rodents and humans [18].
The plasticity of muscle in response to changes in activity is elegantly demonstrated by chronic low-frequency stimulation (CLFS), which is capable of transforming fast-twitch muscle to a slow contractile phenotype [27]. Proteome studies [28, 29] employing CLFS of rabbit tibialis anterior clearly illustrate the spectrum of muscle plasticity and have identified novel biomarkers of muscle transformation. However, voluntary muscle contraction involves ordered recruitment of muscle motorunits [30] corresponding with exercise intensity (as opposed to CLFS, which stimulates the muscle motorunit pool uniformly) and even the most rigorous program of sports training cannot approach the level of activity imposed by CLFS. Nevertheless, voluntary activity results in overt changes in muscle function, which are associated with protection from disease. For example, proteomic analysis [31] of muscle from elderly adults living a physically active lifestyle reveals robust differences in the expression of mitochondrial enzymes and preservation of insulin sensitivity compared to sedentary counterparts. Furthermore, comparative analysis of human upper-and lower-limb muscles [32] reveals that enzymes involved in energy metabolism are more abundant in vastus lateralis than deltoid, despite only moderate differences in MyHC isoform expression between the two muscles. This difference, which is consistent with the role of vastus lateralis in locomotion, clearly demonstrates that the muscle proteome is more closely related to muscle function than is myofibre profile.
Adaptation of the skeletal muscle proteome to exercise
Responsiveness to exercise training can vary widely in humans [33, 34] and this imparts considerable difficulty to investigating exercise-induced adaptations. Moreover, the implausibility of sampling some tissues from healthy humans necessitates the use of animal models to enable access to organs for molecular investigation and, to date, the majority of proteomic investigations of exercise have used laboratory rats and employed exercise models including forced treadmill running, swimming and voluntary wheel-running (Table 1). Muscle adaptation in response to exercise training is specific to the training stimulus, which comprises 3 interrelated variables: (i) intensity, (ii) duration and (iii) frequency. Exercise intensity is arguably the predominant component of the training stimulus, but swimming and free wheel-running models do not afford control of exercise intensity and this imparts difficulties in comparing data across these studies. In contrast, the a rodent model of intensity-controlled treadmill running [35, 36] enables exercise to be prescribed at intensities relative to each animal’s VO2max, which is consistent with best practice in human studies. Proteomics has been used to investigate skeletal muscle responses after either an acute exercise stress (Table 2) or a chronic period of training (Table 3). We will consider these two experiment designs separately because proteins that change immediately after acute exercise may be particularly involved in supporting muscle activity or the processes of recovery and adaptation, whereas, protein differences between trained and untrained muscles may relate more closely with differences in muscle function.
Table 1. Proteomic investigations of exercise.
| Species | Exercise mode | Tissue | Exercise regimen: intensity, duration and frequency. | Ref. |
|---|---|---|---|---|
| Rat | Running | Heart | Progressive endurance training (6 wk): 1-min intervals at treadmill speeds of 24 m/min to 66 m/min, 5 days per week. | [50] |
| Progressive endurance training (6 wk) at 70-75 % VO2max for 30 min, 4 days per week. | [36] | |||
| Endurance training: 30 m/min at 0 % grade, 60 min per day for 5 days. | [63] | |||
| Eight weeks endurance training progressing from 10 m/min, 5 % grade for 10 min to 16 m/min for 50 min performed 5 days per week. | [67] | |||
| Plantaris muscle | Endurance training (5 wk) at 70-75 % VO2max for 30 min, 4 days per week. | [35] | ||
| Gastrocnem ius muscle | Graded exercise test commencing at 12 m/min and encompassing incremental (1-3 min stages) increases in running speed until exhaustion. | [39] | ||
| Hippocamp us | Voluntary wheel running for 5 consecutive nights. | [109] | ||
| Treadmill running, two 20-min bouts per day at 20 m/min or Voluntary wheel running. | [110] | |||
| Cerebellum | Progressive endurance training (3 wk) commencing at 6 m/min and increasing to 15 m/min for 20 min, 7 day per week. | [111] | ||
| Swimming | Gastrocnem ius muscle | Three 30-min bouts interspersed by 10 min rest, total 150 min exercise. | [37] | |
| Single 3-min bout of weighted (10 % body weight) swimming. | [38] | |||
| Epitrochlea ris muscle | Fourteen 20-sec bouts of weighted (14 % body weight) swimming interspersed by 10-sec rest periods. | [43] | ||
| Heart | Progressive training (8 wk): two bouts per day of 15 min to 120 min duration, 6 days per week. | [56] | ||
| Human | Running | Vastus lateralis muscle | High-intensity interval training (6 wk): six 1-min bouts at 100 % VO2max interspersed by 4 min at 50 % VO2max, 3 days per week. | [44] |
| Vibration | Vastus lateralis and soleus muscle | Two 6-min bouts per day (ranging from 19 Hz to 25 Hz) for 89 sessions. | [112] | |
| Mixed | Urine | In-competition doping control samples from athletes participating in endurance, strength or team sports. | [102] | |
| Tai Chi Chuan | Blood | Tai Chi Chuan (tai chi 37 forms) performed 3 times a week for 12 weeks. | [113] | |
| Eccentric resistance | Eccentric contractions of biceps femoris muscle: 3 sets of 10 repetitions using a load equivalent to 8-10 repetition maximum. | [100] | ||
| Horse | Running | Gluteal muscle | Three 5-min bouts of exercise progressing from 60 % to 100 % VO2max over a 15-wk period. | [114] |
| Vastus lateralis muscle | Progressive training (18 wk) from 60 % to 80 % of estimated heart rate maximum, 4 days per week. | [115] | ||
| Rabbit | Running | Renal cortex | Endurance training (12 wk): treadmill speed of 18 m/min, 0 % grade for 60 min periods 5 day per week. | [116] |
| Pig | Running | Blood | Endurance training (14 wk): 65-75 % maximum hear rate for 30 min, 4 days per week. | [99] |
| Mouse | Running | Midbrain | Voluntary wheel running. | [117] |
Table 2. Skeletal muscle proteins modulated by acute exercise.
| Protein description | UniProt accession number | Publication reference |
|---|---|---|
| Glycolysis/ Glycogenolysis | ||
| Triosephosphate isomerase* | P48500 | [39] |
| Glyceraldehyde-3-phosphate dehydrogenase | P04797 | [39] |
| Pyruvate dehydrogenase E1 component beta | P49432 | [39] |
| Sarcomeric proteins | ||
| Troponin T, slow skeletal muscle* | Q7TNB2 | [38] |
| Alpha actin, skeletal muscle | P68136 | [39] |
| High-energy phosphate metabolism | ||
| Muscle creatine kinase* | P00564 | [38] |
| Adenylate kinase 1 | P39069 | [38] |
| Chaperone and heat shock proteins | ||
| Heat shock protein 20 | P97541 | [38] |
| Heat shock cognate 71 kDa protein | P63018 | [39] |
| Lipid metabolism & β-oxidation | ||
| Carnitine O-plamitoyltransferase II | P18886 | [39] |
| Transcriptional regulation | ||
| zinc finger protein 3 | Q8BLB0 | [37] |
Description and UniProt Knowledgebase accession number of proteins identified as differentially expressed in skeletal muscle after an acute bout of exercise.
Proteins that are also modulated by chronic exercise training (Table 3).
Table 3. Skeletal muscle proteins modulated by chronic exercise training.
| Protein description | UniProt accession number | Publication reference |
|---|---|---|
| Sarcomeric and calcium handling proteins | ||
| Troponin T, slow skeletal muscle* | Q7TNB2 | [44] |
| Troponin T, fast skeletal muscle | P50753 | [44] |
| Myosin light chain MLC1-f | P02600 | [43] |
| Myosin light chain, phosphorylatable, fast skeletal muscle | P04466 | [43] |
| Myosin regulatory light chain 2v | P08733 | [35] |
| Parvalbumin | P02625 | [43] |
| Glycolysis/ Glycogenolysis | ||
| Alpha enolase | P04764 | [35] |
| Beta enolase | P13929 | [44] |
| Phosphoglucomutase 1 | P38652 | [35] |
| Triosephosphate isomerase* | P48500 | [35] |
| Muscle glycogen phosphorylase | P09811 | [43] |
| Lactate dehydrogenase A | P04642 | [35] |
| Tricarboxylic acid cycle | ||
| Acontiase 2 | Q63270 | [35] |
| Maltate dehydrogenase, mitochondrial | P04636 | [43] |
| Succinate CoA | B2GV06 | [35] |
| Succinate dehydrogenase [ubiquinone] flavoprotein subunit | P31040 | [44] |
| Oxoglutarate (alpha-ketoglutarate) dehydrogenase | Q5XI78 | [43] |
| Electron transport chain | ||
| ATP synthase alpha subunit | P25705 | [44] |
| ATP synthase beta subunit | P06576/ P10719 | [43, 44] |
| NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75 kDa | Q66HF1 | [43] |
| NADH dehydrogenase (ubiquinone) Fe-S protein 2 | Q641Y2 | [43] |
| Chaperone and heat shock proteins | ||
| Alpha B-crystallin | P23928 | [35] |
| 60 kDa heat shock protein | P10809 | [44] |
| Heat shock protein beta 1, 27 kDa heat shock protein | P04792 | [44] |
| Stress-70 protein, mitochondrial | P48721 | [43] |
| Transport proteins | ||
| Albumin | P02770/P02768 | [35, 44] |
| Transferin | P12346 | [35] |
| myoglobin | Q9QZ76 | [35] |
| Lipid metabolism & β-oxidation | ||
| Trifunctional enzyme subunit alpha | P40939 | [44] |
| High-energy phosphate metabolism | ||
| Muscle creatine kinase* | P06732 | [44] |
Description and UniProt Knowledgebase accession number of proteins identified as differentially expressed in skeletal muscle after a period of endurance training. Proteins are classified and ranked according to biological process.
Proteins that are also modulated after an acute bout of exercise (Table 2).
The first studies in exercise proteomics investigated the acute effects of swimming. For example, Takahashi et al. [37] performed 2-D electrophoresis (2-DE) on muscle pooled from rats killed immediately after 150 minutes swimming and reports lesser expression of a gel spot containing a putative transcription factor, AI854635, which has since been identified as zinc finger protein 3. In C2C12 myoblasts, the mRNA expression of zinc finger protein 3 decreases after serum deprivation [37], implicating this protein in the process of myoblast differentiation. However, as yet, the role of zinc finger protein 3 in the adaptation of differentiated adult muscle has not been elucidated. In a similar study Guelfi et al. [38] isolated gastrocnemius muscles from rats either immediately after or 30 min after a 3-min bout of swimming. Rather than pooled samples, biological replicates were analysed which enabled statistically significant differences to be identified in the expression of troponin T, creatine kinase and a spot containing both adenylate kinase 1 and heat shock protein 20 (hsp20). More recently, Gandra et al [39] investigated changes in rat gastrocnemius muscles isolated either 3 h or 24 h after an incremental treadmill test to exhaustion and reports significant changes in glycolytic enzymes, heat shock cognate protein 70 and carnitine palmitoyltransferase II. These changes observed in proteins involved in high-energy phosphate [38] and glycolytic [39] metabolism are consistent with the energy demands of intense exercise. However, because muscles were isolated at different time points or after differing bouts of acute exercise there is no direct identity among the findings (Table 2) of these studies.
