Abstract
We identified phosphatidylinositol glycan anchor biosynthesis, class X (PIGX), which plays a critical role in the biosynthetic pathway of glycosylphosphatidylinositol (GPI)-anchor motif, to be upregulated highly and frequently in breast cancer cells. Knockdown of PIGX as well as reticulocalbin 1 (RCN1) and reticulocalbin 2 (RCN2), which we found to interact with PIGX and was indicated to regulate calcium-dependent activities, significantly suppressed the growth of breast cancer cells. We also identified PIGX to be a core protein in an RCN1/PIGX/RCN2 complex. Microarray analysis revealed that the expression of two putative tumor suppressor genes, Zic family member 1 (ZIC1) and EH-domain containing 2 (EHD2), were upregulated commonly in cells in which PIGX, RCN1, or RCN2 was knocked down, suggesting that this RCN1/PIGX/RCN2 complex could negatively regulate the expression of these two genes and thereby contribute to human breast carcinogenesis. Our results imply that PIGX may be a good candidate molecule for development of novel anticancer drugs for breast cancer.
Keywords: breast cancer, molecular target, phosphatidylinositol glycan anchor biosynthesis class X, GPI-anchor, reticulocalbin 1, reticulocalbin 2
Introduction
Breast cancer is the most common cancer and the leading cause of cancer death among women worldwide. Global cancer statistics reported 1.7 million newly-diagnosed cases and 521,900 deaths in 2012 (1). While breast cancer incidence and mortality rates in most developing countries have been rapidly increasing (2), in developed countries, the mortality rates of breast cancer have been stable or decreasing due to early detection and development of novel treatment modalities (3–6).
Molecular targeted therapies such as tamoxifen, aromatase inhibitors, and trastuzumab (Herceptin) have markedly improved the clinical outcome of breast cancer treatment (7,8). Tamoxifen and aromatase inhibitors modulate an estrogen-related signaling pathway and trastuzumab is very effective to breast cancers with overexpression of human epidermal growth factor 2 (HER2/ERBB2) (7). Although molecular targeted drugs are available to breast cancer patients with certain molecular characteristics, a subset of patients without the dysfunction of these growth pathways is not able to have the benefit from these treatment options. For example, triple-negative breast cancers (TNBC), which do not express estrogen receptor or progesterone receptor, and do not reveal overexpression of HER2, show poorer prognosis than other subtypes for which established molecular target drugs are available (9). While clinical trials are investigating poly(ADP-ribose) polymerase (PARP) inhibitors as the most promising agents for TNBC, only a subset of patients with TNBC who carry either BRCA1 or BRCA2 mutation expects the treatment benefit (10,11). Therefore, it is required to develop additional molecular target drugs to be applicable to a wide range of breast cancer patients.
Genes upregulated specifically in cancer cells have been considered as potential molecular targets for development of targeted therapy with high efficacy and minimum risk of adverse reactions. However, to effectively develop molecular-targeted drugs, it is critically essential to characterize and understand the precise molecular mechanism how the gene products of interest contribute to development and progression of human cancers.
In this study, we demonstrate that phosphatidylinositol glycan anchor biosynthesis, class X (PIGX), which plays a critical role in the biosynthetic pathway of glycosylphosphatidylinositol (GPI)-anchor motif (12), is upregulated significantly in breast cancer tissues and involved in cancer cell proliferation. We also demonstrate that PIGX forms a complex with reticulocalbin 1 (RCN1) and reticulocalbin 2 (RCN2) proteins, which may regulate calcium-dependent activities in the endoplasmic reticulum (ER) lumen or post-ER compartment, and that the complex regulates gene expression of putative tumor suppressors, Zic family member 1 (ZIC1) and EH-domain containing 2 (EHD2), and thereby contribute to human breast cancer proliferation.
Materials and methods
Cell culture and clinical samples
Human breast cancer cell lines, BT-20, BT-549, HCC-1937, MCF7, MDA-MB-231, SK-BR3, and T-47D, as well as human embryonic kidney 293T cells and human cervical cancer HeLa cells were purchased from the American Type Culture Collection (ATCC, Rockville, MD, USA). HBC4 and HBC5 breast cancer cell lines were kindly provided by Dr T. Yamori (Molecular Pharmacology, Cancer Chemotherapy Center of the Japanese Foundation for Cancer Research). All cell lines were grown in monolayers in appropriate media supplemented with 10% fetal bovine serum and 1% antibiotic/antimycotic solution (Sigma-Aldrich, St. Louis, MO, USA): Dulbecco’s modified Eagle’s medium (D-MEM) for 293T cells; Eagle’s minimal essential medium (E-MEM) for BT-20, MCF7, and HeLa cells; RPMI-1640 for HCC-1937, HBC4, HBC5, T-47D, and BT-549 cells; Leibovitz’s L-15 for MDM-MB-231 cells; McCoy’s 5A for SK-BR3 cells. MDA-MB-231 cells were maintained at 37°C in the atmosphere of humidified air without CO2. Other cells were maintained at 37°C in humid air with 5% CO2 condition. Cells were transfected with FuGENE6 or FuGENE HD (Promega, Madison, WI, USA) following the manufacturer’s protocols. Detailed information of the clinical samples used for the microarray was described previously (13). The use of all clinical materials in this study was approved by Ethics Committees of Institute of Medical Science in the University of Tokyo.
