Abstract
The gel-forming mucins are large glycosylated proteins that are essential components of the mucus layers covering epithelial cells. Using novel methods of identifying mucins based on profile hidden Markov models, we have found a large number of such proteins in Metazoa, aiding in their classification and allowing evolutionary studies. Most vertebrates have 5–6 gel-forming mucin genes and the genomic arrangement of these genes is well conserved throughout vertebrates. An exception is the frog Xenopus tropicalis with an expanded repertoire of at least 26 mucins of this type. Furthermore, we found that the ovomucin protein, originally identified in chicken, is characteristic of reptiles, birds, and amphibians. Muc6 is absent in teleost fish, but we now show that it is present in animals such as ghost sharks, demonstrating an early origin in vertebrate evolution. Public RNA-Seq data were analyzed with respect to mucins in zebrafish, frog, and chicken, thus allowing comparison in regard of tissue and developmental specificity. Analyses of invertebrate proteins reveal that gel-forming-mucin type of proteins is widely distributed also in this group. Their presence in Cnidaria, Porifera, and in Ctenophora (comb jellies) shows that these proteins were present early in metazoan evolution. Finally, we examined the evolution of the FCGBP protein, abundant in mucus and related to gel-forming mucins in terms of structure and localization. We demonstrate that FCGBP, ubiquitous in vertebrates, has a conserved N-terminal domain. Interestingly, this domain is also present as an N-terminal sequence in a number of bacterial proteins.
Keywords: mucin, mucus, bioinformatics, evolution, von Willebrand D domain.
Introduction
Mucins are large glycoproteins that cover the epithelial cell surfaces of the respiratory, digestive, and urogenital tracts. They form gel-like structures, and are thereby able to protect against harmful molecules and microorganisms. Mucins are linked to human disease. For instance, there is an association between the mucin MUC2 and the inflammatory bowel disease ulcerative colitis, presumably because MUC2 protects against bacterial contact (Johansson et al. 2008, 2014). In addition, certain mucins are associated with colon cancer (Hollingsworth and Swanson 2004) and overproduction of gel-forming mucins in the lungs is a key element of both chronic obstructive lung disease and cystic fibrosis (Milla and Moss 2015).
The mucins are classified as membrane-bound or secreted. In mammals, there are five secreted gel-forming mucins, such as MUC2, MUC5AC, MUC5B, MUC6, and MUC19. Experimental studies are restricted to human and mouse (Corfield 2015). In the following nomenclature, human proteins are denoted with uppercase letters, while for other species, the first letter is capitalized. All these proteins contain heavily glycosylated domains that are rich in the amino acids proline, threonine, and serine. These domains, referred to as PTS domains, are often repetitive in nature (Lang et al. 2007; Johansson et al. 2011). Typically, the content of threonine and serine is at least 40% in the PTS domain and proline is often present at a frequency of more than 5%. The threonines and serines are the sites of O-glycan attachment. Through these sugar decorations the PTS domain becomes resistant to proteolysis and adopts an extended, rod-like and stiff conformation, reminiscent of a bottle brush (Johansson et al. 2011; Corfield 2015).
In human, the tissue distribution of mucins is such that MUC2 and MUC5AC are the major mucus components on the surface of the intestine and stomach, respectively (Johansson et al. 2011). The MUC5AC mucin is also found in the respiratory tract together with MUC5B mucin (Thornton et al. 2008). MUC5B is also highly expressed in salivary glands and thus found in saliva. The MUC6 mucin is mainly present in the glands of the stomach and in the pancreas exocrine duct (Corfield 2015). As compared with the other gel-forming mucins, MUC19 is less well characterized in terms of structure and function. It is debated if it is expressed as a protein in humans, but it is the major salivary mucin in pigs, named “pig salivary mucin” (Chen et al. 2004; Rousseau et al. 2008; Yu et al. 2008; Zhu et al. 2011).
A characteristic protein structural domain of the gel-forming mucins is the von Willebrand D (VWD) domain, named after its occurrence in the von Willebrand factor (VWF) (Perez-Vilar and Hill 1999; Zhou et al. 2012). The mucins MUC2, MUC5AC, and MUC5B have a domain architecture which is (VWD-C8-TIL)–(VWD-C8-TIL)–(VWD-C8-TIL)–PTS–(VWD-C8-TIL). (TIL is short for “trypsin inhibitor-like cysteine rich domain” and the “C8” domain has eight conserved cysteines.) The MUC6 and MUC19 mucins have the same domain structure, but lack the C-terminal VWD-C8-TIL unit. In addition, mucins of the types MUC2 or MUC5 are characterized by a domain that we refer to as CysD (with a conserved WxxW motif, in Pfam this domain is known as Mucin2_WxxW) and present within PTS regions (Lang et al. 2007). Toward the C-terminal end of a mucin, there is typically a cystine knot and VWC (VWF type C) domains in some mucins. In addition to the gel-forming mucins, there are other vertebrate proteins that contain multiple units of VWD-C8-TIL. In human, these proteins are known as otogelin, VWF, SCO-spondin, zonadhesin, alpha-tectorin, and FCGBP (Lang et al. 2007). Of these protein families, alpha-tectorin is characterized by NIDO (nidogen-like) and zona pellucida domains, otogelin by AbfB (alpha-l-1arabinofuranosidase B domain), SCO-spondin by LDL_recept_a, VWF by VWA (von Willebrand A) and zonadhesin by MAM (“meprin/A5/mu”) domains (for more information about these domains, see the Pfam database—Finn et al. 2014).
Gel-forming mucins have the ability to form gels which is dependent on their extensive glycosylation and the capacity of monomers to form polymeric structures. The three-dimensional structure of the VWD domain is still poorly characterized but is known to be involved in polymerization of mucin monomers through intermolecular disulfide-bonds (Thornton et al. 2008; Ambort et al. 2012).
The FCGBP protein is abundant in human and mouse mucus and may be functionally related to the gel-forming mucins. Both of these protein classes have multiple VWD domains and they have been shown to be covalently bound to each other (Johansson et al. 2009). This binding probably involves a step where a reactive anhydride is formed by cleavage in VWD domains of FCGBP. The name FCGBP (an acronym for Fc fragment of IgG-binding protein) originates from early studies suggesting that the protein binds to immunoglobulin G (Harada et al. 1997), but we have not been able to reproduce this result (unpublished observations).
In order to better understand the structure and function of mucins and other VWD-domain-containing proteins we have used comparative genomics approaches (Lang et al. 2007). There are many benefits from such investigations. First, it allows more accurate assignment of orthology and paralogy for a protein family of interest. Second, one is able to identify structural elements that are conserved through evolution and that may be considered to be of particular biological and functional importance. For instance, we have identified a highly conserved unit of three VWD-C8-TIL domains that is often associated with PTS domains. Third, it is of functional interest to examine what mucin types are found in different animals and to learn about their tissue specificities. From such studies we will understand what model organisms are relevant for studies of mucin physiology.
We have previously examined the phylogenetic distribution of both gel-forming and membrane-bound mucins. We have examined fish and chicken mucins and bioinformatically characterized chicken ovomucin, a mucin-like protein without a PTS domain (Lang et al. 2006). In a more extensive study of mucin evolution (Lang et al. 2007), we noted that Xenopus tropicalis has a larger number of mucins than other vertebrates. This species is also characterized by a family of secreted mucin-like proteins with alternating SEA (“Sea urchin sperm protein, Enterokinase, Agrin”) and PTS domains. X. tropicalis is also the most deeply branching animal where a protein similar to the mammalian Muc4 is identified. Finally, we noted that proteins related to the gel-forming mucins are present in the cnidarian Nematostella vectensis (Lang et al. 2007). Since these studies were carried out, genome and transcriptome information has recently become available for a large number of species, including ctenophores and choanoflagellates. We have now exploited this novel information to obtain a more accurate and comprehensive account of the evolution of the gel-forming mucins. To make this analysis more effective and accurate, we have used a novel method of identifying mucin-like protein sequences, as well as methods to identify regions in genomes encoding these proteins. In this analysis, we have considered all available metazoan genomes, as well as choanoflagellates and protists to characterize early evolution of gel-forming mucins and their typical protein building blocks. The results provide a very comprehensive collection of protein sequences and demonstrate an early origin for gel-forming mucins as shown by the occurrence of such proteins in Ctenophora. We also examine the evolution of the FCGBP protein, a protein with multiple VWD domains known to colocalize with the gel-forming mucins.
Results
Identification of Gel-Forming Mucins and Related Proteins
We wanted to systematically examine the phylogenetic distribution of gel-forming mucins and related proteins in Metazoa. In order to bioinformatically identify these proteins, we made use of profile hidden Markov models (HMMs) and the hmmer software (http://hmmer.org, last accessed April 11, 2016) (Eddy 2011). Thus, profile HMM models of gel-forming mucin protein sequences were created on the basis of a reliable alignment of previously known full-length mucin sequences (see supplementary dataset 1, Supplementary Material online). The protein sequence databases Genbank and UniProt were searched with this model (see “Analysis with Profile HMMs” for more details). To identify proteins that were not found during genome annotation and thus were lacking in available protein sequence databases, we also analyzed genomic sequences. Thus, selected species with an available genome assembly were analyzed with genewise (Birney et al. 2004). (For more details see –“Prediction of Protein Sequences From Genomic Sequences”.) All proteins identified in this study, including sequences and protein domain structures, are available as supplementary files and at http://www.medkem.gu.se/mucinbiology/mucevo, last accessed April 11, 2016.
