Abstract
Background
Whether increased expression of the tumor suppressor protein p53 indicates a p53 gene mutation in hepatocellular carcinoma (HCC) remains unclear. We conducted a meta-analysis to determine whether p53 protein overexpression detected by immunohistochemistry (IHC) offers a diagnostic prediction for p53 gene mutations in HCC patients.
Methods
Systematic literature searches were conducted with an end date of December 2015. A meta-analysis was performed to estimate the diagnostic accuracy of IHC-determined p53 protein overexpression in the prediction of p53 gene mutations in HCC. Sensitivity, subgroup, and publication bias analyses were also conducted.
Results
Thirty-six studies were included in the meta-analysis. The results showed that the overall sensitivity and specificity for IHC-determined p53 overexpression in the diagnostic prediction of p53 mutations in HCC were 0.83 (95% CI: 0.80–0.86) and 0.74 (95% CI: 0.71–0.76), respectively. The summary positive likelihood ratio (PLR) and negative likelihood ratio (NLR) were 2.65 (95% CI: 2.21–3.18) and 0.36 (95% CI: 0.26–0.50), respectively. The diagnostic odds ratio (DOR) of IHC-determined p53 overexpression in predicting p53 mutations ranged from 0.56 to 105.00 (pooled, 9.77; 95% CI: 6.35–15.02), with significant heterogeneity between the included studies (I2 = 40.7%, P = 0.0067). Moreover, subgroup and sensitivity analyses did not alter the results of the meta-analysis. However, potential publication bias was present in the current meta-analysis.
Conclusion
The upregulation of the tumor suppressor protein p53 was indeed linked to p53 gene mutations. IHC determination of p53 overexpression can predict p53 gene mutations in HCC patients.
Introduction
Hepatocellular carcinoma (HCC) is one of the most prevalent cancers worldwide, and the cancer-related deaths due to this condition are increasing [1,2]. Therefore, elucidating the malignant biological features of HCC is critical for outcome prediction in patients with this disease. Mutations in the tumor suppressor gene p53 are the most common genetic changes in human malignancies. In HCC, the frequency of p53 gene mutations is as high as 50.0% (average 30.0%); therefore, analysis of this gene and its products is of practical importance [3,4]. Several studies have reported that alterations of the p53 gene are correlated with tumor differentiation, vascular invasion, and tumor stage in HCC [5–7]. Moreover, aberrations of the p53 gene have been shown to be prognostic indicators associated with recurrence-free survival and overall survival in HCC patients [3,8].
Wild-type p53 protein is responsible for cell cycle regulation and apoptosis following DNA damage, while mutant p53 protein shows a loss of function [8,9]. Mutational analysis using a variety of techniques, such as direct DNA sequencing, single-strand conformation polymorphism (SSCP) analysis followed by DNA sequencing, and other mutation assays, is the gold standard for the identification of p53 genetic alterations [8–11]. Generally, the transition from wild-type p53 to a mutant phenotype results in mutant p53 protein overexpression due to the resistance to murine double minute gene 2 (MDM2)-mediated degradation and subsequent abnormal stability of the mutant protein; therefore, immunohistochemistry (IHC) can be used to determine the expression and location of mutant p53 protein that has accumulated in the cell nuclei of cancer tissues [12,13]. IHC is an economic and convenient technology; thus, more clinical studies have adopted IHC to identify genetic alterations in the p53 gene rather than using mutational analysis [3]. However, it remains unclear whether a concordance exists between p53 protein overexpression and p53 gene mutations in HCC patients. As reported in a previously published meta-analysis, the association between p53 mutations and p53 overexpression in predicting shorter patient survival times in HCC suggested a correlation between p53 expression and p53 mutations [3]. However, several studies have found that p53 expression determined by IHC assays did not predict p53 mutations [14–16]. Moreover, the accuracy of IHC in measuring p53 protein overexpression for the prediction of p53 mutations in HCC is not clear.
To determine whether p53 protein overexpression is concordant with p53 gene mutations, we performed a diagnostic meta-analysis of relevant observational studies. We evaluated the ability of IHC assessment of p53 protein overexpression to predict p53 mutations identified by mutational analysis as a reference standard in HCC.
Materials and Methods
Literature search
A comprehensive literature search was conducted using the National Center for Biotechnology Information PubMed (MEDLINE) databases with an end date of December 2015 using the following search terms: (liver neoplasm or hepatocellular carcinoma or carcinoma, hepatocellular or HCC) and (tumor suppressor protein p53 or p53) and (immunohistochemistry or IHC or immunostaining or immunoassay or expression or overexpression or up-regulation) and (mutation or mutational analysis or DNA mutational analysis). References in the selected studies and review articles were also manually assessed.
Study selection
Studies were required to meet the following inclusion criteria: (1) provided a confirmed diagnosis of HCC in humans; (2) explicitly reported the detection methods for p53 alterations, including IHC, the specific antibodies used to determine p53 protein overexpression and mutational assays, such as PCR-SSCP and/or DNA sequencing, or other specific approaches for identifying p53 gene mutations; (3) provided data on p53 expression and p53 mutations, with the prevalence of p53 mutations greater than 0%; and (4) written in English, German, or Chinese.
Two investigators (Jiang-Bo Liu and Wei Li) independently read the title and abstract of candidate studies, and irrelevant studies were excluded if they did not meet the inclusion criteria. Then, the two investigators analyzed the full texts of the selected studies and determined whether the studies should be included. If disagreements occurred, the two investigators conducted a discussion or recruited the third investigator (Miao Deng) until a consensus was reached. Additionally, if studies were found to employ overlapping populations after comprehensive evaluation, the one with the largest population or the newest study was usually included.
Data extraction and quality assessment
Two investigators (Jiang-Bo Liu and Wei Li) independently extracted the data, which included the first author, publication year, recruitment period, geographic location, sample size, analytical method (protein/gene), and cut-off values/detected exons. Moreover, the diagnostic data, including the true positive (TP), false positive (FP), false negative (FN), and true negative (TN) values of IHC-determined p53 expression levels and p53 mutations identified by mutational analysis (as a reference standard), were extracted from the relevant articles. The revised version of the Quality Assessment of Diagnostic Accuracy Studies (QUADAS-2) tool, comprising 4 domains (11 items), was used to assess the quality of all included studies [17].
Statistical analysis
The statistical software Meta-DiSc version 1.4 (XI Cochrane Colloquium, Barcelona, Spain) and Stata version 12 (Stata Corporation, College Station, TX, USA) were used in the meta-analysis. Accordingly, TP, TN, FP, and FN were retrieved from each article. The summary sensitivity (SEN), specificity (SPE), positive likelihood ratio (PLR), negative likelihood ratio (NLR), and diagnostic odds ratio (DOR) estimates with 95% confidence intervals (CIs) were analyzed using a random-effects model, and a bivariate summary receiver operating characteristic (SROC) curve was generated. The area under the SROC curve (AUC) represented an analytical summary of the test performance and illustrated the trade-off between SEN and SPE. The between-studies heterogeneity was evaluated with the I2 statistic (range 0% to 100%), and an I2 statistic index greater than 50% indicated substantial heterogeneity [18]. Sensitivity analyses were performed to explore possible heterogeneity, and the influence of individual studies on the meta-analytical results was assessed by applying the leave-one-out method. Deeks’ funnel plots were generated to explore potential publication bias, with P-values less than 0.1 indicating significance [19].
