Abstract
Acute myeloid leukemia (AML) is characterized by the accumulation of immature blood cell precursors in the bone marrow. Pharmacologically overcoming the differentiation block in this condition is an attractive therapeutic avenue, which has only achieved success in a subtype of AML, acute promyelocytic leukemia (APL). Attempts to emulate this success in other AML subtypes have thus far been unsuccessful. Autophagy is a conserved protein degradation pathway with important roles in mammalian cell differentiation, particularly within the hematopoietic system. In this study we demonstrate the functional importance of autophagy in APL cell differentiation. We show that autophagy is increased during ATRA-induced granulocytic differentiation of the APL cell line NB4, and that this is associated with increased expression of LC3-II and GATE-16 proteins involved in autophagosome formation. Autophagy inhibition, using either drugs (chloroquine/3-methyladenine) or short-hairpin (sh)RNA targeting the essential autophagy gene ATG7, attenuates myeloid differentiation. Importantly, we show that enhancing autophagy promotes ATRA-induced granulocytic differentiation of an ATRA-resistant derivative of the non-APL AML HL60 cell line (HL60-Diff-R). These data support the development of strategies to stimulate autophagy as a novel approach to promote differentiation in AML.
Keywords: Differentiation, autophagy, myeloid leukemia, ATRA, promyelocytic leukemia
Graphical Abstract

Introduction
The term ‘acute myeloid leukemia’ (AML) describes a heterogeneous group of clonal disorders of hematopoietic progenitor cells, all of which are characterized by the accumulation of immature blood cell precursors in the bone marrow and peripheral circulation. In addition to environmentally leukemogenic factors, a complex interplay of cytogenetic, genetic and epigenetic abnormalities contributes to AML pathogenesis, conferring on malignant cells an increased proliferation rate, resistance to apoptosis and a differentiation block [1]. AML is the most common acute leukemia in adults, with an incidence that increases with age. The MRC AML trials report an overall survival of 47% for patients under 60 years and 20% for those above 60 years [2]. These figures highlight that despite advances in molecular understanding of the condition, there is a need for improved and tolerable therapeutic strategies. ‘Differentiation therapy’ exploiting pharmacologic override of the cellular differentiation block observed in AML, is one attractive strategy with a favorable toxicity profile. Success with this approach has been achieved with the use of all-trans-retinoic acid (ATRA) for the treatment of acute promyelocytic leukemia (APL) [3].
APL is a distinct form of AML, accounting for approximately 10% of cases. It is distinguished by a characteristic morphology, a potentially fatal coagulation defect at clinical presentation and chromosomal translocations involving the retinoid acid receptor alpha (RARα) gene on chromosome 17 [3]. At least nine driver translocations have been identified, with the most common being at t15;17, which fuses the promyelocytic leukemia (PML) gene with RARα [3]. The resultant PML-RARα fusion oncoprotein represses transcription and blocks granulocyte differentiation at the promyelocyte stage [3]. Therapeutic doses of ATRA, a physiologically active retinoid, restore normal differentiation through two distinct mechanisms: (i) de-repression of transcription [4] and (ii) degradation of the PML-RARα oncoprotein [5, 6]. Arsenic trioxide (ATO) is a second differentiating agent that has now been incorporated into the treatment of APL. When administered in combination with ATRA, ATO eliminates the need for cytotoxic chemotherapy in the treatment of low to intermediate-risk APL [7]. Interestingly, ATO also increases the degradation of the PML-RARα protein [8], and this proteolysis is now thought to be critical for eradicating APL leukemia-initiating cells (LICs) and achieving long-term cures [5, 9].
Autophagy is a degradative cellular process ubiquitously observed across the eukaryotic hierarchy. It allows the disposal of aged, redundant or damaged proteins, aggregates and organelles without steric limitation [10]. Cell contents targeted for autophagic degradation are sequestered within double-membraned autophagic vesicles known as ‘autophagosomes’, which are subsequently delivered to lysosomes and their contents degraded by resident proteases and hydrolases [11]. At a molecular level, autophagy is mediated by a family of AuTophaGy-related (ATG) proteins, which are sequentially recruited to the developing autophagosome membrane [12]. Membrane elongation is specifically dependent on two ubiquitination-like reactions: (i) the formation of an ATG5-ATG12 conjugate and (ii) the conjugation of phosphatidylethanolamine (PE) to the ATG8 homologues LC3, GATE-16 and GABARAP. These lipidated proteins are then incorporated into and stabilize autophagic vesicles at different stages of maturation [11, 13, 14]. Both conjugation reactions are catalyzed by the E1 ligase ATG7 – thought to be an essential component of autophagy machinery [11].
As a pathway involved in protein turnover, autophagy plays an intrinsic role in mammalian development and differentiation [13, 15]. Within the hematopoietic system, autophagy is specifically involved in organelle clearance during reticulocyte differentiation [15], lymphocyte differentiation [15] and plasma cell differentiation [16]. In addition, conditional hematopoietic cell deletion of Focal adhesion kinase (FAK) family interacting protein FIP200 – a key protein involved in autophagy activation, results in severe anemia [17], while ATG7 deletion leads to a fatal myeloproliferation resembling AML [18] - suggesting that autophagy may also be integral to normal myeloid differentiation. Through poorly characterized mechanisms, both ATRA and ATO have been shown to induce autophagy in APL cells [19–22]. This induction is thought to contribute to proteolysis of the PML-RARα oncoprotein [20, 22], similar to the mechanism by which imatinib-induced autophagy degrades the BCR-ABL oncoprotein in chronic myeloid leukemia (CML) [23].
We investigated the role of autophagy in ATRA-mediated granulocytic differentiation of APL cells. We confirm that autophagy is upregulated upon ATRA treatment and demonstrate induction of ATG8 homologues LC3 and GATE-16. Furthermore, we show that inhibition of autophagy with either drugs or targeted ATG7 knockdown attenuates APL cell differentiation. In addition, we show that pharmacologic autophagy induction facilitates the ATRA-mediated differentiation of non-APL, ATRA-resistant HL60-Diff-R AML cells, suggesting that targeting autophagy may represent a means of re-establishing differentiation in this aggressive cancer.
Materials and Methods
Cell lines & culture
NB4 cells were a gift from Prof. Estelle Duprez (INSERM, France). A differentiation-resistant variant of HL60 cells, HL60-Diff-R (HL60 genotype confirmed by LGC standards), were a gift from Prof. Tom Cotter (UCC, Ireland). Cells were maintained at 37°C, 5% CO2 in RPMI 1640 (Sigma, Ireland R8758), 10% heat-inactivated fetal calf serum (Sigma F7524) and 1% Penicillin/Streptomycin (Gibco Ireland 15070-063). Cells were seeded at 2 × 105 cells/ml prior to treatment.
