Abstract
Background
Enterotoxigenic Escherichia coli (ETEC) is a common cause of bacterial infection leading to acute watery diarrhea in infants and young children as well as in travellers to ETEC endemic countries. Ciprofloxacin is a broad-spectrum antimicrobial agent nowadays used for the treatment of diarrhea. This study aimed to characterize ciprofloxacin resistant ETEC strains isolated from diarrheal patients in Bangladesh.
Methods
A total of 8580 stool specimens from diarrheal patients attending the icddr,b Dhaka hospital was screened for ETEC between 2005 and 2009. PCR and Ganglioside GM1- Enzyme Linked Immuno sorbent Assay (ELISA) was used for detection of Heat labile (LT) and Heat stable (ST) toxins of ETEC. Antimicrobial susceptibilities for commonly used antibiotics and the minimum inhibitory concentration (MIC) of nalidixic acid, ciprofloxacin and azithromycin were examined. DNA sequencing of representative ciprofloxacin resistant strains was performed to analyze mutations of the quinolone resistance-determining region of gyrA, gyrB, parC and parE. PCR was used for the detection of qnr, a plasmid mediated ciprofloxacin resistance gene. Clonal variations among ciprofloxacin resistant (CipR) and ciprofloxacin susceptible (CipS) strains were determined by Pulsed-field gel electrophoresis (PFGE).
Results
Among 1067 (12%) ETEC isolates identified, 42% produced LT/ST, 28% ST and 30% LT alone. Forty nine percent (n = 523) of the ETEC strains expressed one or more of the 13 tested colonization factors (CFs) as determined by dot blot immunoassay. Antibiotic resistance of the ETEC strains was observed as follows: ampicillin 66%, azithromycin 27%, ciprofloxacin 27%, ceftriazone 13%, cotrimaxazole 46%, doxycycline 44%, erythromycin 96%, nalidixic acid 83%, norfloxacin 27%, streptomycin 48% and tetracycline 42%. Resistance to ciprofloxacin increased from 13% in 2005 to 34% in 2009. None of the strains was resistant to mecillinam. The MIC of the nalidixic acid and ciprofloxacin of representative CipR strains were 256 μg/ml and 32μg/ml respectively. A single mutation (Ser83-Leu) in gyrA was observed in the nalidixic acid resistant ETEC strains. In contrast, double mutation in gyrA (Ser83-Leu, Asp87-Asn) and a single mutation in parC (Glu84-Ly) were found in ciprofloxacin resistant strains. Mutation of gyrB was not found in either the nalidixic acid or ciprofloxacin resistant strains. None of the ciprofloxacin resistant strains was found to be positive for the qnr gene. Diverse clones were identified from all ciprofloxacin resistant strains by PFGE analysis in both CF positive and CF negative ETEC strains.
Conclusion
Emergence of ciprofloxacin resistant ETEC strains results in a major challenge in current treatment strategies of ETEC diarrhea.
Introduction
Enterotoxigenic E. coli (ETEC) is the most important cause of diarrhea among children under 5 years of age in developing countries and among travellers in ETEC endemic countries [1, 2]. ETEC express one or both of two types of enterotoxins, namely heat stable (ST) and/or heat labile (LT) toxin which are encoded by the same or separate plasmids [3–5], and may produce one or more of several colonization factors (CFs) [6, 7]. In case of human challenge studies where ETEC strains were used [8], ampicillin was the only antibiotic used for the treatment and clearing of bacteria. When ETEC strains were first isolated in Calcutta in 1968, no antimicrobial resistance was reported. Additionally, a completely uniform sensitivity pattern was also observed in ETEC strains isolated from Apache children in Arizona during 1971 [9]. Studying the effects of antimicrobial treatment in children with ETEC disease has been difficult for two reasons. Firstly, childhood diarrhea is caused by ETEC as well as other bacterial and viral agents. Secondly, the clinical presentations are not sufficient to differentiate among the various pathogenic organisms responsible for diarrhea. One retrospective study of ETEC diarrhea in Bangladeshi patients has shown that in adults, only a minimal effect on the severity of diarrhea was evident when tetracycline was used for treatment [10].
Antimicrobial treatment of traveller’s diarrhea, which is caused by ETEC in about 20–50% of cases, has altered over the years due to increasing antimicrobial resistance [11]. The rise in multidrug resistance in organisms, such as ETEC, can be attributed to the widespread and indiscriminate use of chemotherapeutic agents in countries where diarrhea is endemic. Hence, it has become imperative to use newer antimicrobials for treating traveller’s diarrhea. The antibiotics that have been used so far are ciprofloxacin, doxycycline, sulfamethoxazole-trimethoprim, erythromycin, norfloxacin, ofloxacin, azithromycin and rifamycin [12–14]. Fluoroquinolones are powerful broad-spectrum antimicrobial agents for the treatment of a wide variety of community acquired and nosocomial infections [15]. However, resistance to fluoroquinolones has increased markedly since their introduction in the late 1980s [16]. Recently ciprofloxacin resistance Shigella and Salmonella has emerged lately in Bangladesh [17, 18]. In 2001 ciprofloxacin resistant ETEC was isolated from patients in India [19]. A recent study conducted in 2008–2009 demonstrated the presence of ciprofloxacin resistant ETEC in the drinking water of Bangladesh [20]. No studies have yet been carried out on fluoroquinolones resistance mechanism of ETEC in Bangladesh.