Results
Search results
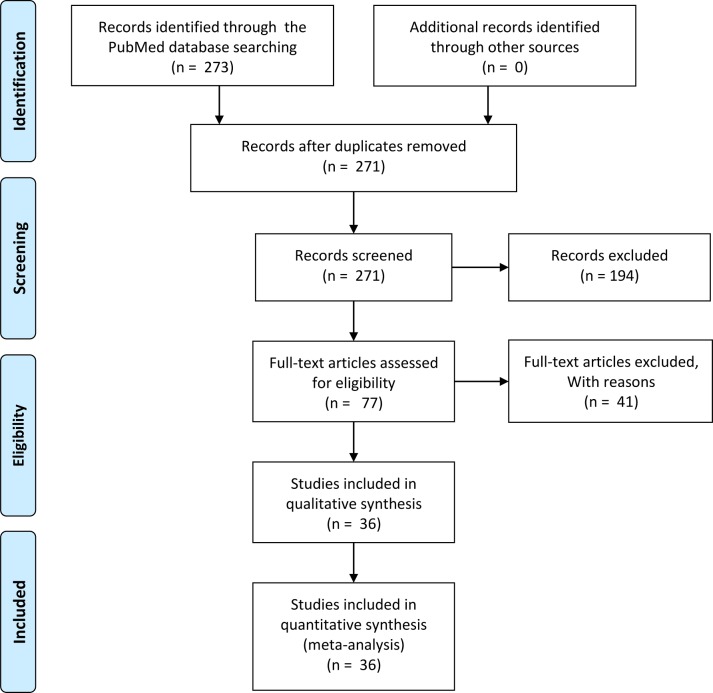
An initial search retrieved 273 published studies. After a careful selection process, thirty-six relevant observational studies (34 in English [4,5,8–12,14–16,20–43] and 2 in Chinese [44,45]) were included in the meta-analysis. Fig 1 shows the literature screening process for the meta-analysis. The included studies had QUADAS-2 scores of 9 to 11 (median = 10).
Fig 1. Flow chart of the selection process for the included studies.
Characteristics of the included studies
The characteristics of each study are shown in Table 1. Of 36 studies, 23 were conducted in high-incidence areas (Asia and Africa) [4,5,8,10–12,23–29,31,34,35,38–40,42–45], and 13 were conducted in low-incidence areas (Europe and USA) [9,14–16,20–22,30,32,33,36,37,41]. The studies included 1,659 HCC patients with a mean sample size of 46 (range 8 to 397). Among the included studies, 584 cases of p53 gene mutations and 765 cases of p53 protein overexpression were found in HCC tissues, with an average mutation and overexpression prevalence of 35.2% (range 2.9% to 60.7%) and 46.1% (range 5.0% to 72.7%), respectively. Thirty-two studies described mutable sites of the p53 gene, reporting 822 mutations in 584 cases of HCC, while another four studies did not report specific sites. Of the 822 reported p53 mutations, the most frequently mutated sites were exons 5 and 7, accounting for 14.2% and 68.9% of the reported mutations, respectively, and codon 249 located in exon 7 had the highest mutation rate of 30.1% (248/822).
Table 1. The analytical results of correlations between p53 mutations and p53 overexpression.
| Reference | Country | Potential mutagen | Sample size | TP | FP | FN | TN | IHC cut-off/Exon | Analytical method (antibody/gene) | QUADAS-2 |
|---|---|---|---|---|---|---|---|---|---|---|
| An et al. 2001 [35] | Japan | HCV | 11 of 41 | 1 | 7 | 0 | 3 | NA/exons 5–8 | IHC (DO-7)/PCR-DNA sequencing | 11 |
| Andersson et al. 1995 [33] | Denmark | alpha-particles | 18 of 36 | 0 | 2 | 1 | 15 | 1%/exons 5,7,8 | IHC (DO-7)/PCR-DGGE, DNA sequencing | 9 |
| Anzola et al. 2004 [15] | Spain | HCV, alcohol | 117 from 78 | 4 | 23 | 8 | 82 | 10%/exons 4–8 | IHC (DO-7)/PCR-SSCP, DNA sequencing | 11 |
| Boix-Ferrero et al. 1999 [20] | Spain | HCV, alcohol | 70 of 129 | 1 | 13 | 1 | 55 | 10%/exons 5–8 | IHC (Bp 53–11)/PCR-DNA sequencing | 11 |
| Bourdon et al. 1995 [14] | France | HBV | 20 | 5 | 5 | 1 | 9 | NA/exons 2–11 | IHC (PAb1801)/PCR-DNA sequencing | 10 |
| Challen et al. 1992 [22] | UK | –* | 19 | 1 | 0 | 1 | 17 | 10%/exons 5–8 | IHC (–)/PCR-DNA sequencing | 9 |
| Chen et al. 2003 [43] | China | HBV | 33 | 16 | 5 | 0 | 12 | NA/exons 2–8 | IHC (Santa)/PCR-DNA sequencing | 10 |
| Cheung et al. 2006 [4] | China | HBV | 55 | 17 | 11 | 9 | 18 | NA/exons 4–9 | IHC (DO-7)/PCR-DNA sequencing | 11 |
| De Benedetti et al. 1996 [41] | USA | Contraceptive | 10 of 11 | 1 | 2 | 0 | 7 | 1%/exons 4–9 | IHC (–)/PCR-DNA sequencing | 9 |
| Greenblatt et al. 1997 [29] | China | HBV | 16 | 1 | 5 | 2 | 8 | 1%/exons 4–8 | IHC (CM-1)/PCR-DNA sequencing | 9 |
| Gross-Goupil et al. 2003 [16] | France | HBV, HCV | 18 | 0 | 4 | 2 | 12 | 10%/exons 2–11 | IHC (DO-7)/PCR-DNA sequencing | 10 |
| Hsia et al. 2000 [24] | China | –* | 28 | 16 | 3 | 1 | 8 | 10%/exons 5–8 | IHC (–)/PCR-DNA sequencing | 9 |
