Abstract
CHK2 is a checkpoint kinase involved in the ATM-mediated response to double strand DNA breaks. Its potential as a drug target is still unclear but inhibitors of CHK2 may increase the efficacy of genotoxic cancer therapies in a p53 mutant background by eliminating one of the checkpoints or DNA repair pathways contributing to cellular resistance. We report here the identification and characterization of a novel CHK2 kinase inhibitor, CCT241533. X-ray crystallography confirmed that CCT241533 bound to CHK2 in the ATP pocket. This compound inhibits CHK2 with an IC50 of 3 nM and shows minimal cross reactivity against a panel of kinases at 1 μM. CCT241533 blocked CHK2 activity in human tumor cell lines in response to DNA damage, as demonstrated by inhibition of CHK2 autophosphorylation at S516, band-shift mobility changes and HDMX degradation. CCT241533 did not potentiate the cytotoxicity of a selection of genotoxic agents in several cell lines. However, this compound significantly potentiates the cytotoxicity of two structurally distinct Poly (ADP-ribose) polymerase (PARP) inhibitors. Clear induction of the pS516 CHK2 signal was seen with a PARP inhibitor alone and this activation was abolished by CCT241533, implying that the potentiation of PARP inhibitor cell killing by CCT241533 was due to inhibition of CHK2. Consequently CHK2 inhibitors may have therapeutic activity in combination with PARP inhibitors.
Keywords: CHK2, PARP, CCT241533, checkpoints, Olaparib
Introduction
The cell responds to genotoxic stress via a complex integrated network of signaling pathways and checkpoints. DNA damage is sensed by either the MRN/ATM/CHK2 pathway (double strand breaks, DSB) or the ATRIP/ATR/CHK1 pathway (single strand breaks, SSB) (1) and there is cross talk at multiple levels. Pathway activation initiates DNA repair, induces cell cycle arrest until repair is completed and primes both the p53-dependent and independent apoptotic pathways (2, 3).
Detection of DNA DSBs by the MRE11–RAD50–NBS1 (MRN) complex leads to activation of ataxia telangiectasia mutated (ATM) kinase which can then phosphorylate CHK2 on threonine 68 (T68). This phosphorylation allows CHK2 to dimerise and autophosphorylate in trans at threonines 383 and 387 resulting in full activation, and autophosphorylation in cis at serine 516 (4, 5). Once active, CHK2 phosphorylates a wide range of targets involved in control of the cell cycle, DNA repair and apoptosis (2, 3). Cell cycle arrest can be achieved through the inhibitory phosphorylation of members of the CDC25 phosphatase family, factors required to activate cyclin dependent kinases and drive the cell cycle forward (6). A main role of CHK2 in DNA repair is to promote the homologous recombination (HR) pathway via BRCA1 (7). Another downstream target of the CHK2 kinase is the tumor suppressor protein p53. When stabilized by phosphorylation, either directly by CHK2 or through intermediate proteins, p53 can transcriptionally activate many factors required for cell cycle arrest, DNA repair and apoptosis (8).
The utility of conventional DNA damaging anticancer drugs is limited by protective mechanisms conferred by cell cycle checkpoints and DNA repair pathways in tumors as well as deleterious side effects on normal tissue. It has been suggested that abrogating the G2 checkpoint may increase the efficacy of genotoxic agents preferentially in p53 deficient backgrounds, as normal tissue would be rescued at the p53 dependent G1/S checkpoint (3, 9, 10). There is increasing evidence that this is the case for inhibitors of CHK1 (11–16) but the case for CHK2 is still unclear and may depend on the cell line and cytotoxic agent employed (17–23). However, in cells that possess a functional p53 pathway, there is evidence that CHK2 inhibitors protect against apoptosis induced by ionizing radiation (IR) and taxol (21, 24–27), presumably by blocking CHK2 dependent activation of p53. This additional protection of normal tissue could make CHK2 inhibitors attractive therapeutic agents.
We report the identification of CCT241533, a potent and selective ATP competitive inhibitor of CHK2. We show that CCT241533 blocks the genotoxic activation of CHK2 in three cell lines exhibiting confirmed functional CHK2 activity. In view of the lack of clarity regarding the role of CHK2 inhibition in p53 mutant cells, a rigorous evaluation of the effects of CCT241533 in p53 compromised tumor cells was performed. Studies with CCT241533 in combination with several different genotoxic anticancer agents, which activate CHK2, showed that inhibition of CHK2 activity did not enhance drug cytotoxicity. By contrast CCT241533 potentiated the cytotoxicity of poly (ADP ribose) polymerase (PARP) inhibitors in p53 defective tumor cells concomitantly with the inhibition of several CHK2 biomarkers.
Materials and Methods
Compounds and ionizing radiation
CCT241533 was synthesized as described (28). Mitomycin C (MMC) and bleomycin were obtained from Kyowa Kirin (Tokyo, Japan). AG14447 was synthesized in house (29). Olaparib was purchased from JS Research Chemicals Trading (Schleswig-Holstein, Germany). All other compounds were obtained from Sigma-Aldrich Chemical Co (Poole, Dorset, UK). Ionizing radiation was generated using an AGO HS 320kV X-ray system (AGO Installations Ltd, Aldermaston, UK) set at 250kV and 10mA and used at 1 or 1.5 Gy/min.
CHK2 enzyme assays
CCT241533 activity against recombinant human CHK2 kinase activity was measured as previously described (28).
Kinome profile
Kinase profiling was performed at The National Centre for Protein Kinase Profiling (MRC Protein Phosphorylation Unit, Dundee, UK) using 20 μM ATP and a fixed concentration of 1 μM test compound.
Crystallography
Expression and purification of the kinase domain of human CHK2 was carried out as previously described (30). Crystals of CHK2 bound to the inhibitor were generated as described in the supplementary material.