Primary APL cells
Following local ethics committee approval and informed consent, peripheral blood samples were obtained from two patients newly-presenting with APL. Whole samples were gradient-separated using Histopaque-1077 (Sigma 10771) and mononuclear cells were extracted comprising a predominantly malignant population.
Drug treatments
ATRA (Sigma R2625) was diluted from a 1 mM stock in 100% ethanol. Lysosomal function was inhibited with E64d (Sigma E8640) and pepstatin A methyl ester (Calbiochem 516485). Autophagy was inhibited with chloroquine (Sigma C6628) and 3-methyladenine (3-MA) (Sigma M9281). Autophagy was induced with lithium chloride (Sigma L9650) and rapamycin (Sigma R8781).
ATG7 knockdown
pLKO.1 plasmid vectors expressing small hairpin (sh)RNAs targeting human ATG7 were purchased from Sigma (TRCN0000007584=shATG7_1 and TRCN0000007587=shATG7_2). Vectors were transfected into HEK293T cells along with lentiviral packaging plasmids pCMVΔ8.9 and pVSVG using Lipofectamine 2000 (Invitrogen 11668). An off-target scrambled vector was transfected as a control (shSCR). Cells were replenished with fresh media following overnight recovery and allowed to produce lentivirus for 48 hours. Lentivirus was harvested by filtering the HEK293T culture supernatant through 0.45 μm filters and supplementing with polybrene. NB4 cells were transduced with viral supernatant in a 1:1 ratio with 2× growth medium. Transduced cells were selected with puromycin (2 μg/mL) for 10 days prior to experiments. ATG7 knockdown was confirmed by western blot.
Morphologic examination
Cells were cytospun onto glass slides and stained with Rapi-Diff (Braidwood Laboratories 22007, 22008, 22009) according to product guidelines. Morphology was examined using an Olympus DP70 digital microscope (Mason Technology, Dublin, Ireland).
Western blotting
Cellular protein extracts were lysed in modified RIPA buffer (50 mM TrisHCl - pH 7.4, 150 mM NaCl, 0.25% sodium deoxycholate, 1% Igepal, 1 mM EDTA, 1× Pefabloc, 1× Protease inhibitor cocktail, 1 mM Na3VO4, 1 mM NaF). Protein samples were separated on NuPAGE 4–12%, Bis-Tris gels (Invitrogen NP0322) and electrophoretically transferred onto PVDF membranes (Invitrogen IB401001). Primary antibodies were: anti-CD11b (Abcam ab52478), anti-LC3A/B (MBL PD014), anti-LC3B (Novus Biologicals 600–1384), anti-GABARAP (Abgent AP1821a), anti-GABARAPL2/GATE-16 (Abgent AP1822d) and anti-ATG7 (Cell Signaling 2631). Proteins were visualized using relevant IR-DYE secondary antibodies and quantified on the Odyssey IR imaging system (Li-Cor, Cambridge, UK).
Flow cytometry assays
Autophagy
Live cells were stained with the Cyto-ID Autophagy Detection Kit (Enzo Life Sciences Exeter, UKENZ-51031) according to product protocol.
Differentiation
CD11b - live cells were incubated for 30 min with PE-conjugated anti-CD11b antibody (eBioscience 12-0118 or Immunotools #21279114) in 1% albumin/phosphate buffered saline (PBS), and washed with PBS prior to analysis.
CellROX Oxidative Stress Assay
To assess reactive oxygen species (ROS) production, cells were stained with CellRox deep red oxidative stress reagent (5 μM) (Molecular probes C10422) at 37 °C for 30 min following stimulation with phorbol 12-myristate 13-acetate (PMA) (50 ng/ml) (Sigma P1585) in complete medium. This cell permeable reagent is non-fluorescent while in a reduced state and upon oxidation, emits fluorescence at 665 nm. Fluorescence was detected using a BD-LSRII flow cytometer (BD Biosciences, Oxford, UK). Data analysis and histogram overlays were performed on FlowJo software.
Colony growth assays
Cells (5.0 × 104 cells/ml) were seeded into methylcellulose media (Stem Cell Technologies - MethoCult H4034). At day 10, cells were resuspended in warm RPMI and counted on a Chemometec nucleocounter.
Quantitative PCR analysis
RNA extraction was performed using the miRCURY RNA Isolation Kit from Exiqon (Vedbaek, Denmark) according to the manufactures description. RNA was reverse-transcribed using the masterscript kit purchased from 5prime (Hilden, Germany). Quantitative PCR analysis of CEBPε and GSF3R mRNA was performed using TaqMan® reagents and the StepOnePlus qPCR system (Applied Biosystems, Zug, Switzerland). Raw Ct values were normalized to HMBS and to the untreated control of day 3 (ΔΔCt method). Gene Expression Assays for CEBPE and CSF3R were HHs00357657_m1 and Hs00167918_m1, respectively (Applied Biosystems, Rotkreuz, Switzerland). HMBS primer and probes have been described previously [24].
Results
Autophagy is upregulated during ATRA-induced APL cell differentiation
We investigated autophagic activity during the myeloid differentiation of APL cells. Human NB4 cells (PML-RARα +ve APL) were induced to differentiate along a granulocytic lineage with therapeutic doses of ATRA (1 μM), administered for 4 days. Morphologically, ATRA treated cells have increased nuclear lobulation and a decreased nuclear to cytoplasmic (N:C) ratio characteristic of maturing granulocytes (Figure S1A). Differentiation was confirmed by the detection of the granulocyte surface marker CD11b by western blotting (Figure S1B) and by flow cytometry (Figure S1C).
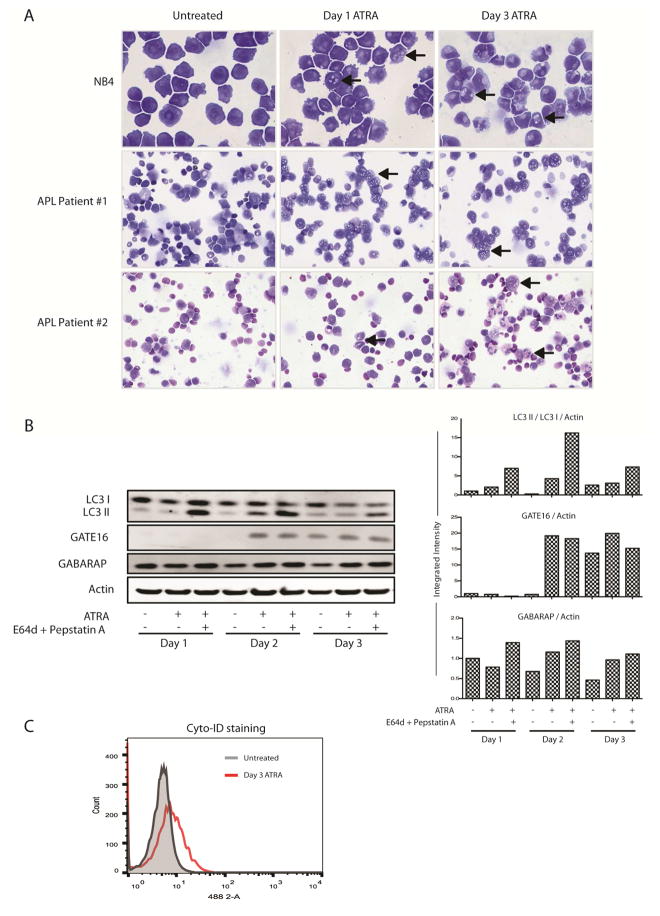
Autophagy is characterized by the accumulation of double-membraned vesicles within the cell cytoplasm. Subsequent fusions events with endosomes and lysosomes can often result in a vesicular morphology, detectable with light microscopy. Coincident with differentiation, we detected a progressive increase in cytoplasmic vesiculation in ATRA treated NB4 cells (Figure 1A, upper panels, arrows). Serial blood samples were collected from two newly diagnosed APL patients receiving in vivo ATRA therapy. Peripheral blood mononuclear cells (PBMCs) obtained from these patients also showed a progressive increase in cytoplasmic vesiculation visible from day 1 of ATRA treatment, suggestive of modulation of trafficking systems or autophagic activity (Figure 1A, middle and lower panels, arrows).
