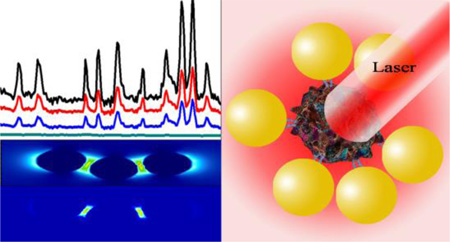
Abstract
Dengue virus (DENV) and West Nile virus (WNV) are two well-documented mosquito-borne flaviviruses that cause significant health problems worldwide. Driven by this need, we have developed a bio-conjugated gold nanoparticle (AuNP)-based surface enhanced Raman spectroscopy (SERS) probe for the detection of both DENV and WNV. Reported data demonstrate anti-flavivirus 4G2 antibody conjugated gold nanoparticle (GNP) SERS probe can be used as a Raman fingerprint for the ultrasensitive detection of DENV and WNV selectively. Experimental data show that due to the plasmon coupling in nano-assembly, antibody conjugated GNP- based SERS is able to detect as low as 10 plaque-forming units (PFU)/ml of DENV-2 and WNV, which is comparable with the sensitivity of quantitative PCR-based assays. Selectivity of our probe was demonstrated using another mosquito-borne chikungunya virus (CHIKV) as a negative control. Experimental data demonstrate a huge enhancement of SERS intensity is mainly due to the strong electric field enhancement, which has been confirmed by the finite-difference time-domain (FDTD) simulation. Reported FDTD simulation data indicate the SERS enhancement factor can be more than 104 times, due to the assembled structure. Reported results suggest that bio-conjugated AuNP-4G2 based SERS probes have great potential to be used to screen viral particles in clinical and research-based laboratories.
Graphical Abstract
Introduction
According to the world health organization (WHO)1–2, over one million people worldwide die from mosquito-borne diseases every year. Dengue virus (DENV) and West Nile virus (WNV) are the leading causative agents of mosquito-borne diseases worldwide1–4. Aside from mosquito transmission, DENV and WNV are also transmitted by blood transfusions and organ transplantation1–4. Importantly, DENV has been identified as a high-priority infectious agent with the potential risk of transfusion-transmission in both the United States and Canada1–4. However, there is currently no routine screening of DENV in clinical settings, which is partially due to lack of a rapid, sensitive and cost-effective detection assay. Driven by the need, we report for the first time the development of an anti-flavivirus 4G2 antibody conjugated gold nanoparticle (AuNP-4G2)-based surface enhanced Raman spectroscopy (SERS) probe that can be used as a cost-effective and rapid detection tool for DENV and WNV selectively.
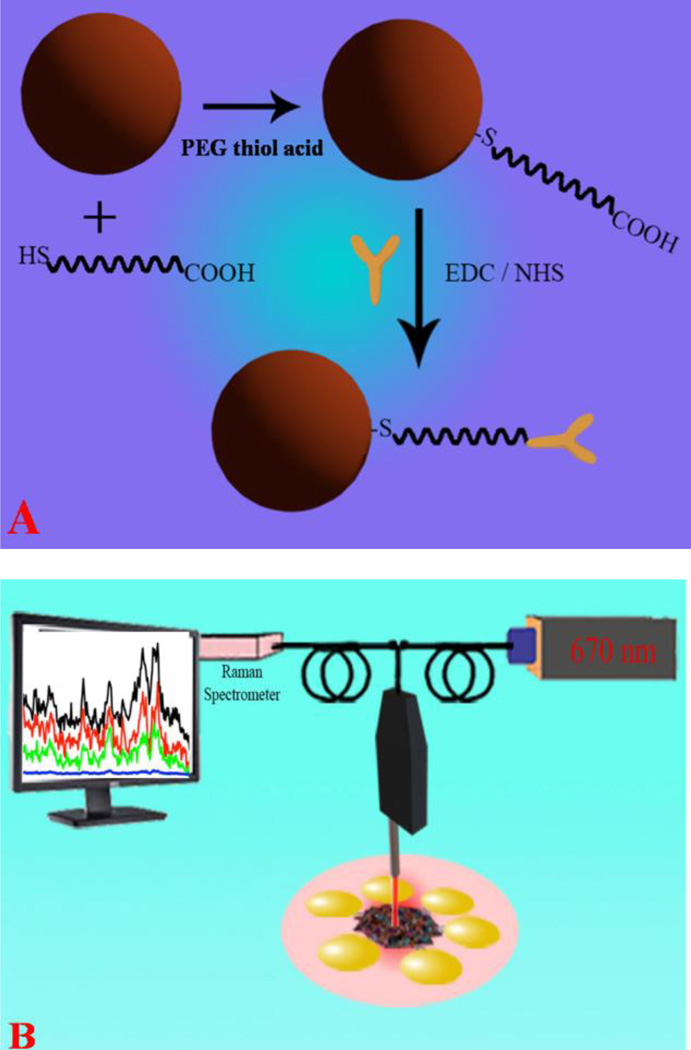
SERS has the ability to rapidly detect microorganisms or biological analytes with chemical specificity intrinsic to vibrational spectroscopy5–14. Since the Raman signal can be enhanced by 108–1014 orders of magnitude in the presence of a metal nanomaterial surface15–23, SERS is emerging as an important tool for identification and classification of microorganisms24–30. Additionally, SERS has the ability to provide detailed information regarding the chemical composition of microorganisms and it can also serve as a fingerprint for detection and identification of microorganisms15–20. In addition to its fingerprinting ability and sensitivity, one of the other important features of the SERS assay is its specificity, which has been achieved here by attaching virus specific antibodies to the gold nanoparticle surface, as shown in Scheme 1.
Scheme 1.
(A) Schematic representation of the construction of anti-flaviviral (4G2) coated gold nanoparticles. (B) Schematic representation shows detection of viruses using Raman fingerprinting. FDTD simulation data indicate a huge enhancement of the SERS intensity is due to strong electric field.