Cells
The human colon cancer cell lines HT-29, HCT-116, breast cancer line MCF7, glioblastoma line U-87 MG, ovarian cancer lines OVCAR-3, OVCAR-5, IGROV and OVCAR-8, osteosarcoma line U-2 OS, lung carcinoma cell line A549, prostate cancer line LNCaP and cervical cancer line HeLa were obtained from ATCC (Manassas, VA, USA). The human ovarian cancer cell line A2780 was obtained from ECACC (Sigma-Aldrich) and the human ovarian cancer cell line CH1 was derived in house (31). Cells were grown in DMEM or RPMI 1640 containing 10% FBS (PAA “Gold”, Pasching, Austria) and 5mM glutamine in a humidified atmosphere of 5% CO2 at 37°C and passaged ≤ 6 months prior to renewal from frozen stocks as indicated. Cells were regularly screened for mycoplasma by PCR (VenorGem, Minerva Biolabs, Berlin, Germany).
Cell growth inhibition assays
Cells were plated in 96 well or 6 well plates and allowed to attach for 36 hours to ensure exponential growth at the time of treatment. Growth inhibition was determined using a 96 hour (four cell-doublings) SRB assay and the growth inhibitory IC50 (GI50) values were derived as previously described (32).
Colony forming assays
Cells were plated out at appropriate concentrations and allowed to attach for at least 16 hours before addition of drugs. After 7-10 days, colonies were fixed, stained and counted by eye or Image J software (NIH, Bethesda, Maryland, USA).
Cell cycle analysis
Ethanol fixed cells were washed with PBS, treated with RNase, stained with propidum iodide and analyzed on a BD LSR II flow cytometer, (Becton Dickinson, Oxford, UK). Cell cycle analysis (Watson pragmatic model) was carried out using FlowJo software (Treestar Inc, Ashland, OR).
Apotosis assay
Cells were harvested using accutase (Sigma) and stained for Annexin V using Annexin V-FITC from eBioscience (San Diego, CA, USA), according to the manufacturers protocol and analyzed on a BD LSR II flow cytometer, (Becton Dickinson, Oxford, UK). Data analysis was performed with FlowJo software (Treestar Inc, Ashland, OR).
Potentiation assays
Cells were exposed to a fixed concentration (GI50) of CCT241533 in combination with increasing concentrations of either PARP inhibitor or cytotoxic drug in a 96 hour SRB assay or 7-10 day colony forming assay. The ability of CCT241533 to enhance cell killing was expressed as a potentiation index (PI) which was the ratio of GI50 for the genotoxic or PARP inhibitor alone : GI50 for the genotoxic or PARP inhibitor in combination with a CHK2 inhibitor. Thus PI >1 indicates potentiation and PI <1 indicates protection.
Immunoblot assays
Western blotting was performed as previously described (13). Antibodies used were: total CHK2 (sc8813, Santa Cruz Biotechnology, Inc., CA, U.S.A.), CHK2-pT68, CHK2-pS516 (2661L, 2669S, NEB, Hitchin, UK), GAPDH (Chemicon, Millipore, Watford, Herts, UK), HDMX (A300-287A , Bethyl Laboratories, TX, USA), p53 (Ab-6, Oncogene Science, Cambridge, MA, USA) p21, (#05-345, Upstate, NY, USA) and RAD50, MRE11, NBS1, (#GTX70228, #GTX70212, #GTX70222 GeneTex inc, Irvine, CA, USA).
Statistics
Statistical significance was determined using unpaired, two-tailed, t-tests as appropriate, using GraphPad Prism 5 software (San Diego, CA, USA). P values indicate the PI was significantly different from unity (PI ≠ 1).
Results
CCT241533 is a potent and selective CHK2 inhibitor
CCT241533 (Figure 1A, (28)) was shown to be an ATP competitive inhibitor of recombinant CHK2 in vitro (Figure 1B) with an IC50 of 3 nM and Ki of 1.16 nM (Figure S1A). Preincubation of CCT241533 with CHK2 had no effect on enzyme kinetics, consistent with a reversible mechanism of action (Figure S1B). The corresponding CHK1 IC50 was 245 nM, giving an 80-fold selectivity. A screen of 85 kinases with 1 μM CCT241533 identified 4 other kinases (PHK, MARK3, GCK and MLK1, highlighted in Table S1) with >80% inhibition. Mouse plasma protein binding was 99% (data not shown).
Figure 1. (A) Chemical structure of CCT241533 B) CCT241533 is an ATP competitive inhibitor of CHK2.
CHK2 enzyme activity was measured at different concentrations of ATP. (C) X-ray crystal structure of CCT241533 bound to CHK2. Key interacting residues surrounding the bound compound are show as ‘ball and stick’ representation. The carbon atoms of CCT241533 are colored in cyan, nitrogens in blue and oxygens in red. The electron density corresponding to an mFo-DFc omit map contoured at 2.5 σ (brown mesh) is shown. The relative positions of the αC helix, glycine-rich loop, DFG motif (magenta) and hinge region are also indicated. (D) Schematic diagram highlighting the key interactions of CCT241533 with the active site residues of CHK2. Dotted lines represent hydrogen bonds whilst solid curved lines represent hydrophobic interactions.
Crystal structure of CCT241533 bound to CHK2
The X-ray crystal structure of CCT241533 bound to the kinase domain of human CHK2 was determined at a resolution of 2.3 Å. The inhibitor occupied the ATP-binding site and was sandwiched between the hydrophobic side chains of valine 234 and leucine 354 (Figures 1 C and D). The hydroxyl of the phenol group was hydrogen-bonded to the backbone amide of methionine 304, providing the “hinge-binding” interaction seen in many kinase inhibitor structures, while also being involved in an intramolecular hydrogen-bond back to the quinazoline core. An additional hydrogen-bond was made between the side-chain of glutamic acid 308 and the 4-amino moiety of the quinazoline, as well as between the backbone oxygen of glutamic acid 251 and the pendant pyrrolidine. The methoxy substituents of the quinazoline core made no direct interactions with the kinase, pointing away from the active site and facing into the solvent.
Interestingly, the glycine-rich loop of the kinase was stabilized through predominantly hydrophobic packing interactions with the dimethylalcohol group of the inhibitor, giving a readily interpretable electron density for the entire loop, with the exception of cysteine 231 which appeared to be conformationally flexible.