Figure 1. Autophagy is increased during the ATRA-mediated differentiation of APL cells.
NB4 cells were treated with 1 μM ATRA for the times indicated in the presence or absence of lysosomal inhibitors: E64d (5 μM) and pepstatin A (1 μM). These experiments were performed a minimum of three times with very similar data. Peripheral blood mononuclear cells (PBMCs) were isolated from serial blood samples drawn from two APL patients undergoing in vivo therapy. Both patients received ATRA at day 0 and idarubicin at day 2. Peripheral white cell counts (WCCs) at diagnosis were 3.2 × 109/L for patient #1 and 1.1 × 109/L for patient #2. In both patients the leukemic clone carried a confirmed t(15;17) translocation and displayed a characteristic APL immunophenotype (HLADR−, CD13+, CD33+, CD117+, CD11c−). (A) Cytoplasmic vesiculation as a morphologic feature of autophagy during granulocytic differentiation was examined by light microscopy in ATRA treated NB4 cells and primary APL patient PBMCs (arrows)(40× magnification). (B) Autophagy and autophagic flux were measured in NB4 cells by evaluating protein levels of LC3-I/II (MBL), GATE-16 (Abgent) and GABARAP (Abgent) in whole cell protein lysates. All bands were quantified using the Odyssey Infrared Imaging System (Li-COR), normalized to β-actin and presented as integrated intensities in the right hand panels. A single representative actin blot is shown. (C) Live NB4 cells were stained using the Cyto-ID autophagy detection kit and levels of autophagy were assessed by flow cytometry. A representative image is shown.
In order to distinguish autophagy, we examined levels of a known marker LC3B. LC3B-I becomes conjugated to phosphatidylethanolamine (PE) to form LC3B-II, which is incorporated into autophagosome membranes. Other members of the ATG8 family of proteins, GABARAP and GATE-16/GABARAPL2, can also be incorporated into autophagosomes [14]. As differentiation proceeded, a decrease in LC3B-I protein levels was observed at day 1 by western blot. An increased level of LC3B-II was observed by day 2 and this was further enhanced in the presence of the lysosomal protease inhibitors E64d and pepstatin, both of which prevent autophagosomal turnover (Figure 1B). We detected increased GABARAP and GATE-16 levels, from day 2, with GATE-16 proving to be the most specific protein marker (Figure 1B), consistent with previously published data [25].
We then quantified autophagic activity by flow cytometry using the Cyto-ID assay, which fluorescently tags autophagosomes [26]. A 1.8-fold increase in mean fluorescence intensity was detected in ATRA treated NB4 cells at day 3 (Figure 1C). Together, these data suggest that autophagic activity is increased during the ATRA-mediated granulocytic differentiation of APL cells.
Pharmacologic inhibition of autophagy attenuates the ATRA-induced differentiation of APL cells
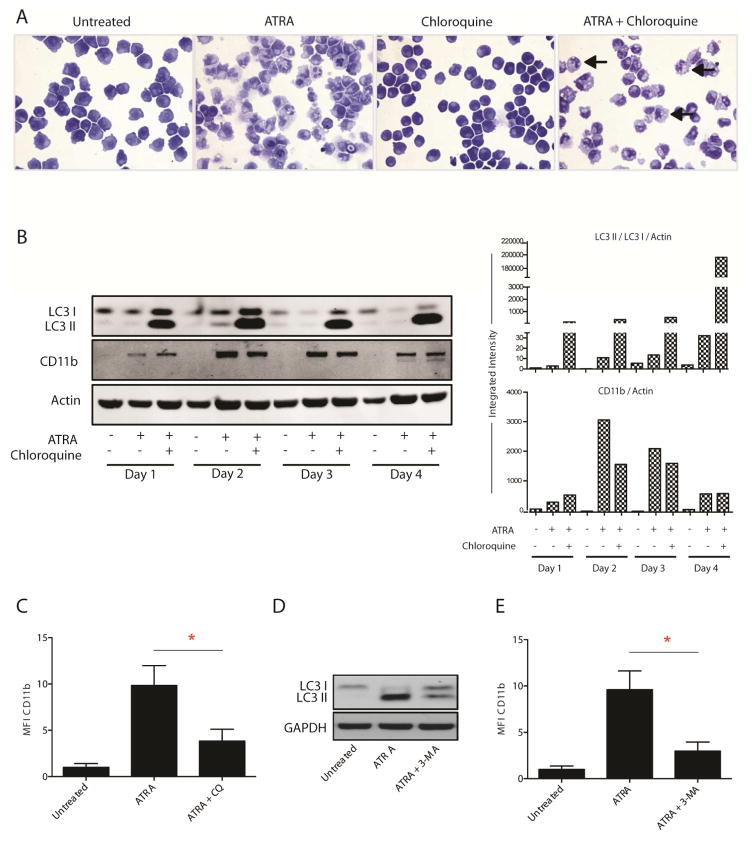
We then examined whether pharmacological inhibition of autophagy would interfere with in vitro leukemic cell differentiation. The anti-malarial compound chloroquine raises lysosomal pH, inhibiting the activity of resident enzymes and thus impeding completion of autophagy [27]. NB4 cells were incubated for four days with chloroquine in the presence or absence of ATRA. While chloroquine alone had little effect on NB4 cells, cells co-treated with ATRA and chloroquine displayed increased cytoplasmic vesiculation (Figure 2A, arrows), along with increased levels of LC3B-II protein (Figure 2B), consistent with impaired autophagosome degradation. Morphologically granulocytic differentiation appeared stunted in co-treated cells (Figure 2A) and protein levels of CD11b were decreased from day 2 compared to cells treated with ATRA alone, consistent with attenuated differentiation (Figures 2B & 2C).