Using the above advantages, we have developed an anti-flaviviral antibody coated AuNP-based SERS assay, as shown in Scheme 1, for rapid and sensitive detection of DENV and WNV selectively. Our reported results demonstrated that antibody conjugated gold nanoparticles can be used as fingerprint spectra for viruses. Since the effective plasmon field generated by nanoparticle assemblies on the viral surface is more intense than individual nanoparticles, our reported experimental data demonstrated that the detection limit is as low as 10 viruses/ml. Our experimental findings on the plasmon coupling enhanced SERS signal was supported by a finite-difference time-domain (FDTD) simulation26–30. FDTD is known to be a powerful tool for modeling electromagnetic near-field enhancement, which is an important parameter for enhancing SERS intensity via nanoparticle assembly. Since the SERS signal enhancement factor is approximately proportional to the fourth power of the electric field enhancement |E|4, we have performed theoretical estimates of |E|4 using FDTD simulation.
Materials and methods
Gold chloride salt, sodium nitrate, sodium citrate, potassium permanganate, polyethylene glycol (PEG) thiol acid (SH-PEG-COOH) and 1-Ethyl-3-[3-dimethylaminopropyl] carbodiimide hydrochloride (EDC) were purchased from Sigma-Aldrich (St. Louis, MO, USA). WNV CT2741 and CHIKV Ross were kindly provided by Dr. John F. Anderson at the Connecticut Agricultural Experiment Station and DENV-2 was purchased from the American Type Culture Collection (ATCC, Rockville, MD).
Pre paration of AuNP-4G2 complexes
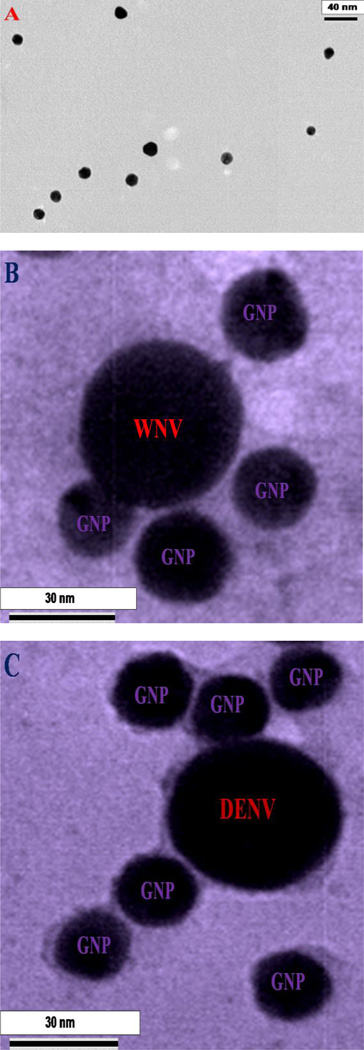
Gold nanoparticle synthesis was achieved through our recently reported method, with additional modifications9,18–20,30. Briefly, AuNPs (10–15 nm in diameter) were synthesized by using 0.01% chloroauric acid and 1% trisodium citrate dehydrate (weight/volume). Gold nanoparticles were then modified with polyethylene glycol (PEG) thiol acid (SH-PEG-COOH, 10−4 M) addition via Au-S chemistry, to prevent aggregation, as shown in Scheme 1. The solution was stirred for 15 minutes and 4G2 antibodies were added to the solution (antibody final concentration 0.1 mg/ml). As shown in Scheme 1, 4G2 antibodies were attached with AuNPs via the carboxy group of PEG using 15 µl (1 mg/ml) of 1-ethyl-3-[3-dimethylaminopropyl] carbodiimide hydrochloride (EDC) and 15 µl (1 mg/ml) of N-hydroxysuccinimide (NHS) to activate the -COOH group. Gold chloride salt, sodium nitrate, sodium citrate, potassium permanganate, polyethylene glycol (PEG) thiol acid (SH-PEG-COOH) and 1-Ethyl-3-[3-dimethylaminopropyl] carbodiimide hydrochloride (EDC) were purchased from Sigma-Aldrich (St. Louis, MO, USA). We used a Zeiss 900 TEM to characterize the gold nanoparticles. Digital images were taken with a Gatan Model 785 ES1000W Erlangshen CCD Camera. As shown in Figure 1A, our data clearly show the size of the antibody attached gold nanoparticle (AuNP-4G2) is approximately 10 nm. Using the KCN dissociation procedure, as previously reported18–20, we estimated that there were about 100–120 4G2 antibodies per AuNP.
Figure 1.
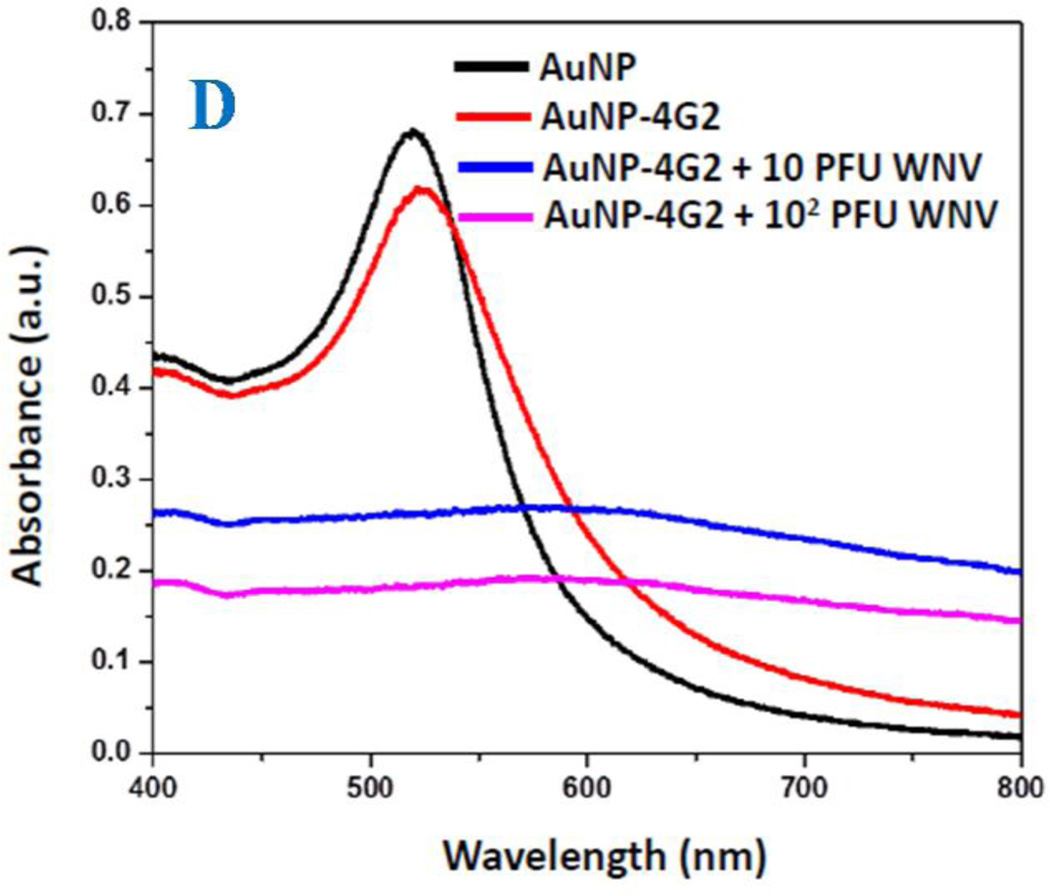
A) TEM image using Zeiss 900 transmission electron microscope shows the morphology of 4G2 antibodies conjugated gold nano-particles. B) TEM picture shows the morphology of 4G2 antibodies modified gold nano-particles attached West Nile virus. To view WNV, virus samples were stained by 2% uranyl acetate for 1 hour. C) TEM picture shows the morphology of 4G2 antibodies modified gold nano-particles attached Dengue virus. To view DENV, virus samples were stained by 2% uranyl acetate for 1 hour, before imaging. D) Extinction spectra of gold nanoparticle, 4G2 antibodies conjugated gold nano-particles, gold nanoparticle attached with WNV. Due to the formation of nanoparticle assembly on WNV surface, excitation spectra is very broad for 4G2 antibodies modified gold nanoparticles after attachment with virus.
