Abstract
Genomics is a promising tool that is becoming more widely available to improve the care and treatment of individuals. While there is much assertion, genomics will most certainly require the use of clinical decision support (CDS) to be fully realized in the routine clinical setting. The National Human Genome Research Institute (NHGRI) of the National Institutes of Health recently convened an in-person, multi-day meeting on this topic. It was widely recognized that there is a need to promote the innovation and development of resources for genomic CDS such as a CDS sandbox. The purpose of this study was to evaluate a proposed approach for such a genomic CDS sandbox among domain experts and potential users. Survey results indicate a significant interest and desire for a genomic CDS sandbox environment among domain experts. These results will be used to guide the development of a genomic CDS sandbox.
Keywords: Clinical Decision Support Systems, Genomics, Medical genetics, Health information technology, Personalized medicine
1. Introduction
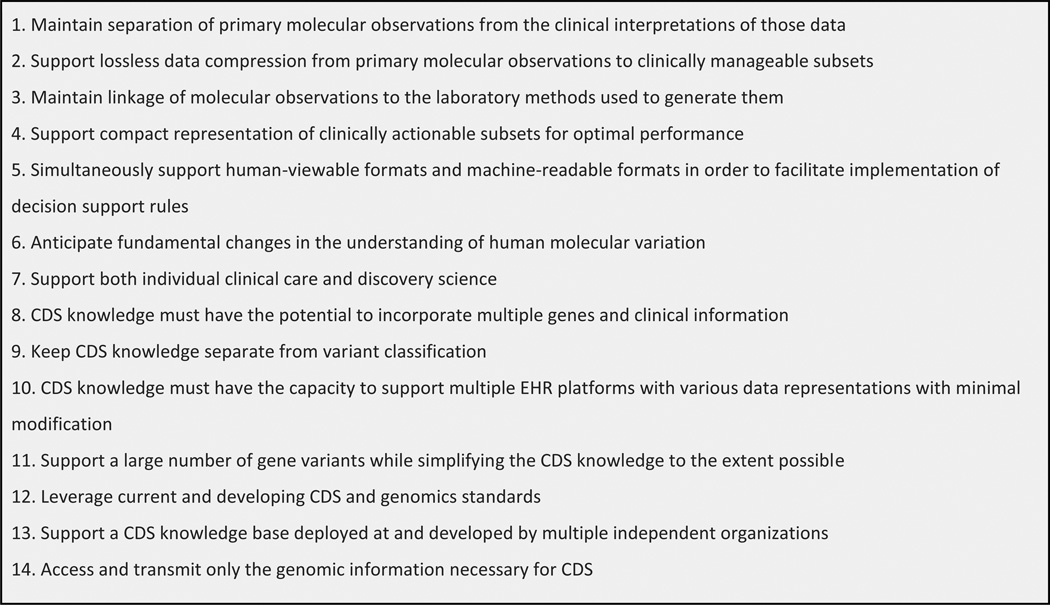
Genomics will have a considerable impact on the future of medicine by personalizing care and treatment of patients.1 However, the application of genomics to routine clinical care will be limited by physician proficiency in genetics, lack of clinical genetics expertise, and most importantly the complex nature of genomic information. Clinical decision support (CDS) has been proposed as a feasible solution to overcome these challenges.2 CDS provides clinicians, patients, and other healthcare stakeholders with pertinent knowledge and person-specific information, intelligently filtered or presented at appropriate times, to enhance health and healthcare.3 CDS can help clinicians manage the complexities of genetics at the point of care and support genetically-guided medicine.2 However, literature and practical implementations of genomic CDS, in particular whole-genome sequence (WGS) information, are currently very nascent.4–6 Before genomic CDS is mature enough to be implemented in routine clinical care, there is much to consider such as health IT architectures and integration, genomic knowledge bases, managing genomic data and its changing interpretations, data models and standards. To guide genomic CDS development, genomics and CDS experts have put forward key technical requirements and considerations.7,8 See Figure 1. Following these technical desiderata, researchers have started developing and exploring novel genomic CDS solutions.9–12
Figure 1.
Technical desiderata for WGS guided CDS from Masys et al. and Welch et al.
