Abstract
To identify cell-surface markers specific to human cardiomyocytes, we screened cardiovascular cell populations derived from human embryonic stem cells (hESCs) against a panel of 370 known CD antibodies. This screen identified the signal-regulatory protein alpha (SIRPA) as a marker expressed specifically on cardiomyocytes derived from hESCs and human induced pluripotent stem cells (hiPSCs), and PECAM, THY1, PDGFRB and ITGA1 as markers of the nonmyocyte population. Cell sorting with an antibody against SIRPA allowed for the enrichment of cardiac precursors and cardiomyocytes from hESC/hiPSC differentiation cultures, yielding populations of up to 98% cardiac troponin T-positive cells. When plated in culture, SIRPA-positive cells were contracting and could be maintained over extended periods of time. These findings provide a simple method for isolating populations of cardiomyocytes from human pluripotent stem cell cultures, and thereby establish a readily adaptable technology for generating large numbers of enriched cardiomyocytes for therapeutic applications.
Generation of cardiovascular cells from human pluripotent stem cells (hPSCs) in culture could provide a powerful model system for investigating cellular interactions and molecular regulators that govern the specification, commitment and maturation of these lineages, as well as a unique and unlimited source of human cardiomyocytes for drug testing and regenerative medicine strategies1–4. Translating this potential into practice, however, will depend on the development of technologies that enable the reproducible generation of highly enriched populations of cardiomyocytes, as contaminating cell types could affect drug responses and other functional properties in vitro and increase the risk of abnormal growth and teratoma formation following transplantation in vivo5. When induced under optimal cardiac conditions, hPSCs efficiently differentiate to generate mixed cardiovascular populations, including cardiomyocytes, smooth muscle cells, fibroblasts and endothelial cells3. Although cardiomyocytes can represent up to 70% of the population for any given hPSC line, the efficiency of generating this lineage varies considerably between different stem cell lines. Manipulation of induction conditions has not yet yielded strategies for the generation of pure populations of cardiomyocytes from a broad range of hPSC lines.
To enrich for cardiomyocytes from differentiation cultures, previous studies have introduced cardiomyocyte-specific fluorescent reporters or drug selectable elements into hPSCs6–8, allowing enrichment by fluorescence-activated cell sorting (FACS) or the addition of appropriate selection drugs. However, these strategies suffer from a major drawback as the introduction of a reporter vector into an hPSC line results in genetically modified cardiomyocytes, reducing their utility for clinical applications. Recently, cardiomyocytes were isolated by FACS based on their high mitochondrial content9. This approach appears useful for isolating mature cardiomyocytes, but cells with fewer mitochondria, such as immature hPSC-derived cardiomyocytes, may be more difficult to distinguish from other cell types.
To overcome these limitations and develop broadly applicable strategies for the enrichment of hPSC-derived cardiomyocytes, we used a high-throughput flow cytometry screen to identify cell-surface markers specific for human cardiomyocytes. Here we report that the cell-surface receptor SIRPA is expressed on hPSC-derived cardiomyocytes as well as on human fetal cardiomyocytes. Using cell sorting with an antibody against SIRPA, we demonstrate that it is possible to isolate populations consisting of up to 98% cardiomyocytes from hPSC differentiation cultures.
RESULTS
Surface of markers expressed on hESC–derived cardiomyocytes
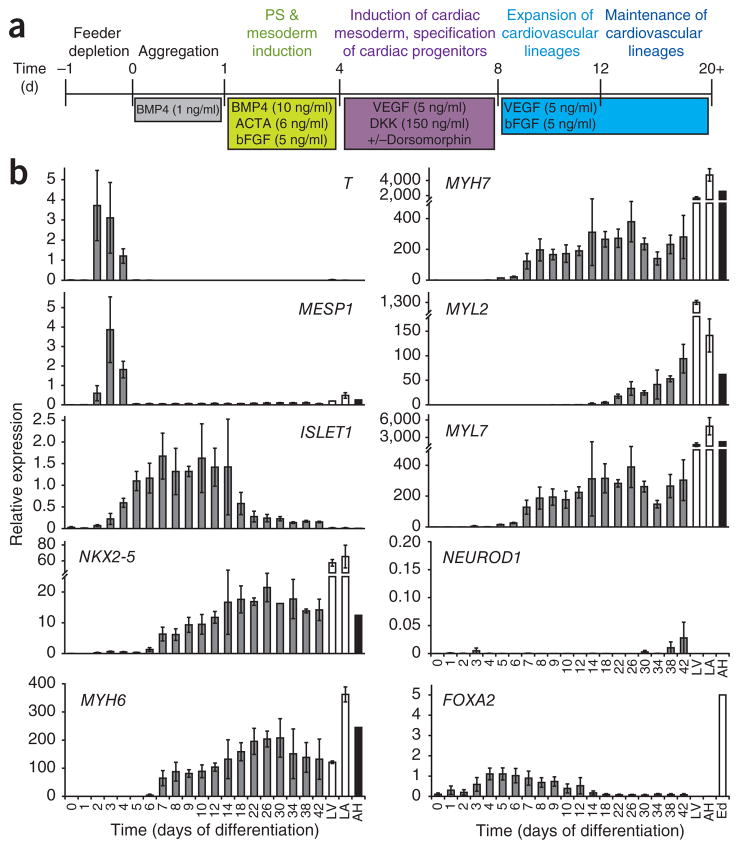
When induced with appropriate concentrations of activin A and BMP4 (Fig. 1a), the HES2 hESC line efficiently and reproducibly differentiates to generate cardiovascular lineage cells2,3. Kinetic analyses of the differentiation cultures revealed a step-wise developmental progression from a primitive streak–like population defined by T (BRACHYURY) expression (days 2–4) to the development of early mesoderm (MESP1; days 3 and 4) and the emergence of NKX2-5− and ISLET1 (also known as ISL1)-positive cardiac precursors (days 4–8). Contracting cardiomyocytes were first detected between days 9 and 12 of differentiation, coincident with the upregulation of MYH6 (also known as αMHC), MYH7 (also known as βMHC) and MYL7 (also known as MLC2A) and later MYL2 (also known as MLC2v) expression (Fig. 1b). The expression levels of some of the cardiac-specific genes in the hESC-derived populations were considerably lower than those in fetal and adult heart tissue. Low levels of NEUROD1 and FOXA2 expression indicated that the cultures were not contaminated with substantial numbers of neuroectoderm or endoderm-derived cells. To monitor cardiomyocyte development in real time, we applied the above protocol to an NKX2-5–GFP reporter hESC line that contains the EGFP cDNA inserted into the NKX2-5 locus of HES3 hESCs10. The first NKX2-5–GFP+ cells developed between days 7 and 8 of differentiation. The size of the NKX2-5–GFP+ population increased with time, reaching a maximum between days 12 and 20 (Supplementary Fig. 1). Epifluorescence analysis of embryoid bodies derived from NKX2-5-GFP hESCs confirmed nuclear GFP expression in the majority of the cells (Supplementary Movie 1). The kinetics of NKX2-5–GFP expression closely paralleled the onset of NKX2-5 expression in the HES2 cultures, indicating that cardiac specification from both hESC lines takes place between days 6 and 8 of differentiation (Fig. 1b and Supplementary Fig. 1). The high proportion of NKX2-5–GFP+ cells in day 20 cultures demonstrates that the differentiation protocol, used efficiently, promotes the generation of cardiomyocytes from this hESC line.