Virus generation, cell culture, and antibody preparation
Dengue virus serotype-2 (DENV-2, ATCC VR-1584, Rockville, MD) and chikungunya virus (CHIKV, Ross strain) were propagated one time in mosquito C6/36 cells (ATCC CRL-1660). WNV isolate (CT2741) was propagated in Vero cells (ATCC CCL-81). All viruses were titered by using a Vero cell plaque assay, as previously described31–32. WNV (CT2741) and CHIKV (Ross strain) were kindly provided by Dr. John F. Anderson at the Connecticut Agricultural Experiment Station. Briefly, C6/36 cells were maintained at 28 °C, 5% CO2 in Eagles minimum essential medium (EMEM, Life Technologies, Grand Island, NY) containing 10% fetal bovine serum (FBS, Atlantic Biologicals, Norcross, GA). Vero cells (ATCC CCL-81) were maintained at 37 °C, 5% CO2 in Dulbecco’s modified Eagle medium (DMEM, Life Technologies, Grand Island, NY) containing 10% FBS, 1% L-glutamine (L-glu, Sigma-Aldrich, St. Louis, MO), and 1% penicillin-streptomycin (Pen/Strep, Sigma-Aldrich, St. Louis, MO).
Mouse IgG monoclonal anti-flaviviral antibodies (4G2) were produced by culturing D1-4G2-4-15 (HB-112, ATCC) hybridoma in DMEM with 10% FBS, 1% Pen/Strep, and 1.5g/l of sodium bicarbonate (Sigma-Aldrich, St. Louis, MO) at 37 °C, 5% CO2. Crude media was collected when cell confluence reached 90–95% and stored at −20°C until purification was performed. The 4G2 antibodies were concentrated using the HiTrap™ Protein G HP columns (GE Healthcare, New Orleans, LA) and purified using Amicon Ultra centrifugal filters (100K MW, Millipore, Billerica, MA), as previously reported31, followed by resuspension in 40% glycerol (Sigma-Aldrich, St. Louis, MO) and storage at −20°C.
SERS sample preparation and Raman experiment
The SERS signals were analyzed by dropping 10 µL of the solution on an eppendorf cap and using a continuous wavelength diode pumped solid state (DPSS) laser (Laser Glow Technology, Toronto, ON) as an excitation light source (670 nm).
As shown in Scheme 1B, we have used a Raman fiber optic probe (InPhotonics, Norwood, MA) for excitation and SERS signal collection. A miniaturized QE65000 Scientific-grade Spectrometer (Ocean Optics, Dunedin, FL) was used for data acquisition. For the SERS experiment, AuNP-4G2 complexes (500 µl, 1:0.1 dilution) were incubated with 50 µl of serially diluted DENV-2, WNV, or CHIKV viruses (10 to 104 PFU/ml) in 1 × PBS, or 1 × PBS only as a negative control, for 15 minutes at room temperature followed by fixation in 4% para-formaldehyde (PFA) and either analyzed immediately or stored at 4°C until further analysis. We have performed TEM and absorption analysis before and after exposure of laser light and we have found out that biological samples remain unchanged during our SERS measurement. For the removal of autofluorescence background signals, baseline correction and spectral smoothing, we have employed the Vancouver Raman algorithms33–34. It is well documented that the Vancouver Raman algorithm is a multi-polynomial fitting algorithm which can be used to improve signal-to-noise ratios by separating autofluorescence background signals33–34. All SERS experiments were performed 10 times separately and the average spectral data are reported.
Quantitative real-time PCR (qPCR)
Viral RNA was extracted from DENV-2 and WNV that were serially diluted (10 to 5 × 103 PFU/ml) in PBS followed by a one-step qPCR assay. Viral RNA was isolated by proteinase K digestion (Qiagen) and AL lysis (Qiagen) of viral particles, followed by RNA purification using the RNase easy isolation kit (Qiagen). The A one-step qPCR method was performed using previously reported primers and probes for the detection of DENV-2 capsid and WNV envelope(E) gene31–32, and were purchased either by Integrated DNA Technologies (Coralville, IA) or Applied Biosystems (Grand Island, NY). All results were expressed as the absolute number of viral RNA copies/100 µl of sample using the iTAQ™ Universal Probes one-step qPCR kit (Bio-Rad, Hercules, CA) and were compared to viral gene standards for absolute copy number quantification.