1.1 NIH genomic CDS conference
Recognizing that more work is necessary to fully support genomic CDS on a widespread scale, the National Human Genome Research Institute (NHGRI) convened a meeting on "Genomic Clinical Decision Support: Developing Solutions for Clinical and Research Implementation" in Bethesda, MD in October of 2014.13 This meeting convened key thought leaders in genomic medicine implementation and application of CDS to (1) compare current state with ideal state of genomic CDS in order to define gaps and strategies to close the gaps; (2) identify and engage US and international health IT initiatives that would support recommended strategies; and (3) define a prioritized research agenda for genomic CDS. A notable outcome was the need for a genomic CDS sandbox to promote the development, evaluation, and testing of genomic CDS.
1.2 Genomic CDS sandbox
During the conference, many issues were highlighted that could be addressed with a genomic CDS sandbox. For example, with a standardized genomic CDS sandbox, researchers will be able to:
1.2.1 Develop and test genomic CDS rules and algorithms
Genomic CDS is still in its infancy, many rules and algorithms for genomics have yet to be developed and tested, and there is still much to be learned. A genomic CDS sandbox provides an ideal environment to develop and test new rules and algorithms, prior to deployment in a real health IT system. Sandbox environments are often part of proprietary EHRs, however they have limited flexibility and control for what users can test or change. In contrast, the proposed genomic CDS sandbox will allow several EHR and health IT components to be used and tested independent of proprietary systems A CDS sandbox environment also promotes rapid development, allowing for success or failure of novel CDS to occur early and often. Furthermore, non-technical domain experts such as pharmacists and clinicians could utilize the sandbox to develop CDS rules.
1.2.2 Design and implement novel architectural approaches
Current CDS approaches are not able to meet the demands of genomic CDS for many reasons.14,7 Architectural approaches need to support disparate data sources and health IT components to effectively provide genomic CDS.15 A genomic CDS sandbox allows these architecture approaches to be developed, evaluated, and refined.
1.2.3 Identify gaps in standards and technologies
To promote interoperability and widespread adoption, genomic CDS should leverage current CDS and genomic CDS standards (Desiderata #12). Oftentimes gaps in health IT standards and capabilities arise during implementation, causing developers to implement non-standard solutions. With a genomic CDS sandbox, such issues can be identified and addressed in the pre-implementation phase, allowing informatics professionals to address standards issues through the proper channels in a formal way.
1.2.4 Conduct usability and human factors research
Usability and human factors often contributes to the success or failure of a CDS intervention.16 As genomic CDS is a new technology, human factors research is critical. However, production EHR systems is not the best place for usability and human factors research. A genomic CDS sandbox would be an ideal environment to create and test innovate user interfaces for genomic CDS, that can ultimately improve the usability and success of genomic CDS.
1.3 Genomic CDS sandbox description
A sandbox environment is a collaborative, non-production, development environment in which novel concepts for emerging technologies can be explored with no risk to clinical systems. Sandbox domains have proven useful in other informatics fields.17,18 The ultimate goal of a genomic CDS sandbox is to provide a common platform which allows domain experts and knowledge translation experts to build and test shareable CDS knowledge base for genomics. Specifically to meet the requirements described above, namely to: 1) develop and test genomic CDS rules and algorithms, (2) design and implement novel architectural approaches, (3) identify gaps in standards and technologies, and (4) conduct usability and human factors research.
To accomplish this, the proposed genomic CDS sandbox would be implemented as an open-source, freely available and distributable virtual machine (VM). The proposed sandbox would consist of several open source health IT and genomics applications (such as OpenMRS, VistA, OpenCDS, SMARTapps), installed and preconfigured on the VM. These components would include synthetic clinical and genomic data, with fully functional genomic CDS examples available to test and modify. Additions and modifications to the genomic CDS sandbox would be limited to the user's virtual environment, but users would be able to share their work with other users. If appropriate, enhancements can be added to the default genomic CDS sandbox. With this approach, a community of technical and clinical domain experts could collaboratively develop and refine genomic CDS solutions and approaches. Additionally, by preconfiguring these applications and distributing them using a VM, it reduces the need for specialized technical expertise to set up and run such components. Additionally the system would include sufficient training resources to help non-technical users build and test genomic CDS approaches. Nevertheless, the intended end users of the genomic CDS sandbox could be both technical and non-technical domain experts. As this genomic CDS sandbox is a community-driven, open source project, any intellectual property generated using the product will belong to the user.