Reverse transcription and real-time PCR
RNA was extracted from the breast cancer cell lines using RNeasy kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions. RNA extracted from a normal mammary gland (BioChain, Newark, CA, USA) was used as a control. The sequences of specific primers are 5′-GCAAATTCCATGGCACCGTC-3′ and 5′-TCGCCCCACTTGATTTTGG-3′ for GAPDH (housekeeping gene), 5′-TTGGCTTGACTCAGGATTTA-3′ and 5′-ATGCTATCACCTCCCCTGTG-3′ for ACTB (housekeeping gene), 5′-GTGAAGATGGAGAAGCCTCG-3′ and 5′-GCAC AGGATTGTAATGAGCA-3′ for PIGX, 5′-GCAGAGAGC AAGTGGAGTTTT-3′ and 5′-CACAAGAGGCAGTAAGCA GAG-3′ for phosphatidylinositol glycan anchor biosynthesis, class M (PIGM), 5′-ATGATGGGGATGGCTTTGT-3′ and 5′-AACCATAGGTGGCTTGTTTGT-3′ for RCN1, 5′-GAT GATACTGTGACTTGGGATG-3′ and 5′-ACTCAAACCGG GACCTGAA-3′ for RCN2, 5′-TGCGAGTTCACGCTTAAC ATC-3′ and 5′-ATCAGCAGCTCCTGCATTTT-3′ for EHD2, 5′-GTCCTACACGCATCCCAGTT-3′ and 5′-GCGATAAG GAGCTTGTGGTC-3′ for ZIC1, and 5′-GGTGACGTGTCTGATGTTGG-3′ and 5′-AGCTCCCAGCTGTAAGACCA-3′ for histone deacetylase 8 (HDAC8). For semi-quantitative reverse transcription PCR (RT-PCR), PCR reactions were performed as previously described (13). The amplification cycle numbers were 22 cycles for ACTB and 27 cycles for PIGX. For quantitative RT-PCR (qRT-PCR), PCR reactions were performed using SYBR Select Master mix and on ViiA 7 real-time PCR system (Life Technologies, Grand Island, NY, USA) following the manufacturer’s protocol. mRNA levels were normalized to GAPDH mRNA expression.
Construction of short hairpin RNA (shRNA)-expressing vectors and cell viability assay
Plasmids designed to express shRNA were prepared by cloning of double-stranded oligonucleotides into psiU6BX vector as described previously (14). The oligonucleotide sequences of target sequences for PIGX are 5′-GATGGAGAAGCCTCGATTG-3′ for shPIGX#1, 5′-GCCAATGGAACAAGATGAAGT-3′ for shPIGX#2, and 5′-CTACAAGTTCCAGTGGGAC-3′ for shPIGX#3. T-47D cells, which expressed PIGX at a high level, were seeded on 10-cm dishes, transfected with psiU6-PIGX or psiU6-siEGFP using FuGENE6 (Promega) according to the manufacturer’s instructions, and then cultured in RPMI-1640 containing 800 μg/ml of geneticin (Sigma-Aldrich) for 14 days. The cells were fixed with 100% methanol, stained with 0.1% of crystal violet-H2O for colony formation assay. In cell viability assay, cell viability was measured using Cell Counting Kit-8 (Dojindo Molecular Technologies, Kumamoto, Japan) 10 days after the transfection. Absorbance was measured at 450 nm as a reference, with a Microplate Reader iMark (Bio-Rad, Hercules, CA, USA). Knockdown effects of these shRNA expression vectors on endogenous PIGX expression were validated 7 days after transfection by qRT-PCR.
Small interfering RNA transfection
siRNA oligonucleotide duplexes were purchased from Sigma-Aldrich for targeting PIGX, PIGM, RCN1, and RCN2 transcripts. siEGFP and siNegative control (siNC, Cosmo Bio, Tokyo, Japan), which is a mixture of three different oligonucleotide duplexes were used as control siRNAs. The siRNA sequences are 5′-GGACAU UCCUGCAGGACUU-3′ for siPIGX, 5′-GUUCCAUCCUGA UUCAAAU-3′ for siPIGM#1, 5′-GGUUUAUAGGGCAG GCCAU-3′ for siPIGM#2, 5′-GAAGCUAACUAAAGAG GAA-3′ for siRCN1#1, 5′-GAUAGACACUCACCAGAAU-3′ for siRCN1#2, 5′-GAAUGGAUACUUGUUGAGA-3′ for siRCN2#1, and 5′-CGGAAUUUGUCAUUCAAGA-3′ for siRCN2#2.siRNA duplexes were transfected with Lipofectamine RNAiMAX (Life Technologies).
Immunoprecipitation
293T cells were lysed 48 h after transfection with Pierce IP lysis buffer (Thermo Scientific, Waltham, MA, USA) containing a complete protease inhibitor cocktail (Roche Life Science, Indianapolis, IN, USA). For FLAG-, HA-, or glutathione S-transferase (GST)-tagged protein, whole-cell extract was incubated with anti-FLAG M2 antibody conjugated agarose beads (Sigma-Aldrich), anti-HA antibody conjugated agarose beads (Sigma-Aldrich), or glutathione sepharose 4b (GE Healthcare, Pittsburgh, PA, USA) at 4°C overnight. Following three times washing with IP lysis buffer, FLAG- or HA-tagged protein bound to the beads was eluted by incubating with FLAG- or HA-peptide at 4°C for 1 h. GST-tagged protein were eluted by boiling in Lane Marker Reducing Sample Buffer (Thermo Scientific).
Mass spectrometry analysis
Immunoprecipitated samples from lysate of 293T cells transfected with mock or PIGX expression vectors were prepared in triplicate. The immunoprecipitant eluted with 3X FLAG peptide was desalted and concentrated with 2D clean-up kit (GE Healthcare). The purified protein samples were lysed in solution of 8 M urea, 50 mM HEPES-NaOH, pH 8.0 and reduced with 10 mM tris(2-carboxyethyl)phosphine (Sigma-Aldrich) at 37°C for 30 min, followed by alkylation with 50 mM iodoacetamide (Sigma-Aldrich) at 25°C in the dark for 45 min. Proteins were digested with Immobilized trypsin (Thermo Scientific) at 37°C for 6 h. The resulting peptides were desalted by Oasis HLB μ-elution plate (Waters, Milford, MA, USA) and analyzed by LTQ-Orbitrap-Velos mass spectrometer (Thermo Scientific) combined with UltiMate 3000 RSLC nano-flow HPLC system (Thermo Scientific). The MS/MS spectra were searched against Homo sapiens protein sequence database in SwissProt using Proteome Discoverer 1.4 software (Thermo Scientific), in which false discovery rate of 1% was set for both peptide and protein identification filters.