Phylogenetic Analysis
With searches of protein and genomic sequences we identified not only gel-forming mucins, but also members of the other protein classes of VWD domain proteins as described above. Further classification required phylogenetic analysis. To generate an accurate multiple alignment we considered first the 5,000 best hits from a search with hmmsearch in the Genbank protein database. These sequences were filtered to remove those that contained less than three VWD domains. Alignment was then made with Clustal Omega (Sievers and Higgins 2014) and edited to leave only the N-terminal part of each protein, containing the three VWD-C8-TIL units. This editing was necessary because the N-terminal region is shared between all mucins and an alignment of PTS domains is not meaningful as a result of strong sequence divergence. The alignment was further edited to remove partial sequences or sequences that contained one or more mispredicted exons. All vertebrate FCGBP proteins were also removed as they contain a large number of VWD domains and cannot be aligned well to the other multiple VWD domain proteins. This procedure resulted in a reliable alignment with 1,260 sequences.
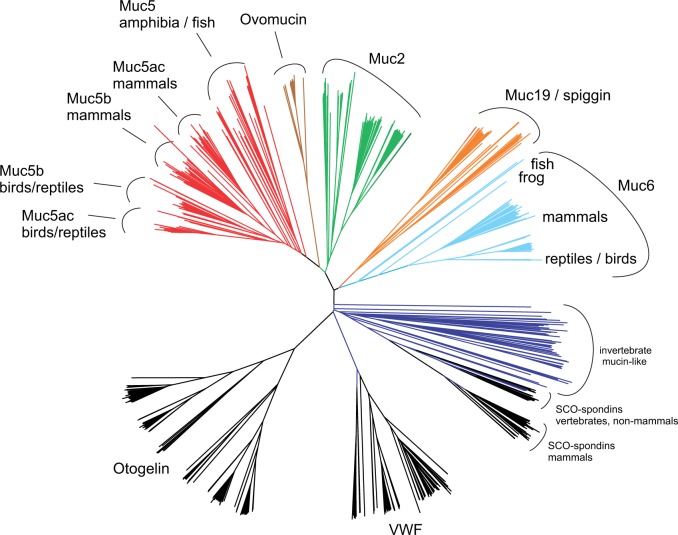
The major purpose of the alignment was to construct phylogenetic trees that would aid in the classification of the different proteins. A neighbor-joining tree constructed from the alignment is shown in figure 1. There are a number of interesting observations from this tree. First, it reveals large clusters containing vertebrate gel-forming mucins, SCO-spondins, otogelins, and VWFs, respectively, showing that we are able to clearly distinguish these groups based on sequence. The tree contains a number of invertebrate mucin-like sequences. These proteins form a fairly homogenous group which is clearly distinct from the vertebrate mucins. Thus, we are not able to associate the invertebrate mucins with any of the vertebrate Muc2, Muc5ac, Muc5b, ovomucin, Muc6, or Muc19 families. Finally, there is an indication that the vertebrate SCO-spondins are the closest relatives of some of the invertebrate mucins, suggesting that they are invertebrate orthologs of SCO-spondin. More details of the tree will be discussed further below.
Fig. 1.
Phylogenetic classification of mucins. Muc2, Muc5, Muc6, Muc19, ovomucin, otogelin, and VWF protein sequences were aligned with Clustal Omega (Sievers and Higgins 2014) and a neighbor-joining tree with 100 bootstrap replicates was obtained with ClustalW (Thompson et al. 2002). Highlighted in color are Muc5 (red), ovomucin (brown), Muc2 (green), Muc19/spiggin (orange), Muc6 (cyan), and invertebrate mucin-like proteins (blue). Classification of Muc5b and Muc5ac is also possible although all bird/reptile Muc5 are in one group and all mammalian Muc5 in another. Invertebrate mucin-like sequences seem to be most similar to the SCO-spondins, proteins previously known to be present in vertebrates only.
Gel-Forming Mucins in Vertebrates—Distribution and Domain Structures
Using a profile HMM-based search of the NCBI protein sequence database we identified a number of vertebrate proteins with at least three VWD domains. Classification of these sequences was possible using a combination of phylogenetic tree construction and analysis of protein domain architecture. Among the sequences identified, a substantial portion was not members of the group of gel-forming mucins; examples are alpha-tectorin, otogelin, VWF, SCO-spondin, and zonadhesins. However, these nonmucin proteins could be filtered out on the basis of their domain structure which is different from the gel-forming mucins. Further analysis of these vertebrate nonmucin proteins revealed that they are all widely distributed among vertebrates. However, none of them had apparent orthologs in invertebrates, with the possible exception of SCO-spondin.
To further characterize the vertebrate proteins that are gel-forming mucins or closely related proteins we restricted our analysis to species where there is a genome assembly available. Filtering out all obvious non-gel-forming mucins (otogelin, VWF, SCO-spondin, zonadhesin, alpha-tectorin, and FCGBP) resulted in a total of 659 sequences from 135 vertebrate species. These were assigned as mucins and annotated as belonging to groups on the basis of the phylogenetic tree in figure 1. According to this classification, there were 133 Muc2-type, 263 Muc5-type, 45 ovomucin-like, 145 Muc6-type, and 73 Muc19-type proteins. The 659 sequences were then analyzed with respect to their predicted protein domain structure (see Analysis with Profile HMMs section). For protein domain structures see http://www.medkem.gu.se/mucinbiology/mucevo/, last accessed April 11, 2016. In addition to proteins identified in Genbank, we analyzed genomic sequences. However, we could only identify two additional vertebrate proteins belonging to the class of gel-forming mucins that did not have a counterpart in GenBank (one Muc5 from the bird Picoides pubescens and one Muc2 from the reptile Python bivitattus).
There are a number of conclusions regarding the domain structure of gel-forming mucins that are consistent with the results of previous studies. Thus, Muc6 and mammalian Muc19 proteins consistently lack the fourth VWD domain characteristic of the other mucins. Furthermore, the Muc2, Muc5ac, and Muc5b proteins from mammals, birds, and reptiles all have CysD domains that are interspersed in the PTS regions.
Ovomucin Is Characteristic of Reptiles and Birds
We previously identified ovomucin as a mucin-like protein which is similar to Muc2 and Muc5ac/5b (Lang et al. 2006). We now demonstrate that ovomucin is present in a variety of birds as well as in the turtles Chelonia mydas and Chrysemys picta bellii and in the reptiles Alligator sinensis and Anolis carolinensis. Therefore, ovomucin seems to be ubiquitous in birds and reptiles. All ovomucin orthologs are different from the gel-forming mucins in that they all lack PTS domains. However, there is a likely ovomucin ortholog in the amphibian X. tropicalis that does have PTS domains (see below).
Muc6 Was Present Early in Vertebrate Evolution
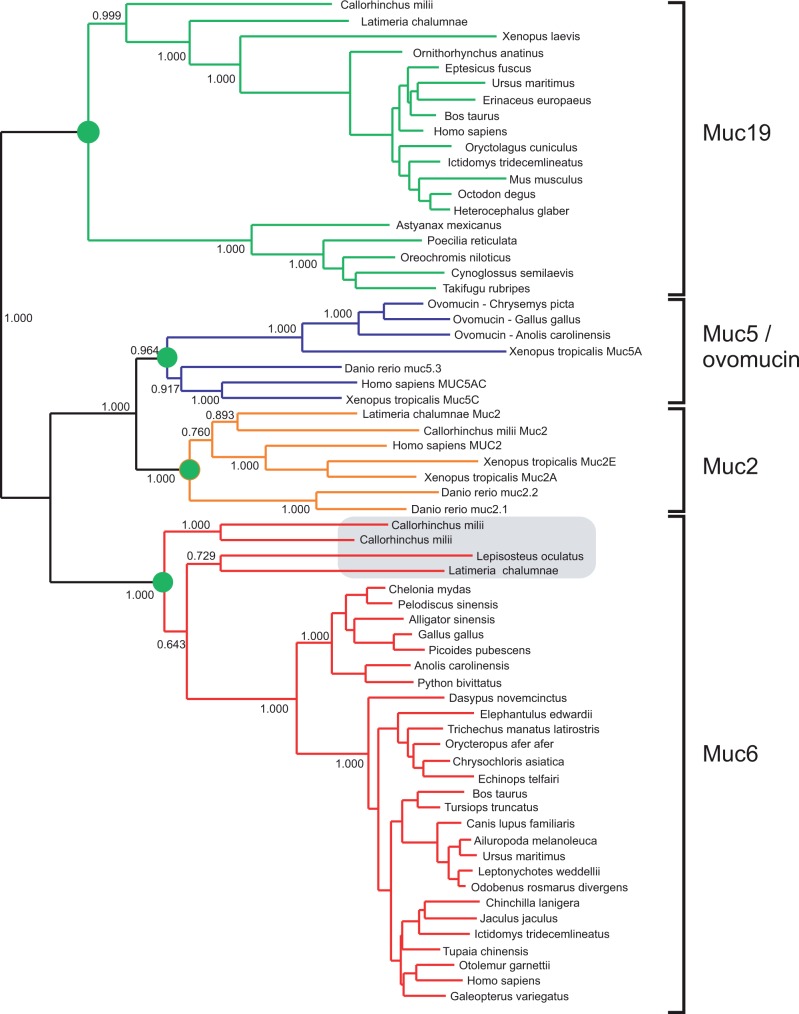
Muc6 is not present in a large majority of teleost fishes, for instance zebrafish. However, we can now show that Muc6 is present in Lepisosteus oculatus (spotted gar), Callorhinchus milii (Australian ghost shark, Chondrichthyes) and Latimeria chalumnae (West Indian Ocean coelacanth), where C. milii has two Muc6 paralogs. The phylogenetic tree obtained with MrBayes in figure 2 demonstrates that these early vertebrate proteins should indeed be classified as Muc6 proteins as they clearly group with other vertebrate Muc6 orthologs and are distinct from Muc19, Muc2, and Muc5. It would therefore seem that Muc6 was present early in vertebrates, but was lost in teleost fishes, a class of the ray-finned fishes (Actinopterygii).