1.4 Purpose of study
To guide the design and development of a genomic CDS sandbox, we engaged potential genomic CDS sandbox end users to assess their level of interest and to determine desired features. Specifically, we sought to (1) validate the proposed genomic CDS sandbox approach among domain experts; (2) identify desired use cases; and (3) determine the best approach for building a community of users.
2. Methods
Using REDCap,19 we distributed an online survey among potential end users of a genomic CDS sandbox. An initial set of 26 Likert-scale questions were developed based on the perceived need of genomic CDS sandbox derived from discussions and comments at the NHGRI meeting. In January 2015, the initial set of questions were distributed to an advisory panel consisting of domain experts in CDS and genomics who attended the NHGRI meeting (see authors). The advisory panel commented and helped revise the questions incorporated into the final survey by AA and BMW. Based on feedback from the advisory panel, the final survey consisted of 22 questions divided into four parts: (1) involvement with genomics and the CDS domain; (2) validation of the genomic CDS sandbox approach; (3) usage of the genomic CDS sandbox; and (4) building a genomic CDS sandbox community. The survey text and questions can be found in Appendix A.
To obtain a diverse representation of domain experts, we distributed the survey to a variety of CDS and genomics professional organizations. Participants included members of the HL7 Clinical Genomics Work Group, the HL7 CDS Work Group, the AMIA Genomics Work Group, the AMIA CDS Workgroup, the ClinGen project, the Clinical Pharmacogenetics Implementation Consortium (CPIC), the Clinical Sequencing Exploratory Research (CSER) consortium EHR working group, the Pharmacogenomics Knowledge Base (PharmGKB), the Pharmacogenomics Research Network (PGRN), the EHR Integration (EHRI) and Phenotyping workgroups of the Electronic Medical Records and Genomics Network (eMERGE), and NHGRI Genomic Medicine meeting attendees. Survey responses were collected in February 2015. The survey was distributed to roughly 600 potential participants via email lists and blog postings for each workgroup or consortia.
3. Results
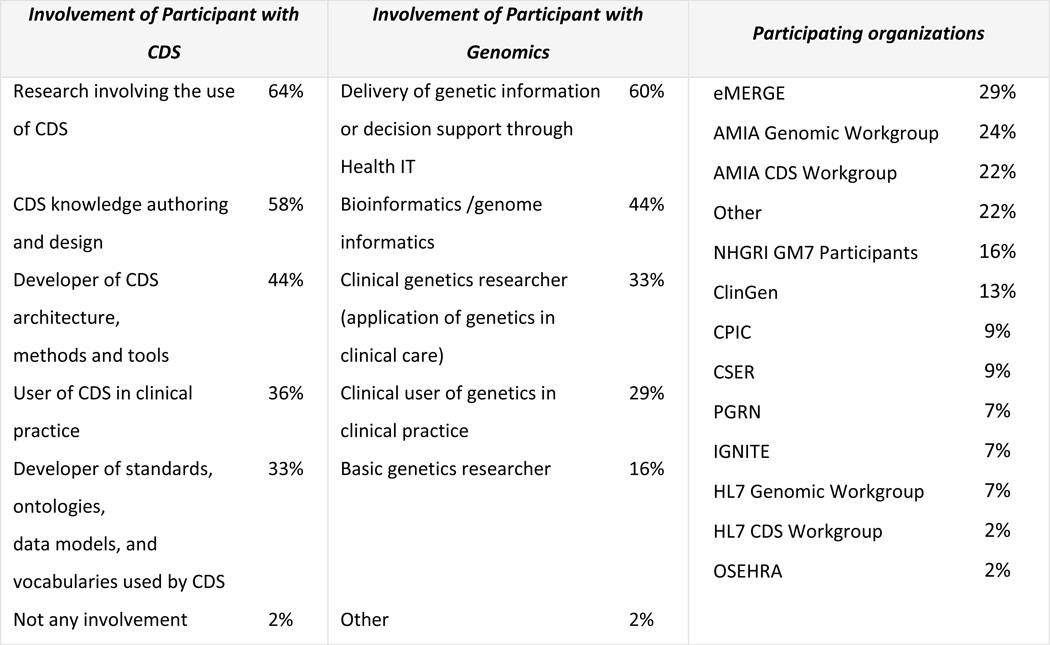
We initially received 54 survey responses, after exclusion of incomplete responses, a final sample of 45 responses was analysed. Respondents had an average of 6 years’ experience in CDS (Min:0, Max:30, SD: 6.93) and an average of 8 years’ experience in genomics (Min:0, Max:30, SD: 5.53). The majority of CDS experience is related to research involving the use of CDS (64%) and CDS knowledge authoring and design (58%) (see Figure 2). Delivery of genetic information or decision support through health IT (60%) and bioinformatics/genome informatics (44%) was the experience most described by those with genomics expertise. Respondents represented a wide range of collaborations and workgroups, with highest participation from the eMERGE Network (29%), the AMIA Genomic Workgroup (24%), the AMIA CDS Workgroup (22%) and ClinGen (13%). Several respondents were involved with multiple groups. 22% described participating in ‘Other’, (see Appendix B). 93% of the respondents do not currently use a CDS sandbox, reinforcing the need to develop a solution.