Several studies showed that fluoroquinolones mainly target DNA gyrase, a type II DNA topoisomerase, in gram-negative organisms [21]. DNA gyrase is composed of two subunits, namely A (gyrA) and B (gyrB). Although the subunit A is highly conserved within bacterial species, most of the mutations reported so far in DNA gyrase are within this subunit [21]. Alterations in subunit A have been documented in a variety of species, including E. coli, Shigella, Salmonella, and Campylobacter [22, 23]. On the contrary, mutations in subunit B, which is associated with quinolone resistance, have been rarely isolated in clinical samples [24]. In addition to targeting DNA gyrase, fluoroquinolones also target topoisomerase IV. Toposiomerase IV is also composed of two subunits, namely parC and parE, which share significant sequence similarity to the subunits A and B of DNA gyrase [25].
The aim of the present study was to compare the antimicrobial resistance pattern, clonal variation, and sequence analysis of Quinolone resistance-determinig regions (QRDR) of gyrA, gyrB, parC and parE mutations in ciprofloxacin resistant ETEC strains.
Material and Methods
Ethics statement
The International Centre for Diarrhoeal Disease Research, Bangladesh (icddr,b) carries out a Surveillance of diarrheal patients attending the Dhaka Hospital where every fiftieth patient’s stool specimens were screened for enteric pathogens [26]. Informed oral consent is obtained from the caregivers or guardians on behalf of the patients for collecting specimens. The 2% surveillance is approved by the Institutional Review Board (IRB) of icddrb. The patients or the guardians are assured about the non-disclosure of information collected from them, but are also informed about the use of data for analysis and keeping identity of the participants anonymous.
Collection and culture of clinical specimens
A total number of 8580 stool specimens were collected from patients, both male (59%) and female (41%), with diarrhea identified from 2% systematic surveillance system [27] at the icddr,b hospital at Dhaka between 2005 and 2009. Two percentage systematic surveillance system means every 50th patient irrespective of age, sex, disease severity or socioeconomic status by administering structured questionnaire. The generated data provides valuable information to hospital clinicians in their decision- making processes and enables the detection of emerging pathogens and early identification of outbreaks and their locations as well as antimicrobial susceptibility pattern. All stool samples were inoculated onto MacConkey agar, Taurocholate-tellurite-gelatin agar and Salmonella–Shigella agar plates (Difco, Becton Dickinson & Company, Sparks, MD, USA). ETEC, Vibrio cholerae, Shigellae and Salmonellae were isolated and identified by using standard microbiological and biochemical methods [28]. Shigella and Salmonella species were confirmed using commercial antisera kits (Denka Seiken, Tokyo, Japan). Vibrio cholerae were confirmed using a combination of biochemical and serological methods as described previously [29, 30]. For the detection of ETEC, fresh stool specimens were inoculated onto MacConkey agar and the plates were incubated for 18h at 37°C. Six lactose fermenting individual colonies morphologically resembling E.coli from MacConkey agar plates were tested immediately for the presence of toxins and CFs as described below.
Detection of toxin types and CFs
The detection of LT and ST was carried out by multiplex PCR and ganglioside GM1 enzyme- linked immunosorbent assays (ELISA) [31, 32]. The colonies tested for toxin production were also cultured on Colonization factor antigen (CFA) agar plates with and without bile salts [7, 33]. Enterotoxin positive E. coli colonies from CFA agar plates were tested for the expression of CFA/I, CS1, CS2, CS3, CS4 and CS6 and colonies from CFA agar plus bile plates were tested for the expression of CS5, CS7, CS8, CS12, CS14 and CS17 by monoclonal antibody-based dot blot assays [31, 34]. Strains grown on TSA were tested for CS21 only [35].