Fig. 2.
The mucin Muc6 was present early in vertebrate evolution. Tree was constructed with MrBayes using an alignment of the N-terminal parts of mucins that include the three VWD–C8–TIL units. Homologs of Muc19, Muc6, Muc5, ovomucin, and Muc2 are shown. Critical nodes are labeled with green circles. The four Muc6 homologs of C. milii, L. chalumnae, and Le. oculatus clearly belong to the group of Muc6.
Evolution of Muc19
The tree in figure 2 also includes Muc19 and confirms a previous report that Muc19 is related to the fish spiggin protein (Kawahara and Nishida 2007). It is also obvious from our analysis that Muc19 is present in fishes, amphibians, and mammals, but seems to have been lost in birds. A schematic view of the genomic context of the Muc19 gene in different animals is shown in supplementary figure S1, Supplementary Material online. The turtle Ch. picta and the lizard An. carolinensis show evidence of a Muc19 gene. However, in the chicken and zebrafinch the Muc19 gene is missing, providing further evidence of the lack of Muc19 in birds. As a rule Muc19 proteins contain a PTS domain, but apparently there are exceptions, like in X. tropicalis. Some of the spiggins are also different from mammalian Muc19 in that they have a PTS domain together with a fourth VWD domain.
An Expansion of Mucin Genes in X. tropicalis
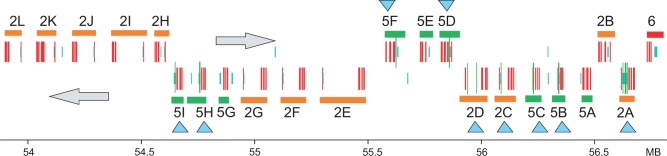
In the analysis of a X. tropicalis genomic scaffold (assembly version 7, accession KB021653.1, region 53,600,000–57,000,000), we discovered a remarkably large cluster with 22 tandemly arranged genes that all encode gel-forming mucins (fig. 3). The mRNA and protein sequences of these genes were obtained either from Genbank or using a prediction based on genewise using the genomic sequence and our HMM model of mucins. The mucins were classified as Muc2 or Muc5 and we named them Muc2A-L and Muc5A-I on the basis of their order in the genomic cluster (note that this is different from our previous nomenclature). In addition, we identified two mucins, Muc5J and Muc5K, on the same scaffold but at a distance several 100,000 nt from the large cluster. Finally, there are singular genes encoding Muc2M and the Muc19 ortholog on other contigs. Therefore, we identified a total of 26 mucin genes in this species. While some of the frog mucins could be classified as Muc5 they could not be associated specifically with either of the Muc5ac or Muc5b of mammals and birds.
Fig. 3.
Genomic organization of X. tropicalis mucins. Genomic region from assembly version 7, accession KB021653.1, region 53,600,000–57,000,000 is shown. Genes are shown for Muc5 paralogs (green) and Muc2 paralogs (orange). Naming of genes was based on their location from right to left. Location of VWD domains, PTS domains, and CysD domains (red, blue, and green vertical bars, respectively) are based on the analysis of the genomic sequence with genewise and an in-house Perl script. Genes where a CysD domain is predicted are indicated with cyan triangles.
All X. tropicalis protein and mRNA sequences were predicted with genewise, except four where the sequences were derived from GenBank entries. The frog proteins were different with respect to their content of CysD domains. Thus, only 11 of the 26 frog mucins contained this domain as judged by analysis of the genome with genewise (see also fig. 3 for the 22 cluster proteins). In this respect they are different from the mammalian, reptile, and bird Muc2, Muc5ac, and Muc5b proteins that all have CysD domains. Another unusual property of some of the X. tropicalis mucins (Muc5A, 5E, 5F, 5G, 5H, and 5I) is that they do not have the fourth VWD domain characteristic of the Muc2 and Muc5 family of mucins in mammals, reptiles, and birds.
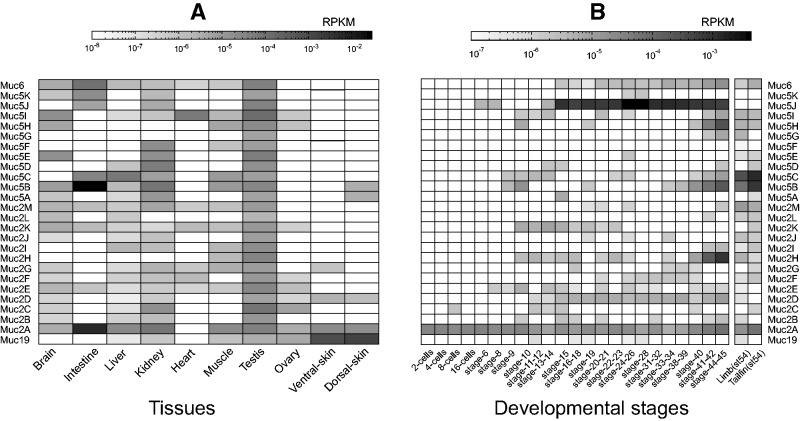
Analysis of publically available RNA-Seq data (Necsulea et al. 2014 and other studies) indicates that the 26 different X. tropicalis mucin genes are expressed in one or more tissues (fig. 4A), suggesting that they are not pseudogenes or the result of misprediction. A number of RNA-Seq samples allowed us to specifically monitor the expression of mucins during embryonic development (Tan et al. 2013). The transcription of a number of mucins is initiated at a specific developmental stage (fig. 4B). For instance, Muc2D appears at stage 11, Muc5J at stage 15, whereas Muc2H, Muc5B, and Muc5H appear relatively late during development. Finally, the Muc2A gene is expressed throughout development, beginning already in the 2-cell stage.
Fig. 4.
X. tropicalis mucin gene transcription. A total of 26 different mucin genes were analyzed with respect to gene expression. Expression levels (RPKM) were estimated from public RNA-Seq data as listed in supplementary dataset S2, Supplementary Material online, and as described in more detail in Materials and Methods section. (A) Expression in different tissues. (B) Expression at different developmental stages.
Eleven Gel-Forming Mucins Are Identified in Zebrafish
The most well-characterized fish genome is that of the zebrafish, Danio rerio. We could identify 11 different gel-forming mucin genes; four Muc2-type genes (Muc2.1–Muc2.4), six mucins of the Muc5-type (Muc5.1–Muc5.6), and one Muc19 gene based on the analysis of the zebrafish genome version 10 (http://www.medkem.gu.se/mucinbiology/mucevo/, last accessed April 11, 2016). The naming of ten of these genes is based on previous nomenclature (Jevtov et al. 2014). Unfortunately, the genome assembly does not contain the full genomic structure of all mucin genes. There is one cluster of six mucins, all situated on the same strand (Muc5.5, Muc5.6, Muc2.1, Muc2.2, Muc5.1, and Muc5.2). All these six genes seem to be full-length. Second, there is another contig with a pair of mucins, Muc5.3 and Muc5.4 that are encoded on in opposite strands and where Muc5.4 is lacking part of its 3′ terminus. Third, there is a yet another contig with genes that we now named Muc2.3 and Muc2.4; they are on the same strand and are both incomplete.
The expression of all ten genes was examined with public RNA-Seq data. The results in supplementary figure S2 (panel C), Supplementary Material online, indicate that all genes are expressed. Exceptions are Muc2.3 and Muc5.4 but it should be noted that our failure to detect these genes may be because neither of them are complete at their 3′-terminal ends. The mucin Muc2.4 seems to be the major intestinal mucin. Also Muc2.1 is present in the intestine but at a lower level as compared with Muc2.4. The Muc2.4 gene is expressed also in liver, spleen, and kidney. A major constituent of gills is Muc5.1, consistent with previous observations (Jevtov et al. 2014). Muc5.2 is present in pectoral fins. Analysis of different embryonic developmental stages indicated that after about 24 h there is an increase in the expression of Muc5.1, Muc5.2, and Muc5.3 (data not shown).