Figure 1.
Specification of the cardiovascular lineage from hESCs. (a) Outline of the protocol used to differentiate hESCs to the cardiac lineage (modified from ref. 3). (b) QPCR analysis of T, MESP1, ISLET1, NKX2-5, MYH6, MYH7, MYL2, MYL7, NEUROD1 and FOXA2 in HES2-derived embryoid bodies at different stages during differentiation. Day 0, hESCs; LV, human fetal left ventricle; LA, human fetal left atria; AH, human adult heart, Ed, hESC-derived endoderm14. Error bars represent s.e.m., n = 3.
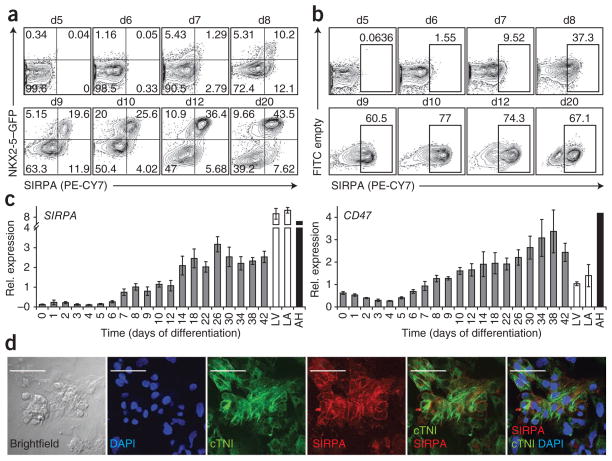
To determine whether the above developmental stages can be distinguished by cell-surface markers, we carried out a screen of 370 known antibodies (http://data.microarrays.ca/AntibodyWeb) using day 8, 12 and 20 populations generated from the NKX2-5–GFP cell line. The initial screen focused on identifying antibodies that recognized antigens present on the NKX2-5–GFP+ population. From this screen, we identified SIRPA (also known as SHPS-1 or CD172a) as a potential cardiac-specific marker, as the anti-SIRPA antibody11 stained the majority of the NKX2-5–GFP+ cells and almost none of the NKX2-5–GFP− cells (Fig. 2a). From the panel of antibodies analyzed, SIRPA was the only one that displayed this cardiomyocyte-specific expression pattern. SIRPA was first detected on emerging GFP-NKX2-5+ cells on day 8 of differentiation, a population considered to represent the cardiac precursor stage of development. Expression was maintained on the GFP-NKX2-5+ population throughout the 20-d time course of the experiment (Fig. 2a and Supplementary Fig. 2a). No SIRPA+ cells were detected in undifferentiated hESC populations or in the day 5 cardiac mesoderm population characterized by co-expression of KDR and PDGFRA (Fig. 2a and data not shown)2. Analyses of embryoid bodies generated from the nongenetically modified HES2 line revealed a similar staining pattern with the anti-SIRPA antibody. SIRPA+ cells were first detected between days 7 and 8 of differentiation and the percentage of positive cells increased strongly over the next 2–4 (Fig. 2b and Supplementary Fig. 2b). Both the directly conjugated (SIRPA-PE-CY7) and the biotinylated (SIRPA-bio) antibodies stained similar portions of the day 20 embryoid body population (Supplementary Fig. 3a–e). Notably, the SIRPA+ cells detected in day 20 embryoid bodies appear to be substantially larger than those in the SIRPA− population (Supplementary Fig. 3f), indicating cardiovascular cell size can be monitored using this antibody. To confirm the specificity of the SIRPA antibody, we carried out western blot analyses and immunoprecipitation for SIRPA (Supplementary Fig. 4). These experiments demonstrated the presence of SIRPA protein in three independent day 20 embryoid body–derived populations, but not in undifferentiated hESCs (Supplementary Fig. 4a). Immunoprecipitation analyses revealed a band the size of that previously described for the SIRPA protein (Supplementary Fig. 4b)12.
Figure 2.
Expression of the cell-surface receptor SIRPA during hESC differentiation. (a) Flow cytometric analysis of SIRPA on embryoid bodies derived from NKX2-5–GFP hESCs. d; day of differentiation. (b) Expression of SIRPA on HES2-derived embryoid body populations at the indicated times. (c) RT-qPCR analysis of expression of SIRPA and its ligand CD47 in HES2-derived embryoid bodies at different times of differentiation. Day 0, ESCs; LV, human fetal left ventricle; LA, human fetal left atrial; AH, human adult heart. Error bars represent s.e.m., n = 4. (d) Immunostaining for SIRPA and cTNI on cardiac monolayer cultures. Monolayers were generated from day 20 HES2-derived embryoid bodies. Scale bars, 50 μm. Rel., relative.
Co-staining of SIRPA and cardiac Troponin T (cTNT) by flow cytometry displayed clear co-expression of the two markers (Supplementary Fig. 5a,b), indicating that SIRPA was specifically expressed on the cardiomyocyte lineage in differentiated populations generated from the nonmodified HES2 cell line. Reverse transcriptase (RT)-quantitative (q)PCR analyses revealed an expression pattern for SIRPA that closely mirrored the flow cytometry antibody staining profile, with an upregulation of SIRPA mRNA between days 6 and 8 of differentiation, followed by persistence of expression over the 42-day time course. Expression of CD47, the ligand for SIRPA, paralleled that observed for SIRPA (Fig. 2c). Flow cytometric analysis of CD47 reflected the gene expression pattern, showing low levels of staining on undifferentiated ESCs and on day 5 differentiation cultures, followed by broad staining of the entire population between days 8 and 20 (data not shown). Immunofluorescence analysis of monolayer cultures derived from day 20 embryoid bodies revealed SIRPA surface expression exclusively on cardiomyocytes, as characterized by co-expression with cardiac TroponinI (cTNI) (Fig. 2d). The respective controls (IgG and secondary antibody only) did not show any staining (data not shown). Collectively, these kinetics studies show that expression of SIRPA uniquely marks the cardiac lineage in hESC differentiation cultures, beginning with the emergence of NKX2-5+ precursor cells and persisting through the development and expansion of contracting populations. In an independent study using genomic analysis, SIRPA was also identified as a marker specific for the cardiac lineage10.