Expression vector construction
PIGX, RCN1, and RCN2 cDNAs were amplified from total human cDNA prepared by reverse transcription from qPCR Human Reference Total RNA (Clontech, Mountainview, CA, USA) using KOD DNA polymerase (Toyobo, Osaka, Japan) and cloned into pCAGGSn3FC or pCAGGSnHC vector. For pull down experiment, GST sequence was cloned together with PIGX.
Antibodies
The following primary antibodies were used: anti-HA (rat, 3F10; Roche Life Science; dilution used in immunocytochemistry (ICC): 1:1,000), anti-KDEL ER Marker (mouse, 10C3; Santa Cruz Biotechnology; dilution used in ICC: 1:50), anti-RCN1 [rabbit, A300-407A-M; Bethyl Laboratories; dilution used in ICC: 1:100, western blotting (WB): 1:1,000], anti-RCN2 (rabbit, 10193-2-AP; Proteintech; dilution used in ICC: 1:100, WB: 1:3,000), anti-FLAG (mouse, M2; Sigma-Aldrich; dilution used in WB: 1:1,000), and anti-HA (rabbit, Y-11; Santa Cruz Biotechnology; dilution used in WB: 1:1,000).
Immunocytochemistry
HeLa cells were fixed 48 h after transfection in 4% paraformaldehyde in PBS at 4°C for 1 h, permeabilized in 0.1% Triton X-100 (Sigma-Aldrich) for 3 min at room temperature and blocked with 3% BSA for 1 h at room temperature. Fixed cells were incubated with each primary antibody overnight at 4°C followed by incubation with Alexa Fluor-conjugated secondary antibody (Life Technologies) for 1 h at room temperature and observed using Leica confocal microscopy (SP5 tandem Scanner Spectral 2-Photon Confocal).
Microarray analysis
Purified RNA was labeled using 3′IVT kit (Affymetrix, Santa Clara, CA, USA) and hybridized onto Human Gene Chip U133 Plus 2.0 arrays (Affymetrix) following the manufacturer’s instructions. Probe signal intensities were normalized by MAS5 method using Expression console software (Affymetrix). Signal intensities with high detection P-value (P>0.05) were eliminated from the analysis. The signal intensities derived from the samples with siRNA treatment targeting PIGX, RCN1, or RCN2 were divided by the ones derived from control siRNA-treated sample. Each experiment was duplicated and average values were used for the analysis.
Statistical analysis
All experiments were performed as triplicate and the data are presented as means ± standard deviation (SD). Statistical significance was calculated using Student’s t-test and the level of significance was set at P<0.05.
Results
PIGX is overexpressed in breast cancer and promotes cell proliferation
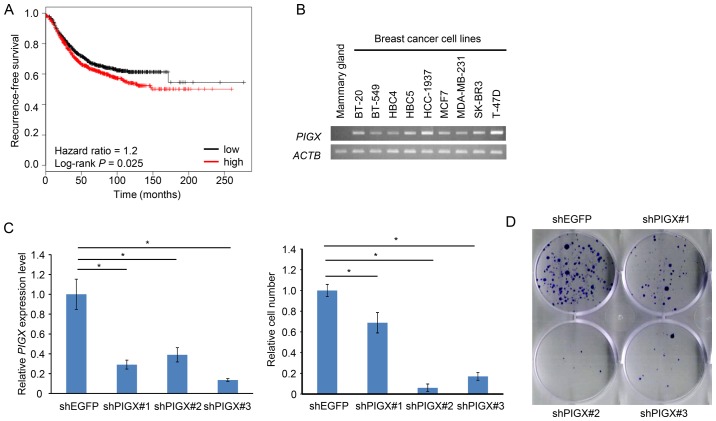
We conducted cDNA microarray analysis to explore molecular targets for development of novel drugs for breast cancer (13), and found that the expression of PIGX was significantly upregulated in breast cancer cells compared to normal breast ductal cells. The datasets from Oncomine database (15) supported the upregulation of PIGX commonly in various subtypes of breast cancer. In addition, the dataset from cBioPortal for Cancer Genomics (16,17) indicated that the amplification of PIGX gene was found in 3.7% of 963 breast cancer samples examined, suggesting that PIGX might have a critical role in human breast carcinogenesis. Furthermore, survival analysis using the KM Plotter, an online tool that incorporates microarray data with survival information (18,19), also suggested a significant association of higher PIGX mRNA expression level with shorter recurrence-free survival (RFS) of breast cancer patients (hazard ratio=1.2, P=0.025; Fig. 1A), implying that PIGX may be related to progression or malignant phenotype of breast cancer. Collectively, these data suggest that PIGX would be a candidate target for the development of novel breast cancer therapeutics.
Figure 1.
Association of PIGX expression with cancer cell growth. (A) The Kaplan-Meier curve showing the recurrence-free survival of breast cancer patients with different expression level of PIGX. Survival information and expression level of PIGX were obtained from the KM plotter (18,19). The Affymetrix ID used for analysis is: 1563111_a_at. (B) Semi-quantitative RT-PCR. PIGX expression was upregulated in various breast cancer cell lines. (C) Cell growth of T-47D with stable knockdown of PIGX was evaluated by MTT assay. All assays were performed in triplicate. The knockdown effect was validated by qRT-PCR. (D) Colony formation assay of T-47D with stable knockdown of PIGX. (E) Cell growth of T-47D with knockdown of PIGM was examined by MTT assay. The knockdown effect was validated by qRT-PCR. Results are the mean ± SD of three independent experiments and P-values were calculated with Student’s t-test (*P<0.05); NS indicates not significant.