Genomic Cluster of Gel-Forming Mucin Genes Is Highly Conserved in Vertebrates
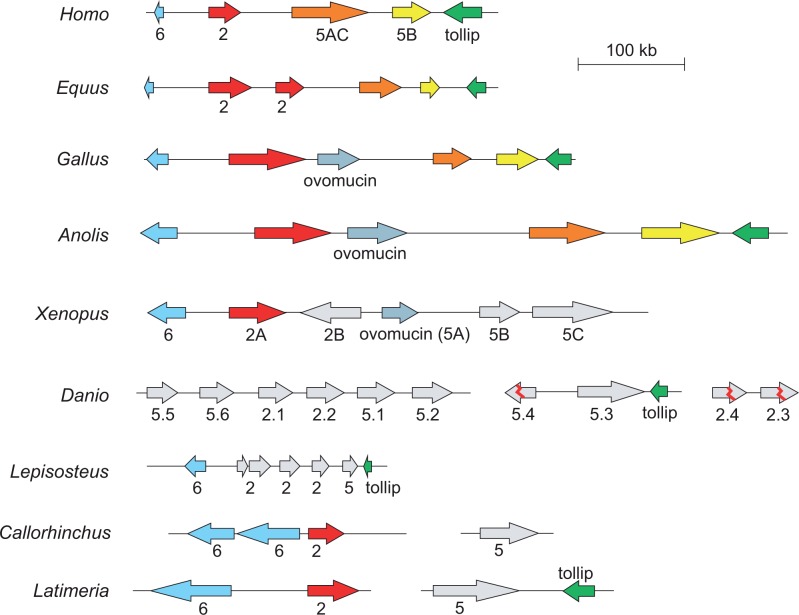
We next examined the genomic organization of mucin genes in different vertebrates (fig. 5). Relevant genomic regions were first identified by matching previously known gel-forming mucins (see Prediction of Protein Sequences From Genomic Sequences section). Then, these regions were analyzed with respect to PTS domains and a profile HMM model of the VWD domain, as well as with full-length profile HMM models of gel-forming mucins. In a majority of vertebrates, the mucins Muc2, Muc6, Muc5ac, and Muc5b are localized to the same genomic region with the protein-coding gene Tollip in a tail to tail arrangement with Muc5b.
Fig. 5.
A conserved structure of mucin genes in vertebrates. Arrows reflect the strand polarity. In the case of X. tropicalis only a part of a larger cluster of mucin genes is shown (see also fig. 3).
Among the mammals, the horse, Equus caballus, presents an unusual case as the Muc2 gene has been duplicated. The horse genome may also have a third copy of the Muc2 gene which is highly similar in sequence to one of the other two Muc2 genes, but not part of the current chromosome assembly. Publically available RNA-Seq data suggest that both of the Muc2 genes being part of the mucin cluster are expressed and the one proximal to Muc5ac seems to be the predominant transcript (data not shown).
Birds and reptiles have a gene arrangement similar to mammals, but with the addition of the ovomucin gene between the Muc2 and Muc5ac genes (fig. 5). In X. tropicalis, the organization of genes at one end of the cluster is similar to that observed in mammals, reptiles, and birds (compare fig. 3), with the Muc6 and Muc2 genes arranged head-to-head. It would also appear that the frog Muc5A is orthologous to bird ovomucin. Thus, Muc5A groups with ovomucins in phylogenetic analysis (supplementary fig. S3, Supplementary Material online) and the Muc5A gene is expressed exclusively in the X. tropicalis oviduct as judged by its presence in EST databases. Furthermore, the frog Muc5B and Muc5C mucins are related in sequence to the bird Muc5ac/Muc5b proteins (data not shown). It therefore seems that this part of the large X. tropicalis cluster corresponds to the cluster in mammals, reptiles, and birds.
Finally, in the ray-finned and cartilaginous fishes as well as in the coelacanth L. chalumnae the gene arrangement is similar to the other birds and mammals (fig. 5). Thus, a Muc6 gene is found in a head-to-head arrangement with Muc2, and Tollip is situated next to the Muc5 gene. These results show that the gene order of the mucin cluster of mammals originates from an arrangement in the early vertebrates. Gene duplication events in amphibians and teleost fishes have apparently given rise to additional paralogs.
Conservation of Tissue-Specific Expression of Mucins Among Vertebrates
Gene expression in zebrafish, frog, and chicken was studied by taking advantage of public RNA-Seq datasets (supplementary fig. S2, Supplementary Material online). Muc2, which is located in the intestine in mammals, was found to be expressed also in the intestine of chicken (Muc2), the frog (Muc2A), and zebrafish (Muc2.1, Muc2.4). Muc2A is the Muc2-type mucin in frog that seems to be closely related to the mammalian Muc2, as discussed above. This relationship is also consistent with the Muc2A expression profile.
In the three animals studied, the testis is unusual as all mucins are expressed at similar levels (fig. 4 and supplementary fig. S2, Supplementary Material online). However, it has previously been shown that the mammalian testis has an unusual mode of gene expression in general (Ramskold et al. 2009; Soumillon et al. 2013; Djureinovic et al. 2014; Fagerberg et al. 2014). In this tissue, transcription of the genome is more widespread than in any other organ. Lower vertebrates may well have the same characteristics. Therefore, our observations regarding the expression of mucins in testis may not be of specific interest to mucin biology.
Human MUC19 transcripts have been detected in mucus cells of the submandibular glands and to tracheal submucosal glands (Chen et al. 2004; Zhu et al. 2011). On the other hand evidence has been presented that the protein product is absent in human saliva (Rousseau et al. 2008). Other studies indicate gene expression in the minor salivary gland and in testis (http://gtexportal.org, last accessed April 11, 2016). A study of Muc19 expression in mouse indicated that it is expressed in both major and minor salivary glands (Das et al. 2010). Our results for chicken and zebrafish do not reveal strong expression in any of the tissues available for analysis. However, the frog Muc19 is expressed in dorsal and ventral skin, suggesting that this is a major mucin on the surface of frog skin.
As discussed above, ovomucin seems to be ubiquitous in birds and reptiles. There are previous reports that ovomucin and Muc6 proteins (also called α- and β-ovomucin, respectively) are found in egg white (Watanabe et al. 2004) and our analysis of chicken RNA-Seq data is in agreement with these results (supplementary fig. S2, Supplementary Material online).
Proteins with Characteristics of Gel-Forming Mucins Were Present Early in Metazoan Evolution
By analyzing both available protein sequences and genomic sequences, we identified a total of 192 proteins with at least three VWD domains from 76 invertebrate species (http://www.medkem.gu.se/mucinbiology/mucevo/, last accessed April 11, 2016). In comparison to vertebrates, invertebrates have fewer gel-forming mucins. However, in the invertebrate species most closely related to the vertebrates (the nonvertebrate Deuterostomia), there are numerous mucin-like proteins. Most notable in this respect are the sea squirts Ciona intestinalis and Oikopleura dioica, each with 17 proteins with multiple VWD domains. Some of these proteins contain PTS domains and seem in this respect clearly related to the vertebrate gel-forming mucins.
Lophotrochozoa and Arthropoda have a lower number of VWD proteins (supplementary fig. S4, Supplementary Material online). This is particularly true for the insects that all have only one protein related to mucins and orthologous to Drosophila melanogaster hemolectin (Goto et al. 2001). This protein is composed of three VWD-C8-TIL units, followed by F5_F8_type_C domains (characteristic of coagulation factors F5 and F8) and yet two more VWD-C8-TIL units (in the following referred to as a 3 + 2 VWD structure). Some of the insect proteins have a very small PTS domain close to the F5_F8_type_C domains. The wasp Microplitis demolitor have two 3 + 2 paralogs, one with and the other without a PTS domain (supplementary fig. S4, Supplementary Material online).
The Lophotrochozoa have a larger number of mucin-like proteins and the F5_F8 domain is characteristic of this phylum. CysD domains have been inserted in the region of the F5_F8 domains in Lophotrochozoa, Chelicerata, Myriapoda, and Crustacea. It would seem that the evolution was such that the CysD domains occurred early in Bilateria evolution, but was lost in the insects. In addition, in Lophotrochozoa we also observed a protein with multiple VWD domains and unknown function (supplementary fig. S4, Supplementary Material online). However, the protein does not have the VWD–C8–TIL structure, and is probably not related to the gel-forming mucins.