| Hsu et al. 1993 [26] | China | HBV, HCV | 78 of 184 | 30 | 9 | 10 | 29 | 10%/exons 2–11 | IHC (DO-7)/PCR-SSCP, DNA sequencing | 9 |
| Jablkowski et al. 2005 [9] | Poland | HBV | 9 of 20 | 4 | 1 | 1 | 3 | 50%/exons 5–8 | IHC (DO-7)/PCR-DNA sequencing | 10 |
| Kang et al. 1998 [39] | Korea | HBV | 8 of 13 | 2 | 2 | 0 | 4 | 20%/exons 5–8 | IHC (DO-7)/PCR-SSCP, DNA sequencing | 9 |
| Kubicka et al. 1995 [32] | Germany | HBV | 20 | 1 | 0 | 1 | 18 | 30%/exons 5–8 | IHC (PAb1801/PAb240) /PCR-DNA sequencing | 9 |
| Lee et al. 2002 [25] | Korea | HBV | 36 from 34 | 6 | 9 | 1 | 20 | 5%/exons 4–10 | IHC (BP53-12)/PCR-SSCP, DNA sequencing | 10 |
| Lunn et al. 1997 [42] | China | HBV, AFB1 | 105 | 22 | 13 | 7 | 63 | 5%/exons 5–9 | IHC (DO-1/Ab-6)/PCR-SSCP, DNA sequencing | 9 |
| Luo et al. 2001 [45] | China | – | 21 | 6 | 5 | 3 | 7 | 10%/exons 5–8 | IHC (DO-7)/PCR-SSCP | 10 |
| Mitsumoto et al. 2004 [31] | Japan | HCV | 50 | 13 | 1 | 8 | 28 | 10%/– | IHC (DO-7)/Yeast p53 Functional Assay, DNA sequencing | 9 |
| Mohamed et al. 2008 [5] | Egypt | HBV, HCV | 30 | 7 | 9 | 4 | 10 | 10%/exons 5–8 | IHC (DO-7)/PCR-SSCP, DNA sequencing | 11 |
| Okada et al. 2003 [27] | Japan | HCV | 10 of 22 | 5 | 1 | 0 | 4 | 10%/exons 5–9 | IHC (DO-7)/PCR-DNA sequencing | 10 |
| Peng et al. 1998 [23] | China | – | 70 | 21 | 9 | 2 | 38 | 5%/exon 7 | IHC (DO-7)/RFLP | 9 |
| Qi et al. 2015 [8] | China | HBV, AFB1 | 397 | 208 | 58 | 15 | 116 | 25%/exons 1–11 | IHC (Abcam)/PCR-DNA sequencing | 11 |
| Qin et al. 1998 [38] | China | HBV | 26 of 31 | 5 | 1 | 1 | 19 | NA/exons 5–9 | IHC (PAb1801/PAb240) /PCR-DNA sequencing | 9 |
| Rashid et al. 1999 [10] | China | HBV | 24 | 10 | 3 | 2 | 9 | NA/exons 2–9 | IHC (DO-7)/PCR-DNA sequencing | 11 |
| Ryder et al. 1996 [37] | Germany | HBV, HCV | 37 of 38 | 15 | 3 | 2 | 17 | 80%/exons 5–8 | IHC (DO-1)/PCR-SSCP, DNA sequencing | 10 |
| Sanefuji et al. 2010 [34] | Japan | HCV | 79 of 82 | 13 | 34 | 0 | 32 | 10%/exons 5–8 | IHC (DO-7)/PCR-DNA sequencing | 11 |
| Shieh et al. 1993 [36] | USA | HBV, HCV | 18 | 1 | 0 | 0 | 17 | NA/exon 7 | IHC (PAb1801)/PCR-DNA sequencing | 9 |
| Soini et al. 1996 [21] | Mexico | AFB1 | 14 of 21 | 2 | 4 | 1 | 7 | 1%/exon 7 | IHC (CM-1)/PCR-DNA sequencing | 9 |
| Stern et al. 2001 [28] | China | HBV, AFB1 | 48 of 64 | 15 | 15 | 3 | 15 | NA/exon 7 | IHC (CM-1)/PCR-DNA sequencing | 10 |
| Szymańska et al. 2004 [11] | Gambia | HBV | 28 of 29 | 9 | 4 | 5 | 10 | 10%/exons 5–8 | IHC (CM-1)/PCR-RFLP, DNA sequencing/SOMA | 9 |
| Volkmann et al. 2001 [30] | Germany | HBV, HCV | 39 | 8 | 3 | 3 | 25 | 10%/exons 5–9 | IHC (DO-1)/PCR-SSCP, DNA sequencing | 10 |
| Zekri et al. 2006 [40] | Egypt | HCV | 25 | 7 | 6 | 3 | 9 | 10%/exons 5–8 | IHC (DO-7)/PCR-SSCP, DNA sequencing | 9 |
| Zhang et al. 2006 [12] | China | HBV, AFB1 | 40 | 9 | 5 | 2 | 24 | 5%/exons 5–8 | IHC (DO-7)/PCR-DNA sequencing | 10 |
| Zhou et al. 1997 [44] | China | HBV | 32 | 2 | 6 | 0 | 24 | NA/exon 7 | IHC (DO-1/Ab-6)/PCR-RFLP | 9 |
HBV/HCV: hepatitis B/C virus; AFB1: aflatoxin B1; IHC: immunohistochemistry; PCR: polymerase chain reaction; SSCP: single-strand conformation polymorphism; RFLP: restriction fragment length polymorphism; SOMA: short oligonucleotide mass spectrometry analysis; DGGE: denaturing gradient gel electrophoresis; QUADAS, Quality Assessment of Diagnostic accuracy studies.