Antimicrobial susceptibility
Antimicrobial agents from Oxoid, Basingstoke, United Kingdom were used to determine bacterial susceptibility by using the guidelines of National Committee for Clinical Laboratory Standards [36]. The antibiotic discs used in the study included ampicillin (10 μg), azithromycin (15 μg), ceftriaxone (30 μg), ciprofloxacin (5 μg), doxycycline (30 μg), erythromycin (15 μg), mecillinam (25 μg), nalidixic acid (30 μg), norfloxacin (10 μg), streptomycin (10 μg), sulfomethoxazole-trimethoprim (25 μg) and tetracycline (30 μg). The minimum inhibitory concentrations (MIC) of nalidixic acid, ciprofloxacin, and azithromycin were determined by the E-test (AB Biodisk, Solna, Sweden) according to the NCCL guideline. E. coli ATCC 25922, susceptible to all antimicrobials was used as a control strain for susceptibility studies. We performed antibiogram of all ETEC strains collected during the study period and randomly selected the representative nalidixic acid and ciprofloxacin resistant strains. These strains were compared with a few representative nalidixic and ciprofloxacin susceptible strains for genotyping.
Pulsed-field gel electrophoresis (PFGE)
Intact chromosomal DNA of clinical ETEC strains was prepared and digested with the XbaI restriction endonuclease (New England Biolabs) according to the PulseNet program descried elsewhere [37]. DNA fragments were separated by pulsed-field gel electrophoresis on a CHEF-MAPPER apparatus (Bio-Rad) under the following conditions: switching time from 2.2 to 54.2 s at 6 V cm−1 for 20 h at 14°C. Analysis of the TIFF image was carried out by the BioNumerics software version 4.5 (Applied Maths, Belgium) with average linkages to generate dendrogram at 1.5% tolerance level using the dice coefficient and unweighted-pair group method.
PCR and sequencing amplification of gyrA, gyrB, parC and parE
Chromosomal DNA was prepared and purified by procedures described earlier [38]. Polymerase chain reaction (PCR) for gyrA, gyrB, parC and parE were done according to the methods described earlier [39–41].
Nucleotide sequencing
GFX PCR DNA and gel band purification kit (Amersham Pharmacia, USA) were used for the purification of gyrA, gyrB, parC and parE PCR amplicons. Sequencing of the purified PCR products were performed by using the dideoxynucleotide chain termination method with an ABI PRISM BigDye Terminator Cycle Sequencing Reaction kit (Perkin-Elmer Applied Biosystems, Foster City, CA, USA) on an automated sequencer (ABI PRISM 310) at the icddr,b core sequencing facility.
DNA and protein sequence analysis
The chromatogram sequencing files were inspected using Chromas 2.23 (Technelysium, Queensland, Australia), and contiguous sequences were prepared using SeqMan II (DNASTAR, Madison, WI, USA). Nucleotide and protein sequences were analyzed using the National Center for Biotechnology Information (NCBI, National Institutes of Health, Bethesda, MD, USA) BLAST (Basic Local Alignment Search Tool) server on GenBank database, release 138, in order to find similarities with previously known sequences [42] Multiple sequence alignments developed using CLUSTALX 1.81 [43] sequences were manually edited in the Gene Doc version 2.6.002 alignment editor.
Results
Isolation of enteric pathogens from stool specimens
A total of 8580 stool specimens were tested from patients at the icddr,b hospital during the period between 2005–2009 for the isolation of Vibrio cholerae, ETEC, Shigella, and Salmonella. Of 8580 specimens, a total of 3194 pathogens were isolated, of them 21% (1784/8580) were V. cholerae O1, 12% (1067/8580) were ETEC, 3.0% (259/8580) were Shigella spp. and rest 1.0% (84/8580) was Salmonella spp. Of V. cholerae O1, 74% were Ogawa and 26% were Inaba serotypes.
Toxin types and CFs on ETEC
Among the ETEC isolates, 448 (42%) were both LT and ST followed by the strains producing LT alone 315 (30%) and ST only 304 (28%) (Table 1). In the present study 523 (49%) of the ETEC strains expressed one or more CFs; CFA/I, CS5+CS6 and CS7 were the predominant phenotypes during the study period.
Table 1. Number of ETEC strains isolated during the study period and their toxin types.
| Year | No. of specimens tested | ETEC No. (%) | ETEC toxin Types | ||
|---|---|---|---|---|---|
| LT | ST | LT+ST | |||
| 2005 | 654 | 91(14) | 24 (26) | 42 (46) | 25 (28) |
| 2006 | 661 | 82 (12) | 27 (33) | 21 (26) | 34 (41) |
| 2007 | 2211 | 291 (13) | 100 (34) | 70 (24) | 121 (42) |
| 2008 | 2369 | 319 (13) | 89 (28) | 87 (27) | 143 (45) |
| 2009 | 2685 | 284 (11) | 75 (26) | 84 (30) | 125 (44) |
| Total (2005–2009) | 8580 | 1067 (12) | 315 (30) | 304 (28) | 448 (42) |
Antibiotic resistance pattern
The majority of the ETEC strains isolated during the study period showed high resistance (Table 2) to the 12 different antibiotics tested, including members of the quinolone (nalidixic acid) and fluoroquinolone groups (ciprofloxacin or norfloxacin). Antibiotic resistance pattern was as follows: ampicillin 66%, azithromycin 27%, ciprofloxacin 27%, ceftriazone 13%, sulfomethoxazole-trimethoprim 46%, doxycycline 44%, erythromycin 96%, nalidixic acid 83%, norfloxacin 27%, streptomycin 48% and tetracycline 42% respectively. Resistance to ciprofloxacin increased from 13% in 2005 to 34% in 2009. However none of the strains was resistant to mecillinam.
