Abstract
Many benthic marine animal populations are established and maintained by free-swimming larvae that recognize cues from surface-bound bacteria to settle and metamorphose. Larvae of the tubeworm Hydroides elegans, an important biofouling agent, require contact with surface-bound bacteria to undergo metamorphosis; however, the mechanisms that underpin this microbially mediated developmental transition have been enigmatic. Here, we show that a marine bacterium, Pseudoalteromonas luteoviolacea, produces arrays of phage tail–like structures that trigger metamorphosis of H. elegans. These arrays comprise about 100 contractile structures with outward-facing baseplates, linked by tail fibers and a dynamic hexagonal net. Not only do these arrays suggest a novel form of bacterium-animal interaction, they provide an entry point to understanding how marine biofilms can trigger animal development.
Environmentally selective settlement of swimming larvae that are the propagules of most benthic invertebrate species is a critical life-cycle stage achieved by recognizing specific physicochemical cues (1, 2). This process is of fundamental importance to the fields of developmental biology and marine benthic community ecology—for example, the recruitment of new larval animals is essential to sustain and disperse coral reef populations (1). Economically, larval settlement is necessary for the supply of products for fisheries and aquaculture industries worldwide (3) and is responsible for millions of dollars of increased fuel consumption per year due to the biofouling of ships (4). Bacteria resident in surface biofilms are now recognized as the sources of metamorphosis-inducing cues for many invertebrates from most phyla (2). Indeed, the importance of microbes to the development and health of diverse animals is becoming increasingly appreciated (5). Yet our understanding of how these microbes interact with their hosts is only in its infancy.
The relation between the marine tubeworm Hydroides elegans and the bacterium Pseudoalteromonas luteoviolacea is a model for the study of invertebrate metamorphosis (2, 6, 7). Bacteria from the genus Pseudoalteromonas are commonly isolated from marine water, sediment, biofilms, or marine eukaryotes (8, 9). P. luteoviolacea strain HI1, used in this study, was isolated from a marine biofilm (8). Recently, Huang et al. (7) identified a set of genes in P. luteoviolacea whose products are essential to metamorphosis of H. elegans. However, the specific cue that triggers this bacterium-mediated developmental transition remained unknown.
In the vicinity of the P. luteoviolacea genes identified as essential to the induction of H. elegans metamorphosis (7) (Fig. 1, A and B), we identified a cluster of open reading frames (ORFs) predicted to encode components of phage tail–like structures, known as bacteriocins (fig. S1). Bacteria typically use bacteriocins to kill other bacteria by puncturing their membrane, causing depolarization (10, 11). R-type bacteriocins resemble contractile phage tails, similar to type VI secretion systems (T6SS) of Gram-negative bacteria (12). Phage tail–like bacteriocins have a contractile sheath, inner tube, baseplate components and tail fibers, but lack a DNA-filled head and are therefore not replicative. Bacteriocin-like structures can mediate several bacterial pathogen-animal interactions, for example, by causing antifeeding activity in grass grubs (13) and insecticidal activity against wax moths (14). No phage tail–like structures are currently known to mediate an interaction that is beneficial for the animal. On the basis of their predicted role in inducing metamorphosis, we named the ORFs surrounding those identified by Huang et al. (7) the metamorphosis-associated contractile structure (mac) genes.