Results and Discussion
To develop the selective SERS probe for WNV and DENV, anti-flaviviral antibodies (4G2) were conjugated to gold nanoparticles (AuNPs) with the detailed synthesis procedures described in methods. Figure 1A shows the image of the antibody attached-gold nanoparticles (AuNP-4G2), which indicates that the size is around 10–15 nm. The excitation spectrum, as shown in Figure 1D indicates that the plasmon band of AuNPs are very slightly shifted (2 nm) after conjugation with the antibody. Due to the lack of antigen-antibody interaction, gold nanoparticles do not form assembly structure in the absence of virus and as a result, we have not observed any broad absorption spectra from antibody conjugated gold nanoparticles.
To determine whether AuNP-4G2 complexes bind to DENV and WNV, one thousand (103) plaque forming units (PFU) of viruses in PBS (50 µl) were incubated for 15 minutes with the AuNP-4G2 complexes (500 µl; 1:0.1 dilution), followed by fixation in 4% PFA. AuNP-4G2 probed with DENV-2 was then spun down (8,000 rpm for 10 minutes) and imaged by TEM. To view the viral particles, all samples were stained with 2% uranyl acetate for 1 h by adding 15–30 µL per grid. The grids were then rinsed with nanowater and air-dried overnight for TEM imaging. As shown in Figures 1B and 1C, AuNP-4G2 complexes bind to WNV and DENV respectively and due to the antigen-antibody interaction, nanoparticles formed assembly structure on the surface of virus. As a result, we have observed very broad plasmon band, which appears as structureless bands in the absorption spectra as shown in Figure 1D.
After binding with DENV and WNV, AuNP-4G2 complexes undergo aggregation on the viral particles and as a result, they formed high intensity “hot spots” that lead to very sensitive and specific viral detection. In the SERS application, formation of these “hot spots” is very important to amplify the SERS signal tremendously10–20. SERS enhancement arises primarily from the enhanced electromagnetic field that is produced by resonant excitation of the surface electrons in the metal nanostructures due to an excitation laser light source10–20.
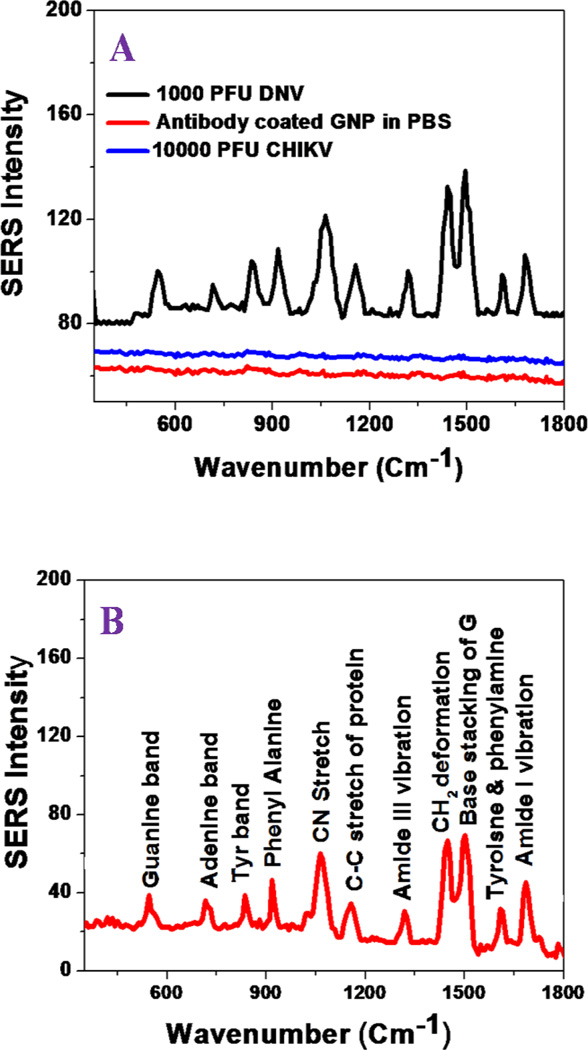
To detect DENV and WNV by the SERS probe, AuNP-4G2 complexes were incubated with serially diluted DENV-2, WNV, or chikungunya virus (CHIKV) as a negative control, starting from 10 to 104 PFU/ml. Figure 2A–2B and 3A show the SERS spectra obtained from WNV and DENV-2, respectively. In contrast, no Raman signal was detected from AuNP-4G2 complexes in the absence of WNV or DENV, which clearly indicates that antibodies attached to AuNPs do not exhibit any Raman band signal in the absence of virus, mainly due to the lack of nanoparticle assembly or aggregate formation. In addition, no Raman signal was detected for CHIKV even at a concentration of 104 PFU/ml, as shown in Figure 2A. All the reported result, clearly show that AuNP-4G2-based SERS assay is highly selective for the accurate detection of WNV and DENV-2.
Figure 2.
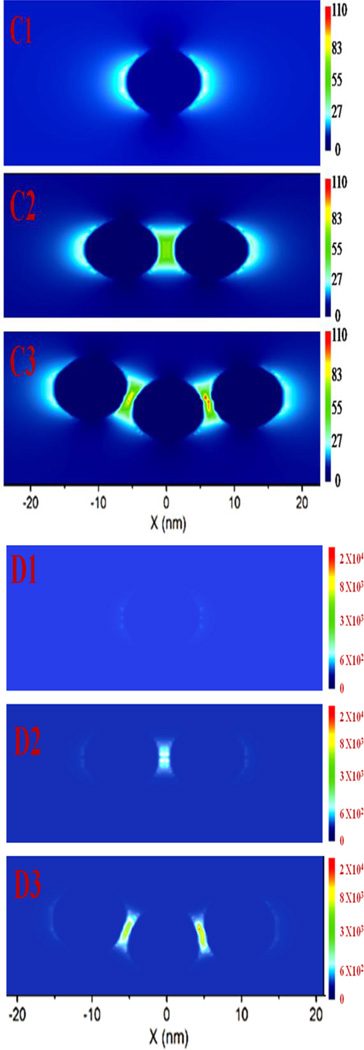
(A) SERS spectra show no Raman signal from 4G2 antibody-conjugated gold nanoparticle. Similarly, no Raman signal was observed in the presence of CHIKV. Conversely, Raman signal was observed in the presence of WNV. It is mainly due to the formation of assembly structure in the presence of WNV. (B) Spectrum shows SERS band assignment from WNV conjugated nanoassembly. Observed SERS signal is directly from the WNV. (C1–C3) Simulated electric field enhancement |E|2 profiles in arbitrary units for monomer and nanoparticle assembly. Calculation has been performed using finite-difference time-domain (FDTD) simulation using the 10 nm particle size and separation distance kept at 2 nm. For trimmer structure we have used bent structure, shown in Figure 1C. (D1–D3) Simulated electric field enhancement |E|4 profiles in arbitrary units for monomer and nanoparticle assembly.