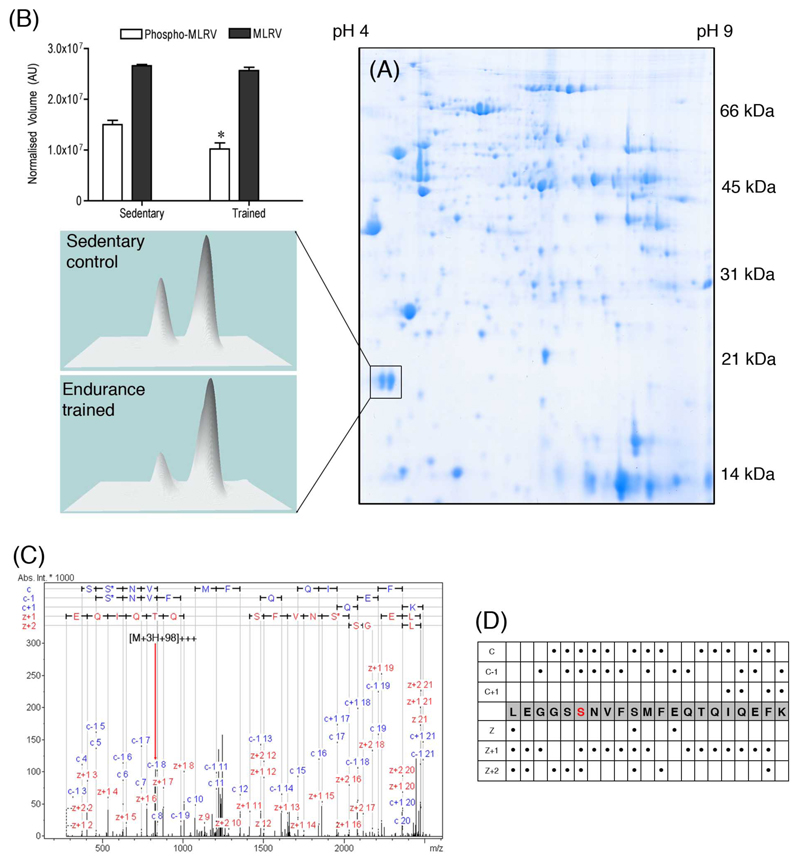
Changes in spot expression in the minutes to hours after exercise may be too rapid to be accounted for by differences in protein synthesis or degradation. Instead, post-translational modification may be the mechanism underlying the changes in spot expression. Covalent modifications, including phosphorylation and acetylation, alter the isoelectric point (pI) of a protein without significantly altering its Mr and result in the appearance of spots with patterns of migration that are markedly different from the non-modified form. Indeed, proteome mining studies consistently identify many muscle proteins as series of spots of similar Mr but different pI. Besides providing separation for MS analysis, this enables shifts in post-translational state to be quantified based on relative changes in spot volume (Figure 1).
Figure 1. Myosin regulatory light chain phosphorylation.
(A) Cardiac samples from sedentary and endurance-trained rats, used in [35], were separated using broad-range (pH 3-10) isoelectric focusing and denaturing polyacrylamide gel electrophoresis. (B) The normalised volume of spot, Mr ~20 kDa, pI ~4.7 was significantly less in endurance-trained hearts, while, no change occurred in the expression of the neighboring spot Mr ~ 20 kDa pI ~4.9. MS analysis of in-gel tryptic digests by nLC-ESI-MS/MS unambiguously identified each of these spots as the ventricular isoform of myosin light chain 2 (MLRV_RAT; P08733). Complementary CID and ETD analysis detected 90 % of the protein sequence and Mascot MOWSE MudPIT scores were 6,933 and 8,187 for the acidic and basic species, respectively. (C) Phosphorylated peptides (residues 10-30: LEGGSSNVFSMFEQTQIQEFK) were only detected in the relatively acidic isoelectric species (pI ~4.7), which was less abundant in endurance-trained hearts. (D) Interpretation of ETD spectra (dots represent observed c, c -1, c +1, z, z +1 or z +2 fragment ions) enabled the specific site of phosphorylation to be mapped to serine 15.
Recently, we [35] found a conspicuous effect of endurance training was to alter the relative expression of individual spots within multi-spot series; thus linking changes in the spot profile of individual proteins with differences in muscle function. Using image analysis we compared the relative expression of 187 protein spots matched across 2-D gels of muscles from 6 endurance-trained and 6 sedentary rats [35]. Proteins were unambiguously identified in 80 gel spots using peptide mass fingerprinting and confirmatory MALDI-ToF MS/MS. Statistical analysis detected significant changes in the expression of 15 gel spots, which represented the products of 11 individual genes. Six of the 11 differentially expressed genes were present as multiple protein spots. Endurance training altered the expression of single spots from each of the multi-spot series identified as transferrin (total of 3 spots), albumin (total of 4 spots) and lactate dehydrogenase A (total of 2 spots), and induced changes in multiple spots from series identified as phosphoglucomutase 1 (PGM1), triosephosphate isomerase and mitochondrial aconitase (ACON2).
When a protein is present in multiple gel spots, it can be erroneous to interpret a change in expression of an individual spot as being indicative of a change in the total abundance of the gene product. For example, ACON2 resolved as 5 discrete spots (Mr ~ 85 kDa, pI 7.5 - 8.0) and endurance training decreased the expression of 2 spots and increased the expression of a third [35]. Despite the marked differences in the spot profile, the sum volume of ACON2 was not different between sedentary and exercised muscle. Nonetheless, the changes in ACON2 expression may be biologically significant. Muscle activity is intimately associated with production of reactive oxygen species and ACON2 is particularly susceptible to metal-catalyzed oxidation. Using 2-DE and western blotting of derivatised proteins we found the shift in ACON2 spot pattern correlated with an increase in carbonylation, which previously has been associated with a loss of enzyme activity. Exercise-induced carbonylation of ACON2 seems inconsistent with its role in the tricarboxylic acid (TCA) cycle but inactivation of aconitase by oxidation may result in its association with mitochondrial DNA nucleoids, which stabilizes the mitochondrial genome during adverse conditions [40].
In the same experiment [35], PGM1 was resolved as 4 spots, and the statistically significant decrease in the normalised volume of 2 spots was accompanied by lesser (non statistically significant) decreases in the 2 other isoelectric species. In this case, the changes in spot expression indicate a general decrease in protein abundance in exercised muscle, and corresponded with a significant decrease in enzyme activity. PGM1 catalyses the conversion of glucose 1-phosphate and glucose 6-phosphate, important in glycogen metabolism, and this finding is consistent with a fundamental adaptation of muscle to endurance training. That is, trained muscle has a greater ability to utilize fatty acids, which spares the muscle’s limited glycogen stores, thus offsetting fatigue and prolonging endurance. Using histochemistry, we found the decreases in enzyme activity and expression of PGM1 were associated with greater glycogen content particularly in SO and FOG myofibres which were hypertrophied in exercise-trained muscle. Interestingly, the effect of exercise on PGM1 is opposite to the increase in PGM1 expression detected by proteomic analysis of muscle from type 2 diabetes patients [41]. So, although, PGM1 is not traditionally considered as a point of control in glycogen metabolism, these findings from unbiased proteomic investigations suggest modulation of PGM1 is a prominent feature that correlates with differences in muscle metabolism and health status.
The incorporation of high-intensity intervals within a training program is associated with more rapid and pronounced improvements in aerobic capacity and enhanced health-related benefits compared to continuous moderate exercise [42]. Yamaguchi et al. [43] investigated the effects in rat epitrochlearis muscle of 5 days of high-intensity interval training incorporating multiple 20-s bouts of weighted swimming interspersed by 10-s recovery periods. Difference in-gel electrophoresis (DIGE) was used to analyse muscle from 4 control and 4 exercise-trained animals. Using DIGE, independent samples labeled with Cy3 or Cy5 canine fluorescent dyes and a pooled internal standard labeled with Cy2 dye are combined equivalently and separated in each gel. This approach minimizes technical variation and affords quantitative expression analysis because each spot is normalised to a consistent internal standard. Yamaguchi et al. [43] reports significant increases in ATP synthase beta and subunits of Complex I in exercise-trained muscle, while, spots containing the soluble calcium-binding protein, parvalbumin, and fast isoforms of myosin regulatory and essential light chains were less abundant, consistent with a shift toward a more fatigue resistance aerobic phenotype.
Recently, we [44] also used high-intensity interval training to conduct the first proteomic investigation of the effects of exercise in human muscle. Healthy adult males completed a 6-wk training programme involving three sessions per week, utilizing six 1-min bouts at VO2max interspersed with 4 min at 50% VO2max. Using 2-DE, we resolved 256 protein spots in small (~15 mg) biopsy samples of vastus lateralis taken before and after the training regimen and, as expected, 2-DE resolved many proteins as multiple isoelectric species, offering the opportunity to learn new information about exercise-induced adaptation in humans. Myosin light chains were clearly resolved but were not differentially expressed as reported [43] in rat muscle. Instead, interval exercise in humans affected both post-transcriptional processing and post-translational modification of slow- and fast-isogenes of troponin T, which may relate to augmented excitation-contraction coupling specifically associated with interval training [42]. Of all myofibrillar proteins, troponin T exhibits the greatest diversity [19] and changes in expression, splice variation and post-translational modification of troponin T isogenes can alter myofibril Ca++-sensitivity and affect muscle contractility [45]. Based on time-series data [46] describing alterations in rat myofibrillar proteins during hindlimb suspension, changes in troponin T are an early event in muscle adaptation and a sensitive indicator of changes in function. Indeed, interval exercise resulted in robust modulation of troponin T proteins in the absence of significant changes in myofibre profile assessed by traditional MyHC isoform analysis [44]. In addition, we discovered interval training increases phosphorylation of muscle creatine kinase (KCRM). Enhanced phosphocreatine metabolism is consistent with the adaptation to sprint training [47], but the significance of post-translational modification of KCRM is not yet clear. During the initial seconds of exercise, the KCRM reaction is the main contributor to ATP re-synthesis and can affect the rate of increase in oxygen consumption [48], as well as modulate the activation of glycogenolysis and glycolysis. The change in post-translational state of KCRM associated with interval training suggests regulatory mechanisms exist that may influence muscle energy metabolism at the onset of exercise by modifying KCRM.