Figure 2. Effects of pharmacologic autophagy inhibition on ATRA-induced APL cell differentiation.
NB4 cells were treated for four days with 1 μM ATRA alone or in combination with an autophagy inhibitor: either chloroquine (10 μM) or 3-methyladenine (3-MA) (5 mM). (A) Differentiation morphology and cytoplasmic vesiculation were examined by light microscopy at day 4 in cells treated with ATRA and chloroquine (40× magnification). ATRA treated cells display increased nuclear lobulation and a decreased nuclear:cytoplasmic (N:C) ratio consistent with differentiation, while these changes are reduced in cells co-treated with ATRA and chloroquine. Increased vesiculation observed in co-treated cells represents an accumulation of autophagosomes due to increased production with a block in terminal autophagosome turnover (arrows). (B) Inhibition of terminal ATRA-induced autophagy by chloroquine was measured by quantifying LC3-I/II protein levels (MBL). Differentiation was assayed by measuring total CD11b protein expression on western blot (Abcam). All bands were quantified using the Odyssey Infrared Imaging System (Li-COR), normalized to β-actin and presented as integrated intensities, in the right hand panel. Data are representative of three independent experiments. (C) Median fluorescence intensity (MFI) of surface CD11b expression at day 4 was used as a marker of differentiation (Immunotools). Data are presented as mean fold induction of CD11b compared to untreated cells for three independent experiments ± S.E.M (*P < 0.05). (D) Western blot analysis of LC3 at day 4 in ATRA alone and ATRA + 3-MA treated cells (Novus Biologicals), with GAPDH used as a loading control. (E) Surface CD11b expression at day 4 was measured by flow cytometry (Immunotools) and analyzed as in (C) (*P < 0.05).
We observed a similar effect on differentiation when NB4 cells were treated with ATRA in the presence of 3-methyladenine (3-MA), a well-established inhibitor of early autophagy. 3-MA inhibits class-III phosphatidylinositol 3-kinase (PI3K) – a positive regulator of autophagy initiation [28]. Treatment of NB4 cells with ATRA alone for 4 days induced LC3B II accumulation. However, the addition of 3-MA to block initiation, attenuated this effect (Figure 2D). Co-treatment resulted in significantly lower CD11b expression than treatment with ATRA alone (Figure 2E). These findings indicate that inhibition of autophagy with pharmacologic agents attenuates the ATRA-mediated differentiation of APL cells and suggest that the process of autophagy may be an important mediator of granulocytic differentiation.
ATG7 is required for effective ATRA-mediated APL cell differentiation
We then assessed whether a known autophagy regulator, ATG7, was important for granulocytic differentiation of APL cells. ATG7 was depleted by lentiviral-mediated shRNA knockdown in NB4 cells. Knockdown in two distinct clones (shATG7_1 & shATG7_2) was evaluated at the protein level under basal conditions and following pharmacologic autophagy induction to ensure that the protein could be effectively depleted when autophagy is induced. Rapamycin directly inhibits mTOR, a master negative regulator of autophagy [29]. Lithium chloride (LiCl) lowers cytosolic inositol 1,4,5-triphosphate (IP3) levels and is a potent pharmacological inducer of autophagy [30]. Following treatment for 24 hours with either lithium or rapamycin, the ATG7-knockdown NB4 cells displayed reduced ATG7 expression (Figure 3A), decreased cytoplasmic vesiculation (Figure S2A), and no evidence of autophagy activation as assessed by flow cytometric Cyto-ID assay (Figure S2B). Thus, genetic inhibition of ATG7 successfully reduced the ability of NB4 cells to induce autophagy.
Figure 3. Effects of genetic ATG7 knockdown on ATRA-induced APL cell differentiation.
ATG7 was knocked down in NB4 cells using lentiviral transduction of target-specific shRNA. (A) Two independent ATG7 knockdown cell lines (shATG7_1 and shATG7_2), wild type NB4 cells (WT), and NB4 cells transduced with an off-target scrambled shRNA (shSCR) were treated for 24 hours with either 20 mM lithium chloride (LiCl) or 400 nM rapamycin (Rap) to induce autophagy. Successful knockdown was confirmed by measurement of ATG7 protein levels by western blot in untreated and treated cells (Cell Signalling), with quantification against β-actin (n=2). (B) Two knockdown NB4 clones and scrambled controls were seeded at 2 × 105 cells/mL and treated with ATRA (1 μM) for 3 days. Differentiation was assessed at day 3 by measurement of surface CD11b expression (eBioscience) (n=2). Grey filled histograms are untreated SCR controls, blue histograms are SCR treated with ATRA and red histograms show reduced differentiation in the ATG7 knockdown clones (C) Morphologic analysis by light microscopy at day 3 in ATG7 knockdown cell lines as assessed by nuclear shape and N:C ratio (40× magnification).
Knockdown of ATG7 significantly impaired the granulocytic differentiation of NB4 cells following three days of ATRA treatment (Figure 3B, red overlays), with reduced CD11b expression when compared to ATRA treated scrambled controls (Figure 3B, blue overlays) and reduced morphological evidence of maturation (Figure 3C). These data suggest that competent autophagy is required for successful ATRA-induced differentiation of APL cells.
Lithium chloride promotes ATRA-induced autophagy in non-APL, differentiation-resistant myeloid leukemic cells
We then evaluated if autophagy induction could be achieved pharmacologically in AML cells that lack the PML-RARα fusion oncogene. HL60-Diff-R cells, an ATRA-resistant variant of the promyeloblastic leukemic cell line HL60, were treated with lithium or rapamycin. Lithium treatment caused a significant increase in GATE-16 protein expression, over four days of treatment, consistent with increased autophagy. In contrast, rapamycin caused a more moderate elevation in GATE16 (Figure 4A).
Figure 4. Effects of lithium or rapamycin alone and combined with ATRA on autophagy in ATRA-resistant non-APL AML cells.
(A) HL60-Diff-R cells were treated with either lithium chloride (LiCl) (10 mM) or rapamycin (Rap) (400 nM). The induction of autophagy by lithium and rapamycin in HL60-Diff-R cells was measured by western blotting for GATE-16 protein levels at indicated times (Abgent). All bands were quantified using the Odyssey Infrared Imaging System (Li-COR), normalized to β-actin and presented as integrated intensities above western blots. This experiment was repeated three times with similar data. (B) HL60-Diff-R cells, treated for three and four days, were stained with Cyto-ID autophagy detection kit and levels of autophagy were quantified by flow cytometry. Single representative overlay histograms of cells treated with lithium (i) and (iii) or rapamycin (ii) and (iv) are shown. Histogram data for both lithium and rapamycin treated cells +/− ATRA (1 μM) are presented as mean fluorescence intensity ± S.E.M to the right. (n=3)(*** P < 0.0001, ** p < 0.01). (C) Induction of autophagy was further confirmed by western blotting for GATE16 (i), GABARAP (ii) and LC3 I/II (iii) levels. All bands were quantified using the Odyssey Infrared Imaging System (Li-COR), normalized to β-actin and presented as integrated intensities, relative to their respective controls.