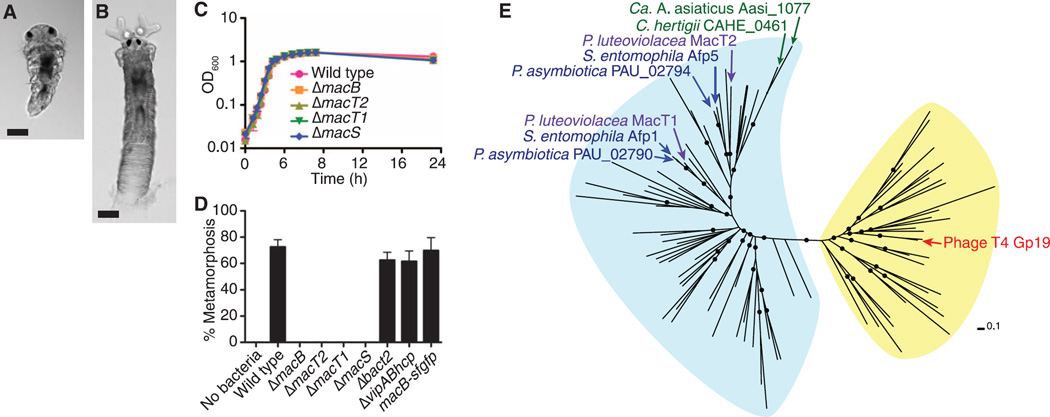
Fig. 1. P. luteoviolacea mac genes are required for metamorphosis of H. elegans and are similar to genes encoding phage tail–like structures.
(A and B) Metamorphically competent H. elegans larva (A) and juvenile adult (B) 12 hours after exposure to a P. luteoviolacea biofilm. Scale bars, 50 µm. (C) Growth of P. luteoviolacea wild type and mutants containing in-frame deletions of mac genes. OD, optical density, a measure of absorbance. (D) Metamorphosis (%) of H. elegans in response to biofilms of P. luteoviolacea wild type; and ΔmacB, ΔmacT1, ΔmacT2, ΔmacS, Δbact2, and ΔvipABhcp mutants; and macB-sfgfp fusion strains. Sterile artificial seawater (no bacteria) was used as a negative control. Error bars represent standard deviations (n = 4). (E) Maximum likelihood unrooted phylogeny of the Gp19 protein family. Gp19-like protein domains originating from bacteria and phages are highlighted in blue and yellow, respectively. Nodes with approximate likelihood-ratio test (aLRT) (29) values ≥0.8 are marked with a black circle. Scale bar indicates amino acid substitutions per site.
To determine whether the mac genes play a role in tubeworm metamorphosis, we made in-frame deletions of genes encoding putative sheath (macS), tube (macT1 and macT2), and baseplate (macB) [previously identified by Huang et al. (7)] proteins (fig. S1). These deletion strains grew identically in rich medium (Fig. 1C) but were unable to induce metamorphosis (Fig. 1D). Complementation of mutant strains in trans with mac genes resulted only in modest restoration of metamorphosis (fig. S2A). When the mac genes were replaced in their native chromosomal loci, metamorphosis induction was restored (fig. S2B). In addition to the mac gene cluster, we identified a second phage tail–like bacteriocin locus (bacteriocin-2) containing two genes predicted to encode putative tube and sheath proteins and a third gene cluster predicted to encode proteins similar to tube (Hcp) and sheath proteins (VipA/B) from a T6SS (table S1). In contrast to the Δmac mutants, strains containing in-frame deletions of genes predicted to encode tube and sheath proteins of bacteriocin-2 (Δbact2) or T6SS (ΔvipABhcp) still induced metamorphosis of H. elegans similarly to wild type (Fig. 1D).
To test whether the mac gene-cluster is responsible for producing phage tail–like structures, we compared negatively stained electron micrographs of cultures of P. luteoviolacea wild type—producing MAC, bacteriocin-2, and T6SS—and mutants producing only MACs (ΔvipABhcpΔbact2), producing only bacteriocin-2 (ΔvipABhcpΔmacSΔmacB), or lacking T6SS, bacteriocin-2, and MACs (ΔvipABhcpΔbact2ΔmacSΔmacB). Contracted and disassembled phage tail−like bacteriocins, as well as aggregated sheaths, were observed in the extracellular space of wild-type cells (Fig. 2A). Cultures producing only MACs (ΔvipABhcpΔbact2) contained dense aggregates of contracted sheaths (length 135 ± 4 nm, n = 13) and possibly tube structures (Fig. 2B), whereas cultures producing only bacteriocin-2 (ΔvipABhcpΔmacSΔmacB) contained contracted and disassembled individual phage tail–like structures, with shorter contracted sheaths (94 ± 3 nm, n = 13) (Fig. 2C). No sheath or phage tail–like structures were detected in the strain lacking MACs, bacteriocin-2, and T6SS (ΔvipABhcpΔbact2ΔmacSΔmacB) (Fig. 2D). Similarly, we observed sheaths or bacteriocins in purifications from the same strains, except that dense aggregates of MACs were not observed (fig. S3, A to D). Mass spectrometry of two bands present in an SDS–polyacrylamide gel electrophoresis (SDS-PAGE) gel of purified proteins from the strain producing only MAC (ΔvipABhcpΔbact2)—not seen in the sample from the strain lacking MAC, bacteriocin-2, and T6SS (ΔvipABhcpΔbact2ΔmacSΔmacB) (fig. S4)—revealed three peptides matching the putative MAC sheath protein (MacS) and one peptide matching a putative MAC tube protein (MacT2).