To date, proteomic analyses of exercise-trained skeletal muscle (Table 3) have mostly detected changes in sarcomeric proteins, metabolic enzymes and molecular chaperones. Some of the proteins differentially expressed after acute exercise (i.e. triosephosphate isomerase, troponin T and muscle creatine kinase; Table 2) are also modulated in exercise-trained muscle (Table 3). In addition, exercise training is associated with modulation of the tricarboxylic acid cycle and electron transport chain enzymes, which is not evident after acute exercise. The metabolic enzymes listed in Table 3 are not regarded as being rate-limiting in their respective metabolic pathways but, nonetheless, these unbiased proteomic findings may point to important information regarding changes in muscle metabolism or other biological processes. In particular, 2 proteins, ATP synthase subunit-β and albumin, have been highlighted in studies of animal and human muscle and this places special significance on these findings. Heat shock proteins (hsp) also represent a notable portion of the proteins identified in proteomic analyses of skeletal (Tables 2 and 3). Exercise-induced changes in hsp expression are commonly observed in skeletal muscle [49] but the exact mechanistic role of such modifications remains to be fully elucidated. The ability of proteomics to measure changes in expression and post-translational modification across numerous hsp simultaneously will hopefully assist in deciphering their specific roles in exercise-induced adaptation.
Adaptation of the cardiac proteome to exercise
Boluyt et al. [50] reports the first MS-based proteomic investigation of the cardiac response to exercise. Rats underwent a 6-week regimen of endurance training incorporating progressive increments in exercise duration, which resulted in a 16 % increase in cardiac mass (i.e. adaptive cardiac hypertrophy). Boluyt et al. [50] investigated hearts isolated 24 h after cessation of exercise and 2-D gel expression profiling detected 26 statistically significant differences between sedentary and exercised animals, including 12 spots that were expressed exclusively in endurance-trained hearts. In particular, endurance training resulted in the expression of hsp20 that was not evident in hearts of sedentary rats. Furthermore, cardiac expression of hsp20 did not occur in response to a bout of unaccustomed exercise in previously sedentary animals, suggesting hsp20 expression is a component of exercise-induced adaptation rather than an acute stress response to treadmill running.
Reproducibility is a fundamental tenet of scientific experimentation and corresponding data from independent work can provide robust evidence that novel discoveries represent important biological phenomena. Using the rat model of intensity-controlled treadmill running we [36] investigated the effects of a 6-week training regimen consisting of 4, 30-minute sessions per week at ~75 % VO2max. To standardize the progression of the training stimulus each animal’s VO2max was measured at 2-week intervals during the training period and their absolute training intensity was increased inline with the increments in performance. This programme of endurance running increased the animals’ VO2max by an average of 23 % and was associated with an 11 % increase in cardiac mass. Consistent with Boluyt et al [50], a spot identified as hsp20 was significantly increased in hearts of endurance-trained animals. However, in our work [36], the hsp20 spot that was increased in endurance-trained hearts exhibited a lower than predicted pI, consistent with post-translational modification. Accordingly, MS/MS analysis unambiguously identified phosphorylation of hsp20 at serine 16, which is important in the interpretation of these findings.
Together, these independent proteomic investigations suggest endurance training is associated with an adaptive increase in cardiac expression of hsp20 [50], and endurance-trained hearts are characterized by greater transient phosphorylation of hsp20 in the hours immediately after exercise [36]. This may be an important mechanism underlying exercise-induced cardioprotection and phosphorylation of hsp20 at serine 16 has been highlighted as a therapeutic target in heart disease [51]. Introduction of a phosphopeptide analogue of hsp20 in vitro increases cardiomyocyte rate of shortening [52], and expression of a constitutively phosphorylated form of hsp20 protects cardiomyocytes from apoptosis induced by the protein kinase A pathway [53]. Moreover, transgenic overexpression of hsp20 enhances basal cardiac function, and can attenuate ischaemia/reperfusion injury [54], whereas, if phosphorylation of hsp20 is prevented the heart is more sensitive to ischaemia/reperfusion injury [55]. Pharmacological manipulation of hsp20 phosphorylation in humans has not yet been reported, nevertheless, moderate endurance exercise may represent a cost-efficient approach to achieving this beneficial molecular adaptation in a clinical setting.
Endurance training is associated with robust changes in the expression of enzymes involved in cardiac energy metabolism, in particular fatty acid oxidation (Table 4). Sun et al [56] first reported changes in the expression of β-oxidation enzymes, including short-chain acyl-CoA dehydrogenase, and components of the TCA cycle in the hearts of rats exposed to high-intensity swimming. Later, we [36] used moderate intensity treadmill running and measured increases in heart fatty acid binding protein (HFABP), short- and long-chain acyl-CoA dehydrogenases and mitochondrial thioesterase-1. Thus despite cardiac muscle being chronically active and characterized by high mitochondrial content, endurance training further increases the heart’s ability to oxidize fatty acids similar to the effect observed in skeletal muscle. Modulation of malate dehydrogenase and aspartate aminotransferase is a second prominent metabolic feature of the exercise-trained cardiac proteome (Table 4). These enzymes form the malate-aspartate shuttle that transports electrons to within the mitochondria and is also enhanced in exercise-trained skeletal muscle [57]. In addition, aspartate aminotransferase is identical to the plasma membrane fatty acid transporter (FABPpm) [58] so changes in this enzyme could also relate to long-chain fatty acid uptake.
Table 4. Cardiac muscle proteins modulated by endurance training.
| Protein description | UniProt accession number | Publication reference |
|---|---|---|
| Fatty acid transport and beta-oxidation | ||
| Heart Fatty acid binding protein | P07483 | [36] |
| Trifunctional enzyme subunit alpha* | P48500 | [63] |
| Long-chain acyl-CoA dehydrogenase | P15650 | [36, 63] |
| Short-chain Acyl-CoA dehydrogenase | P15651 | [36, 56] |
| Enoyl-CoA hydratase | P14604 | [56] |
| Hydroxyacyl-CoA dehydrogenase | QPWVK7 | [63] |
| Delta(3,5)-Delta(2,4)-dienoyl-CoA isomerase | Q62651 | [63] |
| Electron transport chain | ||
| NADH dehydrogenase (ubiquinone) alpha subcomplex | Q561S0 | [36, 56] |
| NADH dehydrogenase (ubiquinone) flavoprotein 1 | Q5XIH3 | [36] |
| ATP synthase D-chain | P31399 | [50] |
| ATP synthase O-subunit | Q06647 | [63] |
| ATP synthase subunit alpha* | P15999 | [56] |
| Electron transfer flavoprotein subunit alpha | P13803 | [36] |
| Electron transfer flavoprotein subunit beta | Q68FU3 | [56] |
| Malate-aspartate shuttle and tricarboxylic acid cycle | ||
| Malate dehydrogenase, cytoplasmic | O88989 | [36, 56, 63] |
| Aspartate aminotransferase | P00507 | [36, 63] |
| Citrate synthase | Q8VHF5 | [63] |
| Aconitase* | Q63270 | [36] |
| Isocitrate dehydrogenase [NAD] subunit alpha | Q99NA5 | [56] |
| Dihydrolipoamide S-succinyltransferase | Q01205 | [50] |
| Antioxidant and heat shock proteins | ||
| Heat shock protein 20 | P97541 | [36, 50] |
| Similar to amine oxidase | P21396 | [63] |
| Stress-70 Protein | P48721 | [56] |
| Antioxidant protein 2 | O35244 | [56] |
| Glutathione peroxidase 1 | P04041 | [56] |
| Thioredoxin-dependent peroxidase reductase | Q9Z0V6 | [63] |
| Sarcomeric proteins | ||
| Alpha actin | P68035 | [36] |
| Alpha cardiac myosin heavy chain | P02563 | [36] |
| Myosin light polypeptide 3 | P16409 | [56] |
| Nebulette | D4A4G1 | [36] |
| Glycolysis | ||
| Triosephosphate isomerase* | P48500 | [50, 56] |
| Similar to glyceraldehyde-3-phosphate dehydrogenase | Q498M9 | [50] |
| High energy phosphate metabolism | ||
| Creatine kinase Basic-type | P07335 | [36] |
| Creatine kinase M-type* | P00564 | [36] |
| Miscellaneous | ||
| 26S proteasome non-ATPase regulatory subunit 1 | O88761 | [36] |
| A kinase (PRKA) anchor protein 3 | Q66HC6 | [56] |
| Aldehyde reductase | P51635 | [56] |
| Alpha-2-HS-glycoprotein precursor (Fetuin-A) | P24090 | [36] |
| Four and a half LIM domains protein 2 | O35115 | [36] |
| Methylmalonate-semialdehyde dehydrogenase [acylating] | Q02253 | [63] |
| Prohibitin | P67779 | [56] |
| Similar to hydroxysteroid dehydrogenase like 2 | Q4V8F9 | [56] |
Description and UniProt Knowledgebase accession number of proteins identified as differentially expressed in cardiac muscle after a period of endurance training. Proteins are classified and ranked according to biological process.
Proteins that are also modulated in skeletal muscle after chronic exercise training (Table 3).
Adaptations in cardiac energy metabolism, in particular mitochondrial fatty acid oxidation, may be a prominent aspect of exercise-mediated cardioprotection. For example, the changes in mitochondrial enzymes induced by endurance training are generally opposite to the changes in the cardiac proteome associated with ageing [59, 60] or heart failure induced by pressure overload [61, 62]. Striated muscle mitochondria can be distinguished as sub-sarcolemmal (SS) and intermyofibrillar (IMF) populations and more recent proteomic work [63] has investigated exercise-induced changes in cardiac SS and IMF mitochondria, separately. iTRAQ analysis of mitochondrial peptides resolved using orthogonal 2-D HPLC-MS/MS detected increases in the expression of mitochondrial enzymes associated with fatty acid oxidation and the TCA cycle specifically in IMF mitochondria. In addition, Kavazis et al [63] discovered endurance training is associated with decreased expression of monoamine oxidase in both SS and IMF populations. Monoamine oxidase generates hydrogen peroxide through oxidation of noradrenaline and other neurotransmitters, and is implicated in pressure-induced maladaptive hypertrophy [64]. The reduction in monoamine oxidase may represent further a potential mechanism of cardioprotection against hypertension relating to enhanced cardiac β-adrenergic receptor responsiveness associated with endurance training [65].
Endurance exercise is beneficial in restoring cardiac function after myocardial infarction induced by coronary artery ligation [66]. Bansal et al [67] reports proteomic analysis of cardiac infarct regions from rats that either remained sedentary or performed a programme of endurance training in the weeks after infarction. Endurance exercise significantly improved cardiac function and was associated with greater expression of glutathione peroxidase-1 and manganese superoxide dismutase, which is consistent with literature linking redox-based mechanisms and exercise-induced cardioprotection [68]. However, heart failure in humans is a progressive and complex condition involving both inherited and environmental factors that co-occur with other complicated risks including hypertension, obesity, and insulin resistance. Thus, the use of acute models of cardiac failure (e.g. myocardial infarction induced by coronary artery ligation) is limited and genetic models involving overexpression or deletion of individual genes do not encompass the complexity of chronic disease. Some years ago, Koch and Britton began artificial selection of rats for low or high aerobic exercise capacity [69]. This work has produced two populations of rats: high capacity runners (HCR) and low capacity runners (LCR) that have the potential to illuminate complex mechanisms underlying diseases associated with low cardiorespiratory fitness. Sedentary HCR and LCR exhibit substantial differences in aerobic capacity, disease susceptibility and mortality rate. For example, LCR exhibit features of metabolic syndrome [70] and are more susceptible to environmental challenges such as ischaemia/reperfusion [71], high-fat diet [72] or hypoxia [73]. In contrast, HCR closely resemble endurance-trained animals and are protected from environmental insults despite living a habitually sedentary lifestyle.