Analysis of autophagy at day 3, using the Cyto-ID assay in HL60-Diff-R cells showed moderate staining with lithium alone (Figure 4B (i), blue overlay). Notably, a striking increase in Cyto-ID staining was seen in cells treated with the combination of ATRA and lithium, indicative of increased levels of autophagosomes (Figure 4B (i), green overlay). In contrast, a more modest increase in Cyto-ID staining was achieved with rapamycin in combination with ATRA (Figure 4B (ii), green overlay). Similar data were collected on day 4 [Figure 4B (iii) & (iv)], with only the lithium & ATRA treatment showing a significant increase in Cyto-ID staining. Analysis of mean fluorescent intensity from three independent experiments is presented to the right of Figure 4B. The effect of lithium and ATRA treatment on additional autophagy markers GATE16, GABARAP and LC3II was also evaluated at days 3 and 4 of differentiation [Figure 4C (i) - (iii)]. The combination of lithium and ATRA shows strong elevation of GATE16. GABARAP is marginally increased on day 3, but not on day 4. LC3II levels are higher in all combination treatments compared to single treatments. Collectively these data suggest augmentation of autophagy in the lithium & ATRA treated cells.
Lithium chloride promotes ATRA-mediated differentiation of non-APL, differentiation-resistant myeloid leukemic cells
We then evaluated if the advancement in autophagy seen with lithium could promote ATRA-mediated differentiation in the non-APL ATRA resistant AML cells. HL60-Diff-R cells failed to successfully differentiate when treated with a therapeutic concentration of ATRA alone for three days (Figure 5A, red overlay). However, co-treatment of these cells with ATRA and lithium resulted in a significant increase in CD11b protein expression at day 3 (Figure 5A, green overlay), with quantitation of triplicate data shown to the right (p = 0.006).
Figure 5. Effects of lithium combined with ATRA on differentiation of ATRA-resistant non-APL AML cells.
(A) HL60-Diff-R cells were treated with ATRA (1 μM) in the presence or absence of lithium chloride (LiCl) (10 mM) for three days. The induction of differentiation was assessed by measuring surface CD11b (eBioscience) by flow cytometry. A single representative overlay histogram is shown, with mean fluorescence intensity ± S.E.M presented to the right (n=4). (B) & (C) GCSF3R and CEBPε qPCR analysis of HL60-Diff-R cells treated as indicated for three days. Raw Ct values were normalized to HMBS and data are shown as n-fold induction compared to the untreated control cells. (D) Reactive oxygen species (ROS) production was assessed with CellROX oxidative stress assay in cells treated for three days with ATRA (1 μM) without and with lithium (10 mM). The percentage of ROS positive cells is presented as mean ± S.E.M to the right. (E) Morphologic features of granulocytic differentiation were assessed at day 4 by light microscopy (40× magnification). Arrows indicate cells with features of early granulocytic differentiation. (F) Cells were removed from their indicated drug treatments at day 2 and cultured in triplicate at a seeding density of 5 × 104 cells/mL in drug-free methylcellulose medium. A cell count was performed at day 10 to assess post-treatment cell proliferation (n=3). (*** p < 0.0005, ** p < 0.005).
Expression of additional differentiation markers, GCSFR and CEBPε was also analyzed by RT-qPCR. Both markers were significantly elevated at day 3 (p = 0.0018 and 0.0004 respectively) in the combined ATRA and lithium treated cells (Figure 5B and C).
Differentiation was also assessed by quantitating the production of reactive oxygen species (ROS) in ATRA or lithium treated cells following stimulation with PMA. This would be indicative of functional maturation in differentiating cells. Figure 5D shows a significant elevation of ROS production in 31 % of cells co-treated with lithium and ATRA for three days, compared to 2.4 and 23.9 % of cells treated with ATRA or lithium alone. Collective mean data is shown to the right.
Analysis of morphology indicated that HL60-Diff-R cells failed to successfully differentiate when treated with therapeutic concentrations of ATRA alone for four days (Figure 5E, lower left panel). In contrast, in the presence of lithium, morphological changes were consistent with early granulocytic differentiation (Figure 5E, lower right panel, arrows indicate lobed/crested nuclei).
The proliferative capacity of HL60-Diff-R cells following drug treatment was evaluated by removing cells from treated media at day 2 and culturing equal cell numbers in methylcellulose media for 10 days to assess cell numbers. At assay completion, decreased cell numbers were observed following co-treatment with ATRA and lithium, consistent with possible arrest or termination of malignant proliferative signaling (Figure 5F).
These data indicate that combination treatment with ATRA and lithium potentiates differentiation of non-APL AML cells and may therefore be of potential therapeutic benefit.
Discussion
In this study, we demonstrate that autophagy is upregulated during ATRA-mediated granulocytic differentiation of APL cells. This is associated with an increase in LC3B-II and GATE-16 levels, consistent with their involvement in autophagosome formation [14]. We show that inhibition of autophagy, either pharmacologically or through targeted genetic knockdown of ATG7, impedes ATRA-mediated differentiation. Thus, we propose that the process of autophagy is an important mediator of ATRA’s effects on APL cells, providing further support to recent data on leukemic cell differentiation [20–22, 31].
ATG7 is a key enzyme in autophagic pathways, acting in both of the conjugation steps involved in autophagosome formation – the conjugation of ATG5 and ATG12, and the conjugation of ATG8 family members to phosphatidylethanolamine (PE) [18]. Previous studies in other cell and in vivo models have shown successful inhibition of autophagy by knockdown/deletion of ATG7 [32]. Within the hematopoietic system, the importance of ATG7 has been demonstrated in normal hematopoietic stem cell renewal, quiescence and remodeling, while also being involved in the differentiation of erythrocytes, lymphocytes and mast cells [13]. The importance of other autophagy regulators such as ULK1, ATG5, PI3KC3, SQSTM1 [20], BECN1 [19], DRAM-1 [31] and GATE-16 [21] for APL cell differentiation has been evaluated; however, the effect of ATG7 knockdown has not previously been examined. Our study thus establishes a novel and important role for ATG7 in APL differentiation and by inference, response to differentiation therapy.