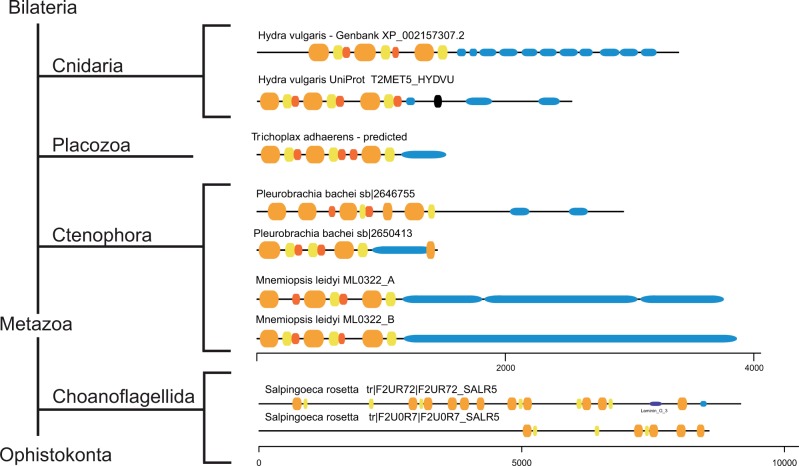
An important conclusion from our analysis of invertebrate proteins is that many mucin-like proteins are observed in deeply branching Metazoa. Gel-forming mucin-like sequences identified in the lower invertebrates are shown in figure 6. In addition to these, the cnidarians N. vectensis and Hydra magnipapillata have mucin-like proteins. It is intriguing to note that there are mucins also in the comb jellies Pleurobrachia bachei and Mnemiopsis leidyi. These are part of the taxon Ctenophora and it is believed that these organisms are our most distant animal relatives (Whelan et al. 2015).
Fig. 6.
Gel-forming mucins evolved early during metazoan evolution. Phylogenetic distribution of proteins in selected basal Metazoa with a domain structure characteristic of gel-forming mucins. VWD, C8, TIL, and PTS domains are shown in orange, yellow, red, and blue, respectively.
The comb jelly M. leidyi has two genes whose sequences are highly similar and that are arranged next to each other in a head-to-head orientation. The authenticity of the Ctenophora mucin genes was further supported by analysis of available RNA-Seq data. According to such data the two P. bachei genes are both expressed. The M. leidyi proteins referred to as A and B in figure 6 are both expressed with the B protein representing the predominant one. The presence of proteins characterized by three VWD8–C8–TIL units and PTS domains (fig. 6) clearly point to a very early origin of gel-forming mucins during metazoan evolution.
There are also proteins with the VWD domain in the choanoflagellates. In fact, our method to predict mucins identified a mucin-like protein present in both Salpingoeca rosetta and Monosiga brevicollis (data not shown). However, these two proteins are likely representing false positives because they are associated with very long introns and they overlap with GenBank proteins that are different from our predicted sequences. Furthermore, the choanoflagellate proteins are lacking TIL domains characteristic of gel-forming mucins. We cannot formally exclude the presence of mucin-like proteins in choanoflagellates, and a definite conclusion can only be reached with improved assemblies of choanoflagellate genomes and transcriptomes.
The Vertebrate FCGBP Protein Is Characterized by an N-Terminal Domain Also Found in Eubacteria
The FCGBP protein colocalizes with mucins in the mucus of epithelial cells. As the function of FCGBP is not understood, we wanted to determine if there is an evolutionary relationship between this protein and the gel-forming mucins and if they have a similar phylogenetic distribution.
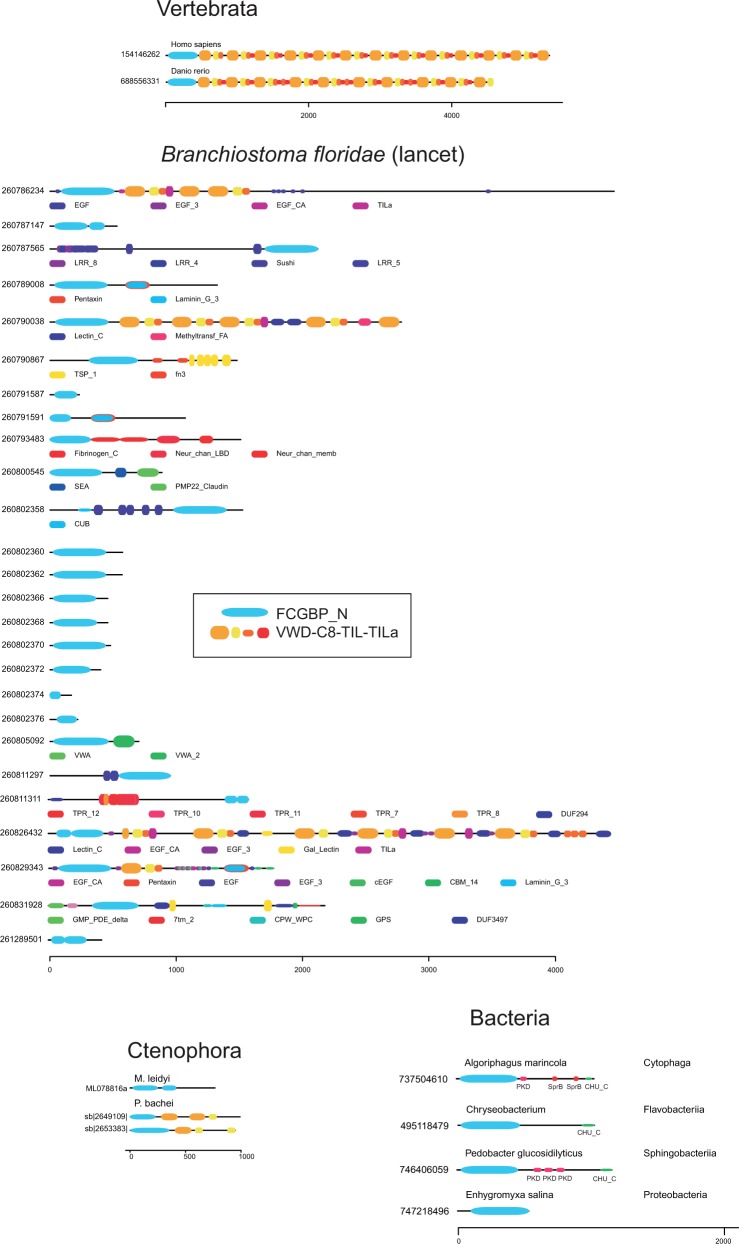
Whereas gel-forming mucins may have up to four VWD–C8–TIL units, the human FCGBP protein is characterized by no less than 13 such units. No other vertebrate protein family has this large number of VWD domains. We found likely orthologs of FCGBP in most vertebrates. In addition to the VWD–C8–TIL units, the human FCGBP protein has an anonymous N-terminal domain. It was earlier suggested that this domain binds to IgG (Harada et al. 1997). We refer to it here as FCGBP_N and this domain was found to be present in all vertebrate FCGBP proteins. With a PSI-BLAST-based approach (Altschul et al. 1997), we identified a total of 974 FCGBP_N-containing proteins from GenBank (supplementary fig. S5, Supplementary Material online). Among these, 235 were from vertebrates, 176 from invertebrates, and 564 were of eubacterial origin. We are confident of the evolutionary relationship between all these proteins as for instance bacterial proteins were identified with very low E-values (1E−90) using human FCGBP as query in a PSI-BLAST search. We were not able to identify the FCGBP_N-domain in fungi, protists, plants, or Archaea. All 974 FCGBP_N-containing proteins were further analyzed with respect to other domains using Pfam information as well as a profile HMM of FCGBP_N. Selected domain structures are shown in figure 7, see also supplementary figure S5, Supplementary Material online.
Fig. 7.
Domain structures of selected proteins with a domain characteristic of FCGBP. In vertebrates, FCGBP proteins have a large number of VWD–C8–TIL units, in addition to the FCGBP_N domain. In the invertebrate B. floridae, there is a large number of proteins with the FCGBP_N domain. All domains that are not FCGBP_N, VWD, C8, TIL, TILa, or PTS domains are explained below each representation of protein domain architecture. For a complete collection of all proteins with the FCGBP domain, see supplementary figure S5, Supplementary Material online.
As Ctenophora proteins are poorly represented in GenBank, we searched specifically for the FCGBP_N domain in the genomes and proteomes of M. leidyi and P. bachei. These searches identified one FCGBP protein in M. leidyi and two in P. bachei. Both of the P. bachei proteins matched transcriptome data from this species, suggesting they are both authentic expressed genes.
The FCGBP protein was found to be well conserved in vertebrates, although an uncertainty is introduced by poor genome assemblies and poor annotation of FCGBP in many vertebrates, due to the presence of multiple and nearly identical VWD domains. It appears though that the FCGBP_N domain is not affected by these assembly and gene prediction problems. Some vertebrate proteins have two copies of the N domain, assuming the protein prediction is correct.
In invertebrates, the FCGBP_N domain occurs in the context of a variety of other protein domain architectures. A remarkable case is Branchiostoma floridae where we identified 26 proteins with the FCGBP_N domain (fig. 7). Some of these have VWD–C8–TIL repeats but the N domain also occurs with other protein domains and in a few proteins in which there is no other domain than FCGBP_N.
In most invertebrates, the FCGBP_N domain does not occur together with VWD domains (supplementary fig. S5, Supplementary Material online). In this respect, it is intriguing to note that there are proteins in Ctenophora that are characterized by both FCGBP_N and VWD domains, reminiscent of proteins of B. floridae and of vertebrates. These results suggest that this combination of domains was invented early in metazoan evolution, just like gel-forming mucins. However, in many other phylas such as Porifera, Placozoa, Cnidaria, Echinodermata, and Hemichordata we do not observe proteins with both FCGBP_N and VWD domains, suggesting that these were lost during evolution and were reinvented at the level of Chordata. The architecture of the FCGBP_N domain combined with VWD–C8–TIL repeats is observed in Cephalochordata and vertebrates implying that this type of protein arose much later in evolution than gel-forming mucins.