Figure 2.
Involvement of survey respondents with CDS and Genomics and their affiliation
3.1. Validation of genomic CDS Sandbox Approach
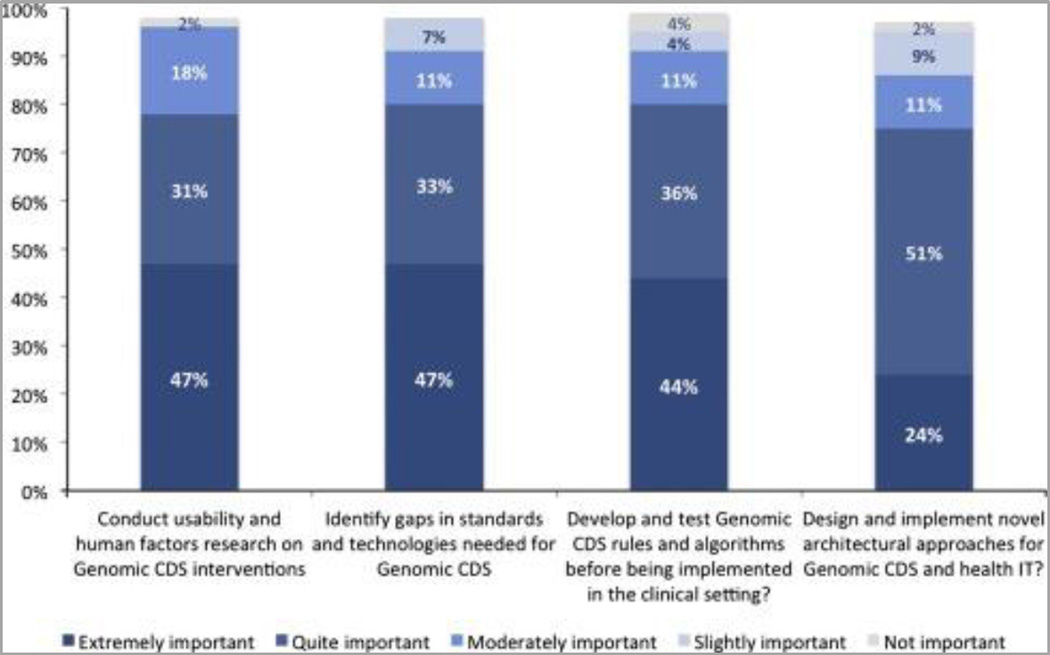
Several questions were asked to validate the proposed genomic CDS sandbox (see Introduction). The responses provide strong support with 93% of respondents agree (51% strongly agree, 42% agree) with the proposed design and features of proposed genomic CDS sandbox. In assessing the uses of a genomic CDS sandbox, roughly 80% of respondents considered it to be ‘extremely important’ or ‘quite important’ to conduct usability and human factors research (47% extremely, 31% quite likely); to identify gaps in standards and technologies (47%); develop and test Genomic CDS rules and algorithms (44%); and a quarter of respondents considered it to be ‘extremely important’ to the design and implement novel architectural approaches (24%). See Figure 3.
Figure 3.