Characterization of the DNA damage response pathway in a panel of cell lines
The MRN complex, which is important for CHK2 activation, has been shown to be defective in several human tumor cell lines (33). In order to identify cell lines with active CHK2 pathways suitable for further studies, we immunoblotted 14 human tumor cell lines for the expression of components of the CHK2 pathway and for biomarkers of CHK2 activation. For this experiment IR treatment was employed to induce DNA DSBs and activate the ATM/CHK2 pathway (34, 35). Samples were prepared 4 hours following IR (5 Gy) and immunoblotted for a range of proteins (Figure 2A). IR activates ATM, which phosphorylates CHK2 on threonine 68 (pT68), and allows dimerisation of the protein. The dimerised CHK2 then autophosphorylates in trans, on threonines 383 and 387 and in cis on serine 516 (pS516). This fully phosphorylated and activated CHK2 gives rise to a change in mobility which can be visualized as a band shift by SDS-PAGE. A CHK2 band shift was clearly observed in 10 of the cell lines (HT-29, U-87 MG, A2780, OVCAR-3, OVCAR-5, IGROV, A459, LNCaP, HeLa, and OVCAR-8) and they all showed induction of pS516, consistent with CHK2 activation. An increase in pS516 signal was seen in the CH1 cell line without a clear band shift. An IR-induced decrease in HDMX, which is targeted for destruction following phosphorylation by CHK2, was seen in 5 of the cell lines (HCT-116, A459, LNCaP, HeLa, MCF7). The expression of the MRN components varied between the different cell lines, with HCT-116 having the lowest levels and this correlated with defective CHK2 activation, as previously published (33). The p53 functional status of the cell lines was assessed by monitoring IR-induced stabilization of p53 protein and subsequent p53-induced transactivation of p21 and agreed with published data (36). The HT-29 and HeLa cell lines were selected for subsequent studies as they demonstrated functional activation of CHK2 in response to IR, as shown by CHK2 band shift and induction of pS516 signal and were functionally defective for p53.
Figure 2. (A) Characterization of the CHK2 activation pathway and p53 status in a panel of human tumor cell lines.
Cells were irradiated (5 Gy) and samples taken 4 hours post treatment. Similar results were obtained in a repeat experiment. (B) Effect of CCT241533 on etoposide induced biomarker changes. HT-29 or HeLa cells were pre-incubated with different concentrations of CCT241533 for 1 hour, followed by the addition of 50 μM etoposide or DMSO control and incubated for a further 5 hours. Similar results were obtained in repeat experiments.
CCT241533 inhibits CHK2 activation in a range of human tumor cell lines
The cytotoxicity of CCT241533 in HT-29, HeLa and MCF-7, measured as the growth inhibitory IC50 (GI50) by SRB assay, was 1.7, 2.2 and 5.1 μM, respectively. Treatment of HT-29 cells with 50 μM etoposide for 5 hours induced phosphorylation of both T68 and S516 on CHK2 and a bandshift in the protein (Figure 2B). While CCT241533 blocked etoposide-induced autophosphorylation of CHK2 on S516 at concentrations as low as 0.5 μM, considerably below the GI50, inhibition of the CHK2 band shift was only seen at ≥5 μM and there was no loss of the ATM dependent T68 phosphorylation in this cell line. Inhibition of the autophosphorylation of S516 in HeLa cells occurred at ≥0.5 μM with inhibition of the band-shift at ≥20 μM (Figure 2B). Moreover, there was clear evidence that T68 phosphorylation was induced or retained following CHK2 inhibition (Figure 2B). There was minimal inhibition of CHK1 autophosphorylation at S296 by CCT241533 (Figure 2B). Cellular inhibition data is summarized in Table 1. CHK2 phosphorylates HDMX in response to DNA damage, which targets this protein for degradation as previously demonstrated in MCF7 cells (37, 38). Figure S2 shows that in this cell line, etoposide treatment caused a clear decrease in HDMX levels and this degradation was inhibited by CCT241533 at ≥0.25 μM, and fully inhibited at ≥1μM. We have therefore established that CCT241533 can inhibit etoposide-induced biomarkers of CHK2 activity in 3 human tumor cell lines at doses below its cellular GI50.
Table 1. Summary of the cytotoxicity, cellular activity and potentiation of genotoxic drugs by the CHK2 inhibitor CCT241533 in vitro.
| Cell Line | HeLa | HT-29 |
|---|---|---|
| GI50 CCT241533 (μM) | 2.2 ± 0.16 (n=3) | 1.7 ± 0.26 (n=3) |
| Full band shift inhibition (μM) | 10 (n=2) | 5 (n=3) |
| Full pS516 inhibition (μM) | 1 (n=2) | 1 (n=3) |
|
P.I. Bleomycin – GI assay 1.5 μM CCT241533 3 μM CCT241533 |
0.8 ± 0.31 (n=5) 2.8 ±2.27 (n=3) |
1.180 ±0.13 (n=7) 0.47 ±0.3 (n=3) |
|
P.I. Bleomycin – CF assay 0.5 μM CCT241533 0.75 μM CCT241533 |
0.86 ± 0.1 (n=3) 0.88 ± 0.07 (n=3) |
N/A |
|
P.I. Etoposide – GI assay 2 μM CCT241533 |
1.16 ± 0.65 (n=3) |
0.67 ± 0.07**(n=3) |
|
P.I. Gemcitabine – GI assay 2 μM CCT241533 |
1.53 ± 0.57 (n=6) |
0.57 ± 0.28 (n=3) |
|
P.I. SN38 – GI assay 2 μM CCT241533 |
1.08 ± 0.02 (n=3) |
0.93 ± 0.35 (n=3) |
|
P.I. MMC – GI assay 2 μM CCT241533 |
1.02 ± 0.28 (n=6) |
0.84 ± 0.77 (n=6) |
|
P.I. AG14447 – GI assay 1.5 μM CCT241533 3 μM CCT241533 |
1.61 ± 0.64* (n=9) 2.31 ± 0.60*** (n=8) |
1.68 ± 0.40** (n=6) 3.01 ± 1.81* (n=6) |
|
P.I. Olaparib – GI assay 1.5 μM CCT241533 3 μM CCT241533 |
2.55 ± 1.1** (n=8) 2.50 ± 1.05* (n=5) |
1.91 ± 1.53 (n=6) 1.43 ± 0.75 (n=3) |
|
P.I. Olaparib – CF assay 0.5 μM CCT241533 0.75 μM CCT241533 |
1.77 ± 0.52** (n=6) 3.66 ± 1.15* (n=3) |
1.4 ± 0.38 (n=3) 2.50 ± 0.31** (n=3) |
Cytotoxicity (GI50) was determined by SRB assay. Band shift inhibition and pS516 inhibition determined by western blot.