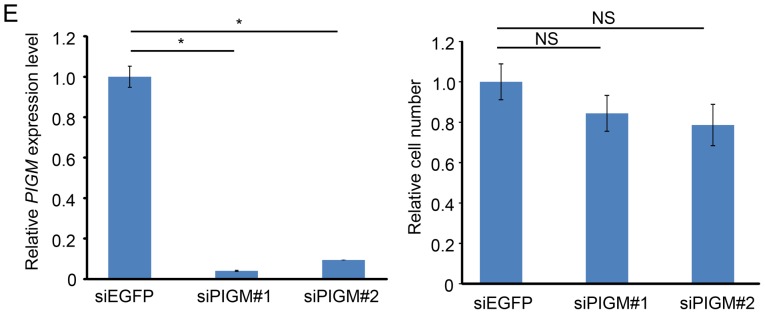
To further assess a possible oncogenic role of PIGX in human breast cancer, we firstly confirmed that PIGX was transactivated in various cell lines regardless of molecular subtypes (BT-20, BT-549, HCC-1937, and MDA-MB-231: TNBC, MCF7, T-47D, and HBC4: estrogen receptor-positive, SK-BR3 and HBC5: HER-2 type) (Fig. 1B). Since T-47D cells revealed the highest level of PIGX expression among the cell lines examined, we chose this cell line for further functional analysis. We transfected T-47D cells with shRNA-expression vector targeting PIGX and examined the growth suppressive effect on these cancer cells. Stable knockdown of PIGX by shRNA resulted in significant suppression of the cancer cell growth (Fig. 1C) and colony formation (Fig. 1D), indicating that PIGX is essential for the growth of T-47D cells. Because a previous report suggested PIGX to stabilize PIGM protein involved in the biosynthetic pathway of GPI-anchor motif (12), we also evaluated knockdown effect of PIGM on cancer cell growth. Knockdown of PIGM did not result in any significant growth suppressive effect (Fig. 1E), suggesting that PIGX is likely to promote cancer cell growth without involvement of PIGM.
RCN1 and RCN2 form a complex with PIGX
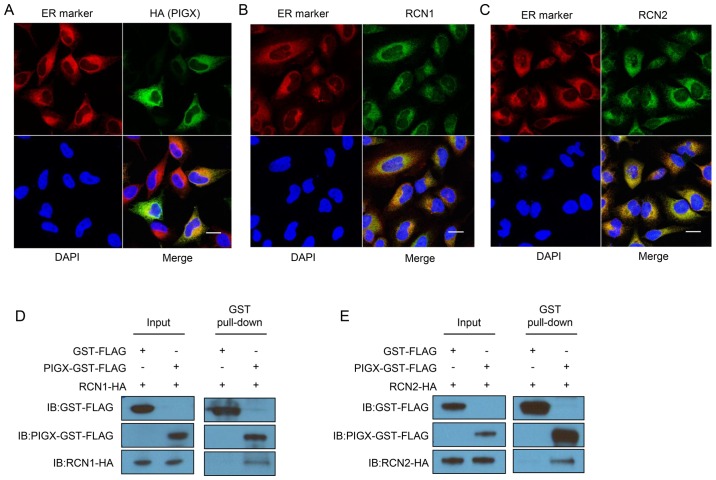
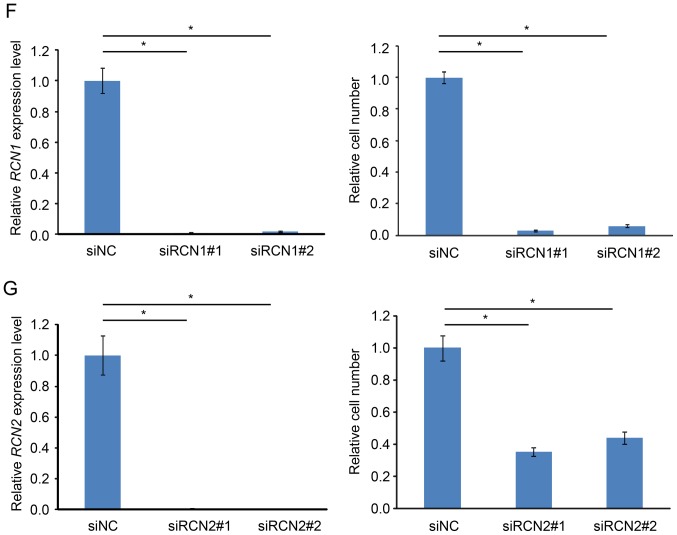
To elucidate the molecular mechanism of PIGX to promote cancer cell proliferation, we attempted to find the functional structural domain using the interproscan program (20,21), but found no characteristic domain structure to lead to speculate its biological function. Hence, we conducted immunoprecipitation followed by mass spectroscopic analysis to identify an interacting protein(s) with PIGX. We prepared samples from cell lysate of PIGX overexpressing cells or control cells, and compared the protein immunoprecipitates. We identified 51 proteins uniquely in all the triplicate samples from PIGX overexpressing cell lysate but not in control cells. Among them, since PIGX is known to be located in ER, we focused on RCN1 and RCN2 as candidate proteins interacting with PIGX. To examine the subcellular localization of these molecules, we transfected HeLa cells with each expression vector and performed immunocytochemical analysis. As shown in Fig. 2A–C, PIGX, RCN1, and RCN2 were localized in ER. Besides, we confirmed the interaction between GST-tagged PIGX and HA-tagged RCN1 or RCN2 by GST-pull down experiment and western blotting (Fig. 2D and E). Importantly, knockdown of RCN1 and RCN2 also significantly suppressed cancer cell growth (Fig. 2F and G). Moreover, survival analysis using the KM Plotter showed that higher RCN1 and RCN2 expression is significantly associated with shorter RFS of breast cancer patients (hazard ratio=1.22, P=0.00072 for RCN1 and hazard ratio=1.33, P=8.8×10−7 for RCN2) (18,19). Altogether these results indicate that the RCN1/PIGX/RCN2 complex has an indispensable role in the growth or survival of cancer cells.
Figure 2.
PIGX and RCN proteins interact with each other in ER. (A) Immunocytochemical analysis of HeLa cells transfected with PIGX expression vector. Signal of exogenous PIGX was merged with that of ER marker, KDEL. Scale bar, 20 μm. (B and C) Immunocytochemical analysis of HeLa cells for RCN1 (B) or RCN2 (C). Endogenous RCN1 or RCN2 was detected and merged with ER marker. Scale bar, 20 μm. (D and E) Co-immunoprecipitation analysis. GST-FLAG tagged PIGX was co-overexpressed with HA-tagged RCN1 or RCN2 and pulled down using GST-tag. (F and G) Cell growth of T-47D with knockdown of RCN1 (F) and RCN2 (G) was examined by MTT assay. The knockdown effect was validated by qRT-PCR. Results are the mean ± SD of three independent experiments and P-values were calculated with Student’s t-test (*P<0.05).