Desired features of Genomic CDS Sandbox
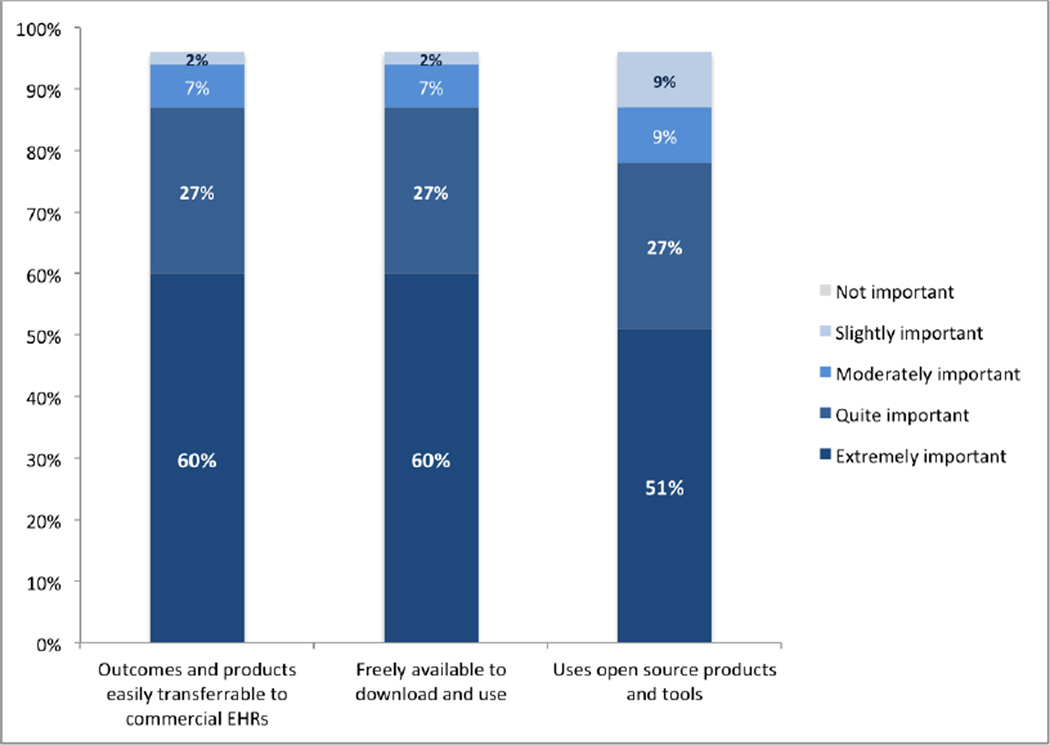
Respondents placed a high importance on the availability of the genetic CDS sandbox, namely: the genomic sandbox is freely available to download and use (60% ‘extremely important’, 27% ‘quite important’); the outcomes and products of the sandbox are easily transferrable to commercial EHRs (60%‘extremely important’, 27% ‘quite important’); and uses open source products and tools (51%‘extremely important’, 27% ‘quite important’). See Figure 4.
Figure 4.
Features relating to Availability and use of genomic CDS sandbox
In addition to these responses, participants proposed additional sandbox features including integration with commercial EHRs, inclusion of existing CDS solutions such as Infobuttons, and making availability of the sandbox as a downloadable solution for personal computers, laptops and servers.20
3.2. Usage of the genomic CDS sandbox
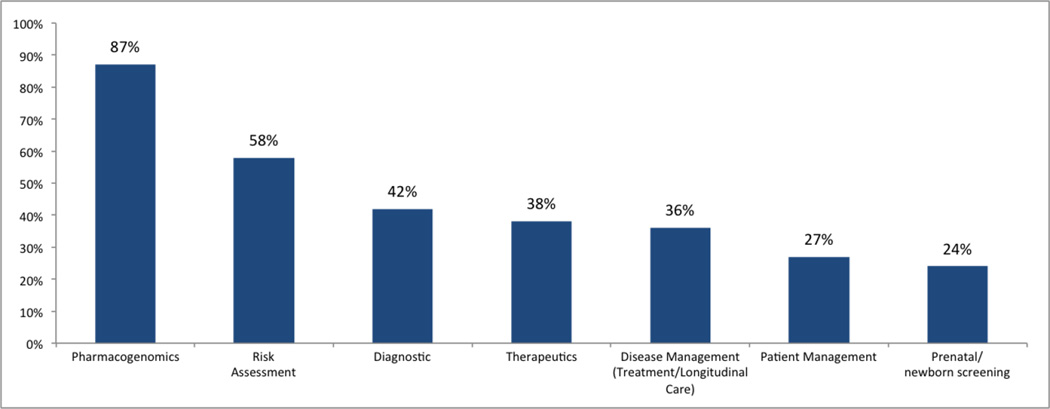
Over two-thirds (67%) of respondents would be ‘extremely likely’ or ‘quite likely’ to use the proposed genomic CDS sandbox. 71% of the respondents are currently working on/or have future projects that will benefit from the genomic CDS sandbox. Examples of such projects include CDS systems for genomically-guided cancer therapy, pharmacogenomics projects, inherited disorder genomics, and ordering of molecular/genomic tests for tumors. Potential genomic CDS sandbox users would use the genomic CDS sandbox for testing use cases in pharmacogenomics (87%), disease risk assessment (58%), diagnostics (42%), therapeutics (38%), disease management (36%), patient management (27%), and prenatal/newborn screening (24%). See Figure 5. Interestingly, all respondents said they would ‘likely’ (31% extremely, quite likely 24%) work with Standards Development Organizations such as HL7, SNOMED, LOINC, etc. to propose new standards if they identified gaps in health IT or genomic standards. For training and technical support for using the genomic CDS Sandbox, of the proposed options, the majority of respondents identified user manuals (71%), community user forums (64%), webinars (60%), video tutorials (58%) and email support (51%), as the preferred media.
Figure 5.
Test Case studies for Sandbox
3.3. Building a Genomic CDS Sandbox Community
We also assessed the importance of developing a community of genomics CDS sandbox users. A majority of respondents (80%) believe that it is ‘extremely important’ or ‘quite important’ to build and support this community of experts to facilitate innovation and enhancements. Approximately two-third of respondents (51% extremely, 29% quite) stated that they would be ‘extremely likely’ or ‘quite likely’ to participate in the community. The majority of respondents said they would participate with the community using email discussions (69%), conferences and publications (69%), and workgroup meetings (53%).
4. Discussion