It is now broadly accepted that ATRA exerts dual effects on APL cells – overcoming the repressive transcriptional effects of the PML-RARα oncoprotein and increasing its degradation [6]. Using synthetic retinoids capable of activating transcription but not protein degradation, these effects have been uncoupled, showing that while differentiation can occur in the absence of proteolysis, PML-RARα elimination is necessary to achieve long-term disease control [5]. Operating without steric limitations, autophagy is the primary pathway available to cells for degrading large aggregated proteins. The chimeric PML-RARα protein (MW 110 kDa), is more prone to aggregation than either PML or RARα alone [22]. This leads to the formation of a large multimeric complex (669 kDa)[33], which requires functioning autophagy for its degradation [20, 22]. While our data would be consistent with autophagy promoting PML-RARα proteolysis, we also show that lithium chloride, an autophagy inducer, potentiates the ATRA-mediated granulocytic differentiation of AML cells that do not carry an abnormal RARα fusion protein. Thus, it is likely that autophagy plays a role in the extensive cellular remodeling that occurs in differentiating cells and is not solely involved in oncoprotein clearance.
There is now increasing data supporting a role for autophagy in cellular differentiation [15]. Within the hematopoietic system, murine studies have elucidated a critical role for autophagy in the removal of intracellular organelles from developing reticulocytes. Indeed, interference with several important autophagy genes (ULK1, ATG7, BNIPL3 or FIP200) results in severe red cell defects and anemia [17, 34]. Autophagy also participates in the differentiation of both B- and T-lymphocytes [35], plasma cells [16] and monocytes [36]. Despite the confounding presence of an abnormal fusion oncoprotein, the ATRA-mediated differentiation of APL cells in culture has long been used as a model for normal myelopoiesis [37]. Our study performed on a leukemic cell line model may have broader application, also suggesting a role for autophagy in normal myelopoiesis. It is possible that defects in autophagy may contribute to the differentiation block seen in malignancies such as AML. Conditional deletion of the ATG7 gene in the murine hematopoietic system results in an abnormal myeloid proliferation with dysplasia which is ultimately fatal – a condition very closely resembling AML [18]. In this study, using the promyeloblastic ATRA-resistant HL60-Diff-R cell line, which does not carry the PML-RARα oncoprotein characteristic of APL, we show that treatment with lithium chloride in combination with ATRA promoted granulocytic differentiation, suggesting that induction of autophagy may overcome the differentiation block in these cells.
Lithium has shown long-standing clinical efficacy in the treatment of mood disorders. Mechanistically, the lithium ion competitively inhibits magnesium binding sites on dependent enzymes [38]. Following observations that patients undergoing lithium therapy developed granulocytosis [39], in vitro studies in the early 1990s showed that combinations of lithium with ATRA induced the differentiation of human myeloid leukemic cell lines [40]. This effect was attributed to either increased RARα pools or to the inhibitory actions of lithium on glycogen synthase kinase 3β (GSK3β), a serine/threonine kinase with well-characterized roles in differentiation [38, 41–43]. Lithium-induced autophagy has been associated with neuroprotective effects in conditions that are attributable to the accumulation of abnormal mutant proteins e.g. huntingtin in Huntington’s disease and α-synuclein in Parkinson’s disease [38, 44]. Lithium’s effects on autophagy in neuronal cells have been reported to be mediated by inhibition of the enzyme inositol monophophatase (IMP) and depletion of free inositol [30]. Lithium has also been reported to augment signaling through the cAMP pathway (reviewed in [45]). Interestingly, a recent study found that augmentation of this pathway with 8-CPT-cAMP, upregulated p62/SQSTM1 and LC3 and promoted ATRA induced differentiation of the resistant cell line NB4-LR1 [46].
It is therefore important to acknowledge that while lithium is an autophagy inducer, the pro-differentiation effects of lithium observed in our experiments are likely to be multi-factorial, with input from several signaling pathways. However, given our data and those of others supporting an important role for autophagy in differentiation, we propose that autophagy induction by lithium, may contribute to its pro-differentiation effects, and this has not been previously recognized in the literature.
Autophagy is a known cellular response to stress - enabling survival in times of nutrient depletion or oxidative stress [47]. However, autophagy has also been linked to the induction of a form of programmed cell death distinct from classical apoptosis [48]. The role of autophagy in cancer is complex and appears to be context-dependent. While in normal cells autophagy has a tumor-suppressive function, cancer cells are often capable of using survival autophagy to sustain their metabolism in hypoxic or nutrient-depleted environments [49]. Indeed, cancer cells have been observed to upregulate autophagy in response to chemotherapeutic stress and studies of several cancer models – including chronic myeloid leukemia [50], have shown that autophagy inhibition increases chemoefficiency [48, 51]. As a result, several clinical trials, combining the autophagy inhibitor chloroquine or its sister drug hydroxychloroquine, with conventional chemotherapy are currently underway in both solid and hematologic malignancies [52]. Conversely, studies in hematologic malignancies have shown that several anti-tumor agents promote autophagic cell death, and that this is critical to their efficacy [48, 53]. Our study supports the concept that autophagy induction may be of clinical benefit through promoting terminal differentiation and subsequent cell death. Further studies with AML clinical samples and other autophagy inducers are warranted.
In summary, this study suggests that autophagy is a critical component of ATRA’s differentiating effects in APL and that pharmacologic autophagy induction in this context may potentiate the ATRA-mediated differentiation of non-APL AML cells. We expect that such a combination therapy would likely be minimally cytotoxic to normal cells and would be well tolerated by elderly patient populations, providing at least transient disease control in the absence of chemotherapy. The exact mechanisms through which ATRA activates autophagy remain to be elucidated and we expect that future studies will address this issue so that we can understand how best to target this process for patient benefit.
Conclusion
This study demonstrates a key role for autophagy in myeloid differentiation. Importantly, these data show that differentiation can be achieved in ATRA resistant AML cells, by co-treatment with lithium. Lithium induces autophagy and diminishes colony regrowth. As pharmacologically overcoming the differentiation block in AML would be a major therapeutic advance, the possibility of achieving this with autophagy inducers is an attractive area for future intervention.
Supplementary Material
Autophagy is increased during ATRA-induced differentiation of NB4 leukemia cells
Inhibition (shRNA) of essential autophagy gene ATG7 attenuates differentiation
Inhibition of autophagy using chloroquine or 3-MA attenuates myeloid differentiation
Lithium and ATRA promote autophagy in HL60-Differentiation resistant (Diff-R) cells
Lithium and ATRA induce differentiation in HL60-Diff-R cells & reduced proliferation
Acknowledgments
We are eternally indebted to our late director of CCRC, Professor Gerry O’Sullivan for his inspiration and support of this study. This work was funded by the Haematology Education and Research Trust (H.E.R.O) and Breakthrough Cancer Research. We acknowledge unrestricted educational support from Pfizer, Novartis, Bristol Myers Squibb, Amgen, and Merck Sharp & Dohme. We are also grateful to the Higher Education Authority of Ireland. Research from the Tschan group in University of Bern was supported by grants from the “Stiftung Klinisch-Experimentelle Tumorforschung” and The Swiss National Science Foundation (31003A_143739) to MPT. We also gratefully acknowledge expert technical assistance by D. Shan-Krauer. We thank the members of the Gudas Laboratory, Weill Cornell Medical College, NY for critical discussion. We gratefully acknowledge the support of the University of Nottingham, Weill Cornell, and NIHRO1CA43796 to LJG.
