Abstract
Periodontitis is an inflammatory disease caused by subgingival microorganisms and their components, such as lipopolysaccharide (LPS). Responses of the host to LPS are mediated by CD14 and LPS-binding protein (LBP). In this study, it was determined that proteases from a periodontal pathogen, Prevotella intermedia, cleave CD14 and LBP, and thereby modulate the virulence of LPS. Culture supernatants from two strains of P. intermedia (ATCC 25611 and 25261) cleaved CD14 and LBP in a concentration-dependent manner. Zymographic and molecular mass analysis revealed the presence of a membrane-associated, 170-kDa, monomeric protease. Class-specific inhibitors and stimulators demonstrated that this enzyme is a metal-requiring, thiol-activated, cysteine protease. The protease was stable over a wide range of temperatures (4–56 °C) and pH values (4.5–8.5). This enzyme also decreased the expression of interleukin-1β (IL-1β)-specific mRNA in the LPS-activated macrophage-like cell lines U937 and THP-1 in a concentration-dependent manner, indicating that it also cleaves membrane-associated CD14. Furthermore, addition of soluble CD14 abrogated protease-mediated inhibition of IL-1 mRNA expression induced by LPS. The observations suggest that proteolysis of CD14 and LBP by P. intermedia protease might modulate the virulence of LPS at sites of periodontal infections.
Keywords: Prevotella intermedia, Cysteine protease, Lipopolysaccharide, Lipopolysaccharide-binding protein, CD14, Periodontitis
Introduction
Periodontitis is an inflammatory process caused by pathogenic microorganisms in the subgingival biofilm, e.g. Porphyromonas gingivalis and Prevotella intermedia (AAP 1996). Subgingival bacteria and their components, e.g. lipopolysaccharide (LPS), induce periodontal breakdown either directly, by releasing tissue-degrading enzymes and toxins, or indirectly, by activating host cells to secrete proinflammatory and catabolic mediators (Gemmell et al. 1997). LPS is a component of the outer membrane of gram-negative bacteria and exerts its stimulatory effects on host cells by binding to a receptor complex comprised of CD14, Toll-like receptor, and other cell surface molecules (Heumann and Roger 2002). LPS binding to CD14 is strongly enhanced in the presence of LPS-binding protein (LBP), which transforms LPS aggregates into monomers and then transfers them to CD14.
CD14 exists as membrane-bound (mCD14) and soluble (sCD14) forms. Soluble CD14 has been implicated in mediating LPS-induced activation of mCD14-negative cells (Pugin et al. 1993). However, it has also been suggested that sCD14 can suppress LPS/LBP-induced activation of mCD14-positive cells by competing with mCD14 for LPS binding (Rokita and Menzel 1997). In addition, sCD14 facilitates the transfer of LPS to lipoproteins, which also results in neutralization of LPS (Wurfel et al. 1994). This shows that LPS responses are tightly controlled by sCD14 and LBP. In addition to LPS, periodontopathogens show a variety of other virulence determinants to avoid host defense mechanisms (Kadowaki et al. 2000). Although very little is known regarding the virulence factors of P. intermedia, a number of membrane-associated endopeptidases are suggested to play important roles in its invasion and colonization. These enzymes are either serine or cysteine proteases and are capable of degrading physiologically significant proteins, e.g. immunoglobulins (Jansen et al. 1997). It has recently been shown that arginine-specific cysteine proteases (gingipains) from P. gingivalis can cleave human monocyte CD14 (Sugawara et al. 2000). The aim of this study was to determine whether proteases from P. intermedia are capable of cleaving CD14 and/or LBP. We show that P. intermedia synthesizes a cysteine protease that cleaves CD14 and LBP dose-dependently. Our findings indicate that this protease might modulate LPS virulence via proteolysis of local CD14 and LBP in periodontal infections.
Materials and methods
Assessment of the sCD14 and LBP proteolytic activity in P. intermedia
Soluble CD14 or LBP (10 μg in 20 μl PBS) were labeled with 0.5 mCi of Na125I using iodogen reagent according to the manufacturer’s recommended protocols. The labeled proteins were analyzed by SDS-PAGE (10% polyacrylamide) (Laemmli 1970) to ascertain the purity and radioiodination. P. intermedia strains ATCC 25261 and ATCC 25611 were grown anaerobically in medium 593 supplemented with 0.05% cysteine. P. intermedia cultures were scraped from agarose slants, transferred to MEM, and incubated at 22 °C for 30 min at a concentration of 108 bacteria/ml medium. Thereafter, bacteria were centrifuged at 3,000×g for 10 min, and the protease activity associated with bacteria or supernatants was analyzed by incubating radioiodinated sCD14 and LBP. Ten μl of 125I-sCD14 (8,000 cpm/ng) or 125I-LBP (6,000 cpm/ng) was incubated with various numbers of P. intermedia cells in a final volume of 20 μl MEM or with supernatants from different numbers of cells in 20 μl MEM.
Following proteolysis of 125I-sCD14 or 125I-LBP, the reaction mixture was subjected to SDS-PAGE (10% polyacrylamide) analysis. Subsequently, the gels were washed, fixed, stained, dried, and autoradiographed by exposure to Reflection NEF 496 film. Additionally, 125I-sCD14 or 125I-LBP bands and their digestion products were cut from each lane of the gels and counted in an LKB-Wallace 1274 γ-counter for quantitative analysis of enzymatic activity.