The purpose of this survey was to demonstrate the need for a genomic CDS sandbox. Our survey results show that there is significant interest and demand for the development of a genomic CDS sandbox. There is a general sentiment that genomic CDS sandbox is an important need, this survey provides that data that confirms this sentiment that was first articulated at the NHGRI conference. Furthermore, we received strong support for our planned approach. Interestingly we found that if such a solution is available to the community of Genomics and CDS users, there is a good chance that they will not only utilize such a system, but also participate in a community of Genomic CDS by sharing their experiences and promoting its use. A majority of respondents would be extremely likely to conduct usability and human factors research on Genomic CDS interventions, and identify gaps in standards and technologies needed for Genomic CDS. Notably, all respondents also stated that they would likely participate in standards development if gaps in standards were found. We identified pharmacogenomics and disease risk assessment as the biggest use cases for users of the genomic CDS sandbox.
4.1 Strengths and Limitations
This survey clearly confirmed and identified needs and desires for a genomic CDS sandbox environment from intended end users. Also the timing of this survey is quite relevant as recent advances in genome sequencing technology have dramatically reduced the cost of obtaining a whole genome sequence, making it less expensive and more accessible. However, it does not reduce the complexity, which will require CDS to manage.9 Furthermore, Meaningful Use is standardizing the health IT infrastructure, making it easier to develop and share CDS in other clinical domains. At this time it is critical to create a collaborative effort among genomics researchers and CDS experts to address issues related to genomic data integration with the EHRs, CDS knowledge base development, genetic test reporting and interpretation. Developing a genomic CDS sandbox is a critical first step toward this goal. The main limitation of the study was its relatively small sample size (n=45). However, we received a diverse representation of responses, and results were sufficient for our purpose of conducting this study. Additionally, results would likely not been significantly different if sample size were increased. While there is potential for response bias because survey respondents were selected to represent the intended users (CDS and genomics experts) of the sandbox, our results generally reflect the current genomic CDS trends in the industry. 14 Another limitation of this study is that the survey instruments did not use more robust survey questionnaire validation methodology.21 However, the questions were derived and refined by a panel of genomics and CDS experts before being distributed to the survey respondents, which we felt was sufficient for this study.