Diagnostic accuracy analysis
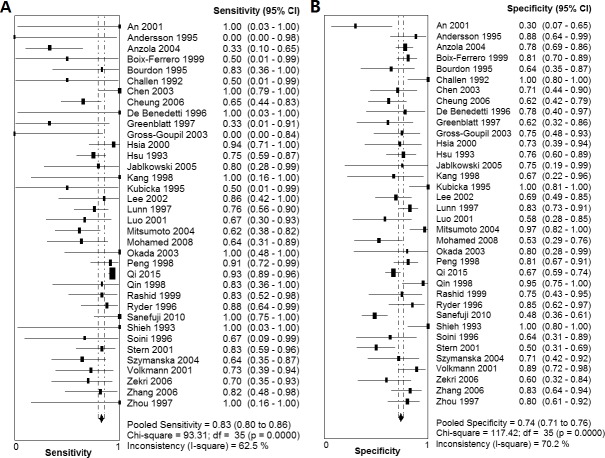
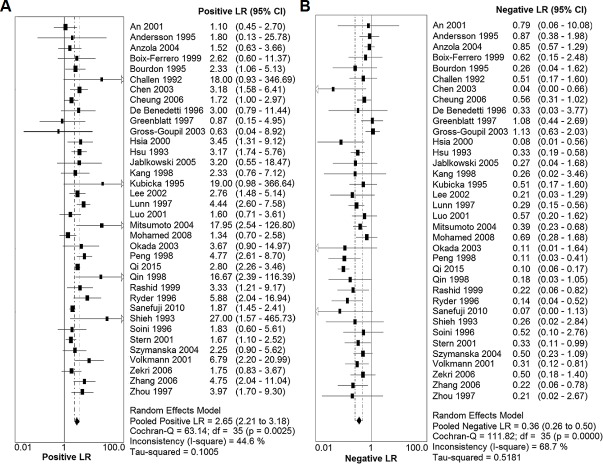
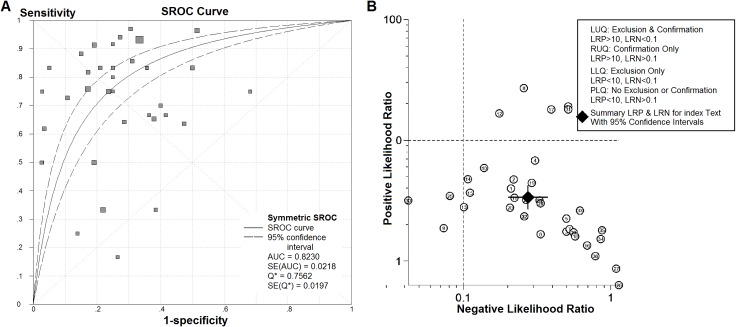
As shown in Fig 2, the summary SEN and SPE for IHC-determined p53 overexpression in the diagnostic prediction of p53 mutations in HCC were 0.83 (95% CI: 0.80–0.86) and 0.74 (95% CI: 0.71–0.76), respectively. Moreover, the summary PLR and NLR were 2.65 (95% CI: 2.21–3.18) and 0.36 (95% CI: 0.26–0.50), respectively (Fig 3). The DOR of IHC-determined p53 overexpression in predicting p53 mutations ranged from 0.56 to 105.00 (pooled, 9.77; 95% CI: 6.35–15.02), with significant heterogeneity among the included studies (I2 = 40.7%, P = 0.0067). Additionally, the estimated accuracy and positive and negative predictive values were 77.0%, 63.3% and 88.8%, respectively. The graph of the symmetric SROC curve showed that the AUC of IHC-determined p53 overexpression was 0.8230 (standard error = 0.0218) with a Q-value of 0.7562 (standard error = 0.0197), indicating that IHC-determined p53 overexpression had an overall moderate level of accuracy in the prediction of p53 mutations in HCC (Fig 4A). The likelihood ratio scattergram shows that IHC-determined p53 overexpression has a limited diagnostic ability to identify p53 mutations in HCC (Fig 4B).
Fig 2. Forest plot of the sensitivity and specificity of IHC-determined p53 overexpression in detecting p53 mutations.
(A) Forest plot showing the sensitivity of IHC-determined p53 overexpression in detecting p53 mutations. (B) Forest plot showing the specificity of IHC-determined p53 overexpression in detecting p53 mutations. Abbreviations: CI, confidence interval.
Fig 3. Forest plot of the positive likelihood ratio (PLR) and the negative likelihood ratio (NLR) of IHC-determined p53 overexpression in detecting p53 mutations.
(A) Forest plot showing the positive LR of IHC-determined p53 overexpression in detecting p53 mutations. (B) Forest plot showing the negative LR of IHC-determined p53 overexpression in detecting p53 mutations.
Fig 4. The summary receiver operating characteristic (SROC) curve and the likelihood ratio scattergram for IHC-determined p53 overexpression in the identification of p53 mutations in HCC for all studies.
(A) The SROC curve summarizes the overall diagnostic accuracy of IHC-determined p53 overexpression for the identification of p53 mutations. The size of the dots for 1-specificity and sensitivity of the single studies in the ROC space reflects the sample size (number of patients) in the study. (B) The likelihood ratio scattergram shows the diagnostic performance of IHC-determined p53 overexpression in the identification of p53 mutations. Q* = point at which sensitivity and specificity were equal.
Subgroup analysis
By grouping studies according to the publication year, geographic location, sample size, different IHC antibodies, mutational analysis methods, or prevalence of p53 alterations, subgroup analysis revealed that the diagnostic accuracy of IHC-determined p53 overexpression in identifying p53 mutations in HCC remained consistent (Table 2). Interestingly, the pooled sensitivities were higher in the studies published after the year 2000, as well as in the studies conducted in Asia and Africa or those with a sample size ≥ 46, but the pooled specificities were much lower compared with those of the corresponding subgroups. In the IHC antibodies subgroup analysis, the highest pooled SEN and SPE were from the studies employing the PAb1801 antibody, while the lowest values were from the studies employing the CM-1 antibody. Moreover, for p53 mutational assays, the studies with all cases detected by direct DNA sequencing yielded much higher sensitivities but much lower SPEs, while the group of partial cases that were abnormal in other mutational assays followed by DNA sequencing presented the reverse of these statistics. Furthermore, the pooled SEN was higher in the studies with a high prevalence of p53 alterations (mutation ≥ 35% or overexpression ≥ 46%), but the pooled SPE was lower compared to the subgroup with a low prevalence of p53 alterations.
Table 2. The results of subgroup analyses.