Cardiomyocytes were recently isolated based on mitochondria content, as measured by retention of MitoTracker dye9, which stains mitochondria. Comparison of MitoTracker dye labeling with SIRPA staining indicated that both procedures mark the same cardiomyocyte population in day 20 embryoid bodies (Supplementary Fig. 6c). The dye retention approach was, however, less useful in tracking the onset of cardiovascular development, as it marked a less distinct population on day 12 of differentiation and almost no cells on day 8 (Supplementary Fig. 6a,b). In contrast, a substantial SIRPA+ population could be clearly resolved at both these time points, indicating that this surface marker allows one to monitor and isolate cells from different stages of cardiac development, whereas labeling with the MitoTracker dye can be used only on populations containing relatively mature cardiomyocytes.
In contrast to the human cells, Sirpa was not detected on mouse ESC–derived cardiomyocytes by antibody staining (Supplementary Fig. 7a). Sirpa+ populations in the culture were cTnT and CD45+, indicating that they represent hematopoietic cells (Supplementary Fig. 7a,b). Gene expression analyses confirmed the flow cytometric data, and showed only low levels of Sirpa mRNA in the mESC-derived cardiomyocytes as well as in adult mouse atrial and ventricular tissues, compared with high expression in the brain (Supplementary Fig. 7c). Expression of the only other known Sirp family member in the mouse, Sirpb, could not be detected in any of these tissues by qPCR (data not shown). Western blot analysis of control and Sirpa-deficient mouse tissue confirmed high Sirpa expression in the brain of control mice, but not in any of the tissues derived from Sirpa-deficient mice (Supplementary Fig. 7d). Most importantly, no Sirpa expression was detected in the heart, kidney or mESC-derived cardiomyocytes from control mice. Differences in SIRPA function and protein homology between mouse and human have been described previously for the interaction of macrophages and red blood cells13.
Purification of cardiomyocytes from hESC-derived populations
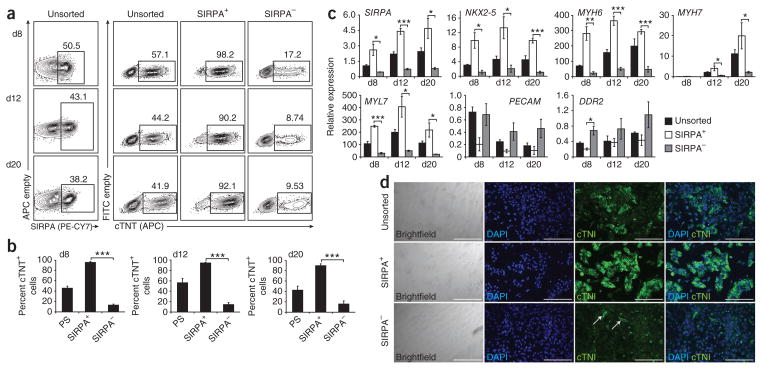
To assess whether expression of the SIRPA surface receptor can be used to generate enriched populations of cardiomyocytes, SIRPA+ and SIRPA− fractions were isolated by cell sorting from HES2-derived embryoid bodies on days 8, 12 and 20 of differentiation and analyzed for expression of cTNT by intracellular flow cytometry (Fig. 3a). Analyses of the unsorted populations demonstrated that cTNT expression closely paralleled that of SIRPA at the corresponding stages during differentiation (days 8, 12 and 20). After sorting, the SIRPA+ fractions from each stage were highly enriched for cTNT+ cardiomyocytes, whereas the SIRPA− fractions were depleted of these cells. It is unclear whether the low numbers of cTNT+ cells present in the SIRPA− fractions are contaminants from the sorting procedure or represent true SIRPA− cardiomyocytes. FACS-based separation in multiple experiments reproducibly yielded significantly enriched populations of cardiomyocytes (SIRPA+: day 8 (95.2% ± 1.9), day 12 (94.4% ± 1.7), day 20 (89.6% ± 3.6); SIRP−: day 8 (13.0% ± 2.1), day 12 (14.3% ± 3.9), day 20 (15.7% ± 6.0)) (Fig. 3b). The purity of the SIRPA+ and SIRPA− sorted populations and the efficiency of cell recovery from the sorting procedure are summarized in Supplementary Figure 8 and Supplementary Table 1.
Figure 3.
Enrichment of cardiomyocytes from hESC-derived cultures by cell sorting based on SIRPA expression. (a) Flow cytometric analysis of SIRPA expression in embryoid bodies at day (d)8, d12 and d20 of differentiation. FACS for SIRPA was performed at d8, d12 and d20. The unsorted, SIRPA+ and SIRPA− fractions from each time point were analyzed for cTNT expression by intracellular flow cytometry. The frequency of cTNT+ cells at d8, d12 and d20 was significantly higher in the SIRPA+ fraction (d8: 95.2% ± 1.9, d12: 94.4 ± 1.7, d20: 89.6 ± 3.6), compared to SIRPA− cells (d8: 13.0 ± 2.1, d12: 14.3 ± 3.9, d20: 15.7 ± 6.0, P ≤ 0.001). (b) Average enrichment of cTNT+ cells from three different cell separation experiments. Error bars represent s.e.m. Asterisks indicate statistical significance as determined by student’s t-test, ***, P ≤ 0.001. (c) QPCR analysis of unsorted, SIRPA+ and SIRPA− cells. Expression of SIRPA, NKX2-5, MYH6, MYH7 and MYL7 was significantly higher in the SIRPA+ fraction compared to SIRPA− fraction at all stages analyzed (d8, d12 and d20). Expression of markers for the noncardiac lineages (PECAM and DDR2) segregated to the SIRPA− fraction. Error bars represent s.e.m. Asterisks indicate statistical significance as determined by student’s t-test. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; n = 3. (d) Immunostaining of cTNI on monolayer cultures generated from unsorted, SIRPA+ and SIRPA− cells sorted on day 20. Scale bars, 200 μm.
Molecular analyses revealed that SIRPA+ cells expressed significantly higher levels of NKX2-5, MYH6, MYH7 and MYL7 compared with the SIRPA− population (Fig. 3c), further demonstrating enrichment of cardiomyocytes. As expected, SIRPA expression segregated to the SIRPA+ population. In contrast to the cardiac markers, nonmyocyte markers, such as the fibroblast markers DDR2 and THY1 (CD90, data not shown) and the endothelial marker PECAM (CD31), were expressed at higher levels in the SIRPA− population (Fig. 3c).
When plated in monolayer cultures, cells from both SIRPA− and SIRPA+ fractions formed viable populations that could easily be maintained for several weeks. Contracting cells were detected in unsorted and SIRPA+-derived populations, but not in the population generated from the SIRPA− cells (Supplementary Movies 2–4). Immunohistochemical analysis revealed broad cTNI expression in the SIRPA+ population, confirming the high proportion of cardiomyocytes in these cultures. Only a few cTNI+ cells were detected in the SIRPA− population (Fig. 3d).
As anticipated from the co-expression of SIRPA and NKX2-5–GFP, it was also possible to isolate populations enriched for cardiac lineage cells from NKX2-5–GFP HES3-derived cultures by sorting with the anti-SIRPA antibody. Cardiac precursors (day 8) and cardiomyocytes (days 12 and 20) defined by gene expression and cTNT staining, segregated to the SIRPA+ fraction, whereas nonmyocyte cells were enriched in the SIRPA− population (Supplementary Fig. 9).