At the transcriptome level, LCR hearts exhibit greater expression of genes associated with glucose metabolism, whereas, genes involved in lipid oxidation are enriched in HCR [74]. This mimics the dichotomy observed between maladaptive and adaptive cardiac hypertrophy [75], but it is important to verify which of these features also occur at the protein level and to this end we recently conducted proteomic analysis of HCR-LCR hearts using DIGE. In many aspects the proteome and transcriptome findings were highly complementary but we found limited direct correspondence between the proteome and transcriptome data. This may be due to the greater resolution of microarray compared to DIGE or the fact that differences detected by DIGE may include post-translational modifications not evident at the transcriptome level. In agreement with the microarray findings each enzyme of the β-oxidation pathway was significantly more abundant in HCR hearts but contrary to the transcriptome dataset there was limited evidence to suggest up-regulation of glycolytic metabolism in LCR. Instead, changes in enzymes associated with ketone body and amino acid metabolism were enriched in LCR. Several features of the HCR cardiac proteome are identical to the effects of endurance training (Table 3) adding further evidence that modulation of cardiac energy metabolism is one of the key benefits of exercise. Interestingly, selection on high running capacity was not associated with elevated expression or phosphorylation of hsp20. Thus, despite similarities in cardiac energy metabolism, differences exist between the hsp response associated with acquired and innate increases in aerobic capacity, and phosphorylation of Hsp20 seems to be a specific training-induced adaptation.
Expert commentary
Exercise proteomics is an emerging discipline, first highlighted in a prospective of research in exercise sciences published in 2000 [76]. Since this time, a small number of original articles have been published reporting proteomic analysis of exercise (Table 1). These pioneering works provide important new information regarding exercise-induced adaptation. In the majority, proteomic analyses of skeletal (Tables 2 and 3) and cardiac (Table 4) muscle have detected changes in metabolic enzymes, myofibrillar components, chaperones and anti-oxidant enzymes. The conclusions from these discovery-based investigations align closely with existing knowledge from hypothesis-led research. Moreover, it is encouraging to note that proteomic analysis of exercise-induced muscle adaptation has uncovered new information whilst providing confirmation, at the protein level, for previously reported changes in gene transcription and simultaneously presenting well-established adaptations that allow the novel findings to be aligned with the wider literature.
Muscle biochemistry has a long history and it may be tempting to assume knowledge regarding abundant muscle proteins is complete. However, shifts in the 2-DE profile of multi-spot proteins are a conspicuous response to exercise and have enabled proteomic investigations to uncover novel information at the level of post-translational modification. The combination of 2-DE and MS-based protein identification, is a powerful investigative tool capable of detecting detailed information without knowledge a priori regarding the identity of the protein or the type of covalent modification. As yet, this approach has not been fully exploited and the identification of state-specific post-translational modifications represents a key area where 2-DE and functional proteomics can contribute knowledge. However, to achieve this goal, sophisticated MS/MS analysis is necessary to map covalent modifications to specific residues, which is crucial to correctly interpret the biological significance of these discoveries.
Phosphorylation is a key regulatory mechanism in striated muscle and is a commonly studied post-translational modification (e.g. [77–82]). Nevertheless, MS/MS mapping of phosphopeptides is challenging because loss of the relatively labile phosphate during fragmentation using collision-induced dissociation (CID) makes it difficult to determine the site of the modification [83]. Electron-transfer dissociation (ETD) is a complementary fragmentation technique that is highly specific for the N-Cα bond and produces MS/MS spectra composed of c- and z-ions. Importantly, covalent modifications to side-chains, such as phosphorylation of serine, threonine and tyrosine residues, remain intact during ETD fragmentation of the peptide backbone and this greatly facilitates unambiguous identification of the specific site of modification. For example, ETD analysis was crucial in mapping phosphorylation of ventricular myosin regulatory light chain (MLRV) to serine 15 (Figure 1). Phosphorylation of MLRV is necessary for normal cardiac function and responsiveness to β1-AR stimulation [84]. Specifically, phosphorylation of MRLV determines the kinetics of force development and Ca++-sensitivity of force production in cardiomyocytes [85]. Proteomic analysis of hearts from animals used in [35] revealed endurance training decreases MLRV phosphorylation, which may relate to the improved myocardial Ca++-handling associated with endurance training [86].
To make full use of the information from functional proteomics studies, new practices and bioinformatic tools are required to integrate 2-DE multi-spot profiles and MS/MS-based identification of site-specific modifications with information regarding changes in biological function. Presently, the majority of 2-D gel investigations only report the identity of spots highlighted as significantly altered by differential image analysis software. In many cases, these individual spots belong to multi-spot series and, as we have discussed, the practice of reporting changes in the expression of individual spots as being synonymous with a change in the total abundance of that protein may be unjustified. Proteome mining and greater use of 2-D gel databases, such as World-2DPAGE; http://world-2dpage.expasy.org/repository/ will help to highlight which spots are commonly observed as multiple isoelectric species and this should guide the interpretation of new findings. Furthermore, integration of 2-D gel maps with available databases of MS-based protein identifications (e.g. PRIDE; http://www.ebi.ac.uk/pride/) and post-translational modifications (e.g. PhosphoSite; http://www.phosphosite.org/) will enable more complete interpretation that will lead to greater insight. Presently, bioinformatic tools specifically suited to functional proteomics are relatively underdeveloped and this has lead to the application of gene pathway analysis tools to proteomic data. However, unlike mRNA expression, 2-D gel data reporting changes across isoelectric subspecies of a gene product may not be dichotomous (i.e. do not depict either an increase or decrease in total protein abundance). Moreover, an increase in a particular gel spot may relate to relatively greater abundance of an inactive form of that protein, and represent a decrease in biological activity. Because of these potential false assumptions, interpretation of proteomics data using current pathway analysis suites is fundamentally flawed and further development of protein ontology and pathway analysis tools (e.g. iProClass; http://pir.georgetown.edu/iproclass/) is required in order to fully exploit functional proteomics data.
Implicit in the term “proteomics” is the desire to analyse the entire protein complement of a cell or tissue. In the case of striated muscle, this objective is not readily achieved and current exercise-proteomics literature is based on separations of about 60 [38] to 1200 [56] protein spots. DIGE, which is considered the gold standard in 2-D gel proteomics and can resolve about 800 [43] to 3500 [87] spots and offers the opportunity to delve deeper in to the proteome. Nonetheless, because 2-DE separates many proteins as multi-spot series the number of non-redundant identifications may be little more than one-half of this number and, at best, the current literature considers only the top 5 % of the most abundant muscle protein species. Besides limited dynamic range, hydrophobic membrane proteins and proteins at the extremes of Mr and pI are poorly resolved using 2-DE. In an attempt to overcome these difficulties we [44, 88, 89] and others [90, 91] have employed size (Mr) fractionation by 1-DE and solution-based separation of tryptic peptides using reverse-phase HPLC. For example, by combining this approach with iTRAQ labelling we [44] were able to detect significant changes in the expression of mitochondrial enzymes in exercise-trained human muscle, which were not observed using 2-DE.
When coupled with high mass-accuracy MS, solution-based approaches can identify almost 5000 proteins in cardiac muscle [92] and about 1000 [90] to 2000 [91] skeletal muscle proteins and provide a tantalizing preview of in-depth muscle proteomics, including regulatory proteins (e.g protein kinase A, calcium calmodulin dependent protein kinase, 5‘AMP activated kinase) important in exercise-induced adaptation. However, while solution-based approaches excel in proteome mining, their reproducibility is relatively poor and, in the majority, peptides that can be detected across multiple samples (i.e. can be included in statistical analyses) represent proteotypic peptides of a protein, which are less likely to provide information regarding post-translational modification. Specifically, solution-based workflows often encounter problems of missing data because, despite analysing multiple replicates, many proteins are identified with low sequence coverage. Thus, when employed in differential expression profiling, less than one-third of the proteins identified can be compared across groups. For example, Hwang et al. [93], identified > 1200 proteins in muscle from lean, obese and type 2 diabetic patients but only 400 of these were present in at least half of the samples investigated and statistical analysis was conducted on only 92 proteins. Nonetheless, Hwang et al. [93] reports novel differences in intermediate filament and microtubule proteins providing new mechanistic insight that may not have been uncovered based on hypothesis-led reasoning. Thus, further development of these techniques is warranted.
In one respect, the limited resolution of striated muscle proteomics is an obstacle to understanding the intricacies of muscle adaptation. However, this feature, along with the reasonable accessibility of skeletal muscle, makes muscle proteomics attractive for biomarker discovery because proteins important in muscle function and whole-body metabolism (i.e. sarcomeric components and metabolic enzymes) are highly enriched. Individually, mechanistic markers such as ‘key’ rate-limiting enzymes may be of limited use, and imbalances within metabolic pathways caused by disease may alter which enzymatic reactions predominate resulting peculiar changes in enzyme stoichiometry. Proteomics affords unbiased and comprehensive analysis extending beyond traditional myofibre phenotyping and proteomic data may highlight new features of interest. Considering the central role of oxygen metabolism in biological complexity [94] and the close correlation between aerobic capacity and health, molecular features associated with endurance exercise should be useful in the selection of biomarkers that distinguish between health and disease.
Tables 2-4 summarize proteins modulated in skeletal and cardiac muscle in response to exercise. Notably few proteins have been identified in more than one investigation, particularly in skeletal muscle, where differences in exercise intensity affect the nature of adaptation more so than in the heart. Nevertheless, some common features emerge and the unbiased nature by which these data were collected provides some validation of their biological importance. These markers may provide key mechanistic insight or be surrogates for mechanisms that are more difficult to detect. For example, changes in subunits of NADH dehydrogenase and F1-ATPase are regularly detected in exercised heart and skeletal muscle. The subunits of NADH dehydrogenase and F1-ATPase highlighted in proteomic studies each protrude in to the mitochondrial matrix and may simply be accessible biomarkers for general changes in electron transport chain proteins, most of which are integral to the inner mitochondrial membrane and may not be resolved using standard 2-DE. Alternatively, these findings may represent new mechanistic insight. For example, ATP synthase is specifically regulated in health vs disease [95] and exercise-induced changes in skeletal muscle ATP synthase-β (ATPB) [43, 44] oppose those reported [41] in proteomic analysis of muscle from diabetes patients. Changes in ATPB expression may correspond with enhanced Ca++-calmodulin-dependent kinase II activity [96] and cardiac preconditioning is associated with specific phosphorylation of ATPB [97, 98], which could regulate the activity of the F1-ATPase. Therefore, post-translational modification of the β-subunit of the F1-ATP synthase may be an important mechanism underling muscle metabolic disease.