| Variables | N | SEN (95% CI), I2 (%) | SPE (95% CI), I2 (%) | PLR (95% CI), I2 (%) | NLR (95% CI), I2 (%) | DOR (95% CI), I2 (%) |
|---|---|---|---|---|---|---|
| Publication year | ||||||
| Before 1999 | 17 | 0.79 (0.72−0.85), 0 | 0.82 (0.78−0.86), 53.1 | 3.66 (2.88−4.64), 0 | 0.37 (0.26−0.53), 28.8 | 13.79 (8.37−22.72), 0 |
| After 2000 | 19 | 0.84 (0.81−0.88), 76.1 | 0.68 (0.65−0.72), 68.0 | 2.23 (1.80−2.76), 46.8 | 0.35 (0.22−0.58), 80.1 | 7.91 (4.18−14.98), 58.6 |
| Geographic location | ||||||
| Asia and Africa | 23 | 0.85 (0.82−0.88), 63.9 | 0.70 (0.66−0.73), 67.6 | 2.57 (2.09−3.15), 54.0 | 0.31 (0.22−0.44), 60.3 | 10.47 (6.32−17.34), 46.8 |
| Europe and America | 13 | 0.66 (0.53−0.77), 40.5 | 0.83 (0.78−0.86), 58.2 | 3.07 (2.02−4.66), 14.8 | 0.54 (0.349−0.82), 48.8 | 8.01 (3.50−18.35), 24.5 |
| Sample size (n, mean) | ||||||
| ≥ 46 | 10 | 0.85 (0.81−0.88), 83.5 | 0.72 (0.68−0.75), 82.3 | 2.60 (1.94−3.48), 69.8 | 0.31 (0.17−0.57), 85.4 | 10.41 (4.97−21.81), 67.9 |
| < 46 | 26 | 0.79 (0.72−0.85), 31.2 | 0.77 (0.73−0.81), 60.5 | 2.70 (2.12−3.44), 24.1 | 0.41 (0.29−0.58), 43.8 | 9.10 (5.39−15.34), 15.9 |
| IHC antibodies | ||||||
| DO-7 antibody | 17 | 0.73 (0.66−0.79), 58.1 | 0.71 (0.67−0.75), 69.0 | 2.27 (1.73−2.99), 47.2 | 0.46 (0.32−0.67), 61.2 | 6.48 (3.53−11.88), 39.0 |
| DO-1 antibody | 4 | 0.80 (0.73−0.89), 0 | 0.84 (0.77−0.89), 0 | 4.74 (3.21−7.01), 0 | 0.26 (0.16−0.43), 0 | 19.75 (8.98−43.42), 0 |
| CM-1 antibody | 4 | 0.71 (0.54−0.85), 17.1 | 0.59 (0.46−0.71), 0 | 1.71 (1.21−2.43), 0 | 0.58 (0.34−0.99), 10.0 | 3.69 (1.46−9.34), 0 |
| PAb1801 antibody | 4 | 0.80 (0.52−0.96), 0 | 0.91 (0.82−0.97), 79.4 | 8.54 (1.82−40.14), 60.3 | 0.34 (0.15−0.75), 0 | 30.79 (6.58−144.13), 0 |
| Mutational assays | ||||||
| All DNA sequencing | 21 | 0.89 (0.85−0.92), 57.3 | 0.70 (0.66−0.74), 75.2 | 2.43 (1.93−3.06), 39.9 | 0.32 (0.19−0.55), 73.6 | 11.10 (5.96−20.68), 36.8 |
| Partial DNA sequencing | 15 | 0.72 (0.65−0.77), 39.4 | 0.78 (0.74−0.81), 53.4 | 2.80 (2.09−3.77), 51.5 | 0.43 (0.31−0.59), 54.5 | 7.81 (4.32−14.10), 49.3 |
| Prevalence of p53 alterations | ||||||
| Mutation ≥ 35% | 15 | 0.85 (0.82−0.88), 71.2 | 0.67 (0.64−0.73), 50.0 | 2.41 (1.92−3.03), 37.0 | 0.30 (0.20−0.47), 66.7 | 9.74 (5.18−18.33), 56.4 |
| Mutation < 35% | 21 | 0.76 (0.68−0.83), 48.0 | 0.77 (0.74−0.80), 75.0 | 2.94 (2.15−4.04), 54.2 | 0.43 (0.28−0.66), 61.3 | 9.91 (5.40−18.18), 25.5 |
| Overexpression ≥ 46% | 18 | 0.87 (0.83−0.90), 63.1 | 0.64 (0.59−0.68), 32.8 | 2.21 (1.84−2.65), 33.0 | 0.29 (0.19−0.45), 60.1 | 8.82 (4.97−15.67), 47.3 |
| Overexpression < 46% | 18 | 0.71 (0.62−0.78), 42.1 | 0.83 (0.79−0.86), 58.3 | 3.71 (2.70−5.11), 27.9 | 0.46 (0.31−0.68), 62.3 | 11.21 (5.63−22.29), 36.8 |
SEN, sensitivity; SPE, specificity; PLR, positive likelihood ratio; NLR, negative likelihood ratio; DOR, diagnostic odds ratio; CI, confidence interval.
Sensitivity analysis
The leave-one-out method sensitivity analysis showed that the results of the meta-analysis were not impacted by individual studies. Overall, the analytical results showed that the pooled SEN ranged from 0.77 (95% CI: 0.72–0.81, I2 = 45.7%), by removing Qi et al. [8], to 0.84 (95% CI: 0.81–0.87, I2 = 56.6%), by removing Anzola et al. [15], and the pooled SPE ranged from 0.73 (95% CI: 0.70–0.76, I2 = 70.1%), by omitting Lunn et al. [42], to 0.76 (95% CI: 0.73–0.78, I2 = 64.9%), by omitting Sanefuji et al. [34].
Publication bias
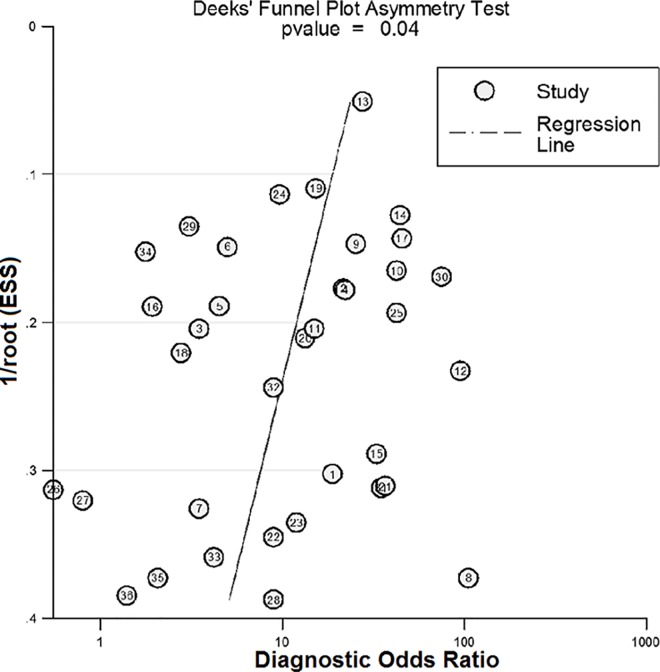
Fig 5 displays the symmetric shape of the funnel plot. However, the P value was less than 0.05 in Deeks’ test, indicating that publication bias may exist in the meta-analysis.
Fig 5. The Deeks’ funnel plot and asymmetry test of the meta-analysis of the 36 included studies.
Discussion
The tumor suppressor gene p53 plays a crucial role in cell cycle control and apoptosis in response to DNA damage, and mutation of the p53 gene has been shown to contribute to carcinogenesis and drug resistance [39,46]. Many studies have reported that p53 mutations are correlated with malignant tumor behaviors in HCC [40,43]. Our previous meta-analysis showed that HCC patients with a mutant p53 gene or p53 protein overexpression had a higher risk of mortality and tumor recurrence than those with wild-type p53 status or low/no p53 expression, which can inform clinical decision-making in HCC [3]. However, it remained unclear whether p53 protein overexpression indicates mutant p53 gene status in HCC. Therefore, the goal of this meta-analysis was to explore the correlation between protein expression and gene mutations of p53 in primary cancer tissues of HCC patients. The results of our meta-analysis, which included 1,659 HCC patients from 36 studies, demonstrated that p53 protein overexpression has a moderate diagnostic concordance to mutational assays in the identification of p53 gene mutations in HCC, with a pooled SEN of 0.83 (95% CI: 0.80–0.86) and SPE of 0.74 (95% CI: 0.71–0.76). Furthermore, the AUC of 0.8230 and the DOR of 9.77 (95% CI: 6.35–15.02) also indicated a moderate overall accuracy.