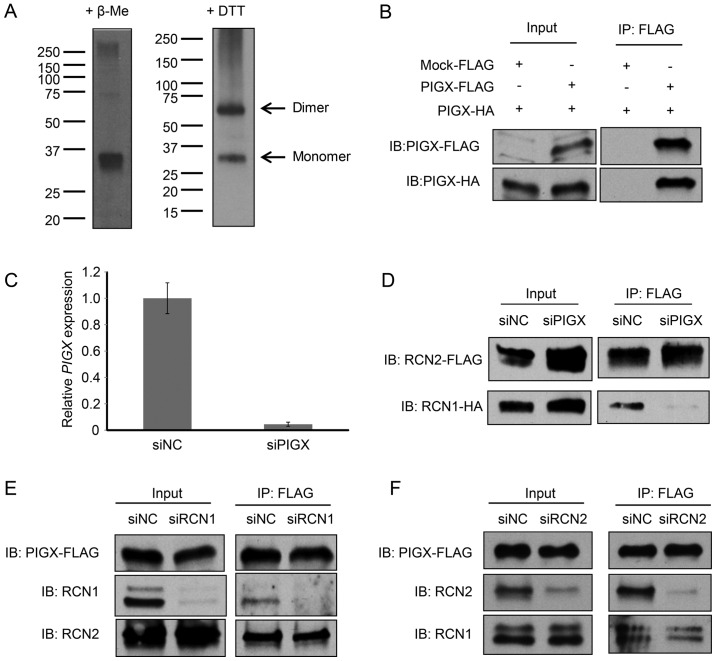
In addition, since we observed two bands in western blotting experiments for PIGX in a mild reducing condition (+DTT) and the molecular weight of a higher band corresponded to the dimer (58 kDa) of PIGX (Fig. 3A), we hypothesized that PIGX would make a dimer possibly through intermolecular disulfide bonds. To address this hypothesis, we co-overexpressed two different PIGX proteins, one with a HA-tag and the other with a FLAG-tag, in 293T cells and conducted immunoprecipitation analysis. As shown in Fig. 3B, HA-tagged PIGX was co-immunoprecipitated with FLAG-tagged PIGX, indicating that PIGX form a dimer. Subsequently, we conducted co-immunoprecipitation analysis with either PIGX, RCN1, or RCN2 knockdown to clarify the complex structure. The results indicated that the binding between RCN1 and RCN2 was diminished by knockdown of PIGX (Fig. 3C and D) whereas knockdown of either RCN1 or RCN2 did not affect the binding of the other RCN protein to PIGX (Fig. 3E and F). Together, these results indicate RCN1 and RCN2 bind directly to PIGX and probably PIGX protein functions as a core protein of this RCN1/PIGX/RCN2 complex although further biochemical analysis of the complex is required to determine the precise molecular stoichiometry in the complex.
Figure 3.
PIGX functions as a core in the complex with RCN1 and RCN2. (A) Western blot analysis of overexpressed PIGX in two different reducing conditions (+β-Me and +DTT). Two different sized bands were observed in mild reducing condition with DTT. β-Me, β-mercaptoethanol; DTT, dithiothreitol. (B) Co-immunoprecipitation analysis of PIGX multimer formation. FLAG- or HA-tagged PIGX was co-overexpressed in 293T cells and immunoprecipitated using FLAG tag. IP, immunoprecipitation; IB, immunoblotting. (C and D) Co-immunoprecipitation analysis of the interaction between RCN1 and RCN2 with PIGX knockdown. Knockdown effect was validated by qRT-PCR. siNC, siNegative control. (E and F) Co-immunoprecipitation analysis between PIGX and RCN1 or RCN2 with knockdown of the other RCN protein. FLAG-tagged PIGX was overexpressed in 293T cells and interaction with endogenous RCN1 or RCN2 were analyzed.
PIGX containing complex regulates EHD2 and ZIC1 transcription
Although previous studies reported that RCN1 and RCN2 might be involved in human cancer (22–24), the detailed molecular function of the proteins how to contribute to cancer remains unclear. Therefore, we conducted cDNA microarray analysis to assess the transcriptional changes caused by the knockdown of each of these three genes. We transfected BT-549 cells with control siRNA or siRNAs targeting PIGX, RCN1, or RCN2 and subjected RNA sample extraction from cells 48 h after transfection. Since PIGX, RCN1, and RCN2 are considered to function as a complex, we explored gene expression changes observed commonly in all knockdown samples. The result revealed that significant upregulation of 17 genes by >2-fold was observed by siRNA treatment targeting PIGX, RCN1, and RCN2 compared to control samples (Fig. 4A), indicating that the RCN1/PIGX/RCN2 complex would regulate the expression of these genes. Among these genes, ZIC1 and EHD2 were recently suggested to have a function related to human tumorigenesis (25–30). The upregulation of these potential downstream genes was validated with qRT-PCR (Fig. 4B and C). Taken together with previous reports that show ZIC1 and EHD2 would have functions as putative tumor suppressors (25–28), our results suggest that the PIGX-containing complex may promote cancer cell proliferation through, at least in part, the modulation of expression of these two genes and thereby contribute to human breast cancer.
Figure 4.
The complex of PIGX, RCN1, and RCN2 regulates gene expression. (A) Heatmap of upregulated genes by PIGX, RCN1, or RCN2 knockdown in common. Gene expression profiles were analyzed by cDNA microarray using Affymetrix HG-U133 Plus 2.0 Array. EHD2 and ZIC1, putative tumor suppressor genes are highlighted in blue. (B and C) Upregulation of EHD2 and ZIC1 was validated by qRT-PCR. Results are the mean ± SD of three independent experiments and P-values were calculated with Student’s t-test (*P<0.05).