Five-year view
Proteomics is a technology-driven discipline and immediate advances in exercise proteomics will come from the further application of DIGE in combination with sub-cellular fractionation, or more sophisticated solution-based proteomics that incorporate enrichment of specific post-translational modifications. For example, phosphorylation events can directly modulate electron transport chain activity and LC-MS/MS analysis of TiO2-enriched fractions [82] can detect more than 200 phosphorylation sites in mitochondrial proteins. Future combination of such enrichment strategies with iTRAQ labelling could be used to address some of the current limitations and enable exercise-induced post-translational modifications to be quantified using solution-based techniques.
The comparison of results between laboratory rodents and humans is confounded by differences in myofibre phenotype and basal metabolic rate [18]. Therefore, while obtaining human skeletal muscle by percutaneous needle biopsy may be regarded as more difficult, the importance of understanding the health benefits of exercise and the complexity of exercise-induced adaptation warrant further carefully conducted studies directly in humans rather than translation of data from laboratory rodents. In that regard, proteomic techniques offer the advantage of comprehensive analysis despite limited sample volume and are a highly efficient means of investigating the protective effects of exercise against human diseases. Likewise, proteomic analysis of human body fluids offers an exciting opportunity to develop accessible biomarkers directly in humans that may be relevant in clinical and sports training scenarios. For example, proteome profiling of blood [99, 100], saliva [101] or urine [102] may reveal novel diagnostic markers indicative of the health-related benefits of physical activity or the effects of doping or underperformance syndrome in elite athletic populations.
Muscle proteome changes in response to ageing and disease have recently been reviewed [16, 17] and herein we have considered responses to exercise. The health benefits of exercise are irrefutable and an important next step will be to compare the effects of different exercise regimens in healthy and diseased populations. To date, proteomic investigations have focused on striated muscle responses to endurance training, which is associated with health benefits underpinned by improvements in aerobic capacity. Training involving resistance exercise also benefits health [103] and helps to preserve muscle function during ageing [104]. As yet, the effects of resistance training have not been investigated using proteomics and this is a clear priority for future work. Proteomic data exist regarding postnatal muscle growth [105] and hypertrophy associated with pharmacological agents [106] or experimental interventions [107] but resistance exercise comprises complex mechanical, metabolic and hormonal signals and is unlikely to be recapitulated by such models. Moreover, many athletic disciplines and recreational activities involve a mixture of both endurance and resistance work that affect muscle differently [108]. The comprehensive nature of proteomic analysis may bring new insight regarding optimization of such concurrent training programs, which remains a current issue in sports physiology and health-based exercise prescription.
Key Issues
Regular exercise brings about adaptations in heart and skeletal muscles that enhance aerobic capacity and are associated with protection against cardio-metabolic diseases. Post-genomic research has illustrated the complexity underlying physiological adaptation; in particular, microarray analysis has uncovered comprehensive changes in muscle gene transcription induced by exercise.
Proteomic techniques, including 2-D electrophoresis and mass spectrometry, have enabled a discovery science approach to be applied at the protein level, and have uncovered novel exercise-induced post-translational modifications that may affect the functional properties of muscle in the absence of changes in gene transcription.
Few of the proteins modulated by exercise have been identified in more than one investigation, particularly in skeletal muscle, where differences in exercise intensity affect the nature of adaptation more so than in the heart. Nonetheless, some common features emerge and the unbiased nature by which these data were collected provides some validation of their biological importance. In skeletal muscle, the most commonly modulated proteins include sarcomeric proteins, glycolytic enzymes and chaperone proteins, whereas cardiac adaptations include comprehensive changes in mitochondrial enzymes involved in fatty acid oxidation.
Proteomic analysis of cardiac muscle discovered that exercise enhances the expression and phosphorylation of heat shock protein 20, which has been associated with enhanced myocardial contractility and protection from cardiac insults such as catecholamine excess and ischemia/reperfusion injury.
Some of the changes highlighted in exercised muscle are opposite to the changes reported from proteomic analysis of muscle metabolic disease or cardiomyopathy, in particular, post-translational modification of F1-ATPase β-subunit is consistent across both cardiac and skeletal muscle.
Exercise proteomics is an emerging discipline and immediate advances in this field will come from further application of advanced analytical techniques including DIGE in combination with sub-cellular fractionation and further development of in-solution techniques such as 2-D LC-MS/MS. To date, proteomic investigations have focused on striated muscle responses to endurance training, which is associated with health benefits underpinned by improvements in aerobic capacity. Training involving resistance exercise also benefits health but, as yet, muscle adaptation in response to resistance training has not been investigated using proteomics and this is a clear priority for future work.
References
*Of interest.
**Of considerable interest.
- 1.Lakka TA, Venäläinen JM, Rauramaa R, Salonen R, Tuomilehto J, Salonen JT. Relation of leisure-time physical activity and cardiorespiratory fitness to the risk of acute myocardial infarction. N Engl J Med. 1994;330:1549–54. doi: 10.1056/NEJM199406023302201. [DOI] [PubMed] [Google Scholar]
- 2.Warburton DE, Nicol CW, Bredin SS. Health benefits of physical activity: the evidence. Cmaj. 2006;174:801–9. doi: 10.1503/cmaj.051351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Myers J, Prakash M, Froelicher V, Do D, Partington S, Atwood JE. Exercise capacity and mortality among men referred for exercise testing. N Engl J Med. 2002;346:793–801. doi: 10.1056/NEJMoa011858. [DOI] [PubMed] [Google Scholar]
- 4.Blair SN, Kampert JB, Kohl WH, 3rd, Barlow CE, Macera CA, Paffenbarger SR, Jr, Gibbons LW. Influences of cardiorespiratory fitness and other precursors on cardiovascular disease and all-cause mortality in men and women. Jama. 1996;276:205–10. [PubMed] [Google Scholar]
- 5.Stepto NK, Coffey VG, Carey AL, Ponnampalam AP, Canny BJ, Powell D, Hawley JA. Global gene expression in skeletal muscle from well-trained strength and endurance athletes. Med Sci Sports Exerc. 2009;41:546–65. doi: 10.1249/MSS.0b013e31818c6be9. [DOI] [PubMed] [Google Scholar]
- 6.Wittwer M, Billeter R, Hoppeler H, Flück M. Regulatory gene expression in skeletal muscle of highly endurance-trained humans. Acta Physiol Scand. 2004;180:217–27. doi: 10.1046/j.0001-6772.2003.01242.x. [DOI] [PubMed] [Google Scholar]
- 7.Chen YW, Hubal MJ, Hoffman EP, Thompson PD, Clarkson PM. Molecular responses of human muscle to eccentric exercise. J Appl Physiol. 2003;95:2485–94. doi: 10.1152/japplphysiol.01161.2002. [DOI] [PubMed] [Google Scholar]
- 8.Liu D, Sartor MA, Nader GA, Gutmann L, Treutelaar MK, Pistilli EE, Iglayreger HB, Burant CF, Hoffman EP, Gordon PM. Skeletal muscle gene expression in response to resistance exercise: sex specific regulation. BMC Genomics. 2010;11:659. doi: 10.1186/1471-2164-11-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mahoney DJ, Parise G, Melov S, Safdar A, Tarnopolsky MA. Analysis of global mRNA expression in human skeletal muscle during recovery from endurance exercise. Faseb J. 2005;19:1498–500. doi: 10.1096/fj.04-3149fje. [DOI] [PubMed] [Google Scholar]
- 10.Gygi SP, Rochon Y, Franza BR, Aebersold R. Correlation between protein and mRNA abundance in yeast. Mol Cell Biol. 1999;19:1720–30. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goldspink DF. Exercise-related changes in protein turnover in mammalian striated muscle. Journal of Experimental Biology. 1991;160:127–48. doi: 10.1242/jeb.160.1.127. [DOI] [PubMed] [Google Scholar]
- 12.Safdar A, Abadi A, Akhtar M, Hettinga BP, Tarnopolsky MA. miRNA in the regulation of skeletal muscle adaptation to acute endurance exercise in C57Bl/6J male mice. PLoS One. 2009;4:e5610. doi: 10.1371/journal.pone.0005610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen YW, Nader GA, Baar KR, Fedele MJ, Hoffman EP, Esser KA. Response of rat muscle to acute resistance exercise defined by transcriptional and translational profiling. J Physiol. 2002;545:27–41. doi: 10.1113/jphysiol.2002.021220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van Hall G, Saltin B, Wagenmakers AJ. Muscle protein degradation and amino acid metabolism during prolonged knee-extensor exercise in humans. Clin Sci (Lond) 1999;97:557–67. doi: 10.1042/cs19980422. [DOI] [PubMed] [Google Scholar]
- 15.Hittel DS, Hathout Y, Hoffman EP. Proteomics and systems biology in exercise and sport sciences research. Exerc Sport Sci Rev. 2007;35:5–11. doi: 10.1097/jes.0b013e31802d744a. [DOI] [PubMed] [Google Scholar]