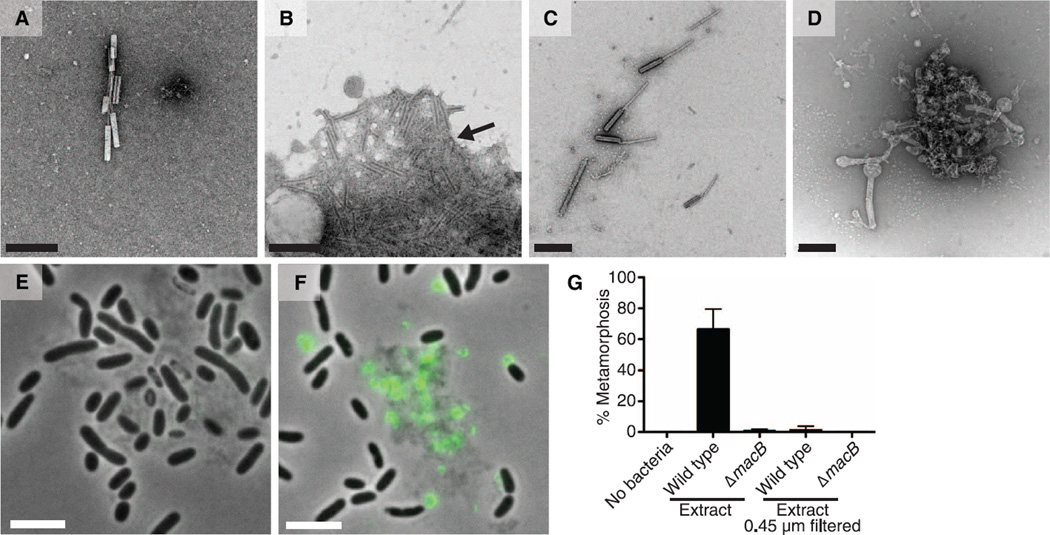
Fig. 2. MACs are phage tail–like structures, are released by cell lysis, and mediate metamorphosis of H. elegans.
(A to D) Negative stain EM of P. luteoviolacea (A) wild type, (B) ΔvipABhcpΔbact2, (C) ΔvipABhcpΔmacSΔmacB, and (D) ΔvipABhcpΔbact2ΔmacSΔmacB. Aggregated sheaths are indicated by an arrow in (B). Scale bars, 200 nm. (E and F) Micrographs of merged phase-contrast and fluorescence images of P. luteoviolacea (E) wild-type and (F) macB-sfgfp strains (see movie S1). Fluorescence of MacB-sfGFP is shown in green. Scale bar, 5 µm. (G) Metamorphosis (%) of H. elegans in response to cell-free extracts from P. luteoviolacea wild type and ΔmacB mutant. Extracts unfiltered and 0.45-µm filtered are indicated. Sterile artificial seawater (no bacteria) was used as a negative control. Error bars represent standard deviation (n = 4).