GI = 96h SRB growth inhibition assay
CF = Colony forming assay.
PI = Potentiation index is the ratio of GI50 : Combination GI50. Values are mean±SD of n independent determinations.
P<0.05
P<0.01
P<0.001 significantly different from unity. See Methods and Materials for further details.
Effects of bleomycin and CCT241533 on CHK2 activation and cell cycle distribution
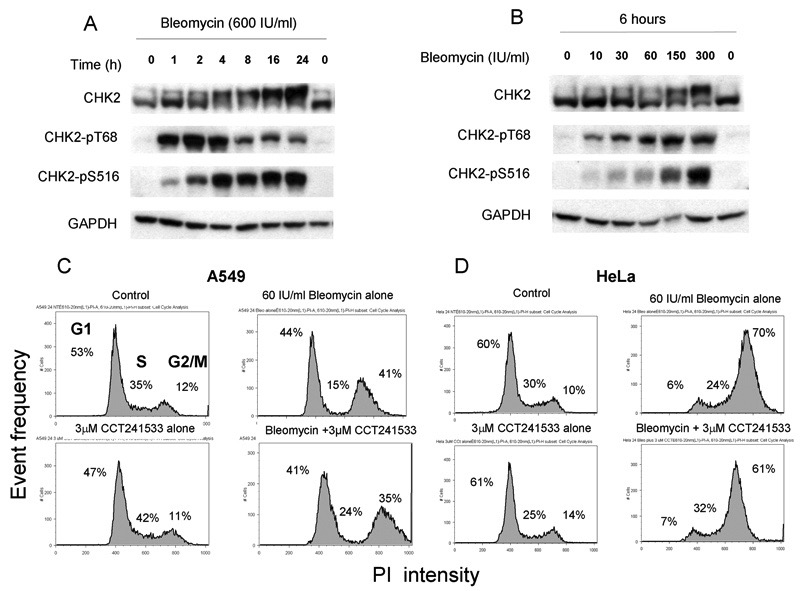
It has been postulated that CHK2 inhibitors may potentiate the action of genotoxic anticancer agents in a functionally defective p53 tumor compared to normal tissue (3, 9). A series of experiments were carried out in HT-29 colon cancer cells (p53 defective) to investigate CHK2 activation in response to different DNA-damaging agents including, bleomycin, IR, etoposide, SN38, 5-fluorodeoxyuridine (5FdU), doxorubicin and mitomycin C (MMC). Figures 3A and B show a time course of CHK2 activation in response to 600 IU/ml bleomycin (GI50= 30 IU/ml), a radiomimetic drug, and a dose response to bleomycin respectively. A band shift and marked pS516 signal were seen after 4 hours treatment with 600 IU/ml bleomycin (Figure 3A) and a marked induction of pS516 occurred after 6 hours at ≥150 IU/ml bleomycin. Similar results were obtained with IR, and other DNA damaging agents (Figure S3). Consequently, bleomycin was selected for further studies as it was a potent activator of CHK2 at therapeutically relevant concentrations.
Figure 3. Effect of Bleomycin treatments on CHK2 activation and cell cycle distribution.
(A) HT-29 cells were treated with 600 IU/ml bleomycin and samples taken at indicated time. (B) HT-29 cells were treated for 4 hours with various concentration of bleomycin. Similar results were obtained in repeat experiments. (C) A549 cells (wild type p53) were treated with either vehicle (control), bleomycin alone, CCT241533 alone or a combincation of bleomycin and CCT241533 (D) HeLa cells (p53 defective) were treated as (C). Cell cycle distribution at 24 h was determined by PI/FACS.
In order to better understand the role of CHK2 in checkpoint control a series of cell cycle experiments were carried out with CCT241533 and bleomycin, in cancer cells expressing functional or defective p53. Figure 3C shows that the human lung adenocarcinoma line A549 (p53+/+) exhibited a G1/S and G2/M arrest 24 hours following 60 IU/ml bleomycin treatment consistent with wild-type p53 status. Treatment of these cells with CCT241533 alone had minimal effects on the cell cycle distribution and bleomycin induced cell cycle arrest. By contrast bleomycin induced a G2/M cell cycle arrest in HeLa (p53 defective) cells with minimal G1/S arrest, consistent with p53 null status (Figure 3D). Once again there were no detectable effects of CCT241533 alone or in combination with bleomycin. Similar results were obtained at lower concentrations of CCT241533 and later time points (48, 72, 96 hours) and in HT-29 (p53 defective) and MCF7 (p53 wild-type) cells (data not shown).
The cytotoxicity of several anticancer drugs is not potentiated by CCT241533
The cytotoxicity of bleomycin in combination with a fixed dose of CCT241533 (GI50) in HT-29 and HeLa cells was investigated. Bleomycin alone had a GI50 of 30 IU/ml and there was no significant potentiation of cell death in the presence of CCT241533 in either cell line (Table 1). Similarly colony forming assays with HeLa cells confirmed the lack of significant effects of CCT241533 on bleomycin cytotoxicity. This lack of potentiation also appeared to be schedule independent in both HT-29 and HeLa cells (Table S2). Similar results were obtained with SN38, gemcitabine, etoposide and MMC (Tables 1 and S2) confirming the lack of potentiation of several different genotoxic agents with CCT241533, despite the clear biomarker evidence of CHK2 target inhibition.