To enable rapid processing of large numbers of cells, we also attempted to isolate SIRPA+ cells by magnetic bead sorting. Isolation of SIRPA+ cells from NKX2-5–GFP differentiation cultures by this approach resulted in populations highly enriched for cardiomyocytes, similar to those derived from FACS experiments (Supplementary Fig. 10a–c). However, with current magnetic bead sorting protocols, a substantial number of cells are lost. Further optimization of these protocols or improvement in the intensity of staining should increase cell enrichment and yield with this approach.
Taken together, the findings from these cell-sorting studies clearly demonstrate that SIRPA expression marks the cardiac lineage in hESC-derived differentiation cultures and that cell sorting with the anti-SIRPA antibody allows for the isolation of populations highly enriched for cardiomyocytes.
Purification of cardiomyocytes from hiPSCs
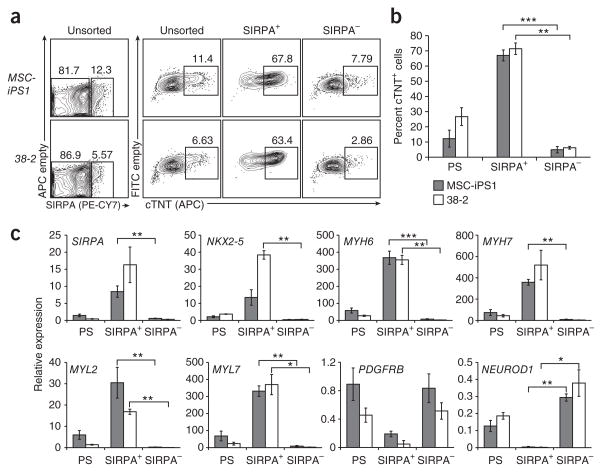
To determine whether SIRPA expression marks the cardiac lineage in other hPSC-derived populations, we next analyzed embryoid bodies generated from two different hiPSC lines, MSC-iPS1 (also known as Y2-1) and 38-2 (refs. 14,15). The efficiency of cardiac differentiation from both lines was low, as demonstrated by the proportion of cTNT+ cells (MSC-iPS1: 12.2% ± 5.6; 38-2: 26.7% ± 5.7; Fig. 4a). Similar low levels of SIRPA expression were detected in both embryoid body populations. FACS of the SIRPA+ cells from both iPSC lines yielded populations significantly enriched for cTNT+ cardiomyocytes (SIRPA+: MSC-iPS1 (67.0% ± 3.6); 38-2 (71.4% ± 3.8); SIRPA−: MSC-iPS1 (4.9% ± 2.1), 38-2 (6.2% ± 0.9)) (Fig. 4a,b). These SIRPA+ populations expressed significantly higher levels of NKX2-5, MYH6, MYH7, MYL2 and MYL7 than did the corresponding SIRPA− cells. As observed with the hESC-derived cells, nonmyocyte markers, including DDR2, PDGFRB, THY1 and NEUROD, segregated to the SIRPA− fraction (Fig. 4b,c and data not shown). These data clearly document the utility of this marker for generating enriched cardiac populations from a range of hPSC lines, including those that do not differentiate efficiently to the cardiac lineage with current protocols.
Figure 4.
Enrichment of cardiomyocytes from hiPSC-derived cultures by cell sorting based on SIRPA expression. (a) Flow cytometric analysis of SIRPA expression at d20 of differentiation on 38-2 and MSC-iPS1 hiPSC-derived cells. FACS for SIRPA was performed on d20 and the unsorted, SIRPA+ and SIRPA− fractions were analyzed for cTNT expression by intracellular flow cytometry. (b) The frequency of cTNT+ cells was significantly higher in the SIRPA+ fraction of both hiPSC-derived cultures (MSC-iPS1: 67.0 ± 3.6, 38-2: 71.4 ± 3.8), compared to SIRPA− cells (MSC-iPS1: 4.9 ± 2.1; 38-2: 6.2 ± 0.9). Error bars represent s.e.m. Asterisks indicate statistical significance as determined by student’s t-test. **, P ≤ 0.01; ***, P ≤ 0.001; n = 3. (c) QPCR analysis of unsorted, SIRPA+ and SIRPA− cells isolated from MSC-iPS1 and 38-2 hiPSC-derived day 20 cultures. Expression of markers specific for the cardiac lineage (SIRPA, NKX2-5, MYH6, MYH7, MYL2 and MYL7) was significantly higher in the SIRPA+ compared to the SIRPA− fraction. Expression of markers for the noncardiac lineages (PDGFRB and NEUROD1) segregated to the SIRPA− fraction and the PS cells. Error bars represent s.e.m. Asterisks indicate statistical significance as determined by student’s t-test. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; n = 5.
SIRPA expression in human fetal and adult heart cells
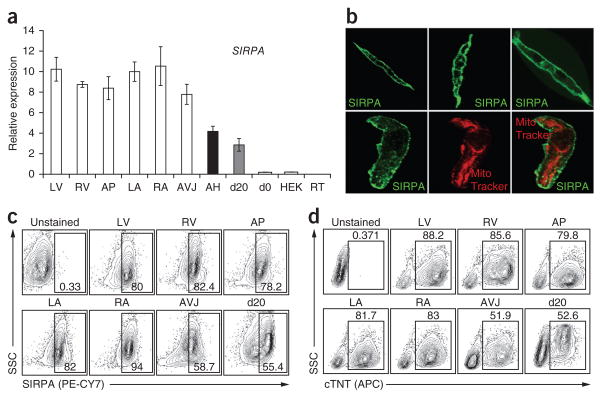
To determine whether SIRPA is expressed on primary human cardiomyocytes, we next analyzed expression patterns in fetal (18–20 weeks of gestation) and adult heart tissue by RT-qPCR. SIRPA transcripts were detected in all fetal-derived heart tissue (left and right atrial cells, left and right ventricle cells, apex and atrioventricular junction; Fig. 5a), at similar or higher levels to those found in day 20 hESC-derived cells (Fig. 3a). SIRPA was not expressed in undifferentiated hESCs (day 0) or in control HEK (human embryonic kidney) cells. Similar to the fetal heart, SIRPA expression was also detected in the adult heart, suggesting that its expression marks cardiomyocytes at different stages of human cardiac development. High levels of SIRPA were also detected in the adult human brain and lung (Supplementary Fig. 11a). Most other human tissues expressed only low levels of SIRPA that could reflect the presence of tissue macrophages that are known to express this receptor16,17. CD47, the SIRPA ligand, was expressed in most tissues, confirming the pattern described in previous studies (Supplementary Fig. 11b)16. Immunofluorescence staining showed that SIRPA was localized on the surface membrane of the fetal ventricular cells but was not present on other membrane fractions such as the mitochondrial membrane, as indicated by the lack of co-staining with MitoTracker Red (Fig. 5b). Flow cytometric analyses revealed a high proportion of SIRPA+ cells in all fetal heart tissues at levels that correlated with the percentage of cTNT+ cells in the respective fractions (Fig. 5c,d).