Table 2. Resistant pattern of ETEC strains isolated during the study periods (2005–2009).
| Name of antibiotics | 2005* (N = 91) (%) | 2006* (N = 82) (%) | 2007* (N = 277) (%) | 2008* (N = 191) (%) | 2009* (N = 262) (%) |
|---|---|---|---|---|---|
| Ampicillin (A) | 53 | 60 | 66 | 70 | 71 |
| Azithromycin (Azm) | 5 | 13 | 22 | 32 | 33 |
| Ciprofloxacin (Cip) | 13 | 26 | 25 | 26 | 34 |
| Ceftriaxone (Cro) | 4 | 6 | 13 | 13 | 17 |
| Doxycline (D) | 33 | 35 | 46 | 49 | 44 |
| Erythromycin (E) | 95 | 95 | 96 | 97 | 100 |
| Nalidixic Acid (Na) | 70 | 76 | 80 | 90 | 87 |
| Norfloxacin (Nor) | 13 | 26 | 25 | 26 | 34 |
| Streptomycin (S) | 35 | 39 | 51 | 47 | 53 |
| Sulfomethoxazole-trimethoprim (Sxt) | 38 | 49 | 49 | 51 | 41 |
| Tetracycline (T) | 31 | 32 | 43 | 49 | 44 |
* Number of ETEC strains tested during the year
Most of the fluoroquinolone resistant ETEC strains tested were CF negative 84% (n = 204). All ETEC strains were tested for 13 colonization factors using 13 specific monoclonal antibodies. A probable reason for the above results may be that other colonization factors were not tested. Moreover, ETEC strains might have possibly lost the plasmids harboring the CF genetic elements because of repeated subculture in vitro or a mutation in genetic locus. Multiple antibiotic resistance patterns and the minimum inhibitory concentrations (MIC) of the representative ciprofloxacin resistant strains are shown in (Table 3). The range of MIC values of the ciprofloxacin was 32 μg/ml and nalidixic acid 64–256 μg/ml.
Table 3. MIC values and amino acids changes in gyrA and ParC in representative ETEC strains isolated in Bangladesh.
| Strain | Toxin types | CFs | Resistance pattern | MIC (μg/ml) | Substitution in QRDRc | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Na | Cip | Azm | gyrA | ParC | ||||||||
| 2533700 | LT+ST | CS5+CS6 | Sensitive | 2 | 0.012 | 0.38 | - | - | - | - | - | - |
| 2960200 | LT+ST | CS1+CS3+CS21 | Sensitive | 0.008 | 1 | 0.75 | - | - | - | - | - | - |
| 2511850 | ST | CFA/I | E Na Sxt T | 256 | 0.19 | 1 | Ser83 | Leu | - | - | - | - |
| 2526600 | LT | Negative | A Cip D E Na Sxt T | 256 | 32 | 4 | Ser83 | Leu | Asp87 | Asn | Glu84 | Ly |
| 2848150 | LT | Negative | A Azm Cip D E Na Sxt T | 256 | 32 | 256 | Ser83 | Leu | Asp87 | Asn | Glu84 | Ly |
| 0008200 | ST | Negative | A Azm Cip Cro D E Na S Sxt T | 256 | 32 | 256 | Ser83 | Leu | Asp87 | Asn | Glu84 | Ly |
Identification of mutations and clonal variations
Further investigation using gene-sequencing QRDR (Quinolone resistance-determining region) of gyrA, gyrB, parC and parE was performed to investigate the mutations of nalidixic acid and ciprofloxacin resistant ETEC strains. A single mutation (Ser83-Leu) in gyrA was observed in the nalidixic acid resistant ETEC strains. In contrast, double mutation in gyrA (Ser83-Leu, Asp87-Asn)) and a single mutation in parC (Glu84-Ly) was found in all of the most recently isolated fluoroquinolone resistant strains in diarrheal patients. Mutation of gyrB was not found either in nalidixic acid or ciprofloxacin resistant strains. No mutation in gyrA, gyrB, parC and parE was observed by gene sequence analyses among the quinolone and fluoroquinolone susceptible ETEC strains.