CCT241533 potentiates the action of PARP inhibitors
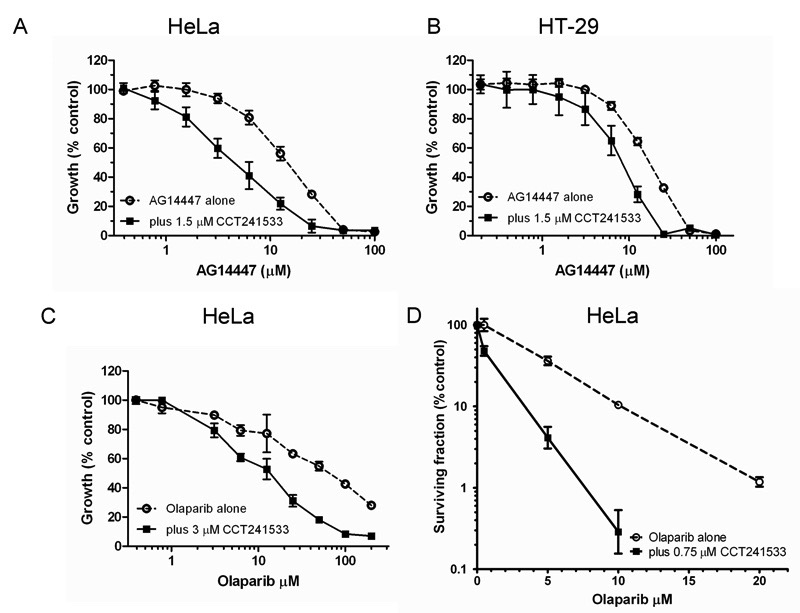
Previously it has been shown that depletion of CHK2 by siRNA increased the sensitivity of HeLa cells to PARP inhibition (20). Our initial studies employed AG14447, a PARP inhibitor currently in clinical trials (29). Both HeLa and HT-29 cells exhibited enhanced sensitivity to AG14447 in the presence of CCT241533 (Figure 4A and B, Table 1). The original studies used a PARP inhibitor from a structurally distinct chemical series of which the clinical candidate is olaparib (20, 39, 40). Significant potentiation between CCT241533 and olaparib was seen in HeLa cells (Figure 4C, Table 1). This relationship was also confirmed in a long term colony forming assay in both cell lines (Figure 4D, Table 1). Once again, neither a 24 hour pretreatment or 24 hour delay in CCT241533 exposure enhanced potentiation above that seen with simultaneous addition of both drugs (Table S2). Although it is not possible to exclude “off-target” effects the observation of potentiation at concentrations of CCT241533 that were non-toxic (Figure S4A) or markedly below the GI50 (Table 1) suggest this is not a major mechanism.
Figure 4. Effect of CCT241533 on PARP inhibitor cytotoxocity in different tumor cell lines.
(A) HeLa or (B) HT-29 cells were treated with AG14447 alone or in combination with a fixed dose (GI50) of CCT241533. (C) HeLa cells treated with either olaparib alone or in combination with 3 μM CCT241533 in a growth delay assay. Values are mean±SD (n=4). Similar results were obtained in repeat experiments and are summarized in Table 1. (D) HeLa cells were treated with olaparib alone or in combination with a fixed dose of CCT241533 in a colony forming assay. Values are mean±SD (n=3). Similar results were obtained in repeat experiments.
In order to investigate the activation and inhibition of CHK2 during the course of the short term growth delay assays, we treated cells with 3 μM CCT241533 together with either 3 or 10 μM olaparib. Figure 5A shows that the pS516 signal increased 72 hours following olaparib treatment alone and was completely inhibited by CCT241533. Therefore olaparib induced phosphorylation of S516 on CHK2 at a biologically relevant concentration and this activation was blocked by CCT241533. Furthermore this combination of olaparib and CCT241533 was shown to enhance apoptosis (Annexin V) in HeLa cells at both 72 and 96 hours post treatment (Figures 5B and S4). These results are consistent with CCT241533 potentiating PARP inhibitor cytotoxicity through the inhibition of CHK2.
Figure 5. (A) Effect of CCT241533 on olaparib cytotoxicity and CHK2 activation in HeLa cells.
HeLa cells were treated with olaparib alone (3 or 10μM) or in combination with 3 μM CCT241533 as indicated. Similar results were obtained in repeat experiments. Et = 50 μM etoposide. (B) Effects of olaparib and CCT241533 on apoptosis in HeLa cells. Cells were treated with olaparib or CCT241533 alone or in combination and apoptosis determined using Annexin V staining. FACS data shown in Figure S4B. Similar results were obtained in repeat experiments. (C) A schematic representation of the possible effects of CHK2 inhibition and PARP inhibition on the DNA damage, repair and survival pathways in cells.
Discussion
We report the first characterization of CCT241533, a novel, potent and selective ATP competitive inhibitor of CHK2 (Figures 1 and S1, Table S1). Crystallographic structural studies showed that CCT241533 binds to the ATP pocket of CHK2. At present there are three other reported selective CHK2 inhibitors, the 2-arylbenzimidazoles (41), VRX0466617 (24) and PV1019 (21) which were all substantially less potent than CCT241533 against CHK2 in vitro.
Many cancer cell lines have compromised DNA repair and several colon tumor cell lines have been reported to lack an active MRN complex or be defective for CHK2 activation (33). Of note is the HCT-116 cell line, which our studies confirm has minimal MRN complex expression and has been used in several previous CHK2 studies (22, 24, 42, 43). In order to overcome this problem, we tested a panel of human cancer cell lines for MRN complex components and IR-induced CHK2 activation. We found a fully functional CHK2 pathway in 10 of the 14 tested cell lines with no evidence for MRN defects in 4/5 ovarian cell lines. In addition there was minimal influence of p53 status on CHK2 pathway activity with half of the 10 functional lines exhibiting wild type p53. The two cancer cell lines picked for further detailed study each expressed all members of the MRN complex and were able to fully activate CHK2 as evidenced by a clear band-shift following genotoxic stress.