Figure 5.
Expression of SIRPA on human fetal cardiomyocytes and in adult human heart. (a) RT-qPCR analysis for SIRPA in human fetal heart tissue and adult heart. LV, left ventricle; RV, right ventricle; AP, Apex; LA, left atria; RA, right atria, AVJ, atrioventricular junction; AH, adult heart; d20, day 20 embryoid bodies, day 0, hESCs; HEK, human embryonic kidney cells; RT, reverse transcriptase control. Error bars represent s.e.m., n = 6. (b) Immunostaining for SIRPA (green) on human fetal ventricular cells and staining with MitoTracker Red. (c) Flow cytometric analysis for SIRPA on human fetal heart tissue. (d) Intracellular flow cytometric analysis for cTNT on human fetal heart tissue.
These findings clearly demonstrate that SIRPA is expressed on fetal cardiomyocytes as well as in adult heart, illustrating that its cardiac-specific expression is not an artifact of hPSC-derived populations.
Monitoring hPSC differentiation by SIRPA expression
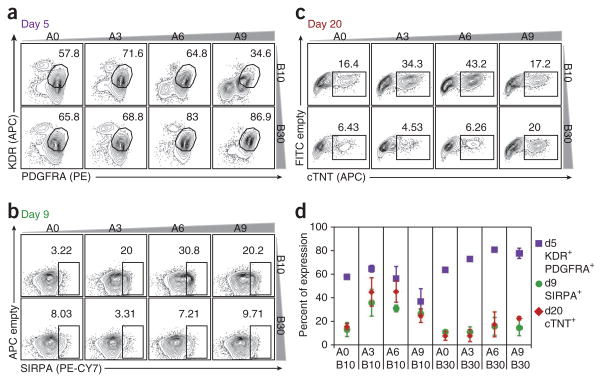
Recently, we reported that co-expression of KDR and PDGFRA provides a reliable method to monitor cardiac mesoderm induction after treatment with BMP4 and activin A2 (Fig. 6a). Although this study showed that induction of a KDR+PDGFRA+ population is an essential first step in the generation of cardiomyocytes, not all KDR+PDGFRA+ populations differentiated to give rise to cardiac lineage cells (an example of this type of population is cells induced with 30 ng/ml BMP4 and no exogenous activin A (A0)). To determine whether SIRPA would more accurately predict the cardiac potential of differentiating populations at an early stage, we monitored its expression in day 9 embryoid bodies induced with different concentrations of activin A and BMP4 (Fig. 6b). The same populations were evaluated on day 5 for expression of KDR and PDGFRA (Fig. 6a) and on day 20 for expression of cTNT (Fig. 6c). Although there was little correlation between the size of the KDR+PDGFRA+ population on day 5 and the proportion of cTNT+ cells on day 20, the cultures with the largest SIRPA+ population on day 9 (activin A, 6 ng/ml; BMP4 10 ng/ml) contained the highest number of cTNT+ cells at the later time point. SIRPA expression correlated well with cTNT output for most conditions tested, and the highest levels of SIRPA predicted the highest cardiomyocyte development on day 20 (Fig. 6d). These data demonstrate that expression of SIRPA on day 9 is a reliable indicator of cardiomyocyte potential, and as such can be used to monitor and optimize induction protocols for directed differentiation of hPSCs to the cardiac lineage.
Figure 6.
Using SIRPA to predict cardiac differentiation efficiency. (a) Day 5 KDR/PDGFRA flow cytometry profiles of cells from cardiac differentiation cultures induced with varying combinations of activin A (A0, 3, 6, 9 ng/ml) and BMP4 (B10, 30 ng/ml). The KDR+PDGFRA+ population has been shown to contain cardiac mesoderm cells2. (b) Day 9 SIRPA flow cytometric analysis expression profiles of the cultures described in a. (c) Day 20 cTNT profiles (intracellular flow cytometric analysis) of the cultures described in a. (d) Quantification of a–c. Close correlation of expression of SIRPA on day 9 (green dots) and cTNT expression on day 20 (red rhombuses) illustrates the predictive potential of SIRPA for cardiac differentiation efficiency.
Depletion of the nonmyocyte lineage cells
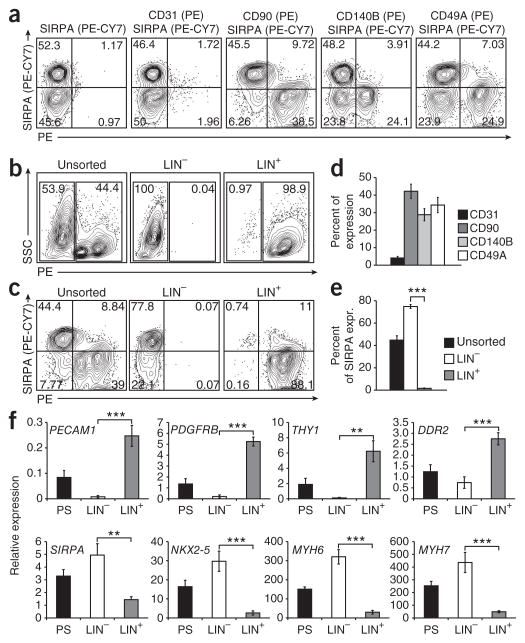
In addition to antibodies that recognize cardiomyocytes, our flow cytometric screen also identified a panel of antibodies that marked the nonmyocyte population in the differentiation cultures. This set of antibodies, including anti-CD90 (also known as THY1, expressed on fibroblast cells), anti-CD31 (also known as PECAM1, expressed on endothelial cells), anti-CD140B (also known as PDGFRB, expressed on smooth muscle cells) and anti-CD49A (also known as ITGA1), all recognized different proportions of the SIRPA− population of day 20 HES2-derived embryoid bodies (Fig. 7a,d). The combination of these antibodies marked the majority of nonmyocyte (SIRPA) cells in the culture (Fig. 7c, unsorted). To determine whether it is possible to enrich for cardiomyocytes by depleting cells expressing the nonmyocyte markers, we combined these antibodies and sorted day 20 embryoid bodies into lineage-positive (LIN+) and lineage-negative (LIN−) fractions (Fig. 7b). This approach has the advantage of generating enriched populations free of any bound antibody or magnetic beads. As expected, the LIN− population was significantly enriched for SIRPA+ cells, whereas the LIN+ population was depleted for the cardiomyocytes (Fig. 7c,e). The efficiency of cell recovery after FACS for LIN− and LIN+ cells is summarized in Supplementary Table 2. Gene expression analyses revealed that nonmyocyte-specific genes, including PECAM1, PDGFRB, THY1 and DDR2, were primarily expressed in the LIN+ fraction, whereas cardiac gene expression was restricted to the LIN− fraction (Fig. 7f). When plated on gelatin-coated dishes or reaggregated as cell clusters, the LIN− fraction generated populations that contained a high proportion of contracting cardiomyocytes (data not shown). The same lineage cocktail of antibodies also marked the nonmyocyte (SIRPA−) fraction of the iPSC (MSC-iPS1)-derived day 20 embryoid body population (Supplementary Fig. 12), indicating that this depletion approach can be applied to different hPSC lines that display variable differentiation efficiencies.