Figure 3.
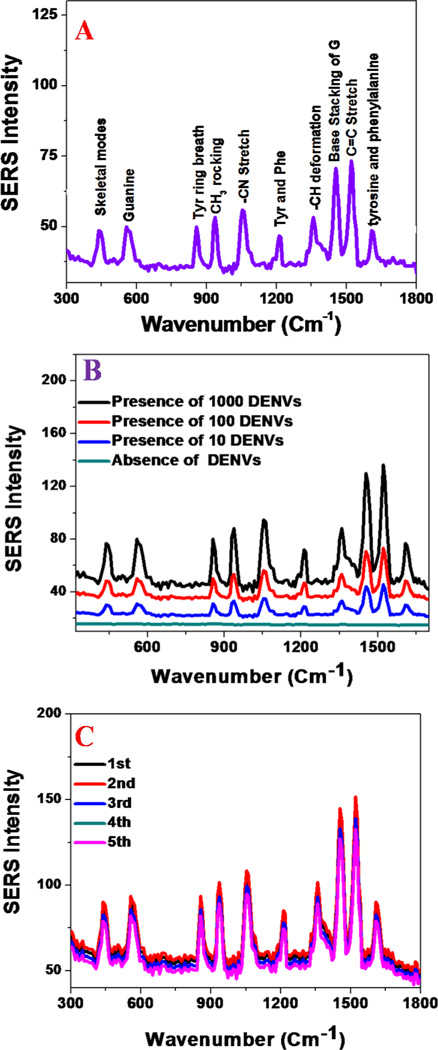
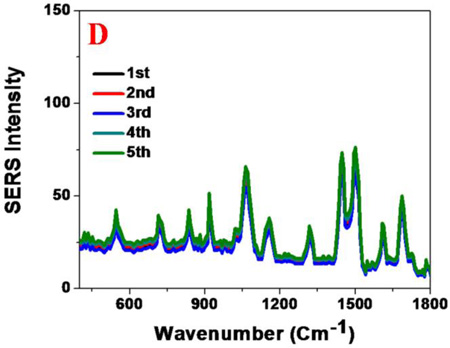
(A) Spectrum shows SERS band assignment from DENV conjugated nano-assembly. Observed SERS signal is directly from the DENV. (B) Concentration dependent study indicates that the SERS probe can be used for the selective and sensitive detection of DENV, as low as 10 PFU DENV/ml. C) SERS spectra from DENV conjugated gold nanoparticle assembly made at different batchs. Reported data shows very good reproducibility. D) SERS spectra from WNV conjugated gold nanoparticle assembly made at different batchs. Reported data shows very good reproducibility.
Tables 1 and 2, show the possible assignment of Raman bands obtained from WNV and DENV-2, respectively. Prominent bands are mainly due to the protein backbone amide band, -COH deformation bands, the guanine band, skeletal vibrations and bands of fatty acids. In the SERS spectrum, the Raman band at ~420 cm−1 is due to the skeleton mode, ~550 cm−1 is due to the guanine band, ~950 cm−1 is due to CH3 rocking, ~1060 cm−1 is due to the –C-N stretch, and ~1160 cm−1 is due to the -C–C stretch of protein. These observed vibrational modes are in good agreement with the Raman bands reported for different microorganism in the literature5.9.25–26. It is interesting to observe that the AuNP-4G2 based SERS assay can be used as a fingerprint for WNV and DENV-2, as shown in Figures 2B and 3A. From the SERS spectra reported in Figures 2 and 3 and Tables 1 and 2, we found that the amide I band at 1680 cm−1 and the -CH2 deformation for lipid and proteins vibrations at 1450 cm−1 are unique for WNV, which were not observed for DENV. Similarly the skeleton mode at 420 cm−1 and the CH3 rocking modes at 950 cm−1 are unique for DENV-2, which were not observed for WNV. These results indicate that the AuNP-4G2-based SERS probe can distinguish between DENV-2 and WNV by generating specific viral fingerprints.
Table 1.
Raman modes analysis observed from WNV. All the bands have been assigned using reported data from different microorganisms5.9.25–26.
| WNV Vibration mode |
WNV Raman peak position (cm−1) |
|---|---|
| Amide I | 1680 |
| Tyrosine and Phenylalanine | 1606 |
| Base stacking of Guanine | 1482 |
| -CH2 deformation for lipid and proteins | 1450 |
| COO− str sym | 1376 |
| -C–C stretch of protein | 1160 |
| -C–N Stretch | 1060 |
| Deoxyribose, backbone | 910 |
| Ring breath Tyr | 828 |
| Adenine | 712 |
| Guanine band | 550 |
Table 2.
Raman modes analysis observed from DENV. All the bands have been assigned using reported data from different microorganisms5.9.25–26.
| DENV-2 Vibration mode |
DENV-2 Raman peak position (cm−1) |
|---|---|
| Tyrosine and Phenylalanine | 1610 |
| C=C stretch | 1495 |
| Base stacking of Guanine | 1482 |
| -CH deformation | 1345 |
| Tyrosine and Phenylalanine | 1190 |
| -C–N Stretch | 1070 |
| CH3 rocking | 950 |
| Tyr ring breath | 858 |
| Trp | 740 |
| Guanine band | 550 |
| Skeletal modes | 420 |
For better understanding of the huge enhancement of SERS signal from mosquito-borne flaviviruses via gold nanoparticle assembly, we have performed full-wave simulations, as reported in the Figure 2C. To do this we have used the finite-difference time-domain (FDTD) simulation platform based on numerical solutions of Maxwell’s equations that others and we have previously reported26–30. As shown in the Figure 2C1–C3, nanoparticles with 10 nm particle sizes were used in our calculation while the separation distance was kept at 2 nm. The multipolar and finite size effects were also included in our calculation. Polarized light at a wavelength of 670 nm was used along the y-axis, for FDTD simulation.