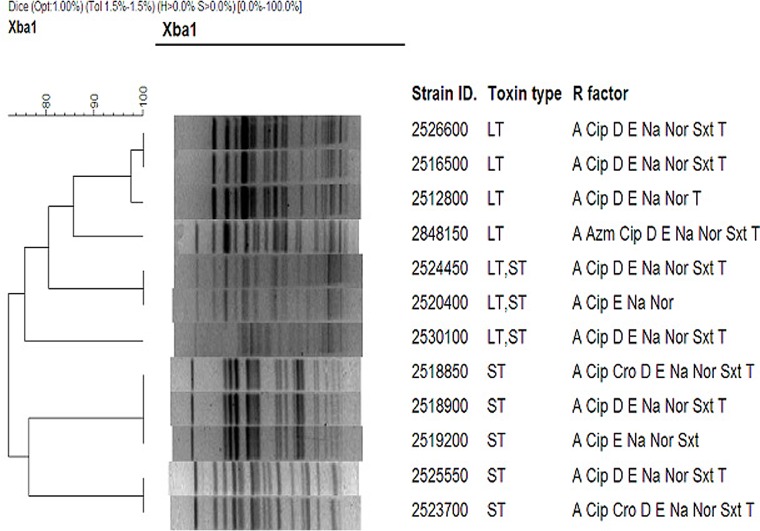
PFGE typing of representative ciprofloxacin resistant strains (n = 12) of different ETEC toxin types (LT, LT/ST and ST ETEC) showed that more than one clone was detected within a specific toxin type (Fig 1).
Fig 1. BioNumeric software generated dendrogram.
The distances shown were calculated by the dice similarity index of PFGE XbaI profiles for clinical ETEC isolates isolated from Bangladesh. The scale shows the degree of similarity (%).
Discussion
Analyses of data from patients hospitalized in diarrheal hospitals, have shown that patients with ETEC diarrhea are often given antibiotic treatment together with rehydration therapy [44]. The treatment is similar to that given to patients with V. cholerae induced diarrhea. This is done to reduce duration of hospital stay as well as decrease of transmission of diarrheal pathogens in the environment. However, careful monitoring of antimicrobial sensitivity in treated patients is generally not carried out. Our results show that ETEC is also following a similar trend of increased antibiotic resistance to drugs commonly used for treatment of V. cholerae O1 and other diarrheal infections [45, 46].
Our present analysis showed that 88% of ETEC strains isolated from stools of diarrheal patients attended at a 2% surveillance system at icddr,b Dhaka Hospital in Bangladesh during five years (2005–2009) were multidrug resistant. Resistance was observed to nalidixic acid and ciprofloxacin as well as reduced sensitivity to azithromycin and ceftriaxone. There have been reports from India and Japan on multidrug resistance to the fluoroquinolones in ETEC strains isolated during outbreaks [19, 46]. In this study no CFs were detected in 84% of the ciprofloxacin resistant strains. Similar finding was also observed in India [19]. A recent study, reported the presence of ciprofloxacin resistant ETEC in environmental samples during 2008–2009 in Bangladesh [20]. This change is trend in Bangladesh is alarming as these antibiotics are used for the treatment of enteric infections.
Quinolones are broad-spectrum antibacterial agents that act by inhibiting the DNA gyrase and topoisomerase IV activities [47]. We showed that the isolated ciprofloxacin resistant ETEC strains on sequencing had a single mutation in gyrA gene in the nalidixic acid resistant strains and double mutation in gyrA in the ciprofloxacin resistant ETEC strains. In addition, a single mutation in parC was found in fluoroquinolone resistant ETEC strains. This suggests that gyrA and parC are intracellular targets of quinolones and fluoroquinolones, and that mutation in these genes are associated with fluoroquinolones and/or quinolones resistance in E. coli, which is also supported by previous studies [40, 48]. No mutation in gyrB was observed in the ciprofloxacin and nalidixic acid resistant ETEC strains examined in our study. Among the quinolone and fluoroquinolone-susceptible ETEC strains, however, no mutations were observed in gyrA, gyrB, parC and parE by sequence analysis. PCR analysis also showed that none of the strains was found to carry the qnr gene. The qnr gene encodes an immune protein which protects E. coli gyrase from quinolone inhibition. Hence, along with these factors, additional factors may also be involved in mediating fluoroquinolone resistance in ETEC. Although in vitro drug resistance pattern not always be related to in vivo activity of the drug, it may indicate the trend of the sensitivity. Our results show that ETEC strains with fluoroquinolone resistance is increasing in Bangladesh and also that the gyrA and parC genes were mutated. There are important informations for therapeutic purposes. However it may mention that in vitro drug resistance pattern has not always been found to correlate with in vivo activity and indicate treatment successor.
Dendogram analysis of representative ETEC strains indicated that Ciprofloxacin resistance in ETEC was not clonally related, since Cip resistance due to gyrA gene mutation was acquired by diverse clones of ETEC. Thus, similar to earlier observations, we did not find any relationship with the toxin phenotype, CFs or antibiotic resistance pattern [6] among the ciprofloxacin resistant strains.