To determine the phylogenetic placement of MACs, we compared sequences of domains from MacT1 andMacT2 to similar domains from the phage T4 Gp19 tail tube (PF06841) protein family (e-value = 2.1e-32 and 3.1e-13, respectively). MacT1 and MacT2 grouped with bacterial Gp19 proteins within distinct clades (Fig. 1E) and are closely related to the phage tail–like bacteriocin tube proteins from Serratia entomophila (13) and Photorhabdus asymbiotica (14). Comparative genomics also revealed that multiple genes in the P. luteoviolacea gene cluster are homologous to genes from the S. entomophila afp locus, with some conservation of synteny (fig. S1, table S1), which suggested a common evolutionary origin. It is noteworthy that MacT1 and MacT2 are related to tube proteins predicted to function as part of T6SSs (15) in the parasitic wasp symbiont, Cardinium hertigii (16), and the amoeba symbiont, Candidatus Amoebophilus asiaticus (17) (Fig. 1E).
Given the relatedness of MACs to phage and phage tail–like structures, we investigated whether P. luteoviolacea produces and releases MACs extracellularly. To track MAC localization in vivo, we constructed a strain encoding a C-terminal fusion of MacB with superfolder green fluorescent protein (sfGFP) (18). When the native chromosomal macB gene was replaced with one encoding MacB-sfGFP, the recombinant strain induced metamorphosis to levels comparable with that of wild type (Fig. 1D). Fluorescence light microscopy revealed that the MacB-sfGFP fusion protein localized extracellularly when broth cultures reached stationary phase and produced approximately 0.5- to 1.0-µm-wide ringlike signals, whereas the untagged strain showed no fluorescence (Fig. 2, E and F). MacB-sfGFP expression was observed by using three different marine media [ASWT, NSWT, and 2216 (fig. S5)], which suggests that extracellular release is dependent neither on soluble factors present in natural seawater nor on a specific nutrient-rich medium. Of the cells in a population, 2.4% (n = 1244) showed intra-cellular GFP expression, many of them seemingly in the process of cell lysis. Time-lapse microscopy revealed that lysis of a subset of cells precedes the appearance of extracellular MacB-sfGFP (movie S1).
Electron cryomicroscopy (19) of a strain producing only MACs (ΔvipABhcpΔbact2) allowed us to visualize these structures at high resolution in a near-native state. Four out of 162 cells (2.5%) showed intracellular phage tail–like bacteriocins. This percentage of MAC-containing cells matches the percentage of cells harboring MacB-sfGFP. Electron cryotomography (ECT) of these four cells revealed that the entire cytoplasm was packed with clusters of MACs, which in some cases appeared to be connected by a filamentous mesh (white arrow in Fig. 3A, movie S2). The shape and membrane structures of MAC-producing cells indicated that they were about to undergo lysis (fig. S6).