- 16.Fu Q, Van Eyk JE. Proteomics and heart disease: identifying biomarkers of clinical utility. Expert Rev Proteomics. 2006;3:237–49. doi: 10.1586/14789450.3.2.237. [DOI] [PubMed] [Google Scholar]
- 17.Ohlendieck K. Proteomics of skeletal muscle differentiation, neuromuscular disorders and fiber aging. Expert Rev Proteomics. 2010;7:283–96. doi: 10.1586/epr.10.2. [DOI] [PubMed] [Google Scholar]
- 18.Schiaffino S. Fibre types in skeletal muscle: a personal account. Acta Physiol (Oxf) 2010;199:451–63. doi: 10.1111/j.1748-1716.2010.02130.x. [DOI] [PubMed] [Google Scholar]
- 19.Schiaffino S, Reggiani C. Molecular diversity of myofibrillar proteins: gene regulation and functional significance. Physiol Rev. 1996;76:371–423. doi: 10.1152/physrev.1996.76.2.371. [DOI] [PubMed] [Google Scholar]
- 20.Li XP, Pleissner KP, Scheler C, Regitz-Zagrosek V, Salnikow J, Jungblut PR. A two-dimensional electrophoresis database of rat heart proteins. Electrophoresis. 1999;20:891–7. doi: 10.1002/(SICI)1522-2683(19990101)20:4/5<891::AID-ELPS891>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 21.Sanchez J-C, Diego C, Converset V, Hoogland C, Binz P-A, Paesano S, Appel RD, Wang S, Sennitt M, Nolan A, Cawthorne MA, et al. The mouse SWISS-2D PAGE database: a tool for proteomics study of diabetes and obesity. Proteomics. 2001;1:136–63. doi: 10.1002/1615-9861(200101)1:1<136::AID-PROT136>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 22.Kim NK, Joh JH, Park HR, Kim OH, Park BY, Lee CS. Differential expression profiling of the proteomes and their mRNAs in porcine white and red skeletal muscles. Proteomics. 2004;4:3422–8. doi: 10.1002/pmic.200400976. [DOI] [PubMed] [Google Scholar]
- 23.Bihan LCM, Hou Y, Harris N, Tarelli E, Coulton GR. Proteomic analysis of fast and slow muscles from normal and kyphoscoliotic mice using protein arrays, 2-DE and MS. Proteomics. 2006;6:4646–61. doi: 10.1002/pmic.200500746. [DOI] [PubMed] [Google Scholar]
- 24.Okumura N, Hashida-Okumura A, Kita K, Matsubae M, Matsubara T, Takao T, Nagai K. Proteomic analysis of slow- and fast-twitch skeletal muscles. Proteomics. 2005;5:2896–906. doi: 10.1002/pmic.200401181. [DOI] [PubMed] [Google Scholar]
- 25.Gelfi C, Vigano A, De Palma S, Ripamonti M, Begum S, Cerretelli P, Wait R. 2-D protein maps of rat gastrocnemius and soleus muscles: a tool for muscle plasticity assessment. Proteomics. 2006;6:321–40. doi: 10.1002/pmic.200501337. [DOI] [PubMed] [Google Scholar]
- 26.Gelfi C, De Palma S, Cerretelli P, Begum S, Wait R. Two-dimensional protein map of human vastus lateralis muscle. Electrophoresis. 2003;24:286–95. doi: 10.1002/elps.200390025. [DOI] [PubMed] [Google Scholar]
- 27.Pette D, Vrbova G. Adaptation of mammalian skeletal muscle fibers to chronic electrical stimulation. Rev Physiol Biochem Pharmacol. 1992;120:115–202. doi: 10.1007/BFb0036123. [DOI] [PubMed] [Google Scholar]
- 28.Donoghue P, Doran P, Dowling P, Ohlendieck K. Differential expression of the fast skeletal muscle proteome following chronic low-frequency stimulation. Biochim Biophys Acta. 2005;1752:166–76. doi: 10.1016/j.bbapap.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 29.Donoghue P, Doran P, Wynne K, Pedersen K, Dunn MJ, Ohlendieck K. Proteomic profiling of chronic low-frequency stimulated fast muscle. Proteomics. 2007;7:3417–30. doi: 10.1002/pmic.200700262. [DOI] [PubMed] [Google Scholar]
- 30.Duchateau J, Semmler JG, Enoka RM. Training adaptations in the behavior of human motor units. J Appl Physiol. 2006;101:1766–75. doi: 10.1152/japplphysiol.00543.2006. [DOI] [PubMed] [Google Scholar]
- 31.Lanza IR, Short DK, Short KR, Raghavakaimal S, Basu R, Joyner MJ, McConnell JP, Nair KS. Endurance exercise as a countermeasure for aging. Diabetes. 2008;57:2933–42. doi: 10.2337/db08-0349. [*Cross-sectional comparison showing comprehensive differences between the muscle proteomes of young and elderly people with either sedentary or habitually active lifestyles.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Capitanio D, Vigano A, Ricci E, Cerretelli P, Wait R, Gelfi C. Comparison of protein expression in human deltoideus and vastus lateralis muscles using two-dimensional gel electrophoresis. Proteomics. 2005;5:2577–86. doi: 10.1002/pmic.200401183. [**Comparison of human upper- and lower-limb muscles that reveals overt differences in metabolic enzymes and demonstrates the muscle proteome more closely relates to muscle function than myofibre phenotype.] [DOI] [PubMed] [Google Scholar]
- 33.Bouchard C, Rankinen T. Individual differences in response to regular physical activity. Med Sci Sports Exerc. 2001;33:S446–51. doi: 10.1097/00005768-200106001-00013. discussion S452-3. [DOI] [PubMed] [Google Scholar]
- 34.Hubal MJ, Gordish-Dressman H, Thompson PD, Price TB, Hoffman EP, Angelopoulos TJ, Gordon PM, Moyna NM, Pescatello LS, Visich PS, Zoeller RF, et al. Variability in muscle size and strength gain after unilateral resistance training. Med Sci Sports Exerc. 2005;37:964–72. [PubMed] [Google Scholar]
- 35.Burniston JG. Changes in the rat skeletal muscle proteome induced by moderate-intensity endurance exercise. Biochimica et Biophysica Acta Proteins and Proteomics. 2008;1784:1077–86. doi: 10.1016/j.bbapap.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 36.Burniston JG. Adaptation of the rat cardiac proteome in response to intensity-controlled endurance exercise. Proteomics. 2009;9:106–15. doi: 10.1002/pmic.200800268. [**Investigated intensity-controlled exercise in rats similar to that advocated for maintaining health in humans and discovered trained hearts exhibit greater phosphorylation of heat shock protein 20 at serine 16, which has been identified as a therapeutic target in heart disease.] [DOI] [PubMed] [Google Scholar]
- 37.Takahashi M, Kubota S. Exercise-related novel gene is involved in myoblast differentiation. Biomed Res. 2005;26:79–85. doi: 10.2220/biomedres.26.79. [DOI] [PubMed] [Google Scholar]
- 38.Guelfi KJ, Casey TM, Giles JJ, Fournier PA, Arthur PG. A proteomic analysis of the acute effects of high-intensity exercise on skeletal muscle proteins in fasted rats. Clin Exp Pharmacol Physiol. 2006;33:952–7. doi: 10.1111/j.1440-1681.2006.04470.x. [DOI] [PubMed] [Google Scholar]
- 39.Gandra PG, Valente RH, Perales J, Pacheco AG, Macedo DV. Proteomic analysis of rat skeletal muscle submitted to one bout of incremental exercise. Scand J Med Sci Sports. 2010 doi: 10.1111/j.1600-0838.2010.01235.x. [DOI] [PubMed] [Google Scholar]
- 40.Shadel GS, Mitochondrial DNA. aconitase 'wraps' it up. Trends Biochem Sci. 2005;30:294–6. doi: 10.1016/j.tibs.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 41.Hojlund K, Wrzesinski K, Larsen PM, Fey SJ, Roepstorff P, Handberg A, Dela F, Vinten J, McCormack JG, Reynet C, Beck-Nielsen H. Proteome analysis reveals phosphorylation of ATP synthase beta -subunit in human skeletal muscle and proteins with potential roles in type 2 diabetes. J Biol Chem. 2003;278:10436–42. doi: 10.1074/jbc.M212881200. [DOI] [PubMed] [Google Scholar]
- 42.Tjonna AE, Lee SJ, Rognmo O, Stolen TO, Bye A, Haram PM, Loennechen JP, Al-Share QY, Skogvoll E, Slordahl SA, Kemi OJ, et al. Aerobic interval training versus continuous moderate exercise as a treatment for the metabolic syndrome: a pilot study. Circulation. 2008;118:346–54. doi: 10.1161/CIRCULATIONAHA.108.772822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamaguchi W, Fujimoto E, Higuchi M, Tabata I. A DIGE proteomic analysis for high-intensity exercise-trained rat skeletal muscle. J Biochem. 2010;148:327–33. doi: 10.1093/jb/mvq073. [DOI] [PubMed] [Google Scholar]
- 44.Holloway KV, O'Gorman M, Woods P, Morton JP, Evans L, Cable NT, Goldspink DF, Burniston JG. Proteomic investigation of changes in human vastus lateralis muscle in response to interval-exercise training. Proteomics. 2009;9:5155–74. doi: 10.1002/pmic.200900068. [**First MS-based proteomic investigation of the effects of exercise training in human muscle, which used gel-based and solution-based separation techniques and detected exercise-induced changes in ATP synthase subunit-β, which is in contrast to diabetic human muscle.] [DOI] [PubMed] [Google Scholar]
- 45.Ogut O, Granzier H, Jin JP. Acidic and basic troponin T isoforms in mature fast-twitch skeletal muscle and effect on contractility. Am J Physiol. 1999;276:C1162–70. doi: 10.1152/ajpcell.1999.276.5.C1162. [DOI] [PubMed] [Google Scholar]
- 46.Yu ZB, Gao F, Feng HZ, Jin JP. Differential regulation of myofilament protein isoforms underlying the contractility changes in skeletal muscle unloading. Am J Physiol Cell Physiol. 2007;292:C1192–203. doi: 10.1152/ajpcell.00462.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ross A, Leveritt M. Long-term metabolic and skeletal muscle adaptations to short-sprint training: implications for sprint training and tapering. Sports Med. 2001;31:1063–82. doi: 10.2165/00007256-200131150-00003. [DOI] [PubMed] [Google Scholar]
- 48.Korzeniewski B, Zoladz JA. Factors determining the oxygen consumption rate (VO2) on-kinetics in skeletal muscles. Biochem J. 2004;379:703–10. doi: 10.1042/BJ20031740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morton JP, Kayani AC, McArdle A, Drust B. The exercise-induced stress response of skeletal muscle, with specific emphasis on humans. Sports Med. 2009;39:643–62. doi: 10.2165/00007256-200939080-00003. [DOI] [PubMed] [Google Scholar]
- 50.Boluyt MO, Brevick JL, Rogers DS, Randall MJ, Scalia AF, Li ZB. Changes in the rat heart proteome induced by exercise training: Increased abundance of heat shock protein hsp20. Proteomics. 2006;6:3154–69. doi: 10.1002/pmic.200401356. [**First MS-based proteomic investigation of the effects exercise in the heart, which discovered exercise training increases the abundance of cardiac heat shock protein 20.] [DOI] [PubMed] [Google Scholar]