Figure 7.
Enrichment of cardiomyocytes through negative selection. (a) Flow cytometric analysis of markers specifically expressed on nonmyocyte (SIRPA-negative) cells in day 20 differentiation cultures (HES2). (b) FACS for the combination of markers specifically expressed on nonmyocyte cells (in PE: CD31, CD90, CD140B, CD49A). (c) Flow cytometric analysis of the unsorted cells, PE-negative (LIN−) and PE-positive (LIN+) samples for SIRPA. (d) Quantification of nonmyocyte markers on day 20 of differentiation (as shown in a), n = 4. (e) Quantification of SIRPA+ cells in PS, LIN− and LIN+ fractions after cell sorting. Asterisks indicate statistical significance as determined by student’s t-test, ***, P ≤ 0.001; n = 3. (f) QPCR analysis of the unsorted, LIN− and LIN+ samples for noncardiac markers (PECAM1, PDGFRB, THY1 and DDR2) and cardiac specific genes (SIRPA, NKX2-5, MYH6 and MYH7). Error bars represent s.e.m. Asterisks indicate statistical significance as determined by student’s t-test, **, P ≤ 0.01; ***, P ≤ 0.001; n = 3. Expr., expression.
Taken together, these data show that cardiomyocytes can be enriched from hPSC-derived differentiation cultures by depletion of the nonmyocte lineages. This method therefore represents an alternative approach to obtaining highly purified cardiomyocyte cultures and as such can be used when purified cardiomyocyte populations free of bound antibodies are required.
DISCUSSION
Advances in our understanding of the signaling pathways that regulate lineage specification have led to strategies for efficient and reproducible directed differentiation of hPSCs to specific cell types1. With respect to cardiac lineage development, protocols have been established that promote the generation of mixed cardiovascular populations containing the major cell types found in the human heart, including cardiomyocytes, endothelial cells, vascular smooth muscle cells and fibroblasts. Cardiomyocytes typically represent 10–70% of such mixed populations2,3, depending on the hPSC line used. Although mixed populations have been used to demonstrate the potential utility of hPSC-derived cells for predictive toxicology5, modeling human disease in vitro18,19 and transplantation-based therapy for heart disease20, highly enriched and well-defined cell populations will ultimately be required to translate this potential into practical applications.
Our identification of SIRPA as a cardiomyocyte-specific marker now enables easy and routine access to highly enriched populations of cardiomyocytes from hESCs and hiPSCs. Such cardiomyocyte-enriched populations can be isolated by FACS or magnetic bead sorting, the latter approach enabling the isolation of large numbers of cells required for in vivo studies. Access to highly enriched populations of cardiomyocytes through simple sorting approaches will enable the development of defined high-throughput drug discovery and toxicology assays, the detailed phenotypic evaluation of cells generated from patient-specific hPSCs and the generation of defined populations safe for transplantation. The fact that SIRPA is expressed on cardiac-lineage cells from the earliest cardiac stage to contracting and more mature cardiomyocytes will allow for comparisons of the in vivo potential of the different populations.
In addition to SIRPA, our screen also identified a panel of markers defining the nonmyocyte fractions of the hPSC-derived cardiovascular population. Although these nonmyocyte populations have not been fully characterized, the markers used suggest that they represent a combination of fibroblasts (CD90)21, vascular smooth muscle cells (CD140B)22 and endothelial cells (CD31). Access to enriched populations of each of these cell types together with cardiomyocytes will allow detailed investigation of interactions between them. Such interactions may alter the drug sensitivity and responsiveness of cardiomyocytes and are likely to influence the survival and maturation of cardiomyocytes in vitro and following transplantation in vivo. Many of the proposed applications for hPSC-derived cardiomyocytes may require three-dimensional engineered tissue to more accurately reflect drug responses and function in the adult heart. Recent studies suggest that appropriate combinations of cardiac cells, endothelial cells and fibroblasts must be incorporated into such tissue constructs for them to function best in vitro or in vivo23–25. Our ability to generate pure myocyte and nonmyocyte populations will allow for the generation of engineered constructs consisting of varying proportions of different cell types, enabling us to determine the optimal combination of each required to form heart tissue with structural and functional properties most similar to that of the human heart.
The specific expression pattern of SIRPA in hPSC-derived populations and in the fetal heart tissue suggests that this receptor plays some functional role in the human cardiomyocyte lineage, perhaps as early as the precursor stage of development. The fact that expression of the ligand, CD47, is upregulated in parallel with SIRPA in embryoid bodies and that CD47 is found on a large proportion of the cells in the culture further supports the interpretation that this ligand/receptor pair plays a role in human cardiomyocyte development and/or function. One thoroughly studied role for SIRPA is on macrophages, where it appears to mediate a signal to eliminate cells from the body that do not express the ligand CD47 (ref. 17). The only other suggested function in human cells is in the smooth muscle lineage, where SIRPA has been shown to play an important role in mediating IGF-1–induced mitogenic signaling26. Given that SIRPA was not detected in mouse cardiomyocytes, its function in human cells may relate to aspects of cardiomyocyte physiology and/or function that differ between the two species. A clearer understanding of its role in the cardiac lineage will require gain- and loss-of-function approaches using the in vitro differentiation model.
In summary, the findings reported here demonstrate that expression of SIRPA uniquely marks the cardiomyocyte lineage in hPSC differentiation cultures. Isolation of SIRPA+ cells by FACS or magnetic bead sorting provides a simple approach for generating highly enriched populations of cardiomyocytes from a broad range of hPSC lines, including those that do not differentiate efficiently to the cardiovascular lineage using current protocols. Although SIRPA appears to mark most cardiomyocytes in the population, future screens may uncover markers that define subpopulations of these cells or specific stages of development within the lineage.
ONLINE METHODS
HPSC maintenance and differentiation
HPSCs were maintained as described27. Embryoid bodies (EBs) were differentiated to the cardiovascular lineage as previously described2,3 (Fig. 1a). In brief, EBs were generated by culture in StemPro-34 (Invitrogen) media containing BMP4 (1 ng/ml). On day 1, EBs were harvested and resuspended in induction medium (StemPro-34, basic fibroblast growth factor (bFGF; 2.5 ng/ml), activin A (6 ng/ml) and BMP4 (10 ng/ml)). On day 4, the EBs were harvested from the induction medium and recultured in StemPro-34 supplemented with vascular endothelial growth factor (VEGF; 10 ng/ml) and DKK1 (150 ng/ml). On day 8, the medium was changed again and the EBs were cultured in StemPro-34 containing VEGF (20 ng/ml) and bFGF (10 ng/ml) for the duration of the experiment. Cultures were maintained in a 5% CO2, 5% O2, 90% N2 environment from for the first 12 d and then transferred into a 5% CO2/air environment for the remainder of the culture period.