In summary, multidrug resistance pattern of ETEC strains especially to fluroquinoles is a matter of great concern for appropriate management of ETEC diarrhea. This also underscores the need for the dissemination of this information not only for control of endemic ETEC diarrheal diseases but also for traveler’s diarrhea. Thus immunoprophylaxis guarantee with effective vaccines is required to prevent ETEC diarrheal cases globally.
Data Availability
All relevant data are within the paper.
Funding Statement
This research was supported by the core grants to the icddr,b, by the Governments of the Australia, Bangladesh, Canada, Sweden, and the UK for providing core/unrestricted support. This work was also supported by a Swedish Sida (SIDA) (grant INT-ICDDR,B-HN-01-AV) and Swedish collaborative grant SIDA-SWE2009-023, as well as by the Commission of the European Communities grant to STOPENTERICS (grant HEALTH-F3-2010-261472).
References
- 1.Qadri F, Svennerholm AM, Faruque AS, Sack RB. Enterotoxigenic Escherichia coli in developing countries: epidemiology, microbiology, clinical features, treatment, and prevention. Clin Microbiol Rev. 2005a;18(3):465–83. 10.1128/CMR.18.3.465-483.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shaheen HI, Kamal KA, Wasfy MO, El-Ghorab NM, Lowe B, Steffen R, et al. Phenotypic diversity of enterotoxigenic Escherichia coli (ETEC) isolated from cases of travelers' diarrhea in Kenya. Int J Infect Dis. 2003;7(1):35–8. . [DOI] [PubMed] [Google Scholar]
- 3.Gill DM, Richardson SH. Adenosine diphosphate-ribosylation of adenylate cyclase catalyzed by heat-labile enterotoxin of Escherichia coli: comparison with cholera toxin. J Infect Dis. 1980;141(1):64–70. . [DOI] [PubMed] [Google Scholar]
- 4.Rao MC. Toxins which activate guanylate cyclase: heat-stable enterotoxins. Ciba Found Symp. 1985;112:74–93. . [DOI] [PubMed] [Google Scholar]
- 5.So M, Dallas WS, Falkow S. Characterization of an Escherichia coli plasmid encoding for synthesis of heat-labile toxin: molecular cloning of the toxin determinant. Infect Immun. 1978;21(2):405–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Begum YA, Talukder KA, Nair GB, Khan SI, Svennerholm AM, Sack RB, et al. Comparison of enterotoxigenic Escherichia coli isolated from surface water and diarrhoeal stool samples in Bangladesh. Can J Microbiol. 2007;53(1):19–26. 10.1139/w06-098 . [DOI] [PubMed] [Google Scholar]
- 7.Gaastra W, Svennerholm AM. Colonization factors of human enterotoxigenic Escherichia coli (ETEC). Trends Microbiol. 1996;4(11):444–52. . [DOI] [PubMed] [Google Scholar]
- 8.DuPont HL, Evans DG, Evans DJ Jr., Satterwhite TK. Antitoxic immunity in cholera and enterotoxigenic Escherichia coli (ETEC) diarrhea. Pharmacol Ther. 1981;13(2):249–55. . [DOI] [PubMed] [Google Scholar]
- 9.Sack RB, Hirschhorn N, Brownlee I, Cash RA, Woodward WE, Sack DA. Enterotoxigenic Escherichia-coli-associated diarrheal disease in Apache children. N Engl J Med. 1975;292(20):1041–5. 10.1056/NEJM197505152922001 . [DOI] [PubMed] [Google Scholar]
- 10.Merson MH, Sack RB, Islam S, Saklayen G, Huda N, Huq I, et al. Disease due to enterotoxigenic Escherichia coli in Bangladeshi adults: clinical aspects and a controlled trial of tetracycline. J Infect Dis. 1980;141(6):702–11. . [DOI] [PubMed] [Google Scholar]
- 11.Ericsson CD. Travellers' diarrhoea. Int J Antimicrob Agents. 2003;21(2):116–24. . [DOI] [PubMed] [Google Scholar]
- 12.Diemert DJ. Prevention and self-treatment of traveler's diarrhea. Clin Microbiol Rev. 2006;19(3):583–94. 10.1128/CMR.00052-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodas C, Mamani R, Blanco J, Blanco JE, Wiklund G, Svennerholm AM, et al. Enterotoxins, colonization factors, serotypes and antimicrobial resistance of enterotoxigenic Escherichia coli (ETEC) strains isolated from hospitalized children with diarrhea in Bolivia. Braz J Infect Dis. 2011;15(2):132–7. . [DOI] [PubMed] [Google Scholar]