For minimum simulation time and maximum field enhancement resolution, the mesh override region was set to 0.1 nm, and the overall simulation time was 500 fs. For the trimmer structure we have used the bent structure, as shown in Figure 1C. As reported in Figure 2C1–C3, the FDTD simulations results indicated the electric field enhancement |E|2 intensity can be greater than 102 times enhancement for gold nano-assembly containing three particles with respect to a monomer gold nanoparticle. Since the SERS signal enhancement factor is directly proportional to the square of the plasmon field enhancement |E|4, theoretically, more than four orders of magnitude of SERS enhancement is expected due to the assembly structure, as shown in Figure 2D1–D3. The observed Raman spectra is mainly from the surface of the virus. Although the size of virus is bigger than nanoparticle, when it forms "hot spot", a portion of virus surface will be in the "hot spot" region. The Raman spectra from the portion of the virus surface which is exposed to "hot spot" are dominated in the reported SERS spectra.
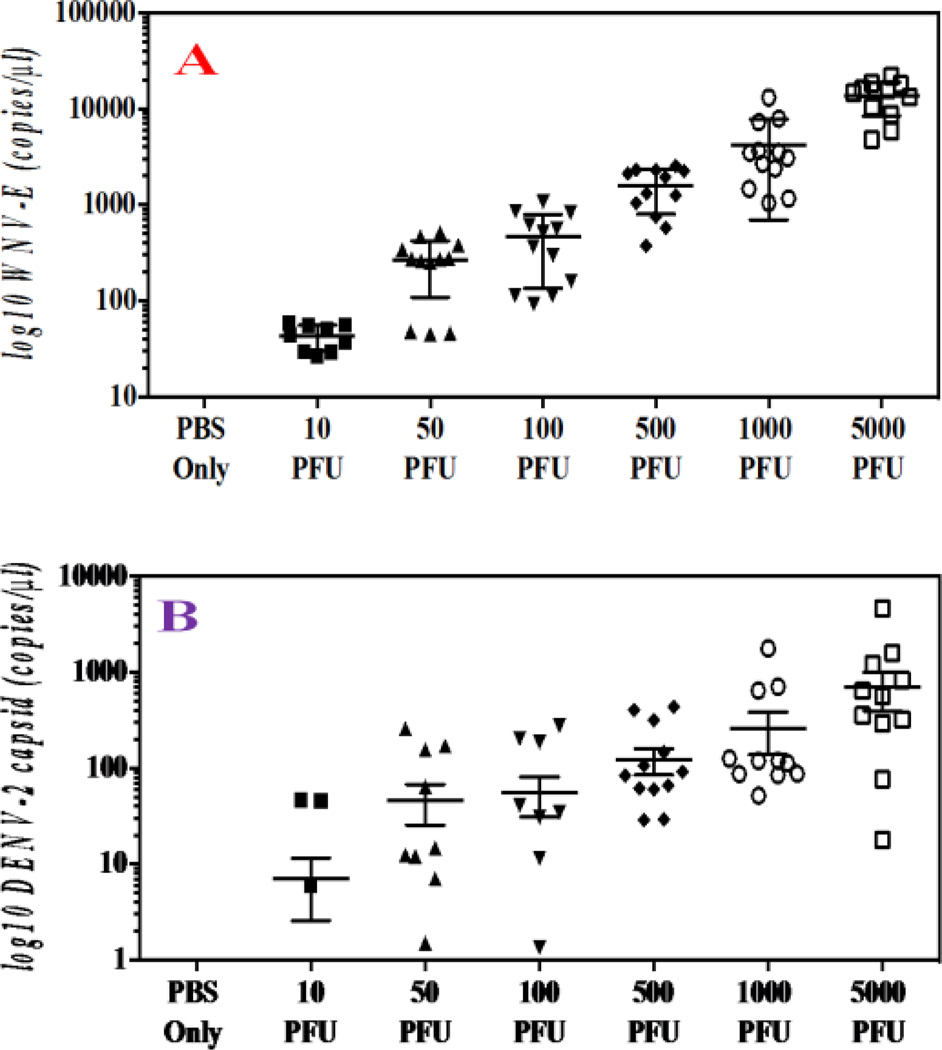
To compare the sensitivity of the SERS-based assay with conventional nucleic-acid testing (NAT), one-step qPCR assays were31–32 performed on serially diluted WNV and DENV-2 (5 × 103 PFU/ml to 10 PFU/ml) in PBS. As shown in the Figure 4, the qPCR results showed that WNV envelope (E) gene (Figure 4A) and DENV-2 capsid gene (Figure 4B) copy numbers (RNA copy number / 100µl sample) increased as the samples contained more viruses, indicating that the qPCR-based assay is able to detect viruses as low as 10 PFU/ml. To compare the sensitivity of the SERS-based assay, serially diluted DENV-2 and WNV were probed with the AuNP-4G2-based SERS probes. Figure 3B showed an unique fingerprint for DENV-2 particles that can be detected even at 10 PFU/ml. All the above data suggest that the SERS-based assay is able to detect low concentrations of flaviviral particles that are comparable to traditional qPCR methods, illustrating the high sensitivity of the SERS-based assays for viral detection.
Figure 4.
(A) q-PCR assay for the detection of DENV-2 in PBS. (B) q-PCR assay for the detection of WNV in PBS.
Since for the real life applications reproducibility and stability of SERS signals are very important criteria, to find out the reproducibility, we have performed SERS spectra from WNV and DENVs with anti-flaviviral antibodies conjugated gold nanoparticlesmade at different batches. Figures 3C and 3D show that SERS reproducibility for DENV and WNV are very good. Our reported data indicate that the Raman frequency shifts are negligible for samples made at different batches, which really confirmed the relative stability of SERS spectral positions. Reported data as shown in Figures 3C and 3D show that the Raman signal intensities change around 5–10% for different batches and it is due to the fact that the spatial distribution of “hot spots” cannot be controlled for different batches of sample.