Usually, wild-type p53 protein is rapidly degraded in a MDM2-dependent manner and is undetectable, while mutant p53 protein can escape from degradation and accumulate to excess levels in the cell nuclei. This p53 protein accumulation has been associated with tumor progression [13,40]; however, studies on p53 protein accumulation have shown inconsistent results. There are several explanations for the differences between the incidence of p53 protein overexpression and p53 genetic alteration: i) other factors, such as the hepatitis virus, may contribute to the transcriptional activation of p53 rather than mutations [5,47]; ii) the presence of a missense mutation [25]; or iii) the threshold values of p53 proteins are different [5,25,30]. Immunoblotting assays revealed that in many tumors, increased p53 was the result of a p53 mutation, but wild-type p53 protein expression was also frequently elevated in HCC. Moreover, elevated wild-type p53 protein expression can upregulate Notch1 (an inhibitor of p53 degradation) in HCC cell lines, resulting in overexpression of wild-type p53 protein [48]. In this meta-analysis, 26.1% (281/1075) of HCC tumor tissues with a wild-type p53 gene exhibited positive staining for p53 protein, while 82.9% (484/584) of specimens with p53 mutations exhibited positive staining. Thus, although the wild-type p53 gene also produced p53 protein upregulation, the association between a p53 mutation and p53 overexpression was easily observable in HCC tissues.
By performing subgroup analysis, we found that the relationship between p53 overexpression and p53 mutations remained unchanged, even when the pooled SENs or SPEs varied due to different stratifications. Notably, the pooled SEN was much higher in high-incidence areas than in low-incidence areas, but the SPE was lower, indicating that in high-incidence areas of HCC, IHC assays for p53 expression accurately predicted p53 alterations with authentic genetic mutations but only showed modest accuracy in identifying wild-type p53 phenotypes with no p53 protein overexpression. However, the pooled SEN and SPE of IHC-determined p53 overexpression in the low-incidence areas showed the opposite results. Specific antibodies for IHC-determined p53 overexpression were critically important in diagnosing p53 mutations. In subgroup analysis, four studies employing IHC PAb1801 antibodies exhibited the best diagnostic performance in identifying p53 mutations compared to the studies using other antibodies, with an SEN of 0.80 (95% CI: 0.52−0.96), SPE of 0.91 (95% CI: 0.82−0.97), and DOR of 30.79 (95% CI: 6.58−144.13), suggesting that the PAb1801 antibody effectively identifies mutant p53 proteins.
In this meta-analysis, significant heterogeneity was observed among the included studies. By excluding each study individually, sensitivity analysis revealed that the diagnostic accuracy of IHC-determined p53 overexpression in identifying p53 mutations in HCC remained consistent. Analytical results showed the lowest pooled SEN (0.77, 95% CI: 0.72–0.81) and the lowest heterogeneity (I2 = 45.7%) by removing the study by Qi et al. [8], and the greatest pooled SEN (0.84, 95% CI: 0.81–0.87) with significant between-studies heterogeneity (I2 = 56.6%) by removing the study by Anzola et al. [15]. However, when the two studies were both removed, the between-studies heterogeneity statistic I2 was reduced to 36.7%, although the effect size remained constant (0.78, 95% CI: 0.73–0.82). In regards to the SPE, by omitting Sanefuji et al. [34], sensitivity analyses yielded the maximal pooled statistics (0.76, 95% CI: 0.73–0.78) and substantial heterogeneity (I2 = 64.9%, the lowest in the sensitivity analyses of SPE).
Although we quantitatively evaluated the association between IHC-determined p53 overexpression and p53 gene mutations, there were some limitations in our meta-analysis. First, due to the wide time span for the included studies, from 1992 to 2015 (17 studies before 1999), the study design and the process of collecting the data on p53 alterations in HCC patients may vary among these studies, resulting in difficulties in controlling relevant clinical and pathological parameters of the patients and a relatively low study quality. Second, our analysis could not clarify the association between the specific characteristics of p53 mutations and p53 overexpression because individual patient data, such as the mutable sites of p53 in each patient and the exposure to hepatitis B/C virus, AFB1, or other potential mutagens, were lacking. Additionally, there could be a potential language bias in this analysis because only studies written in English, German and Chinese were included. Thus, we suggest that the results of the meta-analysis should be interpreted with caution for the above reasons.
Conclusion
In summary, our meta-analysis showed that p53 protein overexpression is indeed correlated with p53 gene mutations, suggesting that IHC-determined p53 overexpression has diagnostic concordance to mutational analysis and the identification of p53 gene mutations. This meta-analysis provides quantitative support for the association of IHC-determined p53 overexpression with p53 genetic alterations in HCC patients, especially in high-incidence areas (Asia and Africa). Furthermore, alterations of the tumor suppressor p53 gene were associated with aggressive malignant behaviors and poor patient survival in HCC. Therefore, to obtain a comprehensive account of p53 alterations, simultaneous evaluation of multiple p53 parameters, including p53 protein expression levels and p53 genetic phenotypes, should be performed in future clinical and pathological or prognostic studies and should present compelling evidence of the clinical and prognostic importance of p53 alterations in HCC patients.
Supporting Information
(DOC)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
Support was provided by 2014-0801 [http://yfy.haust.edu.cn/] from the Scientific Research Foundation for Medical Doctors First Affiliated Hospital, College of Clinical Medicine, Henan University of Science and Technology, JL.