- 14.Sack RB. Travelers' diarrhea: microbiologic bases for prevention and treatment. Rev Infect Dis. 1990;12 Suppl 1:S59–63. . [DOI] [PubMed] [Google Scholar]
- 15.Oethinger M, Conrad S, Kaifel K, Cometta A, Bille J, Klotz G, et al. Molecular epidemiology of fluoroquinolone-resistant Escherichia coli bloodstream isolates from patients admitted to European cancer centers. Antimicrob Agents Chemother. 1996;40(2):387–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aguiar JM, Chacon J, Canton R, Baquero F. The emergence of highly fluoroquinolone-resistant Escherichia coli in community-acquired urinary tract infections. J Antimicrob Chemother. 1992;29(3):349–50. . [DOI] [PubMed] [Google Scholar]
- 17.Ahmed D, D'Costa LT, Alam K, Nair GB, Hossain MA. Multidrug-resistant Salmonella enterica serovar typhi isolates with high-level resistance to ciprofloxacin in Dhaka, Bangladesh. Antimicrob Agents Chemother. 2006;50(10):3516–7. 10.1128/AAC.00667-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Das SK, Ahmed S, Ferdous F, Farzana FD, Chisti MJ, Latham JR, et al. Etiological diversity of diarrhoeal disease in Bangladesh. J Infect Dev Ctries. 2013;7(12):900–9. 10.3855/jidc.3003 . [DOI] [PubMed] [Google Scholar]
- 19.Chakraborty S, Deokule JS, Garg P, Bhattacharya SK, Nandy RK, Nair GB, et al. Concomitant infection of enterotoxigenic Escherichia coli in an outbreak of cholera caused by Vibrio cholerae O1 and O139 in Ahmedabad, India. J Clin Microbiol. 2001;39(9):3241–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Talukdar PK, Rahman M, Rahman M, Nabi A, Islam Z, Hoque MM, et al. Antimicrobial resistance, virulence factors and genetic diversity of Escherichia coli isolates from household water supply in Dhaka, Bangladesh. PLoS One. 2013;8(4):e61090 10.1371/journal.pone.0061090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Everett MJ, Jin YF, Ricci V, Piddock LJ. Contributions of individual mechanisms to fluoroquinolone resistance in 36 Escherichia coli strains isolated from humans and animals. Antimicrob Agents Chemother. 1996;40(10):2380–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Talukder KA, Khajanchi BK, Islam MA, Islam Z, Dutta DK, Rahman M, et al. Fluoroquinolone resistance linked to both gyrA and parC mutations in the quinolone resistance-determining region of Shigella dysenteriae type 1. Curr Microbiol. 2006;52(2):108–11. 10.1007/s00284-005-0140-9 . [DOI] [PubMed] [Google Scholar]
- 23.Thomson CJ. The global epidemiology of resistance to ciprofloxacin and the changing nature of antibiotic resistance: a 10 year perspective. J Antimicrob Chemother. 1999;43 Suppl A:31–40. . [DOI] [PubMed] [Google Scholar]
- 24.Azmi IJ, Khajanchi BK, Akter F, Hasan TN, Shahnaij M, Akter M, et al. Fluoroquinolone resistance mechanisms of Shigella flexneri isolated in Bangladesh. PLoS One. 2014;9(7):e102533 10.1371/journal.pone.0102533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maxwell A. DNA gyrase as a drug target. Trends Microbiol. 1997;5(3):102–9. 10.1016/S0966-842X(96)10085-8 . [DOI] [PubMed] [Google Scholar]
- 26.Stoll BJ, Glass RI, Huq MI, Khan MU, Holt JE, Banu H. Surveillance of patients attending a diarrhoeal disease hospital in Bangladesh. Br Med J (Clin Res Ed). 1982;285(6349):1185–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harris AM, Chowdhury F, Begum YA, Khan AI, Faruque AS, Svennerholm AM, et al. Shifting prevalence of major diarrheal pathogens in patients seeking hospital care during floods in 1998, 2004, and 2007 in Dhaka, Bangladesh. Am J Trop Med Hyg. 2008;79(5):708–14. [PMC free article] [PubMed] [Google Scholar]
- 28.WHO. Programme for control of diarrheal disease Mannual for laboratory investigation of acute enteric infection 1987;CDD/93.3,Rev.1 Geneva, Switzerland:P 9–20.