NKX2-5–GFP hESCs were generated by targeting sequences encoding GFP to the NKX2-5 locus of HES3 cells using previously described protocols10,28.
Work involving human tissue collection and analysis was carried out in accordance with and approved through the Human Ethics Committee at the University Health Network.
Flow cytometry and cell sorting
Dissociation procedure for day 5 to day 12 EBs
EBs generated from hPSC differentiation experiments were dissociated with 0.25% trypsin/EDTA.
Dissociation procedure for day 13 and older EBs and human fetal tissue
EBs generated from hPSC differentiation cultures were incubated in collagenase type II (1 mg/ml; Worthington) in Hanks solution (NaCl, 136 Mm; NaHCO3, 4.16 mM; NaPO4, 0.34 mM; KCl, 5.36 mM; KH2PO4, 0.44 mM; dextrose, 5.55 mM; HEPES, 5 mM) overnight at 25 °C with gentle shaking29. On the following day, the equivalent amount of dissociation solution (in Hanks solution: taurin, 10 mM, EGTA 0.1 mM, BSA 1 mg/ml, collagenase type II 1 mg/ml) was added to the cell suspension and the EBs were pipetted gently to dissociate the cells. After dissociation, cells were centrifuged (1,000 r.p.m., 5 min), filtered and used for analysis. For EBs older than 40 d, additional treatment with 0.25% trypsin/EDTA is often required to obtain complete dissociation to single-cell suspensions.
Cells were stained at a concentration of 2.5 × 106 cells/ml with anti-KDR-allophycocyanin (R&D Systems; 1:10) and anti-PDGFRA–phycoerythrin (R&D Systems; 1:20), anti-SIRPA–IgG-phycoerythrin-Cy7 (clone SE5A5; BioLegend; 1:500)11,30, anti-SIRPA-IgG-biotin (clone SE5A5; BioLegend; 1:500)11, anti-cardiac isoform of cTNT (clone 13-11; NeoMarkers; 1:400), goat anti-mouse IgG– allophycocyanin (BD; 1:200), streptavidin-allophycocyanin (BD: 1:200), anti-IgG1κ-phycoerythrin-Cy7 (clone MOPC-21; BioLegend; 1:500), anti-IgG1κ-biotin (clone MOPC-21; BioLegend; 1:500).
For cell-surface markers, staining was carried out in PBS with 10% FCS. For intracellular proteins, staining was carried out on cells fixed with 4% paraformaldehyde (Electron Microscopy Sciences) in PBS. Staining was done in PBS with 10% FCS and 0.5% saponin (Sigma). Stained cells were analyzed using an LSRII flow cytometer (BD). For FACS, the cells were sorted at a concentration of 106 cells/ml in IMDM/6% FCS using a FACSAriaTMII (BD) cell sorter (SickKids-UHN Flow Cytometry Facility). To prevent cell death due to pressure and sheer stress, all sorts were performed with a 100 μm nozzle. For magnetic bead sorting, the Miltenyi MACS bead sorting system was used according to the manufacturer’s guidelines and sorting conditions. For the high-throughput flow cytometry analysis of the CD antibodies the BD high-throughput sampler (HTS) for the LSRII was used according to the manufacturer’s guidelines. Data were analyzed using FlowJo software (Treestar).
Immunostaining
Immunostaining was performed as previously described14 using the following primary antibodies: rabbit anti-cTNI (Abcam; 1:100), mouse anti-SIRPA (BioLegend; 1:100). Secondary antibodies used were: goat anti-mouse IgGCy3 (Jackson ImmunoResearch; 1:400), donkey anti-mouse IgG-Alexa 488 (Invitrogen; 1:400). DAPI was used to counterstain nuclei. MitoTracker Red (Invitrogen) was used to stain mitochondria. The stained cells were visualized using a fluorescence microscope (Leica CTR6000) and images captured using the Leica Application Suite software.
RT-qPCR
Total RNA was prepared with the RNAqueous-Micro Kit (Ambion) and treated with RNase-free DNase (Ambion). 500 ng to 1 μg of RNA was reverse transcribed into cDNA using random hexamers and Oligo (dT) with Superscript III Reverse Transcriptase (Invitrogen). QPCR was performed on a MasterCycler EP RealPlex (Eppendorf) using QuantiFast SYBR Green PCR Kit (Qiagen) as described previously14. Expression levels were normalized to the housekeeping gene TATA box binding protein (TBP). In addition to TBP for normalization across samples, genomic DNA was used as a DNA standard. The copy number of the target gene present in the genomic DNA can be directly calculated as follows. Human genome size: 2.7 × 109 bp (1.78 × 1012 daltons), corresponds to 6.022 × 1023 copies of a single-copy gene; 1 μg of genomic DNA corresponds to 3.4 × 105 copies of a single-copy gene (on average and assuming no copy number variation occurred). The y axis of RT-qPCR graphs represents copy numbers of the gene of interest divided by copy numbers of TBP, and therefore is an arbitrary but absolute unit, that can be compared between experiments.
The oligonucleotide sequences are summarized in Supplementary Table 3. Total human adult heart RNA was purchased from Ambion and a total human RNA master panel was purchased from Clontech.
Supplementary Material
Acknowledgments
We would like to thank members of the Keller laboratory for discussion and critical reading of the manuscript, G. Daley (Harvard Medical School, Boston) for providing the MSC-iPS1 and 38-2 cell line, B. Neel for providing the Sirpa-mutant mice and for discussion and suggestions, L. Ailles, J. Paterson and E. Hyatt from the antibody core facility, R. Hamilton for assistance in obtaining fetal tissue samples and the Sick Kids/UHN Flow Cytometry Facility for their assistance with cell sorting. This work was supported by funding from VistaGen Therapeutics (San Francisco) and by grants from Canadian Institute of Health Research (CIHR, MOP-84524) and Ontario’s Ministry of Research and Innovation (MRI, GL2-CBD3P3) to G.K. N.C.D. was supported by a Swiss National Foundation postdoctoral fellowship. A.G. and P.S. were supported by the Heart and Stroke Foundation of Ontario (T6281 and NS6636) and CIHR (MOP-106538). A.G.E., E.G.S. and D.A.E. are supported by grants from the Australian Stem Cell Centre, the National Health and Medical Research Council (NHMRC, Australia) and the National Heart Foundation (Australia). E.G.S. and A.G.E. are Senior Research Fellows of the NHMRC (Australia).
Footnotes
Note: Supplementary information is available on the Nature Biotechnology website.
AUTHOR CONTRIBUTIONS
N.C.D. and G.K. designed the study and wrote the paper. N.C.D., A.M.C., P.S. and A.G. designed and performed experiments and analyzed the data. D.A.E., A.G.E. and E.G.S. generated and provided the NKX2-5–GFP hESC line.