References
- 1.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66: 115–132. 10.3322/caac.21338 [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66: 7–30. 10.3322/caac.21332 [DOI] [PubMed] [Google Scholar]
- 3.Liu J, Ma Q, Zhang M, Wang X, Zhang D, Li W, et al. Alterations of TP53 are associated with a poor outcome for patients with hepatocellular carcinoma: evidence from a systematic review and meta-analysis. Eur J Cancer. 2012;48: 2328–2338. 10.1016/j.ejca.2012.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheung ST, Wong SY, Lee YT, Fan ST. GEP associates with wild-type p53 in hepatocellular carcinoma. Oncol Rep. 2006;15: 1507–1511. 10.3892/or.15.6.1507 [DOI] [PubMed] [Google Scholar]
- 5.Mohamed WS, Omar MM, Khayri TM, Fakhr IM. 2008. Assessment of the proliferative marker ki-67 and p53 protein expression in HBV- and HCV-related hepatocellular carcinoma cases in Egypt. Int J Health Sci (Qassim) 2008;2: 27–34. [PMC free article] [PubMed] [Google Scholar]
- 6.Yuan RH, Jeng YM, Chen HL, Lai PL, Pan HW, Hsieh FJ, et al. Stathmin overexpression cooperates with p53 mutation and osteopontin overexpression, and is associated with tumour progression, early recurrence, and poor prognosis in hepatocellular carcinoma. J Pathol. 2006;209: 549–558. 10.1002/path.2011 [DOI] [PubMed] [Google Scholar]
- 7.Park NH, Chung YH, Youn KH, Song BC, Yang SH, Kim JA, et al. Close correlation of p53 mutation to microvascular invasion in hepatocellular carcinoma. J Clin Gastroenterol. 2001;33: 397–401. 10.1097/00004836-200111000-00011 [DOI] [PubMed] [Google Scholar]
- 8.Qi LN, Bai T, Chen ZS, Wu FX, Chen YY, De Xiang B, et al. The p53 mutation spectrum in hepatocellular carcinoma from Guangxi, China: role of chronic hepatitis B virus infection and aflatoxin B1 exposure. Liver Int. 2015;35: 999–1009. 10.1111/liv.12460 [DOI] [PubMed] [Google Scholar]
- 9.Jablkowski M, Bocian A, Bialkowska J, Bartkowiak J. A comparative study of P53/MDM2 genes alterations and P53/MDM2 proteins immunoreactivity in liver cirrhosis and hepatocellular carcinoma. J Exp Clin Cancer Res. 2005;24: 117–125. [PubMed] [Google Scholar]
- 10.Rashid A, Wang JS, Qian GS, Lu BX, Hamilton SR, Groopman JD. Genetic alterations in hepatocellular carcinomas: association between loss of chromosome 4q and p53 gene mutations. Br J Cancer. 1999;80: 59–66. 10.1038/sj.bjc.6690321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Szymańska K, Lesi OA, Kirk GD, Sam O, Taniere P, Scoazec JY, et al. Ser-249TP53 mutation in tumour and plasma DNA of hepatocellular carcinoma patients from a high incidence area in the Gambia, West Africa. Int J Cancer. 2004;110: 374–379. 10.1002/ijc.20103 [DOI] [PubMed] [Google Scholar]
- 12.Zhang YJ, Rossner P Jr, Chen Y, Agrawal M, Wang Q, Wang L, et al. Aflatoxin B1 and polycyclic aromatic hydrocarbon adducts, p53 mutations and p16 methylation in liver tissue and plasma of hepatocellular carcinoma patients. Int J Cancer. 2006;119: 985–991. 10.1002/ijc.21699 [DOI] [PubMed] [Google Scholar]
- 13.Midgley CA, Lane DP. p53 protein stability in tumour cells is not determined by mutation but is dependent on Mdm2 binding. Oncogene. 1997;15: 1179–1189. 10.1038/sj.onc.1201459 [DOI] [PubMed] [Google Scholar]
- 14.Bourdon JC, D'Errico A, Paterlini P, Grigioni W, May E, Debuire B. p53 protein accumulation in European hepatocellular carcinoma is not always dependent on p53 gene mutation. Gastroenterol. 1995;108: 1176–1182. 10.1016/0016-5085(95)90217-1 [DOI] [PubMed] [Google Scholar]
- 15.Anzola M, Saiz A, Cuevas N, Lopez-Martinez M, Martinez de Pancorbo MA, Burgos JJ. High levels of p53 protein expression do not correlate with p53 mutations in hepatocellular carcinoma. J Viral Hepat. 2004;11: 502–510. 10.1111/j.1365-2893.2004.00541.x [DOI] [PubMed] [Google Scholar]
- 16.Gross-Goupil M, Riou P, Emile JF, Saffroy R, Azoulay D, Lacherade I, et al. Analysis of chromosomal instability in pulmonary or liver metastases and matched primary hepatocellular carcinoma after orthotopic liver transplantation. Int J Cancer. 2003;104: 745–751. 10.1002/ijc.11017 [DOI] [PubMed] [Google Scholar]
- 17.Whiting PF, Rutjes AW, Westwood ME, Mallett S, Deeks JJ, Reitsma JB, et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155: 529–536. 10.7326/0003-4819-155-8-201110180-00009 [DOI] [PubMed] [Google Scholar]
- 18.Thompson SG, Higgins JP. How should meta-regression analyses be undertaken and interpreted?. Stat Med. 2002;21: 1559–1573. 10.1002/sim.1187 [DOI] [PubMed] [Google Scholar]
- 19.Deeks JJ, Macaskill P, Irwig L. The performance of tests of publication bias and other sample size effects in systematic reviews of diagnostic test accuracy was assessed. J Clin Epidemiol. 2005;58: 882–893. 10.1016/j.jclinepi.2005.01.016 [DOI] [PubMed] [Google Scholar]
- 20.Boix-Ferrero J, Pellín A, Blesa R, Adrados M, Llombart-Bosch A. Absence of p53 gene mutations in hepatocarcinomas from a Mediterranean area of Spain. A study of 129 archival tumour samples. Virchows Arch. 1999;434: 497–501. 10.1007/s004280050374 [DOI] [PubMed] [Google Scholar]
- 21.Soini Y, Chia SC, Bennett WP, Groopman JD, Wang JS, DeBenedetti VM, et al. An aflatoxin-associated mutational hotspot at codon 249 in the p53 tumor suppressor gene occurs in hepatocellular carcinomas from Mexico. Carcinogenisis. 1996;17: 1007–1012. 10.1093/carcin/17.5.1007 [DOI] [PubMed] [Google Scholar]
- 22.Challen C, Lunec J, Warren W, Collier J, Bassendine MF. Analysis of the p53 tumor-suppressor gene in hepatocellular carcinomas from Britain. Hepatology. 1992;16: 1362–1366. 10.1002/hep.1840160610 [DOI] [PubMed] [Google Scholar]
- 23.Peng XM, Peng WW, Yao JL. Codon 249 mutations of p53 gene in development of hepatocellular carcinoma. World J Gastroenterol. 1998;4: 125–127. 10.3748/wjg.v4.i2.125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsia CC, Nakashima Y, Thorgeirsson SS, Harris CC, Minemura M, Momosaki S, et al. Correlation of immunohistochemical staining and mutations of p53 in human hepatocellular carcinoma. Oncol Rep. 2000;7: 353–356. 10.3892/or.7.2.353 [DOI] [PubMed] [Google Scholar]