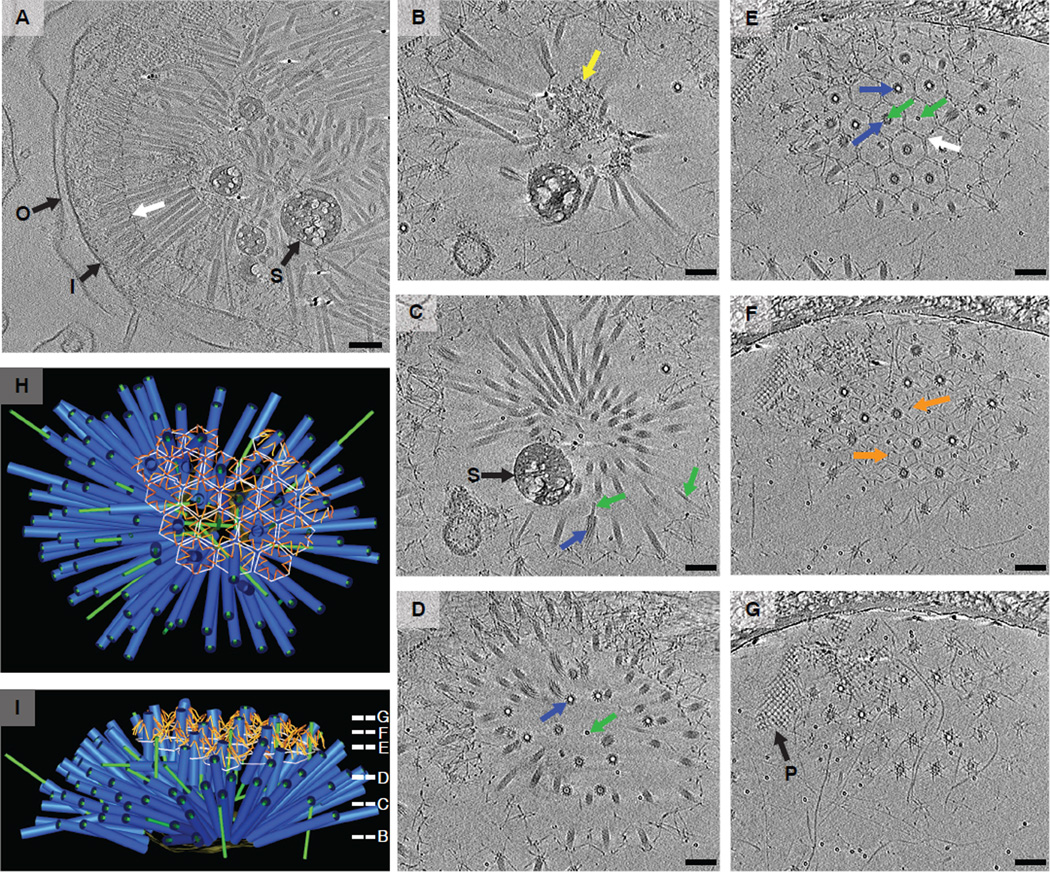
Fig. 3. MACs are assembled intracellularly and expand as an ordered array upon cell lysis.
P. luteoviolacea ΔvipABhcpΔbact2 mutant cells and extracellular aggregates were imaged by ECT (shown are 16.8-nm-thick slices). (A) The cytoplasm of a subset of cells (4 in 162) was packed with MAC aggregates, and cells appeared in the process of lysis (see movie S2). I, inner membrane; O, outer membrane; S, storage granule; white arrow, filamentous connections. (B to G) Extracellular MAC arrays are highly ordered (shown are 2D slices through a tomogram at different z-heights) (see movies S3 and S4). Yellow arrow, amorphous core; green arrow, inner tube; blue arrow, sheath; white arrow, filamentous hexagonal net; orange arrow, tail fibers; P, presumably polymerized sheath protein. (H and I) MAC arrays were hemispherical with MACs coalescing in an amorphous core and the baseplates hexagonally arranged on the surface. Individual MACs were connected by tail fibers and surrounded by a hexagonal net. Different views of a segmented model of the array are shown. Slice z-heights in (B) to (G) are indicated with the corresponding panel letter. Scale bars, 100 nm.
We then asked whether enriched MAC preparations were sufficient to induce metamorphosis in the absence of bacterial cells. MAC preparations obtained by standard bacteriocin purification protocols (12) failed to induce H. elegans metamorphosis. MACs likely contract upon purification, and the arrays fall apart, consistent with our observation that MACs were almost exclusively in a contracted state when visualized by negative-stain electron microscopy (EM). A gentle purification from wild-type P. luteoviolacea resulted in a MAC preparation that induced metamorphosis of H. elegans (Fig. 2G), whereas extracts from a MAC mutant (ΔmacB) did not. Assays were performed with extract concentrations derived from the equivalent of 107 cells/ml (100× dilution). Filtering extracts from wild-type cells through a 0.45-µm filter abolished the metamorphic effect, consistent with the observation that MacB-sfGFP forms >0.45-µm aggregates. Concentrated bacteriocin extracts (derived from the equivalent of 108 cells/ml, 10× dilution) caused 100% larval death after 24 hours, which indicated that MACs or copurifying constituents can have toxic effects at high doses. We do not know the concentration of MACs in laboratory or natural marine biofilms.