COMPETING FINANCIAL INTERESTS
The authors declare competing financial interests: details accompany the full-text HTML version of the paper at http://www.nature.com/naturebiotechnology.
References
- 1.Murry CE, Keller G. Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell. 2008;132:661–680. doi: 10.1016/j.cell.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 2.Kattman SJ, et al. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell. 2011;8:228–240. doi: 10.1016/j.stem.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 3.Yang L, et al. Human cardiovascular progenitor cells develop from a KDR+ embryonic-stem-cell-derived population. Nature. 2008;453:524–528. doi: 10.1038/nature06894. [DOI] [PubMed] [Google Scholar]
- 4.Zwi L, et al. Cardiomyocyte differentiation of human induced pluripotent stem cells. Circulation. 2009;120:1513–1523. doi: 10.1161/CIRCULATIONAHA.109.868885. [DOI] [PubMed] [Google Scholar]
- 5.Braam SR, Passier R, Mummery CL. Cardiomyocytes from human pluripotent stem cells in regenerative medicine and drug discovery. Trends Pharmacol Sci. 2009;30:536–545. doi: 10.1016/j.tips.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 6.Anderson D, et al. Transgenic enrichment of cardiomyocytes from human embryonic stem cells. Mol Ther. 2007;15:2027–2036. doi: 10.1038/sj.mt.6300303. [DOI] [PubMed] [Google Scholar]
- 7.Huber I, et al. Identification and selection of cardiomyocytes during human embryonic stem cell differentiation. FASEB J. 2007;21:2551–2563. doi: 10.1096/fj.05-5711com. [DOI] [PubMed] [Google Scholar]
- 8.Ritner C, et al. An engineered cardiac reporter cell line identifies human embryonic stem cell-derived myocardial precursors. PLoS ONE. 2011;6:e16004. doi: 10.1371/journal.pone.0016004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hattori F, et al. Nongenetic method for purifying stem cell-derived cardiomyocytes. Nat Methods. 2010;7:61–66. doi: 10.1038/nmeth.1403. [DOI] [PubMed] [Google Scholar]
- 10.Elliot DA, et al. Nat Methods. 2011 Oct 23; doi: 10.1038/nmeth.1740. advance online publication. [DOI] [Google Scholar]
- 11.Seiffert M, et al. Signal-regulatory protein alpha (SIRPalpha) but not SIRPbeta is involved in T-cell activation, binds to CD47 with high affinity, and is expressed on immature CD34(+)CD38(−) hematopoietic cells. Blood. 2001;97:2741–2749. doi: 10.1182/blood.v97.9.2741. [DOI] [PubMed] [Google Scholar]
- 12.Timms JF, et al. SHPS-1 is a scaffold for assembling distinct adhesion-regulated multi-protein complexes in macrophages. Curr Biol. 1999;9:927–930. doi: 10.1016/s0960-9822(99)80401-1. [DOI] [PubMed] [Google Scholar]
- 13.Subramanian S, Parthasarathy R, Sen S, Boder ET, Discher DE. Species- and cell type-specific interactions between CD47 and human SIRPalpha. Blood. 2006;107:2548–2556. doi: 10.1182/blood-2005-04-1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nostro MC, et al. Stage-specific signaling through TGFbeta family members and WNT regulates patterning and pancreatic specification of human pluripotent stem cells. Development. 2011;138:861–871. doi: 10.1242/dev.055236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park IH, et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 16.Matozaki T, Murata Y, Okazawa H, Ohnishi H. Functions and molecular mechanisms of the CD47-SIRPalpha signalling pathway. Trends Cell Biol. 2009;19:72–80. doi: 10.1016/j.tcb.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 17.Okazawa H, et al. Negative regulation of phagocytosis in macrophages by the CD47-SHPS-1 system. J Immunol. 2005;174:2004–2011. doi: 10.4049/jimmunol.174.4.2004. [DOI] [PubMed] [Google Scholar]
- 18.Carvajal-Vergara X, et al. Patient-specific induced pluripotent stem-cell-derived models of LEOPARD syndrome. Nature. 2010;465:808–812. doi: 10.1038/nature09005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Itzhaki I, et al. Modelling the long QT syndrome with induced pluripotent stem cells. Nature. 2011;471:225–229. doi: 10.1038/nature09747. [DOI] [PubMed] [Google Scholar]
- 20.Laflamme MA, et al. Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted rat hearts. Nat Biotechnol. 2007;25:1015–1024. doi: 10.1038/nbt1327. [DOI] [PubMed] [Google Scholar]
- 21.Kisselbach L, Merges M, Bossie A, Boyd A. CD90 Expression on human primary cells and elimination of contaminating fibroblasts from cell cultures. Cytotechnology. 2009;59:31–44. doi: 10.1007/s10616-009-9190-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ross R. The pathogenesis of atherosclerosis: a perspective for the 1990s. Nature. 1993;362:801–809. doi: 10.1038/362801a0. [DOI] [PubMed] [Google Scholar]
- 23.Stevens KR, et al. Physiological function and transplantation of scaffold-free and vascularized human cardiac muscle tissue. Proc Natl Acad Sci USA. 2009;106:16568–16573. doi: 10.1073/pnas.0908381106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dvir T, et al. Prevascularization of cardiac patch on the omentum improves its therapeutic outcome. Proc Natl Acad Sci USA. 2009;106:14990–14995. doi: 10.1073/pnas.0812242106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lesman A, et al. Transplantation of a tissue-engineered human vascularized cardiac muscle. Tissue Eng Part A. 2010;16:115–125. doi: 10.1089/ten.TEA.2009.0130. [DOI] [PubMed] [Google Scholar]
- 26.Ling Y, Maile LA, Lieskovska J, Badley-Clarke J, Clemmons DR. Role of SHPS-1 in the regulation of insulin-like growth factor I-stimulated Shc and mitogen-activated protein kinase activation in vascular smooth muscle cells. Mol Biol Cell. 2005;16:3353–3364. doi: 10.1091/mbc.E04-10-0918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kennedy M, D’Souza SL, Lynch-Kattman M, Schwantz S, Keller G. Development of the hemangioblast defines the onset of hematopoiesis in human ES cell differentiation cultures. Blood. 2007;109:2679–2687. doi: 10.1182/blood-2006-09-047704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Costa M, et al. A method for genetic modification of human embryonic stem cells using electroporation. Nat Protoc. 2007;2:792–796. doi: 10.1038/nprot.2007.105. [DOI] [PubMed] [Google Scholar]
- 29.Sharma P, Shathasivam T, Ignatchenko V, Kislinger T, Gramolini AO. Identification of an FHL1 protein complex containing ACTN1, ACTN4, and PDLIM1 using affinity purifications and MS-based protein-protein interaction analysis. Mol Biosyst. 2011;7:1185–1196. doi: 10.1039/c0mb00235f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seiffert M, et al. Human signal-regulatory protein is expressed on normal, but not on subsets of leukemic myeloid cells and mediates cellular adhesion involving its counterreceptor CD47. Blood. 1999;94:3633–3643. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.