- 25.Lee SN, Park CK, Sung CO, Choi JS, Oh YL, Cho JW, et al. Correlation of mutation and immunohistochemistry of p53 in hepatocellular carcinomas in Korean people. J Korean Med Sci. 2002;17: 801–805. 10.3346/jkms.2002.17.6.801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hsu HC, Tseng HJ, Lai PL, Lee PH, Peng SY. Expression of p53 gene in 184 unifocal hepatocellular carcinomas: association with tumor growth and invasiveness. Cancer Res. 1993;53: 4691–4694. [PubMed] [Google Scholar]
- 27.Okada T, Iizuka N, Yamada-Okabe H, Mori N, Tamesa T, Takemoto N, et al. Gene expression profile linked to p53 status in hepatitis C virus-related hepatocellular carcinoma. FEBS Lett. 2003;555: 583–590. 10.1016/S0014-5793(03)01345-0 [DOI] [PubMed] [Google Scholar]
- 28.Stern MC, Umbach DM, Yu MC, London SJ, Zhang ZQ, Taylor JA. Hepatitis B, aflatoxin B(1), and p53 codon 249 mutation in hepatocellular carcinomas from Guangxi, People's Republic of China, and a meta-analysis of existing studies. Cancer Epidemiol Biomarkers Prev. 2001;10: 617–625. [PubMed] [Google Scholar]
- 29.Greenblatt MS, Feitelson MA, Zhu M, Bennett WP, Welsh JA, Jones R, et al. Integrity of p53 in hepatitis B x antigen-positive and -negative hepatocellular carcinomas. Cancer Res. 1997;57: 426–432. [PubMed] [Google Scholar]
- 30.Volkmann M, Schiff J, Hajjar Y, Otto G, Stilgenbauer F, Fiehn W, et al. Loss of CD95 expression is linked to most but not all p53 mutants in European hepatocellular carcinoma. J Mol Med (Berl). 2001;79: 594–600. 10.1007/s001090100244 [DOI] [PubMed] [Google Scholar]
- 31.Mitsumoto Y, Nakajima T, Marutani M, Kashiwazaki H, Moriguchi M, Kimura H, et al. Loss of p53 transcriptional activity in hepatocellular carcinoma evaluated by yeast-based functional assay: comparison with p53 immunohistochemistry. Hum Pathol. 2004;35: 350–356. 10.1016/j.humpath.2003.09.014 [DOI] [PubMed] [Google Scholar]
- 32.Kubicka S, Trautwein C, Schrem H, Tillmann H, Manns M. Low incidence of p53 mutations in European hepatocellular carcinomas with heterogeneous mutation as a rare event. J Hepatol. 1995;23: 412–419. 10.1016/0168-8278(95)80199-5 [DOI] [PubMed] [Google Scholar]
- 33.Andersson M, Jönsson M, Nielsen LL, Vyberg M, Visfeldt J, Storm HH, et al. Mutations in the tumor suppressor gene p53 in human liver cancer induced by alpha-particles. Cancer Epidemiol Biomarkers Prev. 1995;4: 765–770. [PubMed] [Google Scholar]
- 34.Sanefuji K, Taketomi A, Iguchi T, Sugimachi K, Ikegami T, Yamashita Y, et al. Significance of DNA polymerase delta catalytic subunit p125 induced by mutant p53 in the invasive potential of human hepatocellular carcinoma. Oncol. 2010;79: 229–237. 10.1159/000322374 [DOI] [PubMed] [Google Scholar]
- 35.An FQ, Matsuda M, Fujii H, Tang RF, Amemiya H, Dai YM, et al. Tumor heterogeneity in small hepatocellular carcinoma: analysis of tumor cell proliferation, expression and mutation of p53 and beta-catenin. Int J Cancer. 2001;93: 468–474. 10.1002/ijc.1367 [DOI] [PubMed] [Google Scholar]
- 36.Shieh YS, Nguyen C, Vocal MV, Chu HW. Tumor-suppressor p53 gene in hepatitis C and B virus-associated human hepatocellular carcinoma. Int J Cancer. 1993;54: 558–562. 10.1002/ijc.2910540407 [DOI] [PubMed] [Google Scholar]
- 37.Ryder SD, Rizzi PM, Volkmann M, Metivier E, Pereira LM, Galle PR, et al. Use of specific ELISA for the detection of antibodies directed against p53 protein in patients with hepatocellular carcinoma. J Clin Pathol. 1996;49: 295–299. 10.1136/jcp.49.4.295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qin LF, Ng IO, Fan ST, Ng M. p21/WAF1, p53 and PCNA expression and p53 mutation status in hepatocellular carcinoma. Int J Cancer. 1998;79: 424–428. [DOI] [PubMed] [Google Scholar]
- 39.Kang YK, Kim CJ, Kim WH, Kim HO, Kang GH, Kim YI. p53 mutation and overexpression in hepatocellular carcinoma and dysplastic nodules in the liver. Virchows Arch. 1998;432: 27–32. 10.1007/s004280050130 [DOI] [PubMed] [Google Scholar]
- 40.Zekri AR, Bahnassy AA, Madbouly MS, Asaad NY, El-Shehaby AM, Alam El Din HM. p53 mutation in HCV-genotype-4 associated hepatocellular carcinoma in Egyptian patients. J Egypt Natl Canc Inst. 2006;18: 17–29. [PubMed] [Google Scholar]
- 41.De Benedetti VM, Welsh JA, Yu MC, Bennett WP. p53 mutations in hepatocellular carcinoma related to oral contraceptive use. Carcinogenisis. 1996;17: 145–149. 10.1093/carcin/17.1.145 [DOI] [PubMed] [Google Scholar]
- 42.Lunn RM, Zhang YJ, Wang LY, Chen CJ, Lee PH, Lee CS, et al. p53 mutations, chronic hepatitis B virus infection, and aflatoxin exposure in hepatocellular carcinoma in Taiwan. Cancer Res. 1997;57: 3471–3477. [PubMed] [Google Scholar]
- 43.Chen GG, Merchant JL, Lai PB, Ho RL, Hu X, Okada M, et al. Mutation of p53 in recurrent hepatocellular carcinoma and its association with the expression of ZBP-89. Am J Pathol. 2003;162: 1823–1829. 10.1016/S0002-9440(10)64317-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou F, Wang J, Lei B, Zhao L, Tang H, Wang X, et al. Detection of p53 gene mutations in hepatocellular carcinoma. Hua Xi Yi Ke Da Xue Xue Bao. 1997;28: 50–54. [PubMed] [Google Scholar]
- 45.Luo YL, Cheng RX, Feng DY. Role of functional inactivation of p53 from MDM2 overexpression in hepatocarcinogenesis. Hunan Yi Ke Da Xue Xue Bao. 2001;26: 13–16. [PubMed] [Google Scholar]
- 46.Cabelguenne A, Blons H, de Waziers I, Carnot F, Houllier AM, Soussi T, et al. p53 alterations predict tumor response to neoadjuvant chemotherapy in head and neck squamous cell carcinoma: a prospective series. J Clin Oncol. 2000;18: 1465–1473. [DOI] [PubMed] [Google Scholar]
- 47.Otsuka M, Kato N, Lan K, Yoshida H, Kato J, Goto T, et al. Hepatitis C virus core protein enhances p53 function through augmentation of DNA binding affinity and transcriptional ability. J Biol Chem. 2000;275: 34122–34130. 10.1074/jbc.M000578200 [DOI] [PubMed] [Google Scholar]
- 48.Lim SO, Park YM, Kim HS, Quan X, Yoo JE, Park YN, et al. Notch1 differentially regulates oncogenesis by wildtype p53 overexpression and p53 mutation in grade III hepatocellular carcinoma. Hepatology. 2011;53: 1352–1362. 10.1002/hep.24208 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.