4.2 Future Direction
The survey was conducted as a preliminary step towards the development of a genomic CDS sandbox. The results of this survey will guide the next steps and future direction of this effort, including funding opportunities, strategic partners, and an optimal development strategy. The next steps of this effort are to identify potential tools and technologies that would be used to create the genomic CDS sandbox. First, we will need to identify potential EHRs, personal health records, laboratory information systems, CDS tools, genome databases, and technologies that can be used within the genomic CDS sandbox environment. Second, we need to identify necessary resources to design and develop the genomic CDS sandbox including technical and clinical expertise, computational resources, funding, and other types of support. Third, potential case studies to be implemented as primary examples for the genomic CDS sandbox need to be identified and developed. After the genomic CDS sandbox is built, we can then distribute to users and continue to identify opportunities to improve and expand genomic CDS capabilities based on their feedback. We will also be including various Health IT solutions providers in future studies to identify existing components to include in the genomic CDS sandbox. There are certainly many opportunities for future research and development in this space. Finally, just as we used an advisory panel to conduct this needs assessment study, we intend to create a governance committee to guide the development and management of the genomic CDS sandbox.
Conclusion
A genomic CDS sandbox could provide a common platform to develop, test, and evaluate potential genomic CDS approaches and techniques. Results from this study provides strong evidence for the need and desire of a genomic CDS sandbox from among a community of genomics and CDS domain experts. Once built, it is anticipated that genomic CDS approaches developed and tested using this sandbox will be utilized in the clinical setting, improving the ability of clinicians to practice genomically-guided medicine.
Supplementary Material
Acknowledgments
This study was inspired by the outcomes of Genomic Medicine 7 meeting: "Genomic Clinical Decision Support: Developing Solutions for Clinical and Research Implementation" sponsored by the National Human Genome Research Institute (NHGRI) meeting in Bethesda, MD in October of 2014. Co-authors of this manuscript and study were attendees at this conference.
APPENDIX B: Survey Respondent's Affiliation selecting “Other” for their Organizations
Clinical and Translational Science Award (CTSA)
Bioinformatics Australia
Private clinical decision support company that includes pharmacy genomic data in the CDSS
Newborn Sequencing In Genomic medicine and public Health (NSIGHT)
Global Alliance for Genomics and Health (GA4GH)
IOM EHR Action Collaborative
East Carolina University (Academic)
Footnotes
Conflict of Interest Statement
Kensaku Kawamoto (KK) is currently or recently served as a consultant on CDS to the Office of the National Coordinator for Health IT, ARUP Laboratories, McKesson InterQual, ESAC, Inc., JBS International, Inc., Inflexxion, Inc., Intelligent Automation, Inc., Partners HealthCare, the RAND Corporation, and Mayo Clinic. KK receives royalties for a Duke University-owned CDS technology for infectious disease management known as CustomID that he helped develop. KK was formerly a consultant for Religent, Inc. and a co-owner and consultant for Clinica Software, Inc., both of which provide commercial CDS services, including through use of a CDS technology known as SEBASTIAN that KK developed. KK no longer has a financial relationship with either Religent or Clinica Software. KK has no competing interests related to OpenCDS, which is freely available to the community as an open-source resource. MH was previously employed by Cerner Corporation and is an inventor on a number of genomic CDS patents owned by Cerner Corporation but does not receive royalties or other tangible benefits from these inventions. MH owns stock in Cerner Corporation at an amount below the threshold for COI or in inaccessible funds. BHS is a consultant on the Clinical Advisory Board of Avalon Healthcare Solutions. BMW is the founder and shareholder of ItRunsInMyFamily.com, a web-based family health history tool. BMW is also the founder and shareholder of Doxy.me, a telemedicine platform. The remaining authors do not report any conflict of interest.