In response to DNA damage, ATM kinase phosphorylates CHK2 on residue T68. This allows CHK2 to dimerise and autophosphorylate in trans (T383 and T387) and in cis (S516), (Figure 5C). Phosphorylated T68 is therefore a biomarker of the upstream ATM pathway and pS516 a marker of fully activated CHK2 kinase. The hyperphosphorylation of CHK2 also results in its mobility shift on SDS-PAGE. We used these 3 biomarkers (pT68, pS516 and CHK2 bandshift) to investigate the mechanism of action of CCT241533 on CHK2 in cells. When cells were treated with bleomycin, IR, or etoposide, activation of CHK2 was clearly seen, with a complete band shift and presence of both pT68 and pS516 signals. Interestingly, time course studies with these genotoxic agents showed that phosphorylation occurred rapidly on T68 of CHK2 reaching a peak after 1-2 hours and decreasing thereafter, whereas S516 phosphorylation appeared later and increased over the duration of the time course (Figures 3A and S3). In several cell lines the experiments with etoposide and CCT241533 (Figures 2B and S2) showed an increase in pT68 signal as CHK2 kinase activity was inhibited. This is consistent with other studies and suggests that T68 phosphorylation is not required for continued CHK2 kinase activity after the initial ATM dependent activation step (4), but that it remains on CHK2 when it is unable to become fully activated (i.e. in the presence of CCT241533). Consequently T68 phosphorylation alone should not be considered an adequate marker of full CHK2 activation. CCT241533 inhibited the autophosphorylation of S516 on CHK2 at relatively low concentrations (below its single agent GI50) and at higher concentrations the phosphorylation-induced mobility shift was completely blocked, consistent with inhibition of CHK2 activity in a cellular context (Figures 3B and S2). In addition to autophosphorylation on S516, active CHK2 phosphorylates a range of downstream targets, including HDMX which is subsequently degraded, and CCT241533 clearly rescued etoposide-induced HDMX degradation in MCF7 tumor cells. All these robust biomarker changes confirmed that CCT241533 is an active CHK2 inhibitor in cells and these biomarkers may prove useful in the future development and potential clinical evaluation of CHK2 inhibitors.
There is clear evidence that CHK2 inhibition can protect p53 wild type cells from genotoxic agent-induced apoptosis (21, 24–27), however the roles of CHK2 inhibition in p53 deficient cells is complex and unclear. Reduced CHK2 levels have been reported to sensitize HEK293 and HCT-116 cells to IR or camptothecin (17, 22) and the CHK2 inhibitor PV1019 was reported to potentiate the cytotoxicity of topotecan and camptothecin in OVCAR-4 and OVCAR-5 cells (21). However, silencing of CHK2 by RNAi showed no effect or, no additional effect on cell growth over CHK1 siRNA alone, when combined with gemcitabine, FdUrd, fluoruracil, camptothecin and doxorubicin in HeLa, SW620, Panc-1 cells (18, 19) and CHK2 siRNA alone did not potentiate the effect of gemcitabine in p53 deficient pancreatic cell lines (23).
In the present study, CCT241533 did not potentiate the cytotoxic effects of bleomycin or a selection of other cytotoxic agents as assesed by growth delay or colony forming assays (Table 1) using a variety of different schedules (Table S2). CCT241533 had minimal effects on p53 dependent and independent checkpoint control (Figures 3C and D). Endogenous DNA base lesions occur naturally through hydrolysis, reactive oxygen species and alkylation, and if left unrepaired cause the collapse of the replication fork, often leading to DSBs. PARP-1 and 2 ribosylate both themselves and histones in response to DNA base damage (44). This signal recruits other components of the base excision repair pathway (BER) which facilitates DNA repair, thus preventing DSB formation (Figure 5C). The structurally distinct PARP inhibitors olaparib and AG14447 were both shown to activate CHK2 as shown by increased pS516 signal and partial mobility shift, possibly as a result of increased DSB formation following BER inhibition (Figure 5C). CCT241533 significantly enhanced the cytotoxicity of these PARP inhibitors in both 96 hour growth delay and long term colony forming assays and this was associated with a corresponding loss of CHK2 activation and an increase in apoptosis (Figures 5A and B). These results strongly suggest that the cytotoxic effects of the combination of CCT241533 and olaparib are due to inhibition of CHK2 and this may lead to inhibition of BRCA1 phosphorylation and impairment of the homologous recombination pathway, giving rise to unrepairable and lethal DNA DSBs (Figure 5C).
It is interesting that CHK2 inhibition can potentiate the cytotoxicity of PARP inhibitors but not bleomycin despite both treatments generating DNA DSBs. Bleomycin is an acute DNA damaging treatment that activates CHK2 within 1 hour (Figure 3A) whereas PARP inhibition causes DNA damage through endogenous base lesions being converted into DSBs during DNA replication, which will accumulate after each division. Consequently, CHK2 activation in response to olaparib in HeLa cells was slower than with bleomycin, giving an increase in pS516 signal only after 72 hours of treatment. It is likely that these kinetic differences in the creation of damage may result in differences in the signaling context that activates CHK2 and that this in turn influences the pathways downstream of CHK2 and hence sensitivity to CHK2 inhibition.
In conclusion, we have shown that CCT241533 is a potent and selective CHK2 inhibitor in vitro and in cancer cell lines expressing fully functional CHK2. Moreover, we have demonstrated that CCT241533 can enhance the cytotoxicity of PARP inhibitors in p53 deficient cell lines. Consequently the combination of CHK2 inhibitors with PARP inhibitors could provide a new therapeutic avenue for targeted cancer therapy.
Supplementary Material
Financial support
Grant support was provided to VEA, MIW, PDE, KB, LA, JJC, IC and MDG by Cancer Research UK [CRUK] grant number C309/A8274, to IC by Cancer Research UK [CRUK] grant number C309/A8365 and to AWO and LHP by Cancer Research UK [CRUK] grant number C302/A8265. Additional support was provided to MDG by The Institute of Cancer Research. We acknowledge NHS funding to the NIHR Biomedical Research Centre.
Abbreviation List
- PI
potentiation index
- GI50
concentration at which cell growth is inhibited by 50%;
- IR
Ionizing radiation
- PARP
poly (ADP ribose) polymerase
- DSB
double stranded DNA break
- BER
Base excision repair
- MMC
mitomycin C
- 5FdU
5-fluorodeoxyuridine
Footnotes
Conflict of interest statement: The authors are or were all employees of The Institute of Cancer Research which has a commercial interest in CHK2 inhibitors and operates a rewards to inventors scheme.