Discussion
We have demonstrated that PIGX is overexpressed in breast cancer and plays a critical role in cancer cell proliferation. PIGX was identified as a component of a GPI mannosyltransferase I complex, which is essential to transfer mannoses to GPI-anchor precursors in the biosynthesis pathway of GPI-anchor, and its function is suggested to stabilize PIGM, a catalytic component of the complex (12). GPI-anchoring is known as a post-translational glycolipid modification that synthesizes through a multi-step biosynthesis pathway in the ER (31). GPI-anchored proteins are mostly localized to plasma membrane and involved in many biological phenomena such as cell-cell interaction, signal transduction and immune recognition. Abnormalities of GPI-anchoring and elevated expression levels of GPI-anchored proteins have been found in cancer (32,33); overexpression of PIGT and GPAA1, subunits of GPI transamidase that functions in another step of the biosynthesis pathway, plays important roles in tumorigenesis (34). These findings implied that overexpressed PIGX might contribute to development/progression of breast cancer through the activation of the GPI-anchor biosynthesis pathway. Indeed, our result suggested that knockdown of PIGX suppressed breast cancer cell proliferation indicating that PIGX expression is indispensable for the growth or survival of cancer cells. However, knockdown of PIGM, the interacting protein of PIGX, did not cause any growth-suppressive effect on cancer cell growth (Fig. 1E) in this study, suggesting that the PIGX-PIGM interaction might not be a key factor for growth enhancement of breast cancer cells and an interaction with another binding protein(s) could be important in mammary tumorigenesis.
Through the LC-MS/MS analysis, we identified two reticulocalbin (RCN) family proteins, RCN1 and RCN2, which are ER-located calcium-binding proteins possessing EF-hand motifs (35,36), as novel binding partners of PIGX. Although the fundamental roles of RCNs and the functional difference between RCN1 and RCN2 are still unclear, RCN1 is reported to be overexpressed in multiple types of human cancer including breast cancer and was implicated to enhance invasiveness of breast cancer cells (23,37). A previous study indicated that RCN2 is a tumor-associated antigen, whose expression level linearly increased according to the increase of the breast tumor size. In addition, our cell growth analysis using siRNAs targeting RCN1/RCN2 and survival analysis employing public datasets indicate the RCN1/PIGX/RCN2 complex has an indispensable role relating to cancer cell proliferation. Furthermore, our gene expression profile analysis after knocking down of PIGX, RCN1, or RCN2 indicated that the RCN1/PIGX/RCN2 complex deregulates the expression of ZIC1 and EHD2, which are suggested to function as tumor suppressors.
ZIC1 is a zinc finger transcription factor and is known to be involved in vital developmental processes including neural development and body pattern formation through the regulation of a variety of signaling pathways (38–41). ZIC1 was down-regulated in several types of cancer through hypermethylation in its promoter region (27,42,43). Furthermore, Zhong et al showed that ectopic expression of ZIC1 causes the decrease of AKT and ERK1/2 phosphorylation levels, indicating that ZIC1 would serve a tumor suppressive function through regulation of the PI3K-MAPK pathway (28). EHD2, another candidate target gene of the RCN1/PIGX/RCN2 complex, belongs to EH domain-containing protein family and previously reported to be involved in a variety of biological processes such as membrane repair (44), endocytic trafficking (45,46), and myoblast fusion (47). A recent study showed that the EHD2 was downregulated in breast cancer cells and possesses important functions in regulating cancer cell proliferation, migration, and invasion (26). In the Oncomine database (15), downregulation of EHD2 in breast cancer tissues is also reported. In addition, the expression levels of both ZIC1 and EHD2 were positively correlated with that of E-cadherin, downregulation of which is indicated as a hallmark of epithelial-mesenchymal transition (EMT) (25,29). Thus, the RCN1/PIGX/RCN2 complex may contribute to human breast tumorigenesis through not only enhancing cancer cell proliferation, but also promoting EMT by downregulating ZIC1 and EHD2 leading to reduction of E-cadherin expression.
Intriguingly, knockdown of PIGX, RCN1, and RCN2 resulted in upregulation of HDAC8. Given that HDAC8 is suggested to have an oncogenic function and considered as a promising therapeutic target for cancer (30), we assume that HDAC8 upregulation was caused as some kind of feedback mechanism to protect cancer cells from anti-growth inhibitory effect induced by knockdown of PIGX, RCN1, and RCN2.
In conclusion, we identified that PIGX was overexpressed in breast cancer clinical samples as well as breast cancer cell lines, and that PIGX interacted with RCN family member, RCN1 and RCN2, and regulated the expression of tumor suppressors ZIC1 and EHD2. Since several studies reported that ZIC1 negatively regulates the AKT/PI3K pathway, RCN1/PIGX/RCN2 complex may contribute to mammary tumorigenesis through the suppression of ZIC1. Although further analysis is required, our data shed light on the biological role of PIGX in mammary tumorigenesis and suggest that PIGX may be a good candidate molecule for the development of novel anticancer drugs for breast cancer.