Identification of the P. intermedia enzyme responsible for proteolysis of sCD14
The protease responsible for cleavage of sCD14 was characterized as described by Lantz et al. (1991), with minor modifications. P. intermedia (107 cells) suspended in MEM (100 μl) were solubilized in solubilization buffer containing 1% SDS (Laemmli 1970), and the proteins were separated on SDS-PAGE (10% polyacrylamide). The gels were then thoroughly washed with PBS and soaked in 125I-sCD14 (100 ng in 2 ml MEM) or 125I-LBP (100 ng in 2 ml MEM) at 4 °C for 14 h. Subsequently, the gels were briefly washed with PBS at 4 °C, and the molecular mass of the protein that cleaved 125I-sCD14 and 125I-LBP was visualized by autoradiography using Reflection NEF 496 film.
Isolation of CD14 protease from P. intermedia
P. intermedia cells (109 cells/ml) were subjected to lysis with 0.8% deoxycholate and 1.6% NP-40 in buffer A (50 mM HEPES, pH 7.0) containing protease inhibitors (0.5 mM EDTA, 0.25 mM phenylmethyl sulfonyl fluoride, 10 μg leupeptin, 10 μg aprotinin and 2.5 μg benzamidine) and incubated on ice for 15 min. Subsequently, the protein extract was centrifuged at 10,000×g for 10 min at 4 °C. The supernatant was then loaded on a Mono-Q ion-exchange column (Pharmacia, Piscataway, N.J., USA) pre-equilibrated in buffer A. The protease was eluted with a linear concentration gradient of 1 M NaCl prepared in buffer A, and collected in 0.5-ml fractions. Subsequently, each fraction was reacted with 125I-sCD14. Fractions containing proteolytic activity (assessed as described below) were pooled, desalted on Sephadex Quick-spin columns, and immediately frozen on dry ice for concentration. A volume of 200 μl of the pooled concentrated sCD14 protease was then purified sequentially on a Superose 12 and then on an arginyl-Sepharose 4B affinity column pre-equilibrated with 25 mM Tris-HCl, 5 mM CaCl2, 150 mM NaCl, pH 7.5 (Chen et al. 1992). After extensive washing, the purified enzyme was eluted with 50 mM acetate buffer, 5 mM CaCl2, pH 4.5. The fractions were neutralized, reanalyzed for the 125I-sCD14 protease activity, pooled, concentrated, and frozen at −70 °C for further analysis. The molecular mass of the 125I-sCD14 protease was assessed by SDS-PAGE (10% polyacrylamide) followed by silver staining. The presence of LPS in protease preparations was assessed by the limulus amoebocyte lysate assay. Endotoxin levels were below the detection limits in all enzyme preparations.
The activity of the purified enzyme was assessed by its amidolytic activity using Bz-Arg-pNA as a substrate. Briefly, various concentrations of P. intermedia enzyme were added to Bz-Arg-pNA (1.0 mM) in 0.2 M Tris HCl, 5 mM CaCl2, pH 7.5 at 25 °C. The specific activity of the enzyme was expressed as 1 U=ΔA405 nm min−1 ml−1 at 25 °C (Chen et al. 1992).
Characterization of enzymatic properties
To examine the proteolytic activity of the P. intermedia protease, the enzyme was preincubated for 20 min with various concentrations (1–20 μM) of inhibitors, Phe-Pro-Arg-chloromethyl ketone (FPR-cmk) or benzyloxycarbonyl-Phe-Lys-chloromethyl ketone (ZFK-cmk) in 0.1 ml of buffer B (200 mM Tris-HCl, 5 mM CaCl2, pH 7.5). Subsequently, 125I-sCD14 (80,000 cpm) in 10 μl buffer B were added, and the reaction mixture was further incubated at 37 °C for 10 min. The proteolysis of 125I-sCD14 was analyzed by SDS-PAGE (10% polyacrylamide) and autoradiography as described above. For quantitative analysis, the bands were cut from the gels and counted in a LKB-Wallace 1274 γ-counter.
To characterize its enzymatic properties, a total of 2 U of protease in 10 μl buffer B was subjected to various protease modifiers for 10 min. Subsequently, 125I-sCD14 (80,000 CPM in 10 μl buffer B) was added to the reaction mixture and further incubated for 15 min. At the end of incubation, the reaction was stopped by the addition of an equal volume of solubilization buffer (Laemmli 1967), and the reaction mixture was heated at 100 °C for 3 min. The extent of 125I-sCD14 hydrolysis was assessed by SDS-PAGE (10% polyacrylamide) followed by autoradiography. For quantitative analysis, individual bands were analyzed by γ-counting as described above.
To examine the effect of human serum, 125I-sCD14 (80,000 cpm in 10 μl buffer B) was incubated in the presence and absence of 10% human serum (v/v) for 10 min at 37 °C. Subsequently, P. intermedia protease (0.2 U) was added to the reaction mixture and further incubated at 37 °C for various time intervals. The reaction was terminated by addition of equal volumes of solubilization buffer for analysis, as described above.
To examine enzymatic stability, P. intermedia protease (0.2 U) was incubated at 37 °C in the presence of 125I-sCD14 (80,000 cpm in 10 μl buffer B) for 0–120 min. At the end of a required time interval, the reaction was stopped by addition of equal volume of the solubilization buffer, and the extent of 125I-sCD14 proteolysis at each time point was assessed as described above. To examine the effect of different pH values on the enzymatic activity, 0.2 U of enzyme was suspended in 5 μl of Tris-buffered saline (TBS) (1.45 mM NaCl, 100 mM Tris pH 7.5). This was added to reaction mixtures containing 125I-sCD14 (80,000 cpm) in a final volume of 20 μl TBS with pH values ranging between 2.5 and 9.5 (adjusted with 100 mM Tris base or 0.1 N HCl). The reaction mixtures were then incubated at 37 °C for 10 min and the proteolysis of 125I-sCD14 assessed as described above. To examine the thermal stability of the enzyme, 0.2 U of enzyme were suspended in 10 μl of buffer B and reacted with 125I-sCD14 (80,000 cpm in 10 μl buffer B) for 10 min. Thereafter, the reaction mixtures were incubated at various temperatures (0–56 °C) for 10 min. The cleavage of 125I-sCD14 was quantitatively analyzed as described above. All experiments were done at least three times. Mean values and standard error of the mean were calculated for each experiment.
Effect of P. intermedia protease on the LPS-responsiveness of cells
U937 or THP-1 macrophage-like cells (2×106/ml) were incubated for 15 min with various concentrations of P. intermedia protease. Subsequently, cells were washed three times with MEM containing 10% fetal calf serum (FCS) and incubated with 10 μg Escherichia coli J4 LPS ml−1 for 2 h. Cells were pelleted by centrifugation at 1,000×g for 10 min. RNA was extracted with the use of an RNA extraction kit (Qiagen, Santa Clara, Calif., USA), according to the manufacturer’s recommended protocols. One μg of RNA was reverse transcribed with 200 U of MMLV reverse transcriptase at 42 °C for 25 min followed by 65 °C for 5 min. The cDNA thus obtained was amplified with 0.1 μg of specific primers in a reaction mixture (PCR supermix, Invitrogen) containing Taq DNA polymerase (0.5 U), 55 mM KCl, 1.5 mM MgCl2, and 200 μM dNTPs. PCR was carried out in a DNA thermal cycler (Eppendorf) for 30 cycles of 45 s at 94 °C, 45 s at 59 °C, and 60 s at 72 °C. Equivalent cycles were run for all experiments. The sequence of human primers was as follows: GAPDH (346 bp) sense 5′-GCTCTC CAGAACATCATCCCTGCC-3′, antisense 5′-CGTTGTCATACCAGGAAATGAGCTT-3′; interleukin (IL)-1β (388 bp) sense 5′-AAACGAATGAAGTGCTCCTTCAGC-3′ and antisense 5′-ACCTCGTTGTTCACCACAAGAGGT-3′. After amplification, the PCR products were mixed with 0.1 volume of 10× loading buffer (5 ml glycerol, 5 mg xylene cyanol, 5 mg bromophenol blue, 20 μl NaEDTA (0.5 M, pH 8.0), 1 mg ethidium bromide, and 5 ml H2O) and separated on 2% agarose gels prepared in TAE buffer (4.84 g Tris base, 1.14 ml acetic acid, 2 ml EDTA (0.5 M, pH 8.0), 20 ml DEPC water). Following electrophoresis, gels were photographed and PCR products in each band were semiquantitatively analyzed by Fluor-S Imager (Biorad Laboratories, Richmond, Calif., USA).
To confirm the specificity of P. intermedia protease on U937 and THP-1 cell mCD14, the LPS-responsiveness of cells treated with P. intermedia protease in the presence and absence of excess sCD14 was determined. U937 or THP cells (2×106/100 μl) were incubated with various concentrations of P. intermedia protease in 10 μl buffer A with sCD14 (20 μg/ml) for 15 min. Subsequently, cells were washed three times and activated with 10 μg E. coli J4 LPS ml−1 for 2 h; expression of IL-1β mRNA was examined as described above.
Reagents
Soluble CD14 and LBP were used as human recombinant proteins in all experiments and kindly provided by Dr. Peter Tobias (University of California, San Diego). The remaining reagents were purchased as follows: minimal essential medium (MEM) and phosphate-buffered saline (PBS) from Life Technologies (Grand Island, N.Y., USA), P. intermedia strains 25261 and 25611 from ATCC (Rockville, Md., USA), media for P. intermedia cultures from Difco Laboratories (Detroit, Mich., USA), Mono-Q and Superose-12 columns from Pharmacia Biotech Products (Piscataway, N.J., USA), reagents for polyacrylamide gel electrophoresis (PAGE) from Biorad Laboratories (Richmond, Calif., USA), limulus amoebocyte lysate assay kit from Bio Whittaker (Walkersville, Md., USA), radioiodine from New England Nuclear, (Boston, Mass., USA), Iodogen from Pierce (Rockford, Ill., USA), and all other reagents from Sigma (St. Louis, Mo., USA).
Results
Cleavage of sCD14 and LBP by P. intermedia
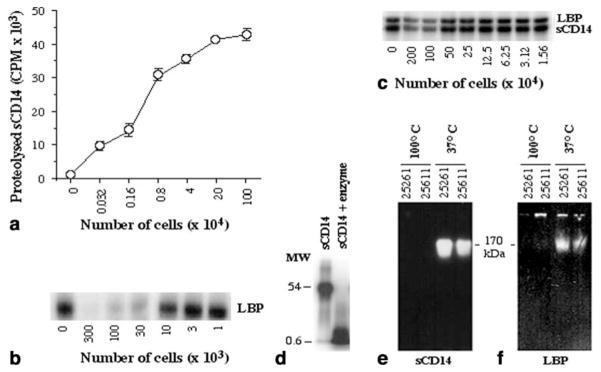
To determine the potential effect of proteolytic enzymes from P. intermedia on LPS-induced periodontal inflammation, the ability of these enzymes to cleave 125I-sCD14 and 125I-LBP was examined. Incubation of 125I-sCD14 or 125I-LBP with suspensions of various numbers of P. intermedia ATCC 25611 cells resulted in a concentration-dependent cleavage of 125I-sCD14 and 125I-LBP, respectively (Fig. 1a, b). By contrast, the supernatant of P. intermedia 25611 revealed only marginal proteolytic activity for both proteins (Fig. 1c). The suspension of P. intermedia ATCC 25611 proteolysed 125I-sCD14 in small fragments of less than 1 kDa, suggesting the presence of multiple cleavage sites on sCD14 for this enzyme (Fig. 1d). Like P. intermedia ATCC 25611, the suspension of strain ATCC 25261 was capable of cleaving 125I-sCD14 and 125I-LBP efficiently in a dose-dependent manner, whereas the supernatant did not reveal notable proteolytic activity for either of the proteins (data not shown). Zymographic analysis of the protein digests of P. intermedia strains 25261 and 25611 revealed that both strains contain a single cell-associated protease of approximately 170 kDa that cleaves 125I-sCD14 and 125I-LBP. The failure of 125I-sCD14 and 125I-LBP cleavage by the bacterial suspensions heated to 100 °C for 5 min indicates that this enzyme is heat sensitive (Fig. 1e, f).
Fig. 1.
Cleavage of 125I-sCD14 and 125I-lipopolysaccharide-binding protein (LBP) by various concentrations of Prevotella intermedia ATCC 25611 cell suspension (a, b). Cleavage of 125I-sCD14 or 125I-LBP by various concentrations of P. intermedia ATCC 25611 cell supernatant (c). SDS-PAGE (10% polyacrylamide) analysis of 125I-sCD14 to show the purity of the sCD14 used in the experiments and its cleavage to less than 1-kDa fragments by P. intermedia strain 25611 enzyme; MW molecular mass (d). Zymographic analysis of the cleavage of 125I-sCD14 by electrophoretically separated proteases from strains 25261 and 25611 and by the heat-inactivated proteases from both strains (e). Zymographic analysis of the cleavage of 125I-LBP by electrophoretically separated proteases from strains 25261 and 25611 and by the heat-inactivated proteases from both strains (f). a All points represent mean and SEM of triplicate values; a–f one out of three separate experiments is shown
Characterization of the purified P. intermedia protease
Molecular mass analysis by SDS-PAGE (10% polyacrylamide) showed that the purified enzyme was a monomeric 170-kDa protein (Fig. 2a). This finding, together with zymographic analysis, suggests that the protease isolated from P. intermedia is a 170-kDa protein. The proteolytic activity of the purified P. intermedia protease was characterized using 125I-sCD14 as substrate in the presence of a variety of protease modifiers, as shown in Table 1. The activity of the 170-kDa protease from P. intermedia was inhibited by N-ethylmaleimide and iodoacetamide, protease modifiers known to inhibit cysteine proteases. Additionally, the cysteine/serine protease inhibitors TLCK, TPCK, leupeptin, and antipain also inhibited its activity. However, the serine protease inhibitors DFP, PMSF, and dichloroisocoumarin were ineffective in inhibiting proteolysis of 125I-sCD14. Proteolysis of 125I-sCD14 was enhanced in the presence of increasing concentrations of cysteine (10 or 30 mM) or reducing agents such as dithiothreitol (15–30 mM) suggesting that this enzyme requires a thiol group to exhibit full activity. Chelating agents, such as EDTA (50 mM), EGTA (10 mM), and phosphoramidon, also inhibited enzyme activity, suggesting a requirement for metal ions. FPR-cmk, an inhibitor of arginine proteases, suppressed the proteolytic activity of P. intermedia enzyme dose-dependently, whereas ZFK-cmk, an inhibitor of lysine proteases, had no inhibitory effect on the enzymatic cleavage of 125I-sCD14 at all concentrations tested (Fig. 2b). The purified 170-kDa protease cleaved 125I-sCD14 and 125I-LBP in a concentration-dependent manner (Fig. 2c).
Fig. 2.
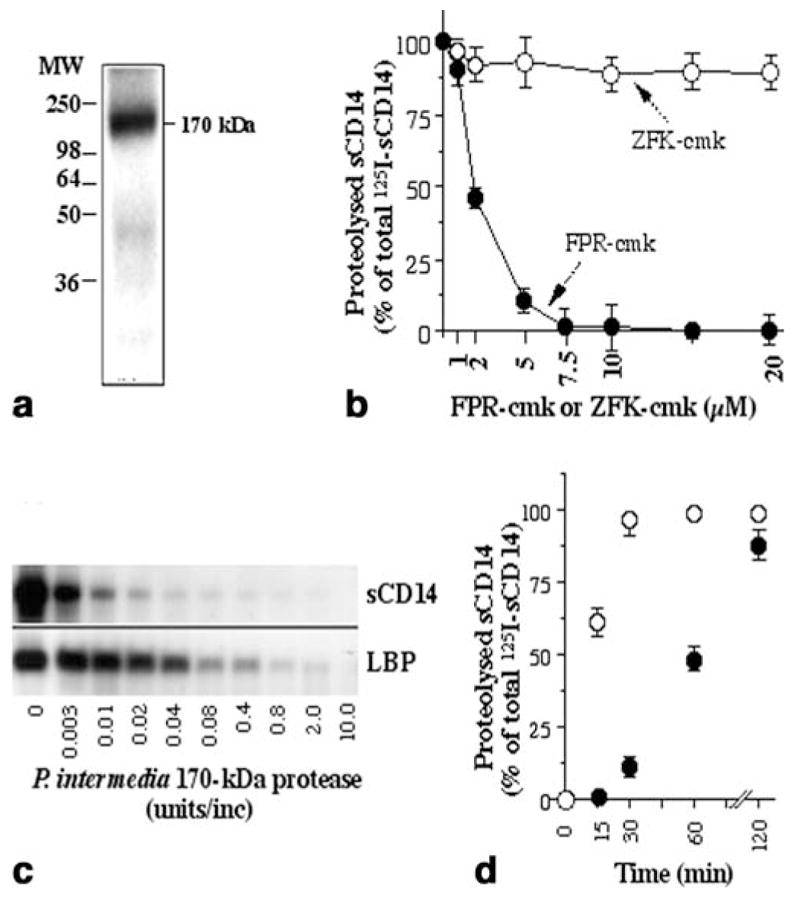
Molecular mass analysis of the radioiodinated purified protease on SDS-PAGE (10% polyacrylamide) (a). Cleavage of 125I-sCD14 by the purified protease in the presence of various concentrations of inhibitors, Phe-Pro-Arg-chloromethyl ketone (FPR-cmk) and benzyloxycarbonyl-Phe-Lys-chloromethyl ketone (ZFK-cmk) (b). Cleavage of 125I-sCD14 or of 125I-LBP by various concentrations of the purified 170-kDa protease (c). Cleavage of 125I-sCD14 by 0.2 U of P. intermedia protease in the absence (○) or presence (●) of 10% human serum (v/v) over a period of 2 h. (d). All points in b and d represent mean and SEM of triplicate values in one out of two experiments. a, c One out of three separate experiments is shown
Table 1.
Effect of various inhibitors and stimulators on the 125I-sCD14 proteolysis by Prevotella intermedia protease. TLCK Tosyl-L-lysine chloromethyl ketone, TPCK tosyl-L-phenylalanine chloromethyl ketone, DFP diisopropylfluorophosphate, PMSF phenylmethylsulfonyl fluoride, EDTA ethylenediamine tetraacetic acid(pH 7.0), EGTA ethyleneglycol-bis-N,N,N′,N′ tetraacetic acid
| Reagent | Class | Concentration | Activity (%) |
|---|---|---|---|
| MEM | – | – | 100 |
| N-ethylmaleimide | Cysteine | 5 mM | <14 |
| Iodoacetamide | Cysteine | 5 mM | <7 |
| TLCK | Serine/cysteine | 0.5 mM | <5 |
| TPCK | Serine/cysteine | 0.5 mM | 84 |
| TPCK | Serine/cysteine | 1 mM | 32 |
| TPCK | Serine/cysteine | 2 mM | <10 |
| Leupeptine | Serine/cysteine | 1 mM | <2 |
| Antipain | Serine/cysteine | 1 mM | <3 |
| DFP | Serine | 0.5 mM | 96 |
| PMSF | Serine | 5 mM | 100 |
| Dichloroisocoumarin | Serine | 0.5 M | 100 |
| L-Cysteine | - | 10 mM | 146 |
| L-Cysteine | - | 30 mM | 206 |
| Dithiothreitol | Reducing agent | 15 mM | 154 |
| Dithiothreitol | Reducing agent | 30 mM | 212 |
| EDTA | Metallo | 10 mM | 72 |
| EDTA | Metallo | 50 mM | <5 |
| EGTA | Metallo | 10 mM | <9 |
| Phosphoramidon | Metallo | 0.2 mM | <12 |
| Poly-L-arginine | Arginine | 1 mg/ml | <35 |
Examination of P. intermedia protease activity in the presence of 10% human serum revealed an inhibition of the enzymatic activity in a time-dependent manner (Fig. 2d).
Optimal conditions for the purified protease activity
A time course analysis showed that the P. intermedia protease cleaves 125I-sCD14 in a time-dependent manner. More than 90% of 125I-sCD14 was cleaved by 0.2 U of purified enzyme in approximately 30 min (Fig. 3a). Incubation of 125I-sCD14 with P. intermedia protease in buffers of different pH values (2.5 and 9.5) for 30 min revealed that the protease cleaves 125I-sCD14 over a wide pH range. Maximal activity was observed between pH values of 4.5–8.5. Below pH 4.5 and above pH 8.5, 125I-sCD14 proteolysis was decreased but not abolished (Fig. 3b). Further analysis of enzyme activity at various temperatures showed that it was stable at temperatures from 4 to 56 °C (Fig. 3c).
Fig. 3.
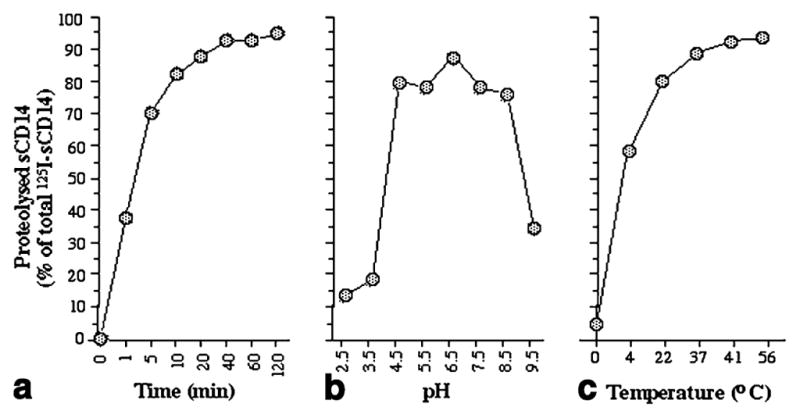
Time course of 125I-sCD14 cleavage by the purified P. intermedia protease (0.2 U) (a). Cleavage of 125I-sCD14 by P. intermedia protease in solutions of different pH values (b). Cleavage of 125I-sCD14 by the purified protease at different temperatures (c). Mean values from one out of three experiments are shown
Effect of the purified protease on the LPS-responsiveness of monocytic cells
To examine whether P. intermedia protease cleaves mCD14, macrophage-like U937 cells were incubated with various concentrations of P. intermedia protease and subsequently stimulated with LPS from E. coli. Exposure of cells to increasing concentrations of the protease resulted in decreased LPS-responsiveness as assessed by the expression of IL-1β-specific mRNA. However, the inhibitory effect of P. intermedia protease on the LPS-responsiveness of U937 cells was abrogated by the addition of sCD14 (Fig. 4a). Similarly, THP-1 macrophage-like cells exhibited a decreased LPS-responsiveness after incubation with P. intermedia protease. Furthermore, an abrogation of this inhibition of LPS-responsiveness was also observed by the addition of sCD14 (Fig. 4b).
Fig. 4.
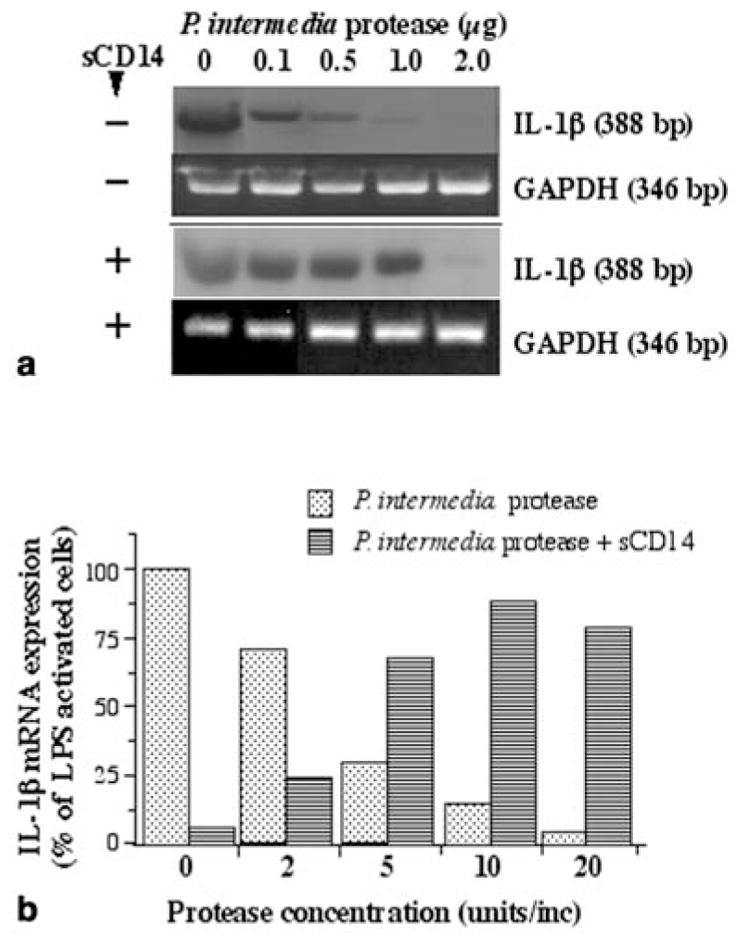
Interleukin (IL)-1β and GAPDH mRNA expression in U937 cells (2×106/ml) treated with various concentrations of P. intermedia protease for 15 min and subsequently activated with 10 μg LPS/ml for 2 h in the presence or absence of sCD14 (20 μg/ml) (a). IL-1β mRNA expression in THP-1 macrophage-like cells (2×106/ml) treated with various concentrations of P. intermedia protease for 15 min and subsequently activated with 10 μg LPS/ml for 2 h in the presence or absence of sCD14 (20 μg/ml) (b). a, b One out of two separate experiments is shown
Discussion
In this study, we have demonstrated the presence of an endoprotease present predominantly on the cell surface of P. intermedia. This enzyme cleaves sCD14 and LBP concentration-dependently. Purification of the proteolytic activity from P. intermedia cells revealed that it resides in a 170-kDa protease.
Although binding of LPS to its receptors is still not fully understood, it has been shown that LPS exerts its stimulatory effects on host cells by binding to a receptor complex comprised of CD14, Toll-like receptor, and other cell surface molecules. Furthermore, LPS binding to CD14 is strongly enhanced in the presence of LBP (Heumann and Roger 2002). When LPS binds to its receptor complex, immunoinflammatory cells are activated to secrete a variety of proinflammatory mediators (Guha and Mackman 2001). This leads to periodontal inflammation and tissue destruction (Birkedal-Hansen 1993; Page 1991). In addition, it is also known that periodontal diseases are exacerbated by the suppression of an adequate immunoinflammatory response necessary to control and eliminate the bacterial infection (Daniel and van-Dyke 1996; Kadowaki et al. 2000). Therefore, a P. intermedia enzyme that can modulate CD14 and LBP levels may be critical in attenuating host responses to this and other pathogens.
This protease has a high affinity for arginine, as shown by our ability to purify it on polyarginine affinity columns. Furthermore, inhibition of its activity by high concentrations of polyarginine or FPR-cmk, an inhibitor of arginine proteases, is also suggestive of its greater affinity for arginine residues. Suppression of the proteolytic activity of the protease by inhibitors of cysteine proteases suggests that this enzyme is a cysteine protease. Since its activity is enhanced in the presence of reducing agents, the enzyme also appears to be a thiol-requiring protease. The observation that EDTA, EGTA, and phosphoramidon inhibit the proteolytic activity of the enzyme suggests that it requires metal ions for activity. Inhibitors of serine- and lysine proteases had little effect on sCD14 cleavage by P. intermedia protease, suggesting that it is unlikely to be a serine or lysine protease. Nevertheless, further analysis is needed to ascertain whether it is a true arginine-specific protease.
Recently, it has been shown that arginine-specific and lysine-specific cysteine proteases (gingipains) from P. gingivalis, one of the most virulent periopathogens, also cleave CD14 and decrease the LPS-responsiveness of monocytes as assessed by the production of tumor necrosis factor (TNF)-α (Sugawara et al. 2000; Tada et al. 2002). It has been suggested that cleavage of CD14 by cysteine proteases from P. gingivalis can result in an attenuation of the cellular recognition of bacteria and thereby sustain chronic inflammation (Sugawara et al. 2000). Even if P. intermedia is not associated with severe periodontal disease as strongly as P. gingivalis, the presence of this cysteine protease may also have significant clinical implications, where P. intermedia prevails. The observation that sCD14 is cleaved very effectively at temperatures above 37 °C by this P. intermedia protease suggests that its escalated activity under hyperthermic conditions may be important in contributing to the reduction of sCD14 levels in the lesions. Additionally, this enzyme hydrolyzes sCD14 over a wide pH range, suggesting the possibility of reduction of sCD14 levels at the nidus of localized periodontal infections, where pH dramatically varies with the presence of various types of bacteria and of inflammatory exudates.
Proteolytic activity of the enzyme in vivo may also be influenced by the presence of serum components. The fact that serum inhibited sCD14 cleavage in vitro supports this assumption. Whether this is due to competitive inhibition by serum LBP, sCD14, other potential enzyme substrates, or endogenous non-specific protease inhibitors remains to be determined.
Soluble CD14 provides three major functions following binding to LPS. Firstly, as the principal LPS receptor in serum and tissue fluids, sCD14 recognizes bacterial LPS in conjunction with LBP and acts as a scavenger by neutralizing circulating LPS (Haziot et al. 1994). Secondly, sCD14 competitively inhibits binding of LPS to mCD14 and retards LPS-mediated activation of CD14-positive cells (Pugin et al. 1993; Rokita and Manzel 1997). Finally, sCD14, when complexed with LPS, can activate CD14-negative cells. Therefore, a dynamic equilibrium exists in which LPS is shuttled to sCD14 or mCD14 resulting in either neutralization or activation of periodontal cells by LPS. As P. intermedia protease inhibited the LPS-responsiveness of macrophage-like cells, it is likely that, in addition to sCD14 and LBP, mCD14 is also cleaved by this protease in vivo. Moreover, the fact that sCD14 protected mCD14 from cleavage by this protease and increased the LPS-responsiveness of macrophage-like cells indicates that sCD14 as well as mCD14 and LBP may be critical to the LPS responsiveness of macrophage-like cells. Since each of these proteins can be proteolysed by P. intermedia, this enzyme may play a significant role in modulating LPS-mediated proinflammatory responses.
Collectively, our findings suggest that the P. intermedia protease represents, in addition to LPS, another critical virulence factor of P. intermedia. We speculate that, through proteolysis of sCD14, mCD14 and LBP, this protease may profoundly influence the virulence of LPS at the site of gram-negative periodontal infections.
Acknowledgments
This work was supported by University of Pittsburgh Central Research Development Fund, and a grant from National Institute of Health DE11010. We thank Dr. Peter Tobias (La Jolla, Calif., USA) for his generosity in providing the recombinant CD14 and LBP, and anti-Cd14 antibodies.
Abbreviations
- CD
Cluster of differentiation
- DFP
Diisopropylfluorophosphate
- IL
Interleukin
- LBP
LPS-binding protein
- LPS
Lipopolysaccharide
- PMSF
Phenylmethylsulfonyl fluoride
- TLCK
Tosyl-L-lysine chloromethyl ketone
- TPCK
Tosyl-L-phenylalanine chloromethyl ketone
Contributor Information
James Deschner, Department of Oral Medicine and Pathology, University of Pittsburgh School of Dental Medicine, 589 Salk Hall, 3501 Terrace Street, Pittsburgh, PA 15261-1964, USA, Tel.: +1-412-648-8951, Fax: +1-412-624-6685.
Anuradha Singhal, Department of Oral Medicine and Pathology, University of Pittsburgh School of Dental Medicine, 589 Salk Hall, 3501 Terrace Street, Pittsburgh, PA 15261-1964, USA, Tel.: +1-412-648-8951, Fax: +1-412-624-6685.
Ping Long, Department of Oral Medicine and Pathology, University of Pittsburgh School of Dental Medicine, 589 Salk Hall, 3501 Terrace Street, Pittsburgh, PA 15261-1964, USA, Tel.: +1-412-648-8951, Fax: +1-412-624-6685.
Chau-Ching Liu, Department of Medicine, University of Pittsburgh School of Medicine, Pittsburgh, PA 15261, USA.
Nicholas Piesco, Department of Oral Medicine and Pathology, University of Pittsburgh School of Dental Medicine, 589 Salk Hall, 3501 Terrace Street, Pittsburgh, PA 15261-1964, USA, Tel.: +1-412-648-8951, Fax: +1-412-624-6685.
Sudha Agarwal, Email: sagar@pitt.edu, Department of Oral Medicine and Pathology, University of Pittsburgh School of Dental Medicine, 589 Salk Hall, 3501 Terrace Street, Pittsburgh, PA 15261-1964, USA, Tel.: +1-412-648-8951, Fax: +1-412-624-6685.
References
- American Academy of Periodontology. Consensus report on periodontal diseases: pathogenesis and microbial factors. Ann Periodontol. 1996;1:926–930. doi: 10.1902/annals.1996.1.1.926. [DOI] [PubMed] [Google Scholar]
- Birkedal-Hansen H. Role of cytokines and inflammatory mediators in tissue destruction. J Periodontal Res. 1993;28:500–510. doi: 10.1111/j.1600-0765.1993.tb02113.x. [DOI] [PubMed] [Google Scholar]
- Chen Z, Potempa J, Polanowski A, Wilkstrom A, Travis J. Purification and characterization of a 50 kDa cysteine proteinase (Gingipain) from Porphyromonas gingivalis. J Biol Chem. 1992;267:18896–18901. [PubMed] [Google Scholar]
- Daniel MA, Van Dyke TE. Alterations in phagocyte function and periodontal infection. J Periodontol. 1996;67:1070–1075. doi: 10.1902/jop.1996.67.10s.1070. [DOI] [PubMed] [Google Scholar]
- Gemmell E, Marshall RI, Seymour GJ. Cytokines and prostaglandins in immune homeostasis and tissue destruction in periodontal disease. Periodontology 2000. 1997;14:112–143. doi: 10.1111/j.1600-0757.1997.tb00194.x. [DOI] [PubMed] [Google Scholar]
- Guha M, Mackman N. LPS induction of gene expression in human monocytes. Cell Signal. 2001;13:85–94. doi: 10.1016/s0898-6568(00)00149-2. [DOI] [PubMed] [Google Scholar]
- Haziot A, Rong GW, Bazil V, Silver J, Goyert SM. Recombinant soluble CD14 inhibits LPS-induced tumor necrosis factor-production by cells in whole blood. J Immunol. 1994;152:5868–5876. [PubMed] [Google Scholar]
- Heumann D, Roger T. Initial responses to endotoxins and gram-negative bacteria. Clin Chim Acta. 2002;323:59–72. doi: 10.1016/s0009-8981(02)00180-8. [DOI] [PubMed] [Google Scholar]
- Jansen HJ, Grenier D, Van der Hoeven JS. Characterization of immunoglobulin G-degrading proteases of Prevotella intermedia and Prevotella nigrescens. Oral Micro Imm. 1997;10:138–145. doi: 10.1111/j.1399-302x.1995.tb00134.x. [DOI] [PubMed] [Google Scholar]
- Kadowaki T, Nakayama K, Okamoto K, Abe N, Baba A, Shi Y, Ratnayake DB, Yamamoto K. Porphyromonas gingivalis proteinases as virulence determinants in progression of periodontal diseases. J Biochem. 2000;128:153–159. doi: 10.1093/oxfordjournals.jbchem.a022735. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lantz MS, Allen RD, Vail TA, Switalski L, Hook M. Specific cell components of Bacteroides gingivalis mediate binding and degradation of human fibrinogen. J Bacteriol. 1991;173:495–504. doi: 10.1128/jb.173.2.495-504.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page RC. The role of inflammatory mediators in the pathogenesis of periodontal disease. J Periodont Res. 1991;26:230–242. doi: 10.1111/j.1600-0765.1991.tb01649.x. [DOI] [PubMed] [Google Scholar]
- Pugin J, Schuer-Maly CC, Leturcq D, Morarity A, Ulevitch RJ, Tobias PS. LPS activation of human endothelial cells is mediated by LPS binding protein and soluble CD14. Proc Nat Acad Sci USA. 1993;90:2744–2748. doi: 10.1073/pnas.90.7.2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokita E, Menzel EJ. Characteristics of CD14 shedding from human monocytes. Evidence for the competition of soluble CD14 (sCD14) with CD14 receptors for lipopolysaccharide (LPS) binding. APMIS. 1997;105:510–518. doi: 10.1111/j.1699-0463.1997.tb05048.x. [DOI] [PubMed] [Google Scholar]
- Sugawara S, Nemoto E, Tada H, Miyake K, Imamura T, Takada H. Proteolysis of human monocyte CD14 by cysteine proteinases (gingipains) from Porphyromonas gingivalis leading to lipopolysaccharide hyporesponsiveness. J Immunol. 2000;165:411–418. doi: 10.4049/jimmunol.165.1.411. [DOI] [PubMed] [Google Scholar]
- Tada H, Sugawara S, Nemoto E, takahashi N, Imamura T, Potempa J, Travis J, Shimauchi H, Takada H. proteolysis of CD14 on human gingival fibroblasts by arginine specific cystein protease from P. gingivalis leading to down regulation of LPS-induced interleukin-8 production. Infect Immun. 2002;70:3304–3307. doi: 10.1128/IAI.70.6.3304-3307.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurfel MM, Kunitake ST, Lichenstein H, Kane JP, Wright SD. Lipopolysaccharide (LPS)-binding protein is carried on lipoproteins and acts as a cofactor in the neutralization of LPS. J Exp Med. 1994;180:1025–1035. doi: 10.1084/jem.180.3.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]