References
- 1.Stracker TH, Usui T, Petrini JH. Taking the time to make important decisions: The checkpoint effector kinases Chk1 and Chk2 and the DNA damage response. DNA Repair (Amst) 2009 doi: 10.1016/j.dnarep.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen Y, Poon RY. The multiple checkpoint functions of CHK1 and CHK2 in maintenance of genome stability. Front Biosci. 2008;13:5016–29. doi: 10.2741/3060. [DOI] [PubMed] [Google Scholar]
- 3.Antoni L, Sodha N, Collins I, Garrett MD. CHK2 kinase: cancer susceptibility and cancer therapy - two sides of the same coin? Nat Rev Cancer. 2007;7:925–36. doi: 10.1038/nrc2251. [DOI] [PubMed] [Google Scholar]
- 4.Ahn JY, Li X, Davis HL, Canman CE. Phosphorylation of threonine 68 promotes oligomerization and autophosphorylation of the Chk2 protein kinase via the forkhead-associated domain. J Biol Chem. 2002;277:19389–95. doi: 10.1074/jbc.M200822200. [DOI] [PubMed] [Google Scholar]
- 5.Schwarz JK, Lovly CM, Piwnica-Worms H. Regulation of the Chk2 protein kinase by oligomerization-mediated cis- and trans-phosphorylation. Mol Cancer Res. 2003;1:598–609. [PubMed] [Google Scholar]
- 6.Reinhardt HC, Yaffe MB. Kinases that control the cell cycle in response to DNA damage: Chk1, Chk2, and MK2. Curr Opin Cell Biol. 2009;21:245–55. doi: 10.1016/j.ceb.2009.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang J, Willers H, Feng Z, et al. Chk2 phosphorylation of BRCA1 regulates DNA double-strand break repair. Mol Cell Biol. 2004;24:708–18. doi: 10.1128/MCB.24.2.708-718.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Efeyan A, Serrano M. p53: guardian of the genome and policeman of the oncogenes. Cell Cycle. 2007;6:1006–10. doi: 10.4161/cc.6.9.4211. [DOI] [PubMed] [Google Scholar]
- 9.Pommier Y, Weinstein JN, Aladjem MI, Kohn KW. Chk2 molecular interaction map and rationale for Chk2 inhibitors. Clin Cancer Res. 2006;12:2657–61. doi: 10.1158/1078-0432.CCR-06-0743. [DOI] [PubMed] [Google Scholar]
- 10.Jiang H, Reinhardt HC, Bartkova J, et al. The combined status of ATM and p53 link tumor development with therapeutic response. Genes Dev. 2009;23:1895–909. doi: 10.1101/gad.1815309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zabludoff SD, Deng C, Grondine MR, et al. AZD7762, a novel checkpoint kinase inhibitor, drives checkpoint abrogation and potentiates DNA-targeted therapies. Mol Cancer Ther. 2008;7:2955–66. doi: 10.1158/1535-7163.MCT-08-0492. [DOI] [PubMed] [Google Scholar]
- 12.Wang GT, Li G, Mantei RA, et al. 1-(5-Chloro-2-alkoxyphenyl)-3-(5-cyanopyrazin-2-yl)ureas [correction of cyanopyrazi] as potent and selective inhibitors of Chk1 kinase: synthesis, preliminary SAR, and biological activities. J Med Chem. 2005;48:3118–21. doi: 10.1021/jm048989d. [DOI] [PubMed] [Google Scholar]
- 13.Walton MI, Eve PD, Hayes A, et al. The preclinical pharmacology and therapeutic activity of the novel CHK1 inhibitor SAR-020106. Mol Cancer Ther. 2010;9:89–100. doi: 10.1158/1535-7163.MCT-09-0938. [DOI] [PubMed] [Google Scholar]
- 14.Tse AN, Rendahl KG, Sheikh T, et al. CHIR-124, a novel potent inhibitor of Chk1, potentiates the cytotoxicity of topoisomerase I poisons in vitro and in vivo. Clin Cancer Res. 2007;13:591–602. doi: 10.1158/1078-0432.CCR-06-1424. [DOI] [PubMed] [Google Scholar]
- 15.Matthews DJ, Yakes FM, Chen J, et al. Pharmacological abrogation of S-phase checkpoint enhances the anti-tumor activity of gemcitabine in vivo. Cell Cycle. 2007;6:104–10. doi: 10.4161/cc.6.1.3699. [DOI] [PubMed] [Google Scholar]
- 16.Blasina A, Hallin J, Chen E, et al. Breaching the DNA damage checkpoint via PF-00477736, a novel small-molecule inhibitor of checkpoint kinase 1. Mol Cancer Ther. 2008;7:2394–404. doi: 10.1158/1535-7163.MCT-07-2391. [DOI] [PubMed] [Google Scholar]
- 17.Yu Q, Rose JH, Zhang H, Pommier Y. Antisense inhibition of Chk2/hCds1 expression attenuates DNA damage-induced S and G2 checkpoints and enhances apoptotic activity in HEK-293 cells. FEBS Lett. 2001;505:7–12. doi: 10.1016/s0014-5793(01)02756-9. [DOI] [PubMed] [Google Scholar]
- 18.Xiao Z, Xue J, Sowin TJ, Zhang H. Differential roles of checkpoint kinase 1, checkpoint kinase 2, and mitogen-activated protein kinase-activated protein kinase 2 in mediating DNA damage-induced cell cycle arrest: implications for cancer therapy. Mol Cancer Ther. 2006;5:1935–43. doi: 10.1158/1535-7163.MCT-06-0077. [DOI] [PubMed] [Google Scholar]
- 19.Morgan MA, Parsels LA, Parsels JD, Lawrence TS, Maybaum J. The relationship of premature mitosis to cytotoxicity in response to checkpoint abrogation and antimetabolite treatment. Cell Cycle. 2006;5:1983–8. doi: 10.4161/cc.5.17.3184. [DOI] [PubMed] [Google Scholar]
- 20.McCabe N, Turner NC, Lord CJ, et al. Deficiency in the repair of DNA damage by homologous recombination and sensitivity to poly(ADP-ribose) polymerase inhibition. Cancer Res. 2006;66:8109–15. doi: 10.1158/0008-5472.CAN-06-0140. [DOI] [PubMed] [Google Scholar]
- 21.Jobson AG, Lountos GT, Lorenzi PL, et al. Cellular inhibition of Chk2 kinase and potentiation of camptothecins and radiation by the novel Chk2 inhibitor PV1019. J Pharmacol Exp Ther. 2009 doi: 10.1124/jpet.109.154997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang M, Miao ZH, Zhu H, Cai YJ, Lu W, Ding J. Chk1 and Chk2 are differentially involved in homologous recombination repair and cell cycle arrest in response to DNA double-strand breaks induced by camptothecins. Mol Cancer Ther. 2008;7:1440–9. doi: 10.1158/1535-7163.MCT-07-2116. [DOI] [PubMed] [Google Scholar]