Footnotes
Disclosures
NO received unrestricted educational support from Pfizer, Novartis, Bristol Myers Squibb, Amgen, and Merck Sharp & Dohme. MRC received support for conference attendance from Roche. All other authors report no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cancer Genome Atlas Research N. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. The New England journal of medicine. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burnett AK. Treatment of acute myeloid leukemia: are we making progress? Hematology / the Education Program of the American Society of Hematology American Society of Hematology Education Program. 2012;2012:1–6. doi: 10.1182/asheducation-2012.1.1. [DOI] [PubMed] [Google Scholar]
- 3.Orfali N, McKenna SL, Cahill MR, Gudas LJ, Mongan NP. Retinoid receptor signaling and autophagy in acute promyelocytic leukemia. Experimental cell research. 2014 doi: 10.1016/j.yexcr.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tang XH, Gudas LJ. Retinoids, retinoic acid receptors, and cancer. Annual review of pathology. 2011;6:345–364. doi: 10.1146/annurev-pathol-011110-130303. [DOI] [PubMed] [Google Scholar]
- 5.Ablain J, Leiva M, Peres L, Fonsart J, Anthony E, de The H. Uncoupling RARA transcriptional activation and degradation clarifies the bases for APL response to therapies. The Journal of experimental medicine. 2013;210:647–653. doi: 10.1084/jem.20122337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dos Santos GA, Kats L, Pandolfi PP. Synergy against PML-RARa: targeting transcription, proteolysis, differentiation, and self-renewal in acute promyelocytic leukemia. The Journal of experimental medicine. 2013;210:2793–2802. doi: 10.1084/jem.20131121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lo-Coco F, Avvisati G, Vignetti M, et al. Retinoic acid and arsenic trioxide for acute promyelocytic leukemia. The New England journal of medicine. 2013;369:111–121. doi: 10.1056/NEJMoa1300874. [DOI] [PubMed] [Google Scholar]
- 8.Jeanne M, Lallemand-Breitenbach V, Ferhi O, et al. PML/RARA oxidation and arsenic binding initiate the antileukemia response of As2O3. Cancer cell. 2010;18:88–98. doi: 10.1016/j.ccr.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 9.Nasr R, Guillemin MC, Ferhi O, et al. Eradication of acute promyelocytic leukemia-initiating cells through PML-RARA degradation. Nature medicine. 2008;14:1333–1342. doi: 10.1038/nm.1891. [DOI] [PubMed] [Google Scholar]
- 10.Mizushima N, Klionsky DJ. Protein turnover via autophagy: implications for metabolism. Annual review of nutrition. 2007;27:19–40. doi: 10.1146/annurev.nutr.27.061406.093749. [DOI] [PubMed] [Google Scholar]
- 11.He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annual review of genetics. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feng Y, He D, Yao Z, Klionsky DJ. The machinery of macroautophagy. Cell research. 2014;24:24–41. doi: 10.1038/cr.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guan JL, Simon AK, Prescott M, et al. Autophagy in stem cells. Autophagy. 2013;9:830–849. doi: 10.4161/auto.24132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weidberg H, Shvets E, Shpilka T, Shimron F, Shinder V, Elazar Z. LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. The EMBO journal. 2010;29:1792–1802. doi: 10.1038/emboj.2010.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mizushima N, Levine B. Autophagy in mammalian development and differentiation. Nat Cell Biol. 2010;12:823–830. doi: 10.1038/ncb0910-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Conway KL, Kuballa P, Khor B, et al. ATG5 regulates plasma cell differentiation. Autophagy. 2013;9 doi: 10.4161/auto.23484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu F, Lee JY, Wei H, et al. FIP200 is required for the cell-autonomous maintenance of fetal hematopoietic stem cells. Blood. 2010;116:4806–4814. doi: 10.1182/blood-2010-06-288589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mortensen M, Soilleux EJ, Djordjevic G, et al. The autophagy protein Atg7 is essential for hematopoietic stem cell maintenance. The Journal of experimental medicine. 2011;208:455–467. doi: 10.1084/jem.20101145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trocoli A, Mathieu J, Priault M, et al. ATRA-induced upregulation of Beclin 1 prolongs the life span of differentiated acute promyelocytic leukemia cells. Autophagy. 2011;7:1108–1114. doi: 10.4161/auto.7.10.16623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Z, Cao L, Kang R, et al. Autophagy regulates myeloid cell differentiation by p62/SQSTM1-mediated degradation of PML-RARalpha oncoprotein. Autophagy. 2011;7:401–411. doi: 10.4161/auto.7.4.14397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brigger D, Torbett BE, Chen J, Fey MF, Tschan MP. Inhibition of GATE-16 attenuates ATRA-induced neutrophil differentiation of APL cells and interferes with autophagosome formation. Biochemical and biophysical research communications. 2013;438:283–288. doi: 10.1016/j.bbrc.2013.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Isakson P, Bjoras M, Boe SO, Simonsen A. Autophagy contributes to therapy-induced degradation of the PML/RARA oncoprotein. Blood. 2010;116:2324–2331. doi: 10.1182/blood-2010-01-261040. [DOI] [PubMed] [Google Scholar]
- 23.Elzinga BM, Nyhan MJ, Crowley LC, O’Donovan TR, Cahill MR, McKenna SL. Induction of autophagy by Imatinib sequesters Bcr-Abl in autophagosomes and down-regulates Bcr-Abl protein. American journal of hematology. 2013;88:455–462. doi: 10.1002/ajh.23428. [DOI] [PubMed] [Google Scholar]
- 24.Britschgi C, Rizzi M, Grob TJ, et al. Identification of the p53 family-responsive element in the promoter region of the tumor suppressor gene hypermethylated in cancer 1. Oncogene. 2006;25:2030–2039. doi: 10.1038/sj.onc.1209240. [DOI] [PubMed] [Google Scholar]
- 25.Chakrama FZ, Seguin-Py S, Le Grand JN, et al. GABARAPL1 (GEC1) associates with autophagic vesicles. Autophagy. 2010;6:495–505. doi: 10.4161/auto.6.4.11819. [DOI] [PubMed] [Google Scholar]
- 26.Lihuan D, Jingcun Z, Ning J, et al. Photodynamic therapy with the novel photosensitizer chlorophyllin f induces apoptosis and autophagy in human bladder cancer cells. Lasers in surgery and medicine. 2014;46:319–334. doi: 10.1002/lsm.22225. [DOI] [PubMed] [Google Scholar]