Conclusion
In this article, we have reported a bio-conjugated gold nanoparticle (AuNP)-based SERS probe for the accurate detection of mosquito-borne flaviviruses. Our experimental data shows that 4G2 antibodies conjugated to the AuNP based SERS probe can be used as a sensitive fingerprinting detection tool for DENV-2 and WNV. Reported data shows that due to the interaction between 4G2 antibody and mosquito-borne flaviviruses, AuNP-4G2 complexes form an assembly structure in the presence of DENV-2 or WNV. FDTD stimulation data indicate that the electric field enhancement |E|2 can be more than 102 times for gold nano-assembly that contains three particles with respect to a monomer gold nanoparticle. Simulation data also suggest that SERS enhancement factor can be greater than 104, due to the assembly structure. As a result, our report has shown that the AuNP-4G2-based SERS probe is an ultrasensitive tool to detect DENV-2 or WNV particles as low as 10 PFU/ml, which is comparable with traditional q-PCR assays. In addition, our experimental results with CHIKV virus clearly indicate that the AuNP-4G2-based SERS assay is highly selective for WNV and DENV. We have shown very good reproducibility for our SERS based DENV and WNV detection techniques. The reported mosquito-borne flaviviruses sensing assay is very fast, it takes less than 30 minutes from virus incubation with antibody conjugated nanoparticles to SERS sensing. After proper engineering, we believe that the reported SERS probe can be further developed into an automated screening assay that is suitable to use in routine blood screening facilities and research-based settings.
Acknowledgments
The authors thank Dr. John F. Anderson at the Connecticut Agricultural Experiment Station for providing WNV and CHIKV. Dr. Ray and Dr. Bai would like to thank the National Institutes of Health for their generous funding (NIH-R15 grant # 1R15A113706-01).
References
- 1. [date of access 01/14/2015]; http://www.who.int/whr/1996/media_centre/executive_summary1/en/index9.html. [Google Scholar]
- 2. [date of access 01/14/2015]; http://www.oxitec.com/health/mosquito-borne-diseases/ [Google Scholar]
- 3.Guzman MG, Halstead SB, Artsob H, Buchy P, Farrar J, Gubler DJ, Hunsperger E, Kroeger A, Margolis HS, Martínez E, Nathan MB, Pelegrino JL, Simmons C, Yoksan S, Peeling RW. Dengue: a continuing global threat. Nat Rev Microbiol. 2010;8:S7–S16. doi: 10.1038/nrmicro2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suthar MS, Diamond MS, Gale M., Jr West Nile virus infection and immunity. Nat. Rev. Microbiol. 2013;11:115–124. doi: 10.1038/nrmicro2950. [DOI] [PubMed] [Google Scholar]
- 5.Shanmukh S, Jones L, Driskell J, Zhao Y, Dluhy R, Tripp RA. Rapid and sensitive detection of respiratory virus molecular signatures using a silver nanorod array SERS substrate. Nano Letters. 2006;6:2630–2636. doi: 10.1021/nl061666f. [DOI] [PubMed] [Google Scholar]
- 6.Saha K, Agasti SS, Kim C, Li X, Rotello VM. Gold Nanoparticles in Chemical and Biological Sensing. Chem. Rev. 2012;112:2739–2779. doi: 10.1021/cr2001178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wanunu M, Dadosh T, Ray V, Jin J, McReynolds L, Drndic M. Rapid Electronic Detection of Probe-Specific MicroRNAs Using Thin Nanopore Sensors. Nat. Nanotechnol. 2010;5:807–814. doi: 10.1038/nnano.2010.202. [DOI] [PubMed] [Google Scholar]
- 8.Qian X, Peng X-H, Ansari DO, Yin-Goen Q, Chen GZ, Shin DM, Yang L, Young AN, Wang MD, Nie S. In Vivo Tumor Targeting and Spectroscopic Detection with Surface-Enhanced Raman Nanoparticle Tags. Nat. Biotechnol. 2008;26:83–90. doi: 10.1038/nbt1377. [DOI] [PubMed] [Google Scholar]
- 9.Fan Z, Yust B, Nellore BOV, Sinha SS, Kanchanapally R, Crouch RA, Pramanik A, Reddy SC, Sardar D, Ray PC. Accurate Identification and Selective Removal of Rotavirus Using a Plasmonic–Magnetic 3D Graphene Oxide Architecture. J. Phys. Chem. Lett. 2014;5:3216–3221. doi: 10.1021/jz501402b. [DOI] [PubMed] [Google Scholar]
- 10.Kleinman SL, Ringe E, Valley N, Wustholz KL, Phillips E, Scheidt KA, Schatz GC, Van Duyne RP. Single-Molecule Surface-Enhanced Raman Spectroscopy of Crystal Violet Isotopologues: Theory and Experiment. J. Am. Chem. Soc. 2011;133:4115–4122. doi: 10.1021/ja110964d. [DOI] [PubMed] [Google Scholar]
- 11.Kim NH, Lee SJ, Moskovits M. Aptamer-Mediated Surface-Enhanced Raman Spectroscopy Intensity Amplification. Nano Lett. 2010;10:4181–4185. doi: 10.1021/nl102495j. [DOI] [PubMed] [Google Scholar]
- 12.Laurence TA, Braun G, Talley C, Schwartzberg A, Moskovits M, Reich N, Huser T. Rapid, Solution-Based Characterization of Optimized SERS Nanoparticle Substrates. J. Am. Chem. Soc. 2008;131:162–169. doi: 10.1021/ja806236k. [DOI] [PubMed] [Google Scholar]
- 13.Barhoumi A, Halas NJ. Label-Free Detection of DNA Hybridization Using Surface Enhanced Raman Spectroscopy. J. Am. Chem. Soc. 2010;132:12792–12793. doi: 10.1021/ja105678z. [DOI] [PubMed] [Google Scholar]
- 14.Demirel MC, Kao P, Malvadkar N, Wang H, Gong X, Poss M, Allara DL. Bio-organism sensing via surface enhanced Raman spectroscopy on controlled metal/polymer nanostructured substrates. Biointerphases. 2009;4:35–41. doi: 10.1116/1.3147962. [DOI] [PubMed] [Google Scholar]