To address whether another factor present in the MAC lysate was responsible for inducing metamorphosis independent of MACs, we constructed macB-sfgfp strains in mutants lacking the MAC sheath protein (ΔmacS), or MAC tube proteins (ΔmacT1 or ΔmacT2). We observed extracellular MacB-sfGFP fluorescence in all mutants (fig. S7, C to E). Levels of MacB-sfGFP were comparable between a macB-sfgfp strain and strains with deletions of macS, macT1, or macT2, as determined by immunoblot analysis with antibodies against GFP (fig. S7, F and G), which suggested that the expression and stability of MacB-sfGFP is not dependent on other MAC components. These results indicate that other biomolecules derived from lysed cells are insufficient to promote metamorphosis of H. elegans in the absence of functional MACs. Although MACs are necessary for metamorphosis, whether they are sufficient remains to be determined. Additional ORFs in the mac gene cluster, such as those identified by Huang et al. (7) (i.e., ORF2, ORF3, and ORF4A/B/C) (fig. S1), might also contribute to the MAC structure or the direct induction of metamorphosis.
We used the same MAC extracts shown to promote metamorphosis of H. elegans in bacteriocidal activity assays with closely related P. luteoviolacea strains and species (P. luteoviolacea strain ATCC 33492, P. piscicida, andP. haloplanktis). These MAC extracts did not kill close relatives of P. luteoviolacea HI1 (fig. S8), unlike bacteriocins produced by other bacteria (20, 21). It remains to be determined whether these or other types of MACs can kill bacterial species under different conditions.
We characterized MAC aggregates using ECT to compare their structure with that of other phage tail–like bacteriocins. In a frozen, near-native state, we observed MACs forming arrays of multiple contractile phage tail–like structures (Fig. 3 and movies S3 and S4). A typical MAC array contained close to 100 individual phagelike tails (96 for the one shown in Fig. 3, B to I), with dimensions matching the size of MacB-sfGFP fluorescence (fig. S9 and table S2). Intracellular MACs were more tightly packed than their extracellular counterparts (table S2), which suggested that MAC arrays expand upon cell lysis. MACs radiate into a hemisphere, originating from an amorphous center (yellow arrow in Fig. 3B). Distal MAC ends were arranged into a regular array, surrounded by a filamentous hexagonal net (white arrow in Fig. 3E). MACs were oriented with their baseplates (“B” in Fig. 4B) outward, with filamentous structures (probably tail fibers; orange arrows in Fig. 3F) emanating from the baseplates and appearing to connect individual MACs to each other (“TF” in Fig. 4B; see also movie S3). To our knowledge, such ordered multitailed arrays have not previously been observed for other types of phagelike structures.