- 27.Kimura T, Takabatake Y, Takahashi A, Isaka Y. Chloroquine in cancer therapy: a double-edged sword of autophagy. Cancer research. 2013;73:3–7. doi: 10.1158/0008-5472.CAN-12-2464. [DOI] [PubMed] [Google Scholar]
- 28.Wu YT, Tan HL, Shui G, et al. Dual role of 3-methyladenine in modulation of autophagy via different temporal patterns of inhibition on class I and III phosphoinositide 3-kinase. The Journal of biological chemistry. 2010;285:10850–10861. doi: 10.1074/jbc.M109.080796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jung CH, Ro SH, Cao J, Otto NM, Kim DH. mTOR regulation of autophagy. FEBS letters. 2010;584:1287–1295. doi: 10.1016/j.febslet.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sarkar S, Floto RA, Berger Z, et al. Lithium induces autophagy by inhibiting inositol monophosphatase. J Cell Biol. 2005;170:1101–1111. doi: 10.1083/jcb.200504035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Humbert M, Mueller C, Fey MF, Tschan MP. Inhibition of damage-regulated autophagy modulator-1 (DRAM-1) impairs neutrophil differentiation of NB4 APL cells. Leukemia research. 2012;36:1552–1556. doi: 10.1016/j.leukres.2012.08.024. [DOI] [PubMed] [Google Scholar]
- 32.Strohecker AM, Guo JY, Karsli-Uzunbas G, et al. Autophagy sustains mitochondrial glutamine metabolism and growth of BrafV600E-driven lung tumors. Cancer discovery. 2013;3:1272–1285. doi: 10.1158/2159-8290.CD-13-0397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shao W, Fanelli M, Ferrara FF, et al. Arsenic trioxide as an inducer of apoptosis and loss of PML/RAR alpha protein in acute promyelocytic leukemia cells. Journal of the National Cancer Institute. 1998;90:124–133. doi: 10.1093/jnci/90.2.124. [DOI] [PubMed] [Google Scholar]
- 34.Yousefi S, Simon HU. Autophagy in cells of the blood. Biochimica et biophysica acta. 2009;1793:1461–1464. doi: 10.1016/j.bbamcr.2008.12.023. [DOI] [PubMed] [Google Scholar]
- 35.Miller BC, Zhao Z, Stephenson LM, et al. The autophagy gene ATG5 plays an essential role in B lymphocyte development. Autophagy. 2008;4:309–314. doi: 10.4161/auto.5474. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Y, Morgan MJ, Chen K, Choksi S, Liu ZG. Induction of autophagy is essential for monocyte-macrophage differentiation. Blood. 2012;119:2895–2905. doi: 10.1182/blood-2011-08-372383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cassinat B, Chomienne C. Biological features of primary APL blasts: their relevance to the understanding of granulopoiesis, leukemogenesis and patient management. Oncogene. 2001;20:7154–7160. doi: 10.1038/sj.onc.1204761. [DOI] [PubMed] [Google Scholar]
- 38.Pasquali L, Busceti CL, Fulceri F, Paparelli A, Fornai F. Intracellular pathways underlying the effects of lithium. Behavioural pharmacology. 2010;21:473–492. doi: 10.1097/FBP.0b013e32833da5da. [DOI] [PubMed] [Google Scholar]
- 39.Focosi D, Azzara A, Kast RE, Carulli G, Petrini M. Lithium and hematology: established and proposed uses. Journal of leukocyte biology. 2009;85:20–28. doi: 10.1189/jlb.0608388. [DOI] [PubMed] [Google Scholar]
- 40.Sokoloski JA, Li J, Nigam A, Sartorelli AC. Induction of the differentiation of HL-60 and WEHI-3B D+ leukemia cells by lithium chloride. Leukemia research. 1993;17:403–410. doi: 10.1016/0145-2126(93)90095-3. [DOI] [PubMed] [Google Scholar]
- 41.Gupta K, Gulen F, Sun L, et al. GSK3 is a regulator of RAR-mediated differentiation. Leukemia. 2012;26:1277–1285. doi: 10.1038/leu.2012.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Si J, Mueller L, Collins SJ. GSK3 inhibitors enhance retinoic acid receptor activity and induce the differentiation of retinoic acid-sensitive myeloid leukemia cells. Leukemia. 2011;25:1914–1918. doi: 10.1038/leu.2011.171. [DOI] [PubMed] [Google Scholar]
- 43.Finch RA, Li J, Chou TC, Sartorelli AC. Maintenance of retinoic acid receptor alpha pools by granulocyte colony-stimulating factor and lithium chloride in all-trans retinoic acid-treated WEHI-3B leukemia cells: relevance to the synergistic induction of terminal differentiation. Blood. 2000;96:2262–2268. [PubMed] [Google Scholar]
- 44.Renna M, Jimenez-Sanchez M, Sarkar S, Rubinsztein DC. Chemical inducers of autophagy that enhance the clearance of mutant proteins in neurodegenerative diseases. The Journal of biological chemistry. 2010;285:11061–11067. doi: 10.1074/jbc.R109.072181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Quiroz JA, Machado-Vieira R, Zarate CA, Jr, Manji HK. Novel insights into lithium’s mechanism of action: neurotrophic and neuroprotective effects. Neuropsychobiology. 2010;62:50–60. doi: 10.1159/000314310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trocoli A, Bensadoun P, Richard E, et al. p62/SQSTM1 upregulation constitutes a survival mechanism that occurs during granulocytic differentiation of acute myeloid leukemia cells. Cell death and differentiation. 2014;21:1852–1861. doi: 10.1038/cdd.2014.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Choi AM, Ryter SW, Levine B. Autophagy in human health and disease. The New England journal of medicine. 2013;368:1845–1846. doi: 10.1056/NEJMc1303158. [DOI] [PubMed] [Google Scholar]
- 48.Nencioni A, Cea M, Montecucco F, et al. Autophagy in blood cancers: biological role and therapeutic implications. Haematologica. 2013;98:1335–1343. doi: 10.3324/haematol.2012.079061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.White E. Deconvoluting the context-dependent role for autophagy in cancer. Nature reviews Cancer. 2012;12:401–410. doi: 10.1038/nrc3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Crowley LC, O’Donovan TR, Nyhan MJ, McKenna SL. Pharmacological agents with inherent anti-autophagic activity improve the cytotoxicity of imatinib. Oncology reports. 2013;29:2261–2268. doi: 10.3892/or.2013.2377. [DOI] [PubMed] [Google Scholar]
- 51.Chen N, Karantza V. Autophagy as a therapeutic target in cancer. Cancer biology & therapy. 2011;11:157–168. doi: 10.4161/cbt.11.2.14622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Townsend KN, Hughson LR, Schlie K, Poon VI, Westerback A, Lum JJ. Autophagy inhibition in cancer therapy: metabolic considerations for antitumor immunity. Immunological reviews. 2012;249:176–194. doi: 10.1111/j.1600-065X.2012.01141.x. [DOI] [PubMed] [Google Scholar]
- 53.Zhang SP, Niu YN, Yuan N, et al. Role of autophagy in acute myeloid leukemia therapy. Chinese journal of cancer. 2013;32:130–135. doi: 10.5732/cjc.012.10073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.