Most bacterial proteins with the FCGBP_N domain were detected within the phyla of Flavobacteria, Deltaproteobacteria, Sphingobacteriia, Cytophaga, and Gammaproteobacteria. A remarkable case is Labilithrix luteola, a member of Myxococcales, where 103 different proteins contain the FCGBP_N domain (supplementary fig. S5, Supplementary Material online). There are a few Pfam domains that sometimes occur in conjunction with the FCGBP_N domain in bacteria. Examples are (1) the Polycystic Kidney Disease (PKD) domain, (2) the structurally and functionally anonymous CHU_C domain (C-terminal domain of CHU protein family, where “CHU” refers to Cytophaga hutchinsonii), and (3) the SprB domain (named after SprB, a cell surface protein).
In human, FCGBP is known to be secreted from goblet cells (Johansson et al. 2008). There are 290 out of 564 bacterial proteins, and 219 out of a total of 411 metazoan sequences that have a signal sequence as predicted by SignalP (Petersen et al. 2011). These results indicate that the FCGBP proteins are secreted like the human ortholog. The FCGBP_N domain typically occurs at the N-terminal end of the protein in both metazoan and bacterial proteins, suggesting the N-terminal location is critical for its function (fig. 7 and supplementary fig. S5, Supplementary Material online).
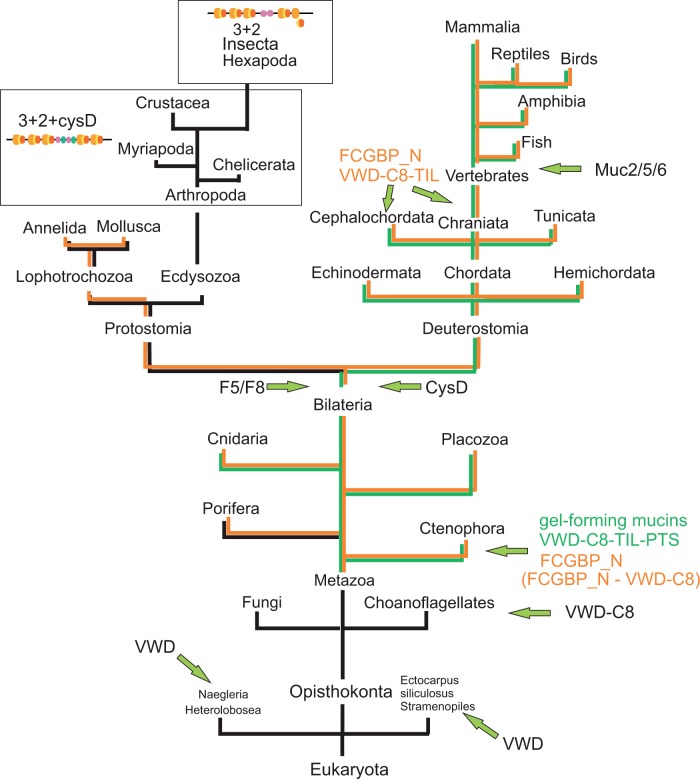
The evolution of gel-forming mucins and the FCGBP protein as well as all the domains characteristic of these proteins is summarized in figure 8 (see Discussion section).
Fig. 8.
Summary of mucin and FCGBP protein evolution. The VWD domain seems to occur in a restricted number of protists, but not in combination with the C8 or TIL domains. The combination of VWD/C8 appears in choanoflagellates and the structure characteristic of gel-forming mucins as well as the FCGBP_N-containing proteins appear in Ctenophora. The F5_F8 and CysD domains occur in Bilateria. A protein with FCGBP_N and multiple VWD–C8–TIL domains appears later in evolution as it is observed only in cephalochordates and vertebrates.
Discussion
We identified gel-forming mucins by searching available genomes with profile HMM models of these proteins. Unless scattered across several contigs, we do not expect to miss any true positive mucins. We eliminated false positive hits through a combination of phylogenetic methods and analysis of protein domain architecture. When considering a complete inventory of mucins in Metazoa an obstacle is that a number of genomes and their corresponding transcriptomes are not fully assembled and for this reason we may lack some homologs from these species. For instance, among the fishes the zebrafish has the most reliable genome assembly, and yet several mucins are not complete in terms of genomic structure. As the mucins often are highly repetitive as they contain PTS domains genome and transcriptome assembly is notoriously difficult. Often one gel-forming mucin gene is mispredicted so as to be represented by two or more different genes. Even in well-characterized genomes, like that of human, there are still genomic pieces being part of mucin genes that are missing. It must also be noted that GenBank and UniProt in general contain a significant portion of erroneous protein sequences. For instance, a protein may represent a pseudogene, it may be based on a partially false exon/intron structure of the corresponding gene, or it may have an incorrect N-terminal end due to misprediction of the translation start site. A protein may also be missing because it was overlooked in a process of genome annotation. Whenever there is uncertainty about the presence of a mucin in a certain phylogenetic clade, we have collected additional information by analyzing the genomic sequence more carefully and we have tried to improve the prediction of a protein by a more careful analysis of the genomic sequence. In addition, we have attempted to find evidence of expression by analyzing RNA-Seq data.
Using a phylogenetic tree based on a large number of sequences we were able to classify gel-forming mucins in a manner that was not previously possible. Using this alignment and tree we are now also able to classify any novel mucins that are discovered. This tree also verified a number of previously known relationships and revealed for the first time that a group of invertebrate gel-forming mucins are similar to vertebrate SCO-spondins.
We have previously reported that a nonPTS mucin-like molecule in chicken corresponds to the alpha subunit of ovomucin whereas the beta subunit is the ortholog to human MUC6 (Lang et al. 2006). We now show that the alpha subunit referred to as “ovomucin” is ubiquitous in reptiles and birds and that the X. tropicalis protein Muc5A seems to be related to ovomucin, although it has a PTS domain. We have not been able to identify a protein resembling ovomucin in fishes, but classification of fish mucins is more difficult as there is a rather significant sequence divergence between fish and amphibians. Chicken ovomucin is known to be a major component of egg white (Watanabe et al. 2004). The occurrence of this protein in birds, reptiles, and in X. tropicalis could be related to the fact that in these species embryonic development takes place external of the female body.
In the frog X. tropicalis, the gel-forming mucin repertoire has been markedly expanded, as judged by our finding of 26 different mucin genes, as compared with only five to six in mammals, reptiles, and birds. Our analysis of public expression data revealed that there is a high degree of diversification also when it comes to tissue distribution and developmental stages. Recently, the genome of the amphibian Nanorana parkeri became available (Sun et al. 2015). A preliminary analysis of its genome (data not shown) indicates that it has approximately the same set of mucins as X. tropicalis. Conclusion about the genomic organization cannot be reached in detail as the current Na. parkeri genome assembly is more fragmented than the X. tropicalis assembly version 7.
The zebrafish D. rerio does not seem to have as many mucins as X. tropicalis and the same is true for ceolacanth (L. chalumnae), spotted gar (Le. oculatus), and Australian ghost shark (C. milii). For instance, in C. milii we identify only one Muc2, one Muc5, two Muc6, and three paralogs of Muc19. It would therefore seem that the early vertebrate development of mucins featured only a small number of mucin genes. It should be kept in mind however that the deeply rooting vertebrates have not been exhaustively sequenced, and we may for this reason have a fragmented view of their mucin repertoire.
Our analysis of RNA-Seq data of zebrafish is by and large consistent with a previous study (Jevtov et al. 2014), although Muc2.4 was not monitored in that particular investigation. We present evidence that in the intestine the Muc2.4 gene is highly expressed, whereas Muc2.1 is expressed at a lower level.
Our results indicate that Muc6 is missing in most teleost fishes and that Muc19 is absent in birds. We cannot formally exclude that these proteins are present in all vertebrates as there for instance could be problems with genome assembly and/or genome annotation. However, we have analyzed a very large number of fish genomes (28 species). We identified Muc6 only in the genome of spotted gar, Le. oculatus, which is the only non-teleost ray-finned fish we investigated. Similarly, we failed to identify Muc19 in any of the bird genomes available, and our analysis of synteny suggests that the Muc19 gene was deleted in birds (supplementary fig. S1, Supplementary Material online).
The FCGBP protein was first characterized in human and mouse (Harada et al. 1997). We noted that an N-terminal domain of FCGBP is unique to this group of proteins and therefore useful in bioinformatics approaches to identify members of the family. It is intriguing that in a large majority of the proteins with the FCGBP_N domain, this domain is located at the N-terminal end. This location may be important for proper function or for correct transport outside the cell.