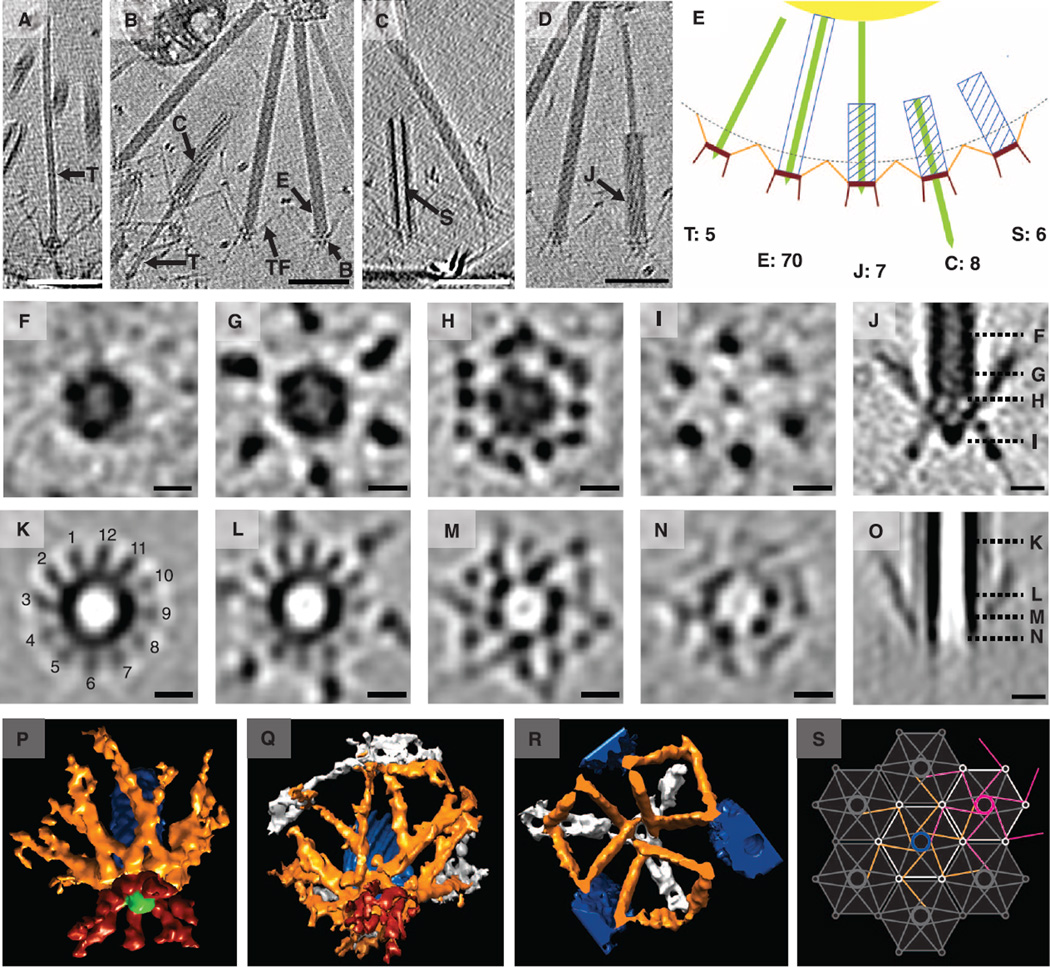
Fig. 4. MACs are observed in different functional states and connected by tail fibers.
(A) Side view of a MAC in the tube only T state. (B) Side views of MACs in extended, E, and contracted, C, states. B, baseplate; TF, tail fibers; T, inner tube. (C) Side view of a MAC in the contracted sheath without tube S state. (D) Side view of a MAC in the contracted state with jammed tube J. Scale bars in (A) to (D), 100 nm. Tomographic slices that are 16.8 nm thick are shown in (A) to (D). (E) Schematic of different functional states. Numbers indicate the quantity of each state found in the MAC array shown in Fig. 3, (B) to (I). (F to J) Subtomogram average of extended MACs. Cross-sectional slices at different z-heights are shown in (F) to (I) and their positions are indicated in the side view (J). Slices that are 8.4 nm thick are shown. Scale bars, 10 nm. (K to O) Subtomogram average of contracted MACs. Cross-sectional slices at different z-heights are shown in (K) to (N), and their positions are indicated in the side view (O). Slices that are 8.4 nm thick are shown. Scale bars, 10 nm. (P to R) Isosurface of subtomogram averages (see movies S6 to S8) of an extended (P) and a contracted (Q) MAC and of the tail fiber junction (R). Tail pins and baseplate, red; tail fibers, orange; inner tube and spike, green; sheath, blue; hexagonal net, white. (S) Schematic model of tail fiber connections in the MAC array. Each MAC unit contributes six tail fibers that connect to neighboring MAC units. The central MAC unit is colored in the color code used in (P) to (R). A second MAC unit is colored magenta.
Individual MACs resembled contractile phage tail–like structures (Fig. 4). Sheaths were observed in extended (“E” in Fig. 4B, table S2) and contracted (“C” in Fig. 4B, table S2) forms, with homogeneous and helical surface patterns, respectively. In some cases, the inner tube (“T” in Fig. 4, A and B, and table S2) was observed. We classified the observed MAC conformations into “tube only” (T), “extended” (E), “contracted with jammed tube” (J), “contracted with fired tube” (C) and “contracted without tube” (S) (Fig. 4, A to E, and movie S5). The “extended” conformation represented 73% of MACs within the aggregate shown (Fig. 3). These different structural classes likely represent different states in a functional sequence. In analogy to phage tail–like structures, