Abstract
In 2012, a new feature of eukaryotic gene expression emerged: ubiquitous expression of circular RNA (circRNA) from genes traditionally thought to express messenger or linear noncoding (nc)RNA only. CircRNAs are covalently closed, circular RNA molecules that typically comprise exonic sequences and are spliced at canonical splice sites. This feature of gene expression was first recognized in humans and mouse, but it quickly emerged that it was common across essentially all eukaryotes studied by molecular biologists. CircRNA abundance, and even which alternatively spliced circRNA isoforms are expressed, varies by cell type and can exceed the abundance of the traditional linear mRNA or ncRNA transcript. CircRNAs are enriched in the brain and increase in abundance during fetal development. Together, these features raise fundamental questions regarding the regulation of circRNA in cis and in trans, and its function.
CircRNAs: The Hidden RNA
Since the discovery that essentially all human protein-coding genes are alternatively spliced, it has been assumed that the major transcriptional product of these genes is mRNA. Similarly, it has been assumed that linear products are the only RNAs processed from the less abundant, but functionally significant, class of long ncRNAs. These assumptions led to a host of models being proposed to explain the function and regulation of splicing as well as transcriptional and post-transcriptional regulation. In 2012, a statistical analysis of RNA-Seq data and subsequent biochemical analysis revealed a complete surprise: circRNA (see Glossary) molecules transcribed and spliced from exons in protein and noncoding genes are ubiquitous in the human and mouse genomes and, thus, are likely to be a pervasive and previously overlooked feature of eukaryotic gene expression and regulation (Figure 1) ([1], reviewed in [2,3]).
Figure 1. The sequence of circular RNA comprises exons from protein and noncoding loci, spliced at canonical splice sites.
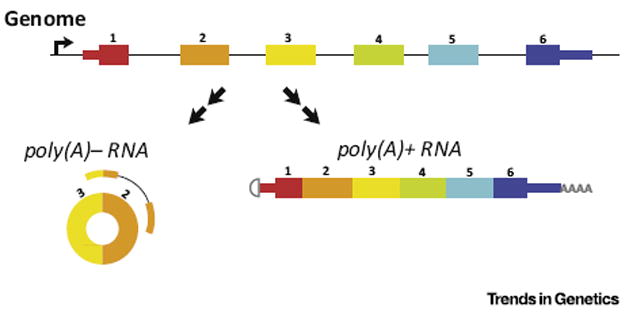
Bi-colored reads represent paired-end RNA-Seq reads used to discover circular RNA that are enriched in polyA-depleted RNA samples.
While circRNAs were first discovered to be ubiquitous in the transcriptomes of humans and mice, they were subsequently found to be abundant in an array of other metazoans, including flies and worms [4,5]. More surprisingly, circRNA expression is a feature of essentially all eukaryotes known today [6-8]. RNA-seq and biochemical studies have revealed that circRNA is expressed across the eukaryotic tree of life, including in Saccharomyces cerevisiae, which has hardly any splicing that involves exon skipping; the parasite that causes malaria, Plasmodium falciparum, amoebae, and plants [6]. In addition, circRNA can also be generated through the U12 spliceosome, which diverged millions of years ago from the major spliceosome that splices most human genes; again suggesting that circRNA expression is ancient [9]. Thus, circRNA production has either been conserved over billions of years or else is a feature that has re-evolved multiple times; either implies a likely functional role for circRNAs in the cell.
Work by many groups has now revealed compelling features of circRNA expression: circular isoforms are themselves differentially spliced and regulated. In hundreds to thousands of human genes, varying by cell type, and in other organisms, circRNA is a more abundant RNA isoform than mRNA [7,8,10]. Recent work in humans and flies has begun to reveal cis and trans regulation of circRNA, and hints at a function for a handful of circRNAs [11-14]. Together, these discoveries suggest greater complexity, regulation, and function of complex eukaryotic gene expression, as well as begging many significant and fundamental questions about the role of circRNA in the cell. Much work remains to be done in this area, which is now very active.
Given its prevalence and the fact that it was overlooked until very recently, circRNA warrants attention from essentially all molecular biologists. This is not only because such attention may reveal functional roles of circRNA, but also because it can confound studies and analysis of linear mRNAs derived from the same locus. For example, depending on experimental design, genome editing and siRNA approaches, among others, can target and impact the abundance of circRNA in addition to the presumed effect on linear messages. Thus, researchers studying linear gene expression must also account for circRNA. Medical researchers are also focusing attention on circRNA because it can be detected in the cell-free components of the blood, and, with its potential for extracellular stability, it shows promise as a biomarker of disease.
History of CircRNA
Although lacking direct biochemical evidence or definitive proof of their existence, the first observation suggesting that human RNAs exist in circular form was made more than 30 years ago by using electron microscopy [15]. Later, it was serendipitously reported that an isoform of the Deleted in Colorectal Cancer (DCC) gene with exons spliced in a scrambled order with respect to the reference genome could be amplified, although it was estimated to comprise less than one one-thousandth of DCC transcripts. The phenomenon responsible for this observation was deemed to be exon scrambling [16]. Over the next 20 years, a few other genes were found to be processed into circRNA isoforms present at low abundance. These included the MLL, ETS-1, Cytochrome P-450 2C18, and Dystrophin genes [17-20].
Thus, generally speaking, circRNAs were thought to be exceptional curiosities or ‘splicing noise’ until 2012. Before this, the only well-studied circRNAs were from the mouse Sex Determining Region Y (Sry) gene, and the Antisense Non-coding RNA in the INK4 locus (ANRIL) and Cerebellar Degeneration-Related protein 1 (CDR1) loci, characterized in 1990, 2010, and 2011, respectively; these were all considered anomalies because of their unusual features, including being transcribed from single exon genes and, in the case of CDR1 and ANRIL, antisense to the messenger RNA [21,22]. The best- characterized circRNA is expressed in mouse from the Sry gene, the sex-determining gene in males. In mice, it comprises a single exon, which, early in development, is transcribed into a linear mRNA transcript that is translated into protein. However, in adults, the RNA exists primarily as a circular product that is predominantly localized to the cytoplasm [23]. The function of the circRNA Sry transcript is not clear, because attempts to show ribosome association have yielded negative results. However, at least one study has found evidence that it acts as a miRNA sink [24,25]. Targeted mutagenesis and RNA biochemical studies of Sry during the 1990s revealed that the circular isoform is processed from a pre-mRNA transcribed from a promoter that is upstream of the promoter used for transcription of the linear transcript. Use of the upstream promoter results in a pre-mRNA that includes inverted repeats flanking the Sry exon that direct transcript circularization [19,26].
The second relatively well-studied circRNA is transcribed from the CDR1 locus, although it is antisense to the CDR1 messenger RNA. Expression of CDR1as circRNA is conserved at least from human to mouse. CDR1as is currently the best-characterized circRNA, with two potential functions: it was first reported to enhance expression of CDR1 by stabilizing the sense CDR1 mRNA [21] and, subsequently, to be a miRNA sponge for mir-7 [4]. Additionally, it was found that mir-671 regulates the Ago-2-dependent cleavage of CDR1as, thereby destabilizing sense CSR1 mRNA [21]. Future research will aim to shed light on co-regulation resulting from the ability of CDR1as to perform its functions.
The sodium transporter NCX1 gene in monkey, the muscleblind (mbl) [27] gene in flies, and the rat Cytochrome P450 2C24 gene are other examples of transcripts that, over the past decade, were tentatively reported as having circular isoforms [20,28,29]. Recently, groups have revisited the finding of high expression of the mbl circle and shown it to regulate the MBL protein in cis [11]. Together, the above genes are part of a decades-long ambiguous history of circRNA that now, in retrospect, is clear: the bias introduced by polyA selection through either RNA purification or oligo-dT priming is likely to be responsible for the serendipitous detection of a few circRNAs and the fact that they were generally thought to be expressed at a very low level.
Original Algorithms to Detect CircRNA Expression
The detection of ubiquitous circRNA expression is due to the RNA-Seq ‘revolution’ that has allowed researchers to easily obtain millions of short sequencing reads representing all RNA isoforms. From the early days of RNA-seq to today, most studies use RNA-Seq reads to quantify gene expression of the known transcriptome or to discover a more complete picture of splicing or transcription that obeys canonical models of its function (e.g., splicing at canonical U2 or U12 spliceosomal boundaries). Development of novel computational methodology has relaxed many of these assumptions, including algorithms that discover de novo splice sites, but even now it remains a significant challenge to distinguish apparently novel splicing or expression events from biochemical noise [30]. Therefore, most algorithms originally designed to detect novel splicing (including ab initio attempts to annotate the transcriptome [31] and even algorithms intended to detect gene fusions in cancer) must impose ad hoc filters due to high false positive rates [32].
A recent study showed that, by using statistical approaches to reduce high rates of false positives generated by algorithms attempting de novo splicing detection, some of the ad hoc bioinformatic filtering steps could be eliminated when cancer and other RNA-seq data sets were analyzed [33], leading to the discovery of the ubiquitous expression of circRNA. Of note, some of the first circular isoforms that were discovered, such as in the MAN1A2 gene, were present in extremely common (i.e., polyA selected) data sets because the circRNA isoforms of MAN1A2 contain exons, which are A-rich. These circRNAs were overlooked in well-studied data sets because of bioinformatic filtering steps. Relaxing these filters allowed the identification of circular RNA in some polyA rich data sets, which led researchers to subsequently look for circRNA more comprehensively. These bioinformatic data were validated using biochemical approaches that included showing that putative circRNA isoforms were resistant to RNaseR, an exonuclease that digests essentially all linear RNAs that have at least seven free 3′ nucleotides [34]. With the continuing improvement of statistical and computational methods, the field will likely gain a clearer picture of the full extent of RNA expression and, in particular, of circRNA. The explosion of interest in circRNA has already led to rapid advances in the field, some of which are described below.
Alternative Splicing of CircRNA
Intron retention is a well-known and common form of alternative splicing regulation in lower eukaryotes [35], as well as in human genes [36], and was recently found to be a widespread and regulated phenomenon [37]. The existence of intron retention in circRNAs would not be surprising, considering that linear intron-retained transcripts are known to be generated by metazoan splicing machinery [38]. Among other examples of cell type-specific alternative splicing of circRNA, sequencing and Northern blotting showed intron retention in circRNA was cell type-specifically regulated [10]. Subsequently, this phenomenon has been more thoroughly characterized and deemed to be exon-intron circRNA (EIcircRNA) [39]. In addition to these examples of bona fide topologically circRNA where all bases have canonical 5′–3′ linkages, recent interest in stable and perhaps functional lariats has emerged [40]. While these molecules are sometimes called circular, the presence of a single unique 2′–5′ linkage in the RNA molecule distinguishes them from a true circRNA, which is topologically invariant under rotations and contains only linkages that are indistinguishable under renaming of bases by cyclic permutation.
In humans and most metazoans, most alternative splicing occurs through alternative inclusion of exons. Dramatic examples of the potential diversity that can be generated from a single gene through this process are highlighted in the nervous systems of flies, where the Dscam gene can give rise to >20 000 different proteins, whose diversity is functionally essential [41]. Essential synaptic genes in humans, such as the best-characterized examples in the Neurexin gene family, have thousands of annotated isoforms [42]. Similar diverse patterns of protein isoform expression regulating synaptic connectivity are found in mice [43].
Similar to linear isoforms, circRNA from both protein-coding and noncoding genes are alternatively spliced through exon skipping and, in some cases, through intron inclusion, and regulated in a cell type-specific manner [10,25,44,45]. This phenomenon will likely be better characterized in the years to come, with some recent work on underlying mechanisms summarized below. Although the processes underlying this regulation are not yet well understood, regulation by cis sequences (such as inverted repeats) alone and passive cell type-invariant decay mechanisms together are not themselves sufficient to explain the observed cell type-specific steady-state abundance of circRNA.
Cis and Trans Regulation of CircRNA
Strong evidence that circRNA production is at least partly regulated in cis came from classical studies of the Sry gene in mouse, where a minimal inverted repeat sequence flanking the circularized RNA was found to be required for circRNA production [46]. Since then, some studies in humans and flies have suggested that inverted repeats, usually derived from transposable elements, are responsible for circRNA biogenesis and that circRNA splicing uses canonical splicing signals [4,5,20,23,47-50]. However, the use of inverted repeats in circRNA biogenesis is not conserved in yeast or plants [7,50]. In the fission yeast S. pombe, for example, it has been shown that circRNA production proceeds through an exon-containing lariat that itself is a ‘pre-mRNA’, giving rise to a circularized exon [50].
However, inverted repeats alone are unlikely to explain the production of circRNA in all metazoan genes. For example, in the Sry gene, the pre-mRNA containing inverted repeats needed for circRNA production requires activation of an upstream promoter [51]. Many human genes with abundant circular isoforms have flanking intron sequences, which lack clear secondary structure, suggesting that inverted repeats are not required for their biogenesis. In addition, in metazoans, the relative expression of linear and circRNA expression from most genes varies by cell type. If this variation is in part due to differences in the production of circRNA, cis motifs alone are not sufficient to explain the dynamic expression of circRNA in different cell types from the same organism.
Moreover, the field has accumulated strong evidence that circRNA expression is not simply correlated with linear RNA expression from the same locus, lending further support to the hypothesis that regulation of circRNA expression is more complex than in cis alone [10,50,52]. Indeed, over the past few years, several trans-acting splicing factors that influence circRNA biogenesis have been, and continue to be, identified, although few studies have conclusively demonstrated a direct interaction between a splicing factor and a pre-mRNA that is subsequently circularized. One set of trans-acting factors are the double-stranded (ds)RNA-binding proteins, discovered for their ability to deaminate adenosine and thereby edit RNA, hence their name: ADARs. One model suggests that, in the presence of ADAR, inverted repeats are destabilized, inhibiting circRNA splicing [47]. However, a model in which circRNA expression is solely regulated by ADAR activity and the presence of inverted repeats, is also too simple to explain cell type-specific expression of circRNA expression because there is no monotonic relation between ADAR expression and circRNA abundance.
The abundance of circRNA in human cells is regulated by the RNA-binding protein Quaking (QKI) through joint protein binding up- and downstream of the circularized exon, which facilitates QKI dimerization by bringing the 3′ and 5′ ends of the circularized exon(s) into close proximity and mediates their splicing [13]. Together with studies of the RNA-binding protein ADAR, these studies provide the first conclusive mechanistic evidence of RNA-binding proteins that affect circRNA abundance in human cells.
In flies, circRNA is regulated by combinatorial relations between many well-studied RNA splicing factors as well as sequences in cis, as has been shown for human cells [11,14,53]. This study and other evidence, for example, that no monotonic relation exists between ADAR expression and circRNA abundance, points to nonlinear relations between each cis- and trans-acting factor and the abundance of the downstream circRNA. Dissecting this relation will likely be both illuminating and difficult; for example, in general, manipulating an endogenous genomic locus (e.g., by CRISPR) will have impacts on both linear and circRNA splicing and, thus, has the potential to confound the impact of these sequences on linear and circular isoforms, especially if they are co-regulated as has been suggested [13,14].
Developmental and Tissue-Specific Regulation: Potential Insight into Function
Several studies have found circRNA enriched in the brain, especially during development and aging, with localization and abundance potentially regulated by synaptic activity in the mouse [9,47,54,55]. Although we do not yet understand all of the mechanisms responsible for generating cell type-specific isoform expression and the regulation of its diversity, it may not be surprising to find circRNA production enriched in the nervous system, where alternative splicing is essential and thousands of isoforms can be generated from a single gene [41-43].
The most convincing evidence for functional circRNAs has been found in the brain. The first example is CDR1as, which has two functions: (i) as a trans-regulator of CDR1 mRNA through a mechanism that is regulated by AGO2- and miR-671-mediated cleavage [21]; and (ii) as a miRNA sink for miR-7, thus regulating neural gene expression [4]. In addition, circRNA has been found to be enriched in the human fetal brain [9] and in synaptosomes [56].
In the fly brain, the abundant circRNA from the mbl gene autoregulates levels of the MBL protein and also binds, and is sequestered by, the circRNA isoform [11]. For example, this study showed that each circRNA molecule of mbl can bind and sequester multiple copies of the MBL protein. In addition, the study showed that binding of the MBL protein to both of the introns flanking the circularized mbl exon is required for circRNA biogenesis. This suggests that circRNA is a key component of a feedback circuit regulating the abundance of this essential RNA-binding protein in the fly brain [57].
Evidence points to the idea that circRNA function through sequestering miRNA or autor-egulating protein levels is the exception, not the rule. First, several studies have rigorously tested the general function for circRNA as a miRNA sponge, through both biochemical and computational analysis, and found negative evidence for a model where circRNA functions to regulate miRNAs [7,25]. Second, circRNA exists in species that lack RNAi pathways, in particular, S. cerevisiae and P. falciparum [6], where, if functional, circRNA must operate through other pathways. Similarly, the function of the mbl circle also seems exceptional: genes hosting highly expressed circRNA isoforms have not been reported as enriched for RNA-binding proteins. Together, this suggests a more general and yet-to-be discovered function for circRNA.
Insights into potential functions of circRNA may come from their developmental regulation, localization, and tissue-specific expression. In particular, the early examples of highly expressed circRNA in the Sry and mbl genes showed developmental regulation, but it was not known whether this was a general feature of circRNA. Recently, the question regarding the generality of developmental regulation has been addressed by several groups in multiple organisms: circRNA expression from hundreds of genes was found to be induced during early development, including in humans, flies, and pigs [9,55,58].
Global Stability of CircRNA and Extracellular Stability
The hypothesis that circRNA is not as efficiently degraded as linear mRNA because it lacks free 5′ or 3′ ends has been tested for a handful of circRNAs [5], although it has not been rigorously tested on a genome-wide scale. If true, cellular levels of circRNA abundance could be reduced through endonucleic activity, and active or passive export of circRNA into extracellular space or dilution out of the cell through mitosis. This is suggested by a study showing that circRNAs are highly enriched in platelets [59]. Additionally, circRNA may be abundant and stable in extracellular space or even secreted by exosomes [60]. In these cases, circRNA, including potentially unique circRNA only expressed by specific cell types or in pathological conditions, could serve as a better biomarker than mRNA for some diseases. Indeed, circRNA may have a role in cancer [61] and, therefore, may be dysregulated in this disease; if so, circRNA may be a useful cancer biomarker.
Concluding Remarks
In summary, circRNA expression is ubiquitous throughout the eukaryotic kingdom and particularly in metazoan genes. Early studies suggested that circRNA expression is combinatorially regulated by multiple splicing factors and cis sequences known to impact the alternative splicing of linear RNAs. A challenge for the field is to determine if and, in which case, how circRNAs are regulated by trans-acting factors and cis sequence elements beyond those known to impact the splicing of pre-mRNA. In addition, functional roles have been assigned to a handful of circRNAs. Determining the function, if any, of the thousands of circRNAs expressed across the tree of life will likely occupy researchers for years to come. The abundance of circRNA and the significant sequence overlap with mRNA or linear ncRNA transcribed from the same locus pose technical and conceptual challenges to studies of circRNA regulation and function that the field will have to address (see Outstanding Questions).
Outstanding Questions.
Are circRNAs ‘noisy’ by-products of splicing that are passively decayed or diluted by cell division?
As expected, known splicing factors regulate circRNA production; but are circRNAs simply secondary products of active linear RNA-regulated splicing or are they themselves actively and specifically regulated?
Many mechanisms for regulating RNA decay and nuclear export exist; could circRNAs be passively diffused or similarly piggyback on nuclear export signals? Or, could their decay be regulated solely by cell division and random nicking and/or degradation processes?
Do most circRNAs have a single or a handful of functions? Or, do they have a rich diversity of functions or redundant functions?
Do circRNA have physiological roles in disease processes and can they be used to diagnose pathology?
Trends.
Many circRNAs have recently been discovered and characterized.
Recently, much light has been shed on the regulation and function of circRNAs
CircRNA has been posited to function as a miRNA or RNA-binding protein sponge. However, a general function has not been identified.
Developmental regulation of circRNA and enrichment in the nervous system are an emerging theme shared by circRNAs across the metazoan lineage, from flies to humans.
CircRNAs can be joined by 3′ –5′ linkages, containing only exonic sequence; 2′ –5′ linkages (intronic lariats); or 3′ –5′ linkages that contain retained intronic sequences.
Acknowledgments
I thank Steven Barrett, Linda Szabo, and Marshae’ Jones for helpful comments. This work was supported by NCI grant R00 CA168987-03, NIGMS grant R01 GM116847, McCormick-Gabilan Fellowship, and a Baxter Family Fellowship to J.S.. J.S. is an Alfred P. Sloan fellow in Computational & Evolutionary Molecular Biology.
Glossary
- Alternative splicing
the process whereby multiple mature RNAs are derived from the same locus by alternative inclusion or exclusion of exons and introns applies to ncRNAs and mRNAs.
- Circular RNA (circRNA)
covalently closed RNA molecule where each nucleotide pair is joined with canonical 3′ –5′ linkages.
- Exon-intron circRNA (EIcircRNA)
circRNA with at least two exons and one intervening intron.
- Exon
RNA sequences retained during the processing of pre-mRNA to a mature transcript.
- Exon skipping
alternative splicing where one isoform differs from another by inclusion or exclusion an exon.
- Intron
RNA sequence excised during the processing of pre-mRNA to a mature transcript.
- Isoform
one of several sequence variants of the mature mRNA or pre-RNAs derived from a locus generating a long ncRNA.
- Lariat
a looped, but not circular byproduct of pre-RNA splicing comprising an intron where a nucleotide near the 3′ end of the intron is linked to the 5′ most nucleotide of the intron through a 2′ –5 ′ linkage.
- Pre-mRNA
incompletely processed RNA copy of the DNA-containing sequences that will be removed during splicing and maturation.
- RNA-binding protein
protein that interacts with, and binds to, RNA, sometimes through structural elements, or double-stranded or single-stranded motifs.
- Splicing factor
RNA-binding protein that regulates splicing.
- RNase R
a highly processive 3′ to 5′ exonuclease that digests essentially all linear RNAs containing at least seven nucleotides of unstructured nucleotides at the 3′ end
- U12 spliceosome
an ancient spliceosome diverged from the major spliceosome responsible for splicing of most metazoan genes that splices a few developmentally regulated transcripts.
References
- 1.Salzman J, et al. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE. 2012;7:e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32:453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lasda E, Parker R. Circular RNAs: diversity of form and function. RNA. 2014;20:1829–1842. doi: 10.1261/rna.047126.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Memczak S, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 5.Jeck WR, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang PL, et al. Circular RNA is expressed across the eukaryotic tree of life. PLoS ONE. 2014;9:e90859. doi: 10.1371/journal.pone.0090859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu T, et al. Transcriptome-wide investigation of circular RNAs in rice. RNA. 2015;21:2076–2087. doi: 10.1261/rna.052282.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Broadbent KM, et al. Strand-specific RNA sequencing in Plasmodium falciparum malaria identifies developmentally regulated long non-coding RNA and circular RNA. BMC Genomics. 2015;16:454. doi: 10.1186/s12864-015-1603-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Szabo L, et al. Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development. Genome Biol. 2015;16:126. doi: 10.1186/s13059-015-0690-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salzman J, et al. Cell-type specific regulation of circular RNA expression. PLoS Genet. 2013;9:e1003777. doi: 10.1371/journal.pgen.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ashwal-Fluss R, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 12.Westholm JM, et al. Genome-wide analysis of Drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep. 2014;9:1966–1980. doi: 10.1016/j.celrep.2014.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conn SJ, et al. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160:1125–1134. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 14.Kramer MC, et al. Combinatorial control of Drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins. Genes Dev. 2015;29:2168–2182. doi: 10.1101/gad.270421.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coca-Prados M, Hsu MT. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature. 1979;280:339–340. doi: 10.1038/280339a0. [DOI] [PubMed] [Google Scholar]
- 16.Nigro JM, et al. Scrambled exons. Cell. 1991;64:607–613. doi: 10.1016/0092-8674(91)90244-s. [DOI] [PubMed] [Google Scholar]
- 17.Cocquerelle C, et al. Splicing with inverted order of exons occurs proximal to large introns. EMBO J. 1992;11:1095–1098. doi: 10.1002/j.1460-2075.1992.tb05148.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saad FA, et al. A 3′ consensus splice mutation in the human dystrophin gene detected by a screening for intra-exonic deletions. Hum Mol Genet. 1992;1:345–346. doi: 10.1093/hmg/1.5.345. [DOI] [PubMed] [Google Scholar]
- 19.Bailleul B. During in vivo maturation of eukaryotic nuclear mRNA, splicing yields excised exon circles. Nucleic Acids Res. 1996;24:1015–1019. doi: 10.1093/nar/24.6.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zaphiropoulos P. Exon skipping and circular RNA formation in transcripts of the human cytochrome P-450 2C18 gene in epidermis and of the rat androgen binding protein gene in testis. Mol Cell Biol. 1997:2985–2993. doi: 10.1128/mcb.17.6.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hansen TB, et al. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 2011;30:4414–4422. doi: 10.1038/emboj.2011.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burd CE, et al. Expression of linear and novel circular forms of an INK4/ARF-associated non–coding RNA correlates with atherosclerosis risk. PLoS Genet. 2010;6:e1001233. doi: 10.1371/journal.pgen.1001233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koopman P, et al. Expression of a candidate sex-determining gene during mouse testis differentiation. Nature. 1990;348:450–452. doi: 10.1038/348450a0. [DOI] [PubMed] [Google Scholar]
- 24.Hansen TB, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 25.Guo JU, et al. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15:409. doi: 10.1186/s13059-014-0409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Capel B, et al. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell. 1993;73:1019–1030. doi: 10.1016/0092-8674(93)90279-y. [DOI] [PubMed] [Google Scholar]
- 27.Nam K, et al. Severe growth defect in a Schizosaccharomyces pombe mutant defective in intron lariat degradation. Mol Cell Biol. 1997;17:809–818. doi: 10.1128/mcb.17.2.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Houseley JM, et al. Noncanonical RNAs from transcripts of the Drosophila muscleblind gene. J Heredity. 2006;97:253–260. doi: 10.1093/jhered/esj037. [DOI] [PubMed] [Google Scholar]
- 29.Lytton J, Li XF. A circularized sodium-calcium exchanger exon 2 transcript. J Biol Chem. 1999;274:8153–8160. doi: 10.1074/jbc.274.12.8153. [DOI] [PubMed] [Google Scholar]
- 30.Hooper JE. A survey of software for genome-wide discovery of differential splicing in RNA-Seq data. Hum Genomics. 2014;8:3. doi: 10.1186/1479-7364-8-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grabherr MG, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peng Z, et al. Hypothesis: artifacts, including spurious chimeric RNAs with a short homologous sequence, caused by consecutive reverse transcriptions and endogenous random primers. J Cancer. 2015;6:555–567. doi: 10.7150/jca.11997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salzman J, et al. ESRRA-C11orf20 is a recurrent gene fusion in serous ovarian carcinoma. PLoS Biol. 2011;9:e1001156. doi: 10.1371/journal.pbio.1001156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vincent HA, Deutscher MP. Substrate recognition and catalysis by the exoribonuclease RNase. R J Biol Chem. 2006;281:29769–29775. doi: 10.1074/jbc.M606744200. [DOI] [PubMed] [Google Scholar]
- 35.Rodriguez-Trelles F, et al. Origins and evolution of spliceosomal introns. Annu Rev Genet. 2006;40:47–76. doi: 10.1146/annurev.genet.40.110405.090625. [DOI] [PubMed] [Google Scholar]
- 36.Dichmann DS, Harland RM. fus/TLS orchestrates splicing of developmental regulators during gastrulation. Genes Dev. 2012;26:1351–1363. doi: 10.1101/gad.187278.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boutz PL, et al. Detained introns are a novel, widespread class of post-transcriptionally spliced introns. Genes Dev. 2015;29:63–80. doi: 10.1101/gad.247361.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lewis BP, et al. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc Natl Acad Sci U S A. 2003;100:189–192. doi: 10.1073/pnas.0136770100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zlotorynski E. Non-coding RNA: circular RNAs promote transcription. Nat Rev Mol Cell Biol. 2015;16:206. doi: 10.1038/nrm3967. [DOI] [PubMed] [Google Scholar]
- 40.Zhang Y, et al. Circular intronic long noncoding RNAs. Mol Cell. 2013;51:792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 41.Schmucker D, et al. Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell. 2000;101:671–684. doi: 10.1016/s0092-8674(00)80878-8. [DOI] [PubMed] [Google Scholar]
- 42.Treutlein B, et al. Cartography of neurexin alternative splicing mapped by single-molecule long-read mRNA sequencing. Proc Natl Acad Sci U S A. 2014;111:E1291–E1299. doi: 10.1073/pnas.1403244111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thu CA, et al. Single-cell identity generated by combinatorial homophilic interactions between alpha, beta, and gamma protocadherins. Cell. 2014;158:1045–1059. doi: 10.1016/j.cell.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lokody I. Alternative splicing: characterizing cell fate. Nat Rev Genet. 2014;15:706. doi: 10.1038/nrg3847. [DOI] [PubMed] [Google Scholar]
- 45.Kelly S, et al. Exon skipping is correlated with exon circularization. J Mol Biol. 2015;427:2414–2417. doi: 10.1016/j.jmb.2015.02.018. [DOI] [PubMed] [Google Scholar]
- 46.Capel B. Sex in the 90s: SRY and the switch to the male pathway. Annu Rev Physiol. 1998;60:497–523. doi: 10.1146/annurev.physiol.60.1.497. [DOI] [PubMed] [Google Scholar]
- 47.Ivanov A, et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 2015;10:170–177. doi: 10.1016/j.celrep.2014.12.019. [DOI] [PubMed] [Google Scholar]
- 48.Liang D, Wilusz JE. Short intronic repeat sequences facilitate circular RNA production. Genes Dev. 2014;28:2233–2247. doi: 10.1101/gad.251926.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Starke S, et al. Exon circularization requires canonical splice signals. Cell Rep. 2015;10:103–111. doi: 10.1016/j.celrep.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 50.Barrett SP, et al. Circular RNA biogenesis can proceed through an exon-containing lariat precursor. Elife. 2015;4:e07540. doi: 10.7554/eLife.07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dolci S, et al. Identification of a promoter region generating Sry circular transcripts both in germ cells from male adult mice and in male mouse embryonal gonads. Biol Reprod. 1997;57:1128–1135. doi: 10.1095/biolreprod57.5.1128. [DOI] [PubMed] [Google Scholar]
- 52.Memczak S, et al. Identification and characterization of circular RNAs as a new class of putative biomarkers in human blood. PLoS ONE. 2015;10:e0141214. doi: 10.1371/journal.pone.0141214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang XO, et al. Complementary sequence-mediated exon circularization. Cell. 2014;159:134–147. doi: 10.1016/j.cell.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 54.You X, et al. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat Neurosci. 2015;18:603–610. doi: 10.1038/nn.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Veno MT, et al. Spatio-temporal regulation of circular RNA expression during porcine embryonic brain development. Genome Biol. 2015;16:245. doi: 10.1186/s13059-015-0801-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rybak-Wolf A, et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell. 2015;58:870–885. doi: 10.1016/j.molcel.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 57.Begemann G, et al. muscleblind, a gene required for photoreceptor differentiation in Drosophila, encodes novel nuclear Cys3His-type zinc-finger-containing proteins. Development. 1997;124:4321–4331. doi: 10.1242/dev.124.21.4321. [DOI] [PubMed] [Google Scholar]
- 58.Westholm JO, et al. Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep. 2014;9:1966–1980. doi: 10.1016/j.celrep.2014.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alhasan AA, et al. Circular RNA enrichment in platelets is a signature of transcriptome degradation. Blood. 2015;127:e1–e11. doi: 10.1182/blood-2015-06-649434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li Y, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25:981–984. doi: 10.1038/cr.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang W, et al. Foxo3 activity promoted by non-coding effects of circular RNA and Foxo3 pseudogene in the inhibition of tumor growth and angiogenesis. Oncogene. 2015 doi: 10.1038/onc.2015.460. Published online December 14, 2015. http://dx.doi.org/10.1038/onc.2015.460. [DOI] [PubMed]