An unexpected observation was that the N-terminal domain is present in a group of bacterial proteins. It seems likely that these bacterial proteins, as well as the eukaryotic proteins with FCGBP_N domain, are secreted. The bacterial proteins have a domain composition different from the eukaryotic proteins. CHU_C, PKD, and SprB are examples of protein domains occurring together with the FCGBP_N domain but they are also found in many other bacterial proteins that do not contain the FCGBP_N domain. The PKD domain often occurs in the extracellular region of membrane-associated bacterial proteins. The function of the CHU domain, originally identified in C. hutchinsonii, is not known.
A majority (425 of 564) of the bacterial proteins with FCGBP_N domains are from the bacterial groups flavobacteria, cytophaga and sphingobacteria. We have noted that a biological function common to these bacteria is a mode of movement known as “gliding” motility (Islam and Mignot 2015). This motility involves neither flagella nor type IV pili. The SprB domain referred to above is found in the SprB protein, a cell surface component that has been implicated in gliding motility (Nakane et al. 2013). The bacterium C. hutchinsonii mentioned above has been studied because of its gliding motility, but the molecular basis of this type of bacterial motility is in general poorly understood. One exception is the mechanism characterizing Myxococcus xanthus, a species where we also identified FCGBP_N proteins. According to one model the motility in M. xanthus is achieved by focal adhesion that involves slime secretion, where the slime acts as a glue for surface adhesion (Islam and Mignot 2015). There could possibly be a role for the FCGBP_N domain in gliding motility in bacteria that is somehow related to its mucus association in higher animals.
The evolution of mucin-like proteins and individual domains characteristic of these proteins is summarized in figure 8. A key component of mucins is the VWD domain. In our analysis, we identify this element in a restricted number of protists (such as Naegleria and Ectocarpus), suggesting that it was an early invention during eukaryotic evolution. In choanoflagellates, it appears in combination with the C8 domain. In ctenophores, there is a substantial change as we now observe three repeats of the VWD–C8–TIL unit as well as PTS domains, that is, the hallmarks of gel-forming mucins.
The CysD domain appears at the level of Bilateria. This domain then persists in both the Deuterostomia and Protostomia branches except for Hexapoda. In Deuterostomia, it appears together with PTS. The F5/F8 domain has a similar evolution as it appears in Bilateria. However, while it is present in Protostomia, Hemichordata, and Cephalochordata, it is lost from the gel-forming mucins in vertebrates.
Finally, we can trace the eukaryotic origin of the FCGBP_N domain to the Ctenophora. This domain is present in all major metazoan branches, but seems to have been lost in Arthropoda.
In conclusion, we have identified proteins with properties of gel-forming mucins in Ctenophora, a group shown to be sister to all other animals (Whelan et al. 2015). This demonstrates a very early origin of this family of proteins. We hypothesize that the gel-forming mucin-like proteins in Ctenophora play a role in the digestive system of these animals. A protein resembling the vertebrate FCGBP protein appears later in evolution as it is observed only in cephalochordates and vertebrates. In human, the FCGBP protein colocalizes with mucins in goblet cells and in the mucus. Experimental work is now required to clarify the biological role of this protein and of its N-terminal domain.
Materials and Methods
Analysis with Profile HMMs
In order to bioinformatically identify gel-forming mucins we made use of the hmmer software (http://hmmer.org, last accessed April 11, 2016, Eddy 2011). Profile HMM models of gel-forming mucin protein sequences were created on the basis of a reliable alignment of previously described and well-characterized full-length mucin sequences. In this alignment, we replaced the authentic PTS region with a region containing a random sequence of proline, threonine, and serine but with the same composition as a PTS domain. The length of the PTS domain differed between the sequences in the alignment and was in the range 100–2,000. The protein sequence databases GenBank, release 205 (Benson et al. 2015) and UniProt, release 2014_09 (2015) were searched with this model using hmmsearch of the hmmer package and hits were filtered so that only these with E-value below 1E−07 were retrieved.
Protein domain analysis was performed with hmmscan of the hmmer package using protein domains in the Pfam database version 27.0 (Finn et al. 2014). For construction of the profile HMM of the N-terminal domain of FCGBP, we used an alignment with 637 sequences (available at http://www.medkem.gu.se/mucinbiology/mucevo/, last accessed April 11, 2016). For analysis of the domain composition of the retrieved protein sequences, the HMM of this domain was added to the Pfam HMM database. In the next release of Pfam (30.0), the FCGBP_N domain will be included in Pfam A. Protein domain structure was graphically presented using in-house R scripts.
PSI-BLAST (Altschul et al. 1997) was used for identification of proteins containing the N-terminal domain characteristic of vertebrate FCGBP. The human FCGBP N-terminal domain was used as original query and all hits after six rounds above default E-value cutoff were retrieved.
Analysis of PTS Domains
PTS domains were predicted using a previously described method using an in-house Perl script (Lang et al. 2004).
Prediction of Protein Sequences From Genomic Sequences
To identify gel-forming mucins we also analyzed genomic sequences. The genome assemblies used are listed in the supplementary dataset 2, Supplementary Material online. Selected species (194 vertebrates and 79 invertebrates) with an available genome assembly were analyzed with genewise (Wise2, Birney et al. 2004). In this method we used a similar profile HMM model as for protein sequence database searches. We tested different lengths of the PTS domain and found that an optimal length of the modeled PTS domain for the purpose of gene prediction with genewise was ∼2,000 amino acids. To improve on the speed of this step based on genewise, we first identified the relevant regions of the genome, either using tblastn (Altschul et al. 1990, 1997) with selected mucins as queries or using genewise (version wise2-2-0) with the Pfam VWD domain profile HMM. These identified regions were then analyzed with genewise using the mucin hmmer model, thus generating peptide sequences for the predicted genes. The parameters used with genewise were “-hmmer -pretty -silent -both -pep”. We only considered for further analysis predicted proteins with at least three VWD domains.
To test the efficiency of this prediction based on genomic sequence there are few mucin sequences available where the complete structure of the gene and protein is known. Testing with available human, mouse, and cat mucin sequences, showed that the method predicted the correct domain structure of Muc2, Muc5ac, Muc5b, and Muc6 with an overall sequence identity of ∼95% for the nonPTS portion of the protein.
In order to compare all our predicted sequences to sequences already present in GenBank and UniProt we carried out blastp searches using the predicted sequence as queries. The top hit for every query was aligned to the query using ClustalW (Thompson et al. 2002). The resulting alignment was analyzed and whenever the two sequences were from the same species and there was a region of at least 30 amino acids being identical between the two sequences, we inferred that this GenBank/UniProt sequence corresponded to the query. Predicted sequences that did not fulfill this criterion were considered as novel proteins.
Availability of Sequence Data
All proteins identified in this study, including sequences and domain structures, are available as supplementary files and at http://www.medkem.gu.se/mucinbiology/mucevo, last accessed April 11, 2016.
Phylogeny
Phylogenetic analyses were carried out using neighbor-joining as performed by ClustalW (Thompson et al. 2002), Clustal Omega (Sievers and Higgins 2014), or with MrBayes (Ronquist et al. 2012). For the MrBayes tree in figure 2 we used 400,000 generations and the parameter prset aamodelpr = mixed.
Analysis of Public RNA-Seq data
RNA-Seq data were downloaded from the Sequence Read Archive, NCBI. In addition, RNA-Seq data of P. bachei and M. leidyi were obtained from http://neurobase.rc.ufl.edu/pleurobrachia, last accessed April 11, 2016 (Moroz et al. 2014), and the National Human Genome Research Institute (http://research.nhgri.nih.gov/mnemiopsis, last accessed April 11, 2016). For X. tropicalis, D. rerio, and Gallus gallus, available annotation in GFF3 format was modified to produce a more accurate annotation of mucin genes. This annotation was based on our careful analysis of mucin gene structure in these species. Reads from an RNA-Seq dataset were aligned to the relevant reference genome using HISAT (Kim et al. 2015). Information about the RNA-Seq datasets used are in the supplementary dataset 2, Supplementary Material online. The number of reads mapping to each gene as listed in the annotation table was determined with htseq-count (Anders et al. 2015). Normalization of gene expression was obtained by calculating RPKM values for each RNA-Seq sample. The heat maps were generated in MATLAB by displaying the logarithmic value of the normalized expression. Each sample type is comprised of the mean value of the expression for all samples from that particular tissue type or developmental stage.