- 51.Fan GC, Chu G, Kranias EG. Hsp20 and its cardioprotection. Trends Cardiovasc Med. 2005;15:138–41. doi: 10.1016/j.tcm.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 52.Pipkin W, Johnson JA, Creazzo TL, Burch J, Komalavilas P, Brophy C. Localization, macromolecular associations, and function of the small heat shock-related protein HSP20 in rat heart. Circulation. 2003;107:469–76. doi: 10.1161/01.cir.0000044386.27444.5a. [DOI] [PubMed] [Google Scholar]
- 53.Fan GC, Chu G, Mitton B, Song Q, Yuan Q, Kranias EG. Small heat-shock protein Hsp20 phosphorylation inhibits beta-agonist-induced cardiac apoptosis. Circ Res. 2004;94:1474–82. doi: 10.1161/01.RES.0000129179.66631.00. [DOI] [PubMed] [Google Scholar]
- 54.Fan GC, Ren X, Qian J, Yuan Q, Nicolaou P, Wang Y, Jones WK, Chu G, Kranias EG. Novel cardioprotective role of a small heat-shock protein, Hsp20, against ischemia/reperfusion injury. Circulation. 2005;111:1792–9. doi: 10.1161/01.CIR.0000160851.41872.C6. [DOI] [PubMed] [Google Scholar]
- 55.Qian J, Ren X, Wang X, Zhang P, Jones WK, Molkentin JD, Fan GC, Kranias EG. Blockade of Hsp20 phosphorylation exacerbates cardiac ischemia/reperfusion injury by suppressed autophagy and increased cell death. Circ Res. 2009;105:1223–31. doi: 10.1161/CIRCRESAHA.109.200378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sun B, Wang JH, Lv YY, Zhu SS, Yang J, Ma JZ. Proteomic adaptation to chronic high intensity swimming training in the rat heart. Comparative Biochemistry and Physiology-Part D. 2008;3:108–17. doi: 10.1016/j.cbd.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 57.Hittel DS, Kraus WE, Tanner CJ, Houmard JA, Hoffman EP. Exercise training increases electron and substrate shuttling proteins in muscle of overweight men and women with the metabolic syndrome. J Appl Physiol. 2005;98:168–79. doi: 10.1152/japplphysiol.00331.2004. [DOI] [PubMed] [Google Scholar]
- 58.Roepstorff C, Helge JW, Vistisen B, Kiens B. Studies of plasma membrane fatty acid-binding protein and other lipid-binding proteins in human skeletal muscle. Proc Nutr Soc. 2004;63:239–44. doi: 10.1079/PNS2004332. [DOI] [PubMed] [Google Scholar]
- 59.Dai Q, Escobar GP, Hakala KW, Lambert JM, Weintraub ST, Lindsey ML. The left ventricle proteome differentiates middle-aged and old left ventricles in mice. J Proteome Res. 2008;7:756–65. doi: 10.1021/pr700685e. [DOI] [PubMed] [Google Scholar]
- 60.Grant JE, Bradshaw AD, Schwacke JH, Baicu CF, Zile MR, Schey KL. Quantification of protein expression changes in the aging left ventricle of Rattus norvegicus. J Proteome Res. 2009;8:4252–63. doi: 10.1021/pr900297f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Faber MJ, Dalinghaus M, Lankhuizen IM, Bezstarosti K, Dekkers DH, Duncker DJ, Helbing WA, Lamers JM. Proteomic changes in the pressure overloaded right ventricle after 6 weeks in young rats: correlations with the degree of hypertrophy. Proteomics. 2005;5:2519–30. doi: 10.1002/pmic.200401313. [DOI] [PubMed] [Google Scholar]
- 62.Bugger H, Schwarzer M, Chen D, Schrepper A, Amorim PA, Schoepe M, Nguyen TD, Mohr FW, Khalimonchuk O, Weimer BC, Doenst T. Proteomic remodelling of mitochondrial oxidative pathways in pressure overload-induced heart failure. Cardiovasc Res. 2010;85:376–84. doi: 10.1093/cvr/cvp344. [DOI] [PubMed] [Google Scholar]
- 63.Kavazis AN, Alvarez S, Talbert E, Lee Y, Powers SK. Exercise training induces a cardioprotective phenotype and alterations in cardiac subsarcolemmal and intermyofibrillar mitochondrial proteins. Am J Physiol Heart Circ Physiol. 2009;297:H144–52. doi: 10.1152/ajpheart.01278.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kaludercic N, Takimoto E, Nagayama T, Feng N, Lai EW, Bedja D, Chen K, Gabrielson KL, Blakely RD, Shih JC, Pacak K, et al. Monoamine oxidase A-mediated enhanced catabolism of norepinephrine contributes to adverse remodeling and pump failure in hearts with pressure overload. Circ Res. 2010;106:193–202. doi: 10.1161/CIRCRESAHA.109.198366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.MacDonnell SM, Kubo H, Crabbe DL, Renna BF, Reger PO, Mohara J, Smithwick LA, Koch WJ, Houser SR, Libonati JR. Improved myocardial beta-adrenergic responsiveness and signaling with exercise training in hypertension. Circulation. 2005;111:3420–8. doi: 10.1161/CIRCULATIONAHA.104.505784. [DOI] [PubMed] [Google Scholar]
- 66.Kemi OJ, Hoydal MA, Haram PM, Garnier A, Fortin D, Ventura-Clapier R, Ellingsen O. Exercise training restores aerobic capacity and energy transfer systems in heart failure treated with losartan. Cardiovasc Res. 2007;76:91–9. doi: 10.1016/j.cardiores.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 67.Bansal A, Dai Q, Chiao YA, Hakala KW, Zhang JQ, Weintraub ST, Lindsey ML. Proteomic analysis reveals late exercise effects on cardiac remodeling following myocardial infarction. J Proteomics. 2010 doi: 10.1016/j.jprot.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ascensao A, Ferreira R, Magalhaes J. Exercise-induced cardioprotection - biochemical, morphological and functional evidence in whole tissue and isolated mitochondria. Int J Cardiol. 2007;117:16–30. doi: 10.1016/j.ijcard.2006.04.076. [DOI] [PubMed] [Google Scholar]
- 69.Koch LG, Britton SL. Artificial selection for intrinsic aerobic endurance running capacity in rats. Physiol Genomics. 2001;5:45–52. doi: 10.1152/physiolgenomics.2001.5.1.45. [DOI] [PubMed] [Google Scholar]
- 70.Wisloff U, Najjar SM, Ellingsen O, Haram PM, Swoap S, Al-Share Q, Fernstrom M, Rezaei K, Lee SJ, Koch LG, Britton SL. Cardiovascular risk factors emerge after artificial selection for low aerobic capacity. Science. 2005;307:418–20. doi: 10.1126/science.1108177. [DOI] [PubMed] [Google Scholar]
- 71.Lujan HL, Britton SL, Koch LG, DiCarlo SE. Reduced susceptibility to ventricular tachyarrhythmias in rats selectively bred for high aerobic capacity. Am J Physiol Heart Circ Physiol. 2006;291:H2933–41. doi: 10.1152/ajpheart.00514.2006. [DOI] [PubMed] [Google Scholar]
- 72.Noland RC, Thyfault JP, Henes ST, Whitfield BR, Woodlief TL, Evans JR, Lust JA, Britton SL, Koch LG, Dudek RW, Dohm GL, et al. Artificial selection for high-capacity endurance running is protective against high-fat diet-induced insulin resistance. Am J Physiol Endocrinol Metab. 2007;293:E31–41. doi: 10.1152/ajpendo.00500.2006. [DOI] [PubMed] [Google Scholar]
- 73.Palpant NJ, Szatkowski ML, Wang W, Townsend D, Bedada FB, Koch LG, Britton SL, Metzger JM. Artificial selection for whole animal low intrinsic aerobic capacity co-segregates with hypoxia-induced cardiac pump failure. PLoS One. 2009;4:e6117. doi: 10.1371/journal.pone.0006117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bye A, Langaas M, Hoydal MA, Kemi OJ, Heinrich G, Koch LG, Britton SL, Najjar SM, Ellingsen O, Wisloff U. Aerobic capacity-dependent differences in cardiac gene expression. Physiol Genomics. 2008;33:100–9. doi: 10.1152/physiolgenomics.00269.2007. [DOI] [PubMed] [Google Scholar]
- 75.Strom CC, Aplin M, Ploug T, Christoffersen TE, Langfort J, Viese M, Galbo H, Haunso S, Sheikh SP. Expression profiling reveals differences in metabolic gene expression between exercise-induced cardiac effects and maladaptive cardiac hypertrophy. Febs J. 2005;272:2684–95. doi: 10.1111/j.1742-4658.2005.04684.x. [DOI] [PubMed] [Google Scholar]
- 76.Baldwin KM. Research in the exercise sciences: where do we go from here? J Appl Physiol. 2000;88:332–6. doi: 10.1152/jappl.2000.88.1.332. [DOI] [PubMed] [Google Scholar]
- 77.Boja ES, Phillips D, French SA, Harris RA, Balaban RS. Quantitative Mitochondrial Phosphoproteomics Using iTRAQ on an LTQ-Orbitrap with High Energy Collision Dissociation. J Proteome Res. 2009;8:4665–75. doi: 10.1021/pr900387b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gannon J, Staunton L, O'Connell K, Doran P, Ohlendieck K. Phosphoproteomic analysis of aged skeletal muscle. Int J Mol Med. 2008;22:33–42. [PubMed] [Google Scholar]
- 79.Lefort N, Yi Z, Bowen B, Glancy B, De Filippis EA, Mapes R, Hwang H, Flynn CR, Willis WT, Civitarese A, Højlund K, et al. Proteome profile of functional mitochondria from human skeletal muscle using one-dimensional gel electrophoresis and HPLC-ESI-MS/MS. J Proteomics. 2009;72:1046–60. doi: 10.1016/j.jprot.2009.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yin X, Cuello F, Mayr U, Hao Z, Hornshaw M, Ehler E, Avkiran M, Mayr M. Proteomic analysis of the cardiac myofilament subproteome reveals dynamic alterations in phosphatase subunit distribution. Mol Cell Proteomics. 2009 doi: 10.1074/mcp.M900275-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yuan C, Sheng Q, Tang H, Li Y, Zeng R, Solaro RJ. Quantitative comparison of sarcomeric phosphoproteomes of neonatal and adult rat hearts. Am J Physiol Heart Circ Physiol. 2008;295:H647–56. doi: 10.1152/ajpheart.00357.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Deng N, Zhang J, Zong C, Wang Y, Lu H, Yang P, Wang W, Young GW, Wang Y, Korge P, Lotz C, et al. Phosphoproteome analysis reveals regulatory sites in major pathways of cardiac mitochondria. Mol Cell Proteomics. 2011;10:M110.000117. doi: 10.1074/mcp.M110.000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Boersema PJ, Mohammed S, Heck AJ. Phosphopeptide fragmentation and analysis by mass spectrometry. J Mass Spectrom. 2009;44:861–78. doi: 10.1002/jms.1599. [*Overview comparing different mass spectrometry techniques that can be used to identify site-specific phosphorylation of proteins.] [DOI] [PubMed] [Google Scholar]