Acknowledgements
We thank Dr Ryuji Hamamoto for helpful discussion and critical reading of the manuscript. We also thank the members of Nakamura laboratory for helpful discussion. This study was supported partly by the funding from OncoTherapy Science, Inc. Y. Nakamura is a stock holder and a scientific advisor of OncoTherapy Science, Inc.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Center MM, DeSantis C, Ward EM. Global patterns of cancer incidence and mortality rates and trends. Cancer Epidemiol Biomarkers Prev. 2010;19:1893–1907. doi: 10.1158/1055-9965.EPI-10-0437. [DOI] [PubMed] [Google Scholar]
- 3.Althuis MD, Dozier JM, Anderson WF, Devesa SS, Brinton LA. Global trends in breast cancer incidence and mortality 1973–1997. Int J Epidemiol. 2005;34:405–412. doi: 10.1093/ije/dyh414. [DOI] [PubMed] [Google Scholar]
- 4.Canfell K, Banks E, Moa AM, Beral V. Decrease in breast cancer incidence following a rapid fall in use of hormone replacement therapy in Australia. Med J Aust. 2008;188:641–644. doi: 10.5694/j.1326-5377.2008.tb01821.x. [DOI] [PubMed] [Google Scholar]
- 5.Parkin DM. Is the recent fall in incidence of post-menopausal breast cancer in UK related to changes in use of hormone replacement therapy? Eur J Cancer. 2009;45:1649–1653. doi: 10.1016/j.ejca.2009.01.016. [DOI] [PubMed] [Google Scholar]
- 6.Séradour B, Allemand H, Weill A, Ricordeau P. Changes by age in breast cancer incidence, mammography screening and hormone therapy use in France from 2000 to 2006. Bull Cancer. 2009;96:E1–E6. doi: 10.1684/bdc.2009.0869. [DOI] [PubMed] [Google Scholar]
- 7.Schlotter CM, Vogt U, Allgayer H, Brandt B. Molecular targeted therapies for breast cancer treatment. Breast Cancer Res. 2008;10:211. doi: 10.1186/bcr2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gasparini G, Longo R, Torino F, Morabito A. Therapy of breast cancer with molecular targeting agents. Ann Oncol. 2005;16(Suppl 4):iv28–iv36. doi: 10.1093/annonc/mdi905. [DOI] [PubMed] [Google Scholar]
- 9.Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N Engl J Med. 2010;363:1938–1948. doi: 10.1056/NEJMra1001389. [DOI] [PubMed] [Google Scholar]
- 10.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 11.Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, Mortimer P, Swaisland H, Lau A, O’Connor MJ, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361:123–134. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 12.Ashida H, Hong Y, Murakami Y, Shishioh N, Sugimoto N, Kim YU, Maeda Y, Kinoshita T. Mammalian PIG-X and yeast Pbn1p are the essential components of glycosylphosphatidylinositol-mannosyltransferase I. Mol Biol Cell. 2005;16:1439–1448. doi: 10.1091/mbc.E04-09-0802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nishidate T, Katagiri T, Lin ML, Mano Y, Miki Y, Kasumi F, Yoshimoto M, Tsunoda T, Hirata K, Nakamura Y. Genome-wide gene-expression profiles of breast-cancer cells purified with laser microbeam microdissection: Identification of genes associated with progression and metastasis. Int J Oncol. 2004;25:797–819. [PubMed] [Google Scholar]
- 14.Tamura K, Furihata M, Tsunoda T, Ashida S, Takata R, Obara W, Yoshioka H, Daigo Y, Nasu Y, Kumon H, et al. Molecular features of hormone-refractory prostate cancer cells by genome-wide gene expression profiles. Cancer Res. 2007;67:5117–5125. doi: 10.1158/0008-5472.CAN-06-4040. [DOI] [PubMed] [Google Scholar]
- 15.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: A cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/S1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBio-Portal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Györffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, Szallasi Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123:725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 19.Győrffy B, Surowiak P, Budczies J, Lánczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One. 2013;8:e82241. doi: 10.1371/journal.pone.0082241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goujon M, McWilliam H, Li W, Valentin F, Squizzato S, Paern J, Lopez R. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 2010;38(Web Server):W695–W699. doi: 10.1093/nar/gkq313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zdobnov EM, Apweiler R. InterProScan - an integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001;17:847–848. doi: 10.1093/bioinformatics/17.9.847. [DOI] [PubMed] [Google Scholar]
- 22.Giribaldi G, Barbero G, Mandili G, Daniele L, Khadjavi A, Notarpietro A, Ulliers D, Prato M, Minero VG, Battaglia A, et al. Proteomic identification of Reticulocalbin 1 as potential tumor marker in renal cell carcinoma. J Proteomics. 2013;91:385–392. doi: 10.1016/j.jprot.2013.07.018. [DOI] [PubMed] [Google Scholar]
- 23.Liu Z, Brattain MG, Appert H. Differential display of reticulocalbin in the highly invasive cell line, MDA-MB-435, versus the poorly invasive cell line, MCF-7. Biochem Biophys Res Commun. 1997;231:283–289. doi: 10.1006/bbrc.1997.6083. [DOI] [PubMed] [Google Scholar]
- 24.Chen JJ, Reid CE, Band V, Androphy EJ. Interaction of papillomavirus E6 oncoproteins with a putative calcium-binding protein. Science. 1995;269:529–531. doi: 10.1126/science.7624774. [DOI] [PubMed] [Google Scholar]
- 25.Shi Y, Liu X, Sun Y, Wu D, Qiu A, Cheng H, Wu C, Wang X. Decreased expression and prognostic role of EHD2 in human breast carcinoma: Correlation with E-cadherin. J Mol Histol. 2015;46:221–231. doi: 10.1007/s10735-015-9614-7. [DOI] [PubMed] [Google Scholar]
- 26.Yang X, Ren H, Yao L, Chen X, He A. Role of EHD2 in migration and invasion of human breast cancer cells. Tumour Biol. 2015;36:3717–3726. doi: 10.1007/s13277-014-3011-9. [DOI] [PubMed] [Google Scholar]