Supplementary Material
Supplementary figures S1–S5 and datasets S1 and S2 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors acknowledge the grants to T.L. from the State Scholarship Fund of the China Scholarship Council (file number 201404910355) and the National Natural Science Foundation of China (grant number 61271447). This work was supported by the Swedish Research Council, The Swedish Cancer Foundation, The Knut and Alice Wallenberg Foundation, National Institute of Allergy and Infectious Diseases (U01AI095473, the content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH), and The Swedish Foundation for Strategic Research - The Mucus-Bacteria-Colitis Center (MBC) of the Innate Immunity Program.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215:403–410. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambort D, Johansson ME, Gustafsson JK, Ermund A, Hansson GC. 2012. Perspectives on mucus properties and formation—lessons from the biochemical world. Cold Spring Harb Perspect Med. 2:a014159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W. 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson DA, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. 2015. GenBank. Nucleic Acids Res. 43:D30–D35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, Clamp M, Durbin R. 2004. GeneWise and Genomewise. Genome Res. 14:988–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Zhao YH, Kalaslavadi TB, Hamati E, Nehrke K, Le AD, Ann DK, Wu R. 2004. Genome-wide search and identification of a novel gel-forming mucin MUC19/Muc19 in glandular tissues. Am J Respir Cell Mol Biol. 30:155–165. [DOI] [PubMed] [Google Scholar]
- Corfield AP. 2015. Mucins: a biologically relevant glycan barrier in mucosal protection. Biochim Biophys Acta 1850:236–252. [DOI] [PubMed] [Google Scholar]
- Das B, Cash MN, Hand AR, Shivazad A, Grieshaber SS, Robinson B, Culp DJ. 2010. Tissue distibution of murine Muc19/smgc gene products. J Histochem Cytochem. 58:141–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djureinovic D, Fagerberg L, Hallstrom B, Danielsson A, Lindskog C, Uhlen M, Ponten F. 2014. The human testis-specific proteome defined by transcriptomics and antibody-based profiling. Mol Hum Reprod. 20:476–488. [DOI] [PubMed] [Google Scholar]
- Eddy SR. 2011. Accelerated profile HMM searches. PLoS Comput Biol. 7:e1002195.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagerberg L, Hallstrom BM, Oksvold P, Kampf C, Djureinovic D, Odeberg J, Habuka M, Tahmasebpoor S, Danielsson A, Edlund K, et al. 2014. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics 13:397–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, et al. 2014. Pfam: the protein families database. Nucleic Acids Res. 42:D222–D230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto A, Kumagai T, Kumagai C, Hirose J, Narita H, Mori H, Kadowaki T, Beck K, Kitagawa Y. 2001. A Drosophila haemocyte-specific protein, hemolectin, similar to human von Willebrand factor. Biochem J. 359:99–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada N, Iijima S, Kobayashi K, Yoshida T, Brown WR, Hibi T, Oshima A, Morikawa M. 1997. Human IgGFc binding protein (FcgammaBP) in colonic epithelial cells exhibits mucin-like structure. J Biol Chem. 272:15232–15241. [DOI] [PubMed] [Google Scholar]
- Hollingsworth MA, Swanson BJ. 2004. Mucins in cancer: protection and control of the cell surface. Nat Rev Cancer. 4:45–60. [DOI] [PubMed] [Google Scholar]
- Islam ST, Mignot T. 2015. The mysterious nature of bacterial surface (gliding) motility: a focal adhesion-based mechanism in Myxococcus xanthus. Semin Cell Dev Biol. 46:143–154. [DOI] [PubMed] [Google Scholar]
- Jevtov I, Samuelsson T, Yao G, Amsterdam A, Ribbeck K. 2014. Zebrafish as a model to study live mucus physiology. Sci Rep. 4:6653.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson ME, Ambort D, Pelaseyed T, Schutte A, Gustafsson JK, Ermund A, Subramani DB, Holmen-Larsson JM, Thomsson KA, Bergstrom JH, et al. 2011. Composition and functional role of the mucus layers in the intestine. Cell Mol Life Sci. 68:3635–3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson ME, Gustafsson JK, Holmen-Larsson J, Jabbar KS, Xia L, Xu H, Ghishan FK, Carvalho FA, Gewirtz AT, Sjovall H, et al. 2014. Bacteria penetrate the normally impenetrable inner colon mucus layer in both murine colitis models and patients with ulcerative colitis. Gut 63:281–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson ME, Phillipson M, Petersson J, Velcich A, Holm L, Hansson GC. 2008. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci U S A. 105:15064–15069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson ME, Thomsson KA, Hansson GC. 2009. Proteomic analyses of the two mucus layers of the colon barrier reveal that their main component, the Muc2 mucin, is strongly bound to the Fcgbp protein. J Proteome Res. 8:3549–3557. [DOI] [PubMed] [Google Scholar]
- Kawahara R, Nishida M. 2007. Extensive lineage-specific gene duplication and evolution of the spiggin multi-gene family in stickleback. BMC Evol Biol. 7:209.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL. 2015. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12:357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang T, Alexandersson M, Hansson GC, Samuelsson T. 2004. Bioinformatic identification of polymerizing and transmembrane mucins in the puffer fish Fugu rubripes. Glycobiology 14:521–527. [DOI] [PubMed] [Google Scholar]
- Lang T, Hansson GC, Samuelsson T. 2006. An inventory of mucin genes in the chicken genome shows that the mucin domain of Muc13 is encoded by multiple exons and that ovomucin is part of a locus of related gel-forming mucins. BMC Genomics 7:197.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang T, Hansson GC, Samuelsson T. 2007. Gel-forming mucins appeared early in metazoan evolution. Proc Natl Acad Sci U S A. 104:16209–16214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milla CE, Moss RB. 2015. Recent advances in cystic fibrosis. Curr Opin Pediatr. 27:317–324. [DOI] [PubMed] [Google Scholar]
- Moroz LL, Kocot KM, Citarella MR, Dosung S, Norekian TP, Povolotskaya IS, Grigorenko AP, Dailey C, Berezikov E, Buckley KM, et al. 2014. The ctenophore genome and the evolutionary origins of neural systems. Nature 510:109–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakane D, Sato K, Wada H, McBride MJ, Nakayama K. 2013. Helical flow of surface protein required for bacterial gliding motility. Proc Natl Acad Sci U S A. 110:11145–11150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Necsulea A, Soumillon M, Warnefors M, Liechti A, Daish T, Zeller U, Baker JC, Grutzner F, Kaessmann H. 2014. The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature 505:635–640. [DOI] [PubMed] [Google Scholar]
- Perez-Vilar J, Hill RL. 1999. The structure and assembly of secreted mucins. J Biol Chem. 274:31751–31754. [DOI] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786. [DOI] [PubMed] [Google Scholar]
- Ramskold D, Wang ET, Burge CB, Sandberg R. 2009. An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. PLoS Comput Biol. 5:e1000598.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61:539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseau K, Kirkham S, Johnson L, Fitzpatrick B, Howard M, Adams EJ, Rogers DF, Knight D, Clegg P, Thornton DJ. 2008. Proteomic analysis of polymeric salivary mucins: no evidence for MUC19 in human saliva. Biochem J. 413:545–552. [DOI] [PubMed] [Google Scholar]
- Sievers F, Higgins DG. 2014. Clustal Omega, accurate alignment of very large numbers of sequences. Methods Mol Biol. 1079:105–116. [DOI] [PubMed] [Google Scholar]
- Soumillon M, Necsulea A, Weier M, Brawand D, Zhang X, Gu H, Barthes P, Kokkinaki M, Nef S, Gnirke A, et al. 2013. Cellular source and mechanisms of high transcriptome complexity in the mammalian testis. Cell Rep. 3:2179–2190. [DOI] [PubMed] [Google Scholar]
- Sun YB, Xiong ZJ, Xiang XY, Liu SP, Zhou WW, Tu XL, Zhong L, Wang L, Wu DD, Zhang BL, et al. 2015. Whole-genome sequence of the Tibetan frog Nanorana parkeri and the comparative evolution of tetrapod genomes. Proc Natl Acad Sci U S A. 112:E1257–E1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan MH, Au KF, Yablonovitch AL, Wills AE, Chuang J, Baker JC, Wong WH, Li JB. 2013. RNA sequencing reveals a diverse and dynamic repertoire of the Xenopus tropicalis transcriptome over development. Genome Res. 23:201–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Higgins DG. 2002. Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics Chapter 2:Unit 2.3. [DOI] [PubMed] [Google Scholar]
- Thornton DJ, Rousseau K, McGuckin MA. 2008. Structure and function of the polymeric mucins in airways mucus. Annu Rev Physiol. 70:459–486. [DOI] [PubMed] [Google Scholar]
- UniProt Consortium. 2015. UniProt: a hub for protein information. Nucleic Acids Res. 43:D204–D212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Shimoyamada M, Onizuka T, Akiyama H, Niwa M, Ido T, Tsuge Y. 2004. Amino acid sequence of alpha-subunit in hen egg white ovomucin deduced from cloned cDNA. DNA Seq. 15:251–261. [DOI] [PubMed] [Google Scholar]
- Whelan NV, Kocot KM, Moroz LL, Halanych KM. 2015. Error, signal, and the placement of Ctenophora sister to all other animals. Proc Natl Acad Sci U S A. 112:5773–5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu DF, Chen Y, Han JM, Zhang H, Chen XP, Zou WJ, Liang LY, Xu CC, Liu ZG. 2008. MUC19 expression in human ocular surface and lacrimal gland and its alteration in Sjogren syndrome patients. Exp Eye Res. 86:403–411. [DOI] [PubMed] [Google Scholar]
- Zhou YF, Eng ET, Zhu J, Lu C, Walz T, Springer TA. 2012. Sequence and structure relationships within von Willebrand factor. Blood 120:449–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Lee P, Yu D, Tao S, Chen Y. 2011. Cloning and characterization of human MUC19 gene. Am J Respir Cell Mol Biol. 45:348–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.