- 29.Qadri F, Azim T, Chowdhury A, Hossain J, Sack RB, Albert MJ. Production, characterization, and application of monoclonal antibodies to Vibrio cholerae O139 synonym Bengal. Clin Diagn Lab Immunol. 1994;1(1):51–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rahman M, Sack DA, Mahmood S, Hossain A. Rapid diagnosis of cholera by coagglutination test using 4-h fecal enrichment cultures. J Clin Microbiol. 1987;25(11):2204–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sjoling A, Wiklund G, Savarino SJ, Cohen DI, Svennerholm AM. Comparative analyses of phenotypic and genotypic methods for detection of enterotoxigenic Escherichia coli toxins and colonization factors. J Clin Microbiol. 2007;45(10):3295–301. 10.1128/JCM.00471-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Svennerholm AM, Wikstrom M, Lindblad M, Holmgren J. Monoclonal antibodies against Escherichia coli heat-stable toxin (STa) and their use in a diagnostic ST ganglioside GM1-enzyme-linked immunosorbent assay. J Clin Microbiol. 1986;24(4):585–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McConnell MM, Chart H, Field AM, Hibberd M, Rowe B. Characterization of a putative colonization factor (PCFO166) of enterotoxigenic Escherichia coli of serogroup O166. J Gen Microbiol. 1989;135(5):1135–44. . [DOI] [PubMed] [Google Scholar]
- 34.Qadri F, Das SK, Faruque AS, Fuchs GJ, Albert MJ, Sack RB, et al. Prevalence of toxin types and colonization factors in enterotoxigenic Escherichia coli isolated during a 2-year period from diarrheal patients in Bangladesh. J Clin Microbiol. 2000a;38(1):27–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qadri F, Giron JA, Helander A, Begum YA, Asaduzzaman M, Xicohtencatl-Cortes J, et al. Human antibody response to longus type IV pilus and study of its prevalence among enterotoxigenic Escherichia coli in Bangladesh by using monoclonal antibodies. J Infect Dis. 2000b;181(6):2071–4. 10.1086/315507 . [DOI] [PubMed] [Google Scholar]
- 36.National Committee for Clinical Laboratory Standards 2000. Performance standard for antimicrobial disc susceptibility test approved standard M2 National committee for clinical laboratory standards, villanova, Pa, USA.
- 37.Pichel M, Brengi SP, Cooper KL, Ribot EM, Al-Busaidy S, Araya P, et al. Standardization and international multicenter validation of a PulseNet pulsed-field gel electrophoresis protocol for subtyping Shigella flexneri isolates. Foodborne Pathog Dis. 2012;9(5):418–24. 10.1089/fpd.2011.1067 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Talukder KA, Islam MA, Dutta DK, Hassan F, Safa A, Nair GB, et al. Phenotypic and genotypic characterization of serologically atypical strains of Shigella flexneri type 4 isolated in Dhaka, Bangladesh. J Clin Microbiol. 2002;40(7):2490–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rahman M, Mauff G, Levy J, Couturier M, Pulverer G, Glasdorff N, et al. Detection of 4-quinolone resistance mutation in gyrA gene of Shigella dysenteriae type 1 by PCR. Antimicrob Agents Chemother. 1994;38(10):2488–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vila J, Ruiz J, Goni P, De Anta MT. Detection of mutations in parC in quinolone-resistant clinical isolates of Escherichia coli. Antimicrob Agents Chemother. 1996;40(2):491–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang H, Dzink-Fox JL, Chen M, Levy SB. Genetic characterization of highly fluoroquinolone-resistant clinical Escherichia coli strains from China: role of acrR mutations. Antimicrob Agents Chemother. 2001;45(5):1515–21. 10.1128/AAC.45.5.1515-1521.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–10. 10.1016/S0022-2836(05)80360-2 . [DOI] [PubMed] [Google Scholar]
- 43.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25(24):4876–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qadri F, Khan AI, Faruque AS, Begum YA, Chowdhury F, Nair GB, et al. Enterotoxigenic Escherichia coli and Vibrio cholerae diarrhea, Bangladesh, 2004. Emerg Infect Dis. 2005b;11(7):1104–7. 10.3201/eid1107.041266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Khan WA, Saha D, Rahman A, Salam MA, Bogaerts J, Bennish ML. Comparison of single-dose azithromycin and 12-dose, 3-day erythromycin for childhood cholera: a randomised, double-blind trial. Lancet. 2002;360(9347):1722–7. 10.1016/S0140-6736(02)11680-1 . [DOI] [PubMed] [Google Scholar]
- 46.Matsushita S, Kawamura M, Takahashi M, Yokoyama K, Konishi N, Hatakeyama K, et al. [Increasing fluoroquinolone low-sensitivity in enterotoxigenic Escherichia coli isolated from diarrhea of overseas travelers in Tokyo]. Kansenshogaku Zasshi. 2001;75(9):785–91. . [DOI] [PubMed] [Google Scholar]
- 47.Hooper DC. Quinolone mode of action—new aspects. Drugs. 1993;45 Suppl 3:8–14. . [DOI] [PubMed] [Google Scholar]
- 48.Vila J, Ruiz J, Marco F, Barcelo A, Goni P, Giralt E, et al. Association between double mutation in gyrA gene of ciprofloxacin-resistant clinical isolates of Escherichia coli and MICs. Antimicrob Agents Chemother. 1994;38(10):2477–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.