- 23.Azorsa DO, Gonzales IM, Basu GD, et al. Synthetic lethal RNAi screening identifies sensitizing targets for gemcitabine therapy in pancreatic cancer. J Transl Med. 2009;7:43. doi: 10.1186/1479-5876-7-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carlessi L, Buscemi G, Larson G, Hong Z, Wu JZ, Delia D. Biochemical and cellular characterization of VRX0466617, a novel and selective inhibitor for the checkpoint kinase Chk2. Mol Cancer Ther. 2007;6:935–44. doi: 10.1158/1535-7163.MCT-06-0567. [DOI] [PubMed] [Google Scholar]
- 25.Chabalier-Taste C, Racca C, Dozier C, Larminat F. BRCA1 is regulated by Chk2 in response to spindle damage. Biochim Biophys Acta. 2008;1783:2223–33. doi: 10.1016/j.bbamcr.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 26.Hirao A, Kong YY, Matsuoka S, et al. DNA damage-induced activation of p53 by the checkpoint kinase Chk2. Science. 2000;287:1824–7. doi: 10.1126/science.287.5459.1824. [DOI] [PubMed] [Google Scholar]
- 27.Takai H, Naka K, Okada Y, et al. Chk2-deficient mice exhibit radioresistance and defective p53-mediated transcription. Embo J. 2002;21:5195–205. doi: 10.1093/emboj/cdf506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Collins I, Caldwell JJ, Oliver AW, Raynham TM, Welsh EJ, CAJ M. Preparation of oxy-phenyl-aryl compounds as CHK2 kinase inhibitors for treating proliferative disorders and for radioprotection. Intl Patent Appl WO2009053694 2009. Chem Abs. 2009;150:447975. [Google Scholar]
- 29.Thomas HD, Calabrese CR, Batey MA, et al. Preclinical selection of a novel poly(ADP-ribose) polymerase inhibitor for clinical trial. Mol Cancer Ther. 2007;6:945–56. doi: 10.1158/1535-7163.MCT-06-0552. [DOI] [PubMed] [Google Scholar]
- 30.Oliver AW, Paul A, Boxall KJ, et al. Trans-activation of the DNA-damage signalling protein kinase Chk2 by T-loop exchange. Embo J. 2006;25:3179–90. doi: 10.1038/sj.emboj.7601209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hills CA, Kelland LR, Abel G, Siracky J, Wilson AP, Harrap KR. Biological properties of ten human ovarian carcinoma cell lines: calibration in vitro against four platinum complexes. Br J Cancer. 1989;59:527–34. doi: 10.1038/bjc.1989.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Skehan P, Storeng R, Scudiero D, et al. New colorimetric cytotoxicity assay for anticancer-drug screening. J Natl Cancer Inst. 1990;82:1107–12. doi: 10.1093/jnci/82.13.1107. [DOI] [PubMed] [Google Scholar]
- 33.Takemura H, Rao VA, Sordet O, et al. Defective Mre11-dependent activation of Chk2 by ataxia telangiectasia mutated in colorectal carcinoma cells in response to replication-dependent DNA double strand breaks. J Biol Chem. 2006;281:30814–23. doi: 10.1074/jbc.M603747200. [DOI] [PubMed] [Google Scholar]
- 34.Chaturvedi P, Eng WK, Zhu Y, et al. Mammalian Chk2 is a downstream effector of the ATM-dependent DNA damage checkpoint pathway. Oncogene. 1999;18:4047–54. doi: 10.1038/sj.onc.1202925. [DOI] [PubMed] [Google Scholar]
- 35.Matsuoka S, Huang M, Elledge SJ. Linkage of ATM to cell cycle regulation by the Chk2 protein kinase. Science. 1998;282:1893–7. doi: 10.1126/science.282.5395.1893. [DOI] [PubMed] [Google Scholar]
- 36.O'Connor PM, Jackman J, Bae I, et al. Characterization of the p53 tumor suppressor pathway in cell lines of the National Cancer Institute anticancer drug screen and correlations with the growth-inhibitory potency of 123 anticancer agents. Cancer Res. 1997;57:4285–300. [PubMed] [Google Scholar]
- 37.Chen L, Gilkes DM, Pan Y, Lane WS, Chen J. ATM and Chk2-dependent phosphorylation of MDMX contribute to p53 activation after DNA damage. Embo J. 2005;24:3411–22. doi: 10.1038/sj.emboj.7600812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pereg Y, Lam S, Teunisse A, et al. Differential roles of ATM- and Chk2-mediated phosphorylations of Hdmx in response to DNA damage. Mol Cell Biol. 2006;26:6819–31. doi: 10.1128/MCB.00562-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fong PC, Boss DS, Yap TA, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361:123–34. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 40.Menear KA, Adcock C, Boulter R, et al. 4-[3-(4-cyclopropanecarbonylpiperazine-1-carbonyl)-4-fluorobenzyl]-2H-phth alazin-1-one: a novel bioavailable inhibitor of poly(ADP-ribose) polymerase-1. J Med Chem. 2008;51:6581–91. doi: 10.1021/jm8001263. [DOI] [PubMed] [Google Scholar]
- 41.Arienti KL, Brunmark A, Axe FU, et al. Checkpoint kinase inhibitors: SAR and radioprotective properties of a series of 2-arylbenzimidazoles. J Med Chem. 2005;48:1873–85. doi: 10.1021/jm0495935. [DOI] [PubMed] [Google Scholar]
- 42.Jin J, Ang XL, Ye X, Livingstone M, Harper JW. Differential roles for checkpoint kinases in DNA damage-dependent degradation of the Cdc25A protein phosphatase. J Biol Chem. 2008;283:19322–8. doi: 10.1074/jbc.M802474200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Y, Wiltshire T, Senft J, Reed E, Wang W. Irofulven induces replication-dependent CHK2 activation related to p53 status. Biochem Pharmacol. 2007;73:469–80. doi: 10.1016/j.bcp.2006.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Farmer H, McCabe N, Lord CJ, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–21. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.