- 15.Brown LV, Zhao K, King N, Sobhani H, Nordlander P, Halas NJ. Surface-Enhanced Infrared Absorption Using Individual Cross Antennas Tailored to Chemical Moieties. J. Am. Chem. Soc. 2013;135:3688–3695. doi: 10.1021/ja312694g. [DOI] [PubMed] [Google Scholar]
- 16.Lim DK, Jeon KS, Hwang JH, Kim H, Kwon S, Suh YD, Nam JM. Highly Uniform and Reproducible Surface-Enhanced Raman Scattering from DNA-Tailorable Nanoparticles with 1-nm Interior Gap. Nat. Nanotechnol. 2011;6:452–460. doi: 10.1038/nnano.2011.79. [DOI] [PubMed] [Google Scholar]
- 17.Sivapalan ST, DeVetter BM, Yang TK, van Dijk T, Schulmerich MV, Carney PS, Bhargava R, Murphy CJ. Off-Resonance Surface-Enhanced Raman Spectroscopy from Gold Nanorod Suspensions as a Function of Aspect Ratio: Not What We Thought. ACS Nano. 2013;7:2099–2210. doi: 10.1021/nn305710k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dassary SR, Sing AK, Senapati D, Yu H, Ray PC. Gold Nanoparticle Based Label-Free SERS Probe for Ultrasensitive and Selective Detection of Trinitrotoluene. J. Am. Chem. Soc. 2009;131:13806–13812. doi: 10.1021/ja905134d. [DOI] [PubMed] [Google Scholar]
- 19.Lu W, Singh AK, Khan SA, Senapati D, Yu H, Ray PC. Gold Nano-Popcorn-Based Targeted Diagnosis, Nanotherapy Treatment, and In Situ Monitoring of Photothermal Destruction Response of Prostate Cancer Cells Using Surface-Enhanced Raman Spectroscopy. J. Am. Chem. Soc. 2010;132:18103–18114. doi: 10.1021/ja104924b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singh AK, Khan SA, Fan Z, Demeritte T, Senapati D, Kanchanapally R, Ray PC. Development of a Long-Range Surface-Enhanced Raman Spectroscopy Ruler. J. Am. Chem. Soc. 2012;134:8662–8669. doi: 10.1021/ja301921k. [DOI] [PubMed] [Google Scholar]
- 21.Malvadkar NA, Demirel G, Poss M, Javed A, Dressick WJ, Demirel MC. Fabrication and use of electroless plated polymer surface-enhanced Raman spectroscopy substrates for viral gene detection. J. Phys. Chem. C. 2010;114:10730–10738. [Google Scholar]
- 22.Panikkanvalappil SR, Mackey MA, El-Sayed MA. Probing the Unique Dehydration-Induced Structural Modifications in Cancer Cell DNA using Surface Enhanced Raman Spectroscopy. J. Am. Chem. Soc. 2013;135:4815–4821. doi: 10.1021/ja400187b. [DOI] [PubMed] [Google Scholar]
- 23.Murphy S, Huang L, Kamat PV. Reduced Graphene Oxide–Silver Nanoparticle Composite as an Active SERS Material. J. Phys. Chem. C. 2013;117:4740–4747. [Google Scholar]
- 24.Angulo AM, Noguez C, Schatz GC. Electromagnetic Field Enhancement for Wedge-Shaped Metal Nanostructures. J. Phys. Chem. Lett. 2011;2:1978–1983. [Google Scholar]
- 25.Premasiri WR, Moir DT, Klempner MS, Krieger N, Jones G, Ziegler LD. Characterization of the surface enhanced Raman scattering (SERS) of bacteria. J. Phys. Chem. B. 2005;109:312–320. doi: 10.1021/jp040442n. [DOI] [PubMed] [Google Scholar]
- 26.Wang Y, Lee K, Irudayaraj J. Silver nano-sphere SERS probes for sensitive identification of pathogens. J. Phys. Chem. C. 2010;114:16122–16128. [Google Scholar]
- 27.Demeritte T, Fan Z, Sinha SS, Duan J, Pachter R, Ray PC. Gold Nanocage Assembly for Selective Second Harmonic Generation Imaging of Cancer Cell. Chem.—Eur. J. 2014;20:1017–1022. doi: 10.1002/chem.201303306. [DOI] [PubMed] [Google Scholar]
- 28.Zhao J, Pinchuk AO, McMahon JM, Li S, Ausman LK, Atkinson AL, Schatz GC. Methods for Describing the Electromagnetic Properties of Silver and Gold Nanoparticles. Acc. Chem. Res. 2008;41:1710–1720. doi: 10.1021/ar800028j. [DOI] [PubMed] [Google Scholar]
- 29.Wells SM, Merkulov IA, Kravchenko II, Lavrik NV, Sepaniak MJ. Silicon Nanopillars for Field-Enhanced Surface Spectroscopy. Acs Nano. 2012;6:2948–2959. doi: 10.1021/nn204110z. [DOI] [PubMed] [Google Scholar]
- 30.Sinha SS, Paul DK, Kanchanapally R, Pramanik A, Chavva SR, Nellore BPV, Jones SJ, Ray PC. Long-range Two-photon Scattering Spectroscopy Ruler for Screening Prostate Cancer Cells. Chem. Sci. 2015;4:2411–2418. doi: 10.1039/c4sc03843f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bai F, Wang T, Pal U, Bao F, Gould LH, Fikrig E. Use of RNA interference to prevent lethal murine west nile virus infection. J. Infec. Dis. 2005;191:1148–1154. doi: 10.1086/428507. [DOI] [PubMed] [Google Scholar]
- 32.Waggoner J, Abeynayake J, Sahoo MK, Gresh L, Tellez Y, Gonzalez K, Ballesteros G, Balmaseda A, Karunaratne K, Harris E, Pinsky BA. Development of an internally controlled, real-time RT-PCR for pan-dengue virus detection and comparison of four molecular dengue assays. J. Clin. Microbiol. 2013;51:2172–2181. doi: 10.1128/JCM.00548-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao J, Lui H, Mclean D, Zeng H. Automated autofluorescence background subtraction algorithm for biomedical Raman spectroscopy. Appl. spectrosc. 2007;61:1225–1232. doi: 10.1366/000370207782597003. [DOI] [PubMed] [Google Scholar]
- 34.Beier BD, Berger AJ. Method for automated background subtraction from Raman spectra containing known contaminants. Analyst. 2009;134:1198–1202. doi: 10.1039/b821856k. [DOI] [PubMed] [Google Scholar]