- 27.Gan L, Chen S, Zhong J, Wang X, Lam EK, Liu X, Zhang J, Zhou T, Yu J, Si J, et al. ZIC1 is downregulated through promoter hypermethylation, and functions as a tumor suppressor gene in colorectal cancer. PLoS One. 2011;6:e16916. doi: 10.1371/journal.pone.0016916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhong J, Chen S, Xue M, Du Q, Cai J, Jin H, Si J, Wang L. ZIC1 modulates cell-cycle distributions and cell migration through regulation of sonic hedgehog, PI(3)K and MAPK signaling pathways in gastric cancer. BMC Cancer. 2012;12:290. doi: 10.1186/1471-2407-12-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qiang W, Zhao Y, Yang Q, Liu W, Guan H, Lv S, Ji M, Shi B, Hou P. ZIC1 is a putative tumor suppressor in thyroid cancer by modulating major signaling pathways and transcription factor FOXO3a. J Clin Endocrinol Metab. 2014;99:E1163–E1172. doi: 10.1210/jc.2013-3729. [DOI] [PubMed] [Google Scholar]
- 30.Chakrabarti A, Oehme I, Witt O, Oliveira G, Sippl W, Romier C, Pierce RJ, Jung M. HDAC8: A multifaceted target for therapeutic interventions. Trends Pharmacol Sci. 2015;36:481–492. doi: 10.1016/j.tips.2015.04.013. [DOI] [PubMed] [Google Scholar]
- 31.Ferguson MAJ, Kinoshita T, Hart GW. Glycosylphosphatidylinositol anchors. In: Varki A, Cummings RD, Esko JD, editors. Essentials of Glycobiology. Cold Spring Harbor, NY: 2009. [PubMed] [Google Scholar]
- 32.Gamage DG, Hendrickson TL. GPI transamidase and GPI anchored proteins: Oncogenes and biomarkers for cancer. Crit Rev Biochem Mol Biol. 2013;48:446–464. doi: 10.3109/10409238.2013.831024. [DOI] [PubMed] [Google Scholar]
- 33.Zhao P, Nairn AV, Hester S, Moremen KW, O’Regan RM, Oprea G, Wells L, Pierce M, Abbott KL. Proteomic identification of glycosylphosphatidylinositol anchor-dependent membrane proteins elevated in breast carcinoma. J Biol Chem. 2012;287:25230–25240. doi: 10.1074/jbc.M112.339465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu G, Guo Z, Chatterjee A, Huang X, Rubin E, Wu F, Mambo E, Chang X, Osada M, Sook Kim M, et al. Overexpression of glycosylphosphatidylinositol (GPI) transamidase subunits phosphatidylinositol glycan class T and/or GPI anchor attachment 1 induces tumorigenesis and contributes to invasion in human breast cancer. Cancer Res. 2006;66:9829–9836. doi: 10.1158/0008-5472.CAN-06-0506. [DOI] [PubMed] [Google Scholar]
- 35.Ozawa M, Muramatsu T. Reticulocalbin, a novel endoplasmic reticulum resident Ca(2+)-binding protein with multiple EF-hand motifs and a carboxyl-terminal HDEL sequence. J Biol Chem. 1993;268:699–705. [PubMed] [Google Scholar]
- 36.Weis K, Griffiths G, Lamond AI. The endoplasmic reticulum calcium-binding protein of 55 kDa is a novel EF-hand protein retained in the endoplasmic reticulum by a carboxyl-terminal His-Asp-Glu-Leu motif. J Biol Chem. 1994;269:19142–19150. [PubMed] [Google Scholar]
- 37.Yu LR, Zeng R, Shao XX, Wang N, Xu YH, Xia QC. Identification of differentially expressed proteins between human hepatoma and normal liver cell lines by two-dimensional electrophoresis and liquid chromatography-ion trap mass spectrometry. Electrophoresis. 2000;21:3058–3068. doi: 10.1002/1522-2683(20000801)21:14<3058::AID-ELPS3058>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 38.Aruga J. The role of Zic genes in neural development. Mol Cell Neurosci. 2004;26:205–221. doi: 10.1016/j.mcn.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 39.Nagai T, Aruga J, Takada S, Günther T, Spörle R, Schughart K, Mikoshiba K. The expression of the mouse Zic1, Zic2, and Zic3 gene suggests an essential role for Zic genes in body pattern formation. Dev Biol. 1997;182:299–313. doi: 10.1006/dbio.1996.8449. [DOI] [PubMed] [Google Scholar]
- 40.Merzdorf CS, Sive HL. The zic1 gene is an activator of Wnt signaling. Int J Dev Biol. 2006;50:611–617. doi: 10.1387/ijdb.052110cm. [DOI] [PubMed] [Google Scholar]
- 41.Maurus D, Harris WA. Zic-associated holoprosencephaly: Zebrafish Zic1 controls midline formation and forebrain patterning by regulating Nodal, Hedgehog, and retinoic acid signaling. Genes Dev. 2009;23:1461–1473. doi: 10.1101/gad.517009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang LJ, Jin HC, Wang X, Lam EK, Zhang JB, Liu X, Chan FK, Si JM, Sung JJ. ZIC1 is downregulated through promoter hypermethylation in gastric cancer. Biochem Biophys Res Commun. 2009;379:959–963. doi: 10.1016/j.bbrc.2008.12.180. [DOI] [PubMed] [Google Scholar]
- 43.Wang YY, Jiang JX, Ma H, Han J, Sun ZY, Liu ZM, Xu ZG. Role of ZIC1 methylation in hepatocellular carcinoma and its clinical significance. Tumour Biol. 2014;35:7429–7433. doi: 10.1007/s13277-014-1971-4. [DOI] [PubMed] [Google Scholar]
- 44.Marg A, Schoewel V, Timmel T, Schulze A, Shah C, Daumke O, Spuler S. Sarcolemmal repair is a slow process and includes EHD2. Traffic. 2012;13:1286–1294. doi: 10.1111/j.1600-0854.2012.01386.x. [DOI] [PubMed] [Google Scholar]
- 45.Benjamin S, Weidberg H, Rapaport D, Pekar O, Nudelman M, Segal D, Hirschberg K, Katzav S, Ehrlich M, Horowitz M. EHD2 mediates trafficking from the plasma membrane by modulating Rac1 activity. Biochem J. 2011;439:433–442. doi: 10.1042/BJ20111010. [DOI] [PubMed] [Google Scholar]
- 46.Naslavsky N, Caplan S. C-terminal EH-domain-containing proteins: Consensus for a role in endocytic trafficking, EH? J Cell Sci. 2005;118:4093–4101. doi: 10.1242/jcs.02595. [DOI] [PubMed] [Google Scholar]
- 47.Posey AD, Jr, Pytel P, Gardikiotes K, Demonbreun AR, Rainey M, George M, Band H, McNally EM. Endocytic recycling proteins EHD1 and EHD2 interact with fer-1-like-5 (Fer1L5) and mediate myoblast fusion. J Biol Chem. 2011;286:7379–7388. doi: 10.1074/jbc.M110.157222. [DOI] [PMC free article] [PubMed] [Google Scholar]