- 84.Scruggs SB, Hinken AC, Thawornkaiwong A, Robbins J, Walker LA, de Tombe PP, Geenen DL, Buttrick PM, Solaro RJ. Ablation of ventricular myosin regulatory light chain phosphorylation in mice causes cardiac dysfunction in situ and affects neighboring myofilament protein phosphorylation. J Biol Chem. 2009;284:5097–106. doi: 10.1074/jbc.M807414200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Olsson MC, Patel JR, Fitzsimons DP, Walker JW, Moss RL. Basal myosin light chain phosphorylation is a determinant of Ca2+ sensitivity of force and activation dependence of the kinetics of myocardial force development. Am J Physiol Heart Circ Physiol. 2004;287:H2712–8. doi: 10.1152/ajpheart.01067.2003. [DOI] [PubMed] [Google Scholar]
- 86.Kemi OJ, Ellingsen O, Smith GL, Wisloff U. Exercise-induced changes in calcium handling in left ventricular cardiomyocytes. Front Biosci. 2008;13:356–68. doi: 10.2741/2685. [DOI] [PubMed] [Google Scholar]
- 87.Moriggi M, Cassano P, Vasso M, Capitanio D, Fania C, Musicco C, Pesce V, Gadaleta MN, Gelfi C. A DIGE approach for the assessment of rat soleus muscle changes during unloading: effect of acetyl-L-carnitine supplementation. Proteomics. 2008 doi: 10.1002/pmic.200701176. [DOI] [PubMed] [Google Scholar]
- 88.Mintz M, Vanderver A, Brown KJ, Lin J, Wang Z, Kaneski C, Schiffmann R, Nagaraju K, Hoffman EP, Hathout Y. Time series proteome profiling to study endoplasmic reticulum stress response. J Proteome Res. 2008;7:2435–44. doi: 10.1021/pr700842m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Burniston JG, Connolly JB. Characterisation of isoform-specific tryptic peptides of rat cardiac myosin heavy chains using automated liquid chromatography-matrix assisted laser desorption ionisation (LC-MALDI) mass spectrometry. Int J Cardiol. 2010;140:363–6. doi: 10.1016/j.ijcard.2008.11.058. [DOI] [PubMed] [Google Scholar]
- 90.Hojlund K, Yi Z, Hwang H, Bowen B, Lefort N, Flynn CR, Langlais P, Weintraub ST, Mandarino LJ. Characterization of the human skeletal muscle proteome by one-dimensional Gel electrophoresis and HPLC-ESI-MS/MS. Mol Cell Proteomics. 2007;7:257–67. doi: 10.1074/mcp.M700304-MCP200. [*Comprehensive catalogue of human muscle proteins that includes each enzyme of the major metabolic pathways in muscle and regulatory proteins known to be involved in mediating exercise-induced adaptations.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Parker KC, Walsh R, Salajegheh M, Amato A, Krastins B, Sarracino D, Greenberg SA. Characterization of human skeletal muscle biopsy samples using shotgun proteomics. J Proteome Res. 2009 doi: 10.1021/pr800873q. [*Comparison of total analysis-time vs number of human skeletal muscle proteins identified using different solution-based separation techniques.] [DOI] [PubMed] [Google Scholar]
- 92.Bousette N, Kislinger T, Fong V, Isserlin R, Hewel JA, Emil A, Gramolini AO. Large-scale characterization and analysis of the murine cardiac proteome. J Proteome Res. 2009;8:1887–901. doi: 10.1021/pr800845a. [DOI] [PubMed] [Google Scholar]
- 93.Hwang H, Bowen BP, Lefort N, Flynn CR, De Filippis EA, Roberts C, Smoke CC, Meyer C, Højlund K, Yi Z, Mandarino LJ. Proteomics analysis of human skeletal muscle reveals novel abnormalities in obesity and type 2 diabetes. Diabetes. 2010;59:33–42. doi: 10.2337/db09-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Koch LG, Britton SL. Evolution, atmospheric oxygen, and complex disease. Physiol Genomics. 2007;30:205–8. doi: 10.1152/physiolgenomics.00043.2007. [DOI] [PubMed] [Google Scholar]
- 95.Das AM. Regulation of the mitochondrial ATP-synthase in health and disease. Mol Genet Metab. 2003;79:71–82. doi: 10.1016/s1096-7192(03)00069-6. [DOI] [PubMed] [Google Scholar]
- 96.Rose AJ, Frosig C, Kiens B, Wojtaszewski JF, Richter EA. Effect of endurance exercise training on Ca2+ calmodulin-dependent protein kinase II expression and signalling in skeletal muscle of humans. J Physiol. 2007;583:785–95. doi: 10.1113/jphysiol.2007.138529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Arrell DK, Elliott ST, Kane LA, Guo Y, Ko YH, Pedersen PL, Robinson J, Murata M, Murphy AM, Marbán E, Van Eyk JE. Proteomic analysis of pharmacological preconditioning: novel protein targets converge to mitochondrial metabolism pathways. Circ Res. 2006;99:706–14. doi: 10.1161/01.RES.0000243995.74395.f8. [DOI] [PubMed] [Google Scholar]
- 98.Kane LA, Van Eyk JE. Post-translational modifications of ATP synthase in the heart: biology an function. J Bioenerg Biomembr. 2009;41:145–50. doi: 10.1007/s10863-009-9218-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Richardson MR, Lai X, Dixon JL, Sturek M, Witzmann FA. Diabetic dyslipidemia and exercise alter the plasma low-density lipoproteome in Yucatan pigs. Proteomics. 2009;9:2468–83. doi: 10.1002/pmic.200800613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sietsema KE, Meng F, Yates NA, Hendrickson RC, Liaw A, Song Q, Brass EP, Ulrich RG. Potential biomarkers of muscle injury after eccentric exercise. Biomarkers. 2010;15:249–58. doi: 10.3109/13547500903502802. [DOI] [PubMed] [Google Scholar]
- 101.Hu S, Xie Y, Ramachandran P, Ogorzalek Loo RR, Li Y, Loo JA, Wong DT. Large-scale identification of proteins in human salivary proteome by liquid chromatography/mass spectrometry and two-dimensional gel electrophoresis-mass spectrometry. Proteomics. 2005;5:1714–28. doi: 10.1002/pmic.200401037. [DOI] [PubMed] [Google Scholar]
- 102.Kohler M, Franz S, Regeniter A, Ikonen A, Walpurgis K, Thomas A, Schänzer W, Thevis M. Comparison of the urinary protein patterns of athletes by 2D-gel electrophoresis and mass spectrometry-a pilot study. Drug Test Anal. 2009;1:382–6. doi: 10.1002/dta.80. [DOI] [PubMed] [Google Scholar]
- 103.Maiorana A, O'Driscoll G, Cheetham C, Collis J, Goodman C, Rankin S, Taylor R, Green D. Combined aerobic and resistance exercise training improves functional capacity and strength in CHF. J Appl Physiol. 2000;88:1565–70. doi: 10.1152/jappl.2000.88.5.1565. [DOI] [PubMed] [Google Scholar]
- 104.Goldspink DF. Ageing and activity: their effects on the functional reserve capacities of the heart and vascular smooth and skeletal muscles. Ergonomics. 2005;48:1334–51. doi: 10.1080/00140130500101247. [DOI] [PubMed] [Google Scholar]
- 105.Doherty MK, McLean L, Hayter JR, Pratt JM, Robertson DH, El-Shafei A, Gaskell SJ, Beynon RJ. The proteome of chicken skeletal muscle: changes in soluble protein expression during growth in a layer strain. Proteomics. 2004;4:2082–93. doi: 10.1002/pmic.200300716. [DOI] [PubMed] [Google Scholar]
- 106.Burniston JG, McLean L, Beynon RJ, Goldspink DF. Anabolic effects of a non-myotoxic dose of the β2-adrenergic receptor agonist clenbuterol on the rat plantaris muscle. Muscle Nerve. 2007;35:217–23. doi: 10.1002/mus.20684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Isfort RJ, Wang F, Greis KD, Sun Y, Keough TW, Farrar RP, Bodine SC, Anderson NL. Proteomic analysis of rat soleus muscle undergoing hindlimb suspension-induced atrophy and reweighting hypertrophy. Proteomics. 2002;2:543–50. doi: 10.1002/1615-9861(200205)2:5<543::AID-PROT543>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 108.Nader GA. Concurrent strength and endurance training: from molecules to man. Med Sci Sports Exerc. 2006;38:1965–70. doi: 10.1249/01.mss.0000233795.39282.33. [DOI] [PubMed] [Google Scholar]
- 109.Ding Q, Vaynman S, Souda P, Whitelegge JP, Gomez-Pinilla F. Exercise affects energy metabolism and neural plasticity-related proteins in the hippocampus as revealed by proteomic analysis. Eur J Neurosci. 2006;24:1265–76. doi: 10.1111/j.1460-9568.2006.05026.x. [DOI] [PubMed] [Google Scholar]
- 110.Kirchner L, Chen WQ, Afjehi-Sadat L, Viidik A, Skalicky M, Höger H, Lubec G. Hippocampal metabolic proteins are modulated in voluntary and treadmill exercise rats. Exp Neurol. 2008;212:145–51. doi: 10.1016/j.expneurol.2008.03.014. [DOI] [PubMed] [Google Scholar]
- 111.Mizutani K, Sonoda S, Hayashi N, Takasaki A, Beppu H, Saitoh E, Shimpo K. Analysis of protein expression profile in the cerebellum of cerebral infarction rats after treadmill training. Am J Phys Med Rehabil. 2010;89:107–14. doi: 10.1097/PHM.0b013e3181b3323b. [DOI] [PubMed] [Google Scholar]
- 112.Moriggi M, Vasso M, Fania C, Capitanio D, Bonifacio G, Salanova M, Blottner D, Rittweger J, Felsenberg D, Cerretelli P, Gelfi C. Long term bed rest with and without vibration exercise countermeasures: effects on human muscle protein dysregulation. Proteomics. 2010;10:3756–74. doi: 10.1002/pmic.200900817. [DOI] [PubMed] [Google Scholar]
- 113.Yang KD, Chang WC, Chuang H, Wang PW, Liu RT, Yeh SH. Increased complement factor H with decreased factor B determined by proteomic differential displays as a biomarker of tai chi chuan exercise. Clin Chem. 2010;56:127–31. doi: 10.1373/clinchem.2009.126615. [DOI] [PubMed] [Google Scholar]
- 114.Ichibangase T, Imai K. Application of Fluorogenic Derivatization-Liquid Chromatography-Tandem Mass Spectrometric Proteome Method to Skeletal Muscle Proteins in Fast Thoroughbred Horses. J Proteome Res. 2009 doi: 10.1021/pr801004s. [DOI] [PubMed] [Google Scholar]
- 115.Bouwman FG, van Ginneken MM, Noben JP, Royackers E, de Graaf-Roelfsema E, Wijnberg ID, van der Kolk JH, Mariman EC, van Breda E. Differential expression of equine muscle biopsy proteins during normal training and intensified training in young standardbred horses using proteomics technology. Comp Biochem Physiol Part D Genomics Proteomics. 2010;5:55–64. doi: 10.1016/j.cbd.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 116.de Moraes R, Valente RH, León IR, Trugilho MR, Nóbrega AC, Perales J, TibiriÇá E. Chronic dynamic exercise increases apolipoprotein A-I expression in rabbit renal cortex as determined by proteomic technology. Br J Sports Med. 2008;42:386–8. doi: 10.1136/bjsm.2007.038646. [DOI] [PubMed] [Google Scholar]
- 117.Gerecke KM, Jiao Y, Pani A, Pagala V, Smeyne RJ. Exercise protects against MPTP-induced neurotoxicity in mice. Brain Res. 2010;1341:72–83. doi: 10.1016/j.brainres.2010.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]