References
- 1.Abrahams E, Ginsburg GS, Silver M. The Personalized Medicine Coalition: goals and strategies. Am. J. Pharmacogenomics. 2005;5:345–355. doi: 10.2165/00129785-200505060-00002. [DOI] [PubMed] [Google Scholar]
- 2.Welch BM, Kawamoto K. The need for clinical decision support integrated with the electronic health record for the clinical application of whole genome sequencing information. J Pers Med. 2013;3:306–325. doi: 10.3390/jpm3040306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Osheroff JA, et al. A roadmap for national action on clinical decision support. J. Am. Med. Inform. Assoc. 2007;14:141–145. doi: 10.1197/jamia.M2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Welch BM, Kawamoto K. Clinical decision support for genetically guided personalized medicine: a systematic review. J. Am. Med. Inform. Assoc. 2013;20:388–400. doi: 10.1136/amiajnl-2012-000892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Overby CL, et al. Opportunities for genomic clinical decision support interventions. Genet. Med. 2013;15:817–823. doi: 10.1038/gim.2013.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kho AN, et al. Practical challenges in integrating genomic data into the electronic health record. Genet. Med. 2013;15:772–778. doi: 10.1038/gim.2013.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Welch BM, Eilbeck K, Del Fiol G, Meyer LJ, Kawamoto K. Technical desiderata for the integration of genomic data with clinical decision support. J. Biomed. Inform. 2014;51:3–7. doi: 10.1016/j.jbi.2014.05.014. [DOI] [PubMed] [Google Scholar]
- 8.Masys DR, et al. Technical desiderata for the integration of genomic data into Electronic Health Records. J. Biomed. Inform. 2012;45:419–422. doi: 10.1016/j.jbi.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Welch BM, Loya SR, Eilbeck K, Kawamoto K. A proposed clinical decision support architecture capable of supporting whole genome sequence information. J Pers Med. 2014;4:176–199. doi: 10.3390/jpm4020176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Welch BM, Rodriguez-Loya S, Eilbeck K, Kawamoto K. Clinical decision support for whole genome sequence information leveraging a service-oriented architecture: a prototype. AMIA Annu. Symp. Proc. 2014;2014:1188–1197. [PMC free article] [PubMed] [Google Scholar]
- 11.Herr A conceptual model for translating omic data into clinical action. J. Pathol. Inform. 2015;6:46. doi: 10.4103/2153-3539.163985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shirts BH, et al. CSER and eMERGE: current and potential state of the display of genetic information in the electronic health record. J. Am. Med. Inform. Assoc. 2015 doi: 10.1093/jamia/ocv065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Genomic Clinical Decision Support: Developing Solutions for Clinical and Research Implementation. National Human Genome Research Institute (NHGRI) at < http://www.genome.gov/27558904>.
- 14.Kannry JL, Williams MS. The undiscovered country: the future of integrating genomic information into the EHR. Genet. Med. 2013;15:842–845. doi: 10.1038/gim.2013.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tarczy-Hornoch P, et al. A survey of informatics approaches to whole-exome and whole-genome clinical reporting in the electronic health record. Genet. Med. 2013;15:824–832. doi: 10.1038/gim.2013.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ohno-Machado L. Health IT and clinical decision support systems: human factors and successful adoption. J. Am. Med. Inform. Assoc. 2014;21:e180. doi: 10.1136/amiajnl-2014-003279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ambler SW, Sadalage PJ. In: Refactoring databases: Evolutionary database design. Ambler SW, Sadalage PJ, editors. Pearson Education; 2006. at < https://books.google.com/books?hl=en&lr=&id=fN5Aa_m38vAC&oi=fnd&pg=PT4&dq=refactoring%2Bdatabases&ots=jHof1yM-MI&sig=tkBptwDv5lgFYRKfkKDOhqeaUyE>. [Google Scholar]
- 18.Fenwick M, et al. An Open-Source Sandbox for Increasing the Accessibility of Functional Programming to the Bioinformatics and Scientific Communities. Proc. Int. Conf. Inf. Technol. New Gener. 2012;2012:89–94. doi: 10.1109/ITNG.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harris PA, et al. Research electronic data capture (REDCap)--a metadata-driven methodology and workflow process for providing translational research informatics support. J. Biomed. Inform. 2009;42:377–381. doi: 10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.HL7. Product Brief - Context-Aware Knowledge Retrieval (Infobutton) HL7 Inc.; at < http://wiki.hl7.org/index.php?title=Product_Infobutton>. [Google Scholar]
- 21.Portney LG, Watkins MP, et al. Foundations of clinical research: applications to practice. Vol. 2. Upper Saddle River, NJ: Prentice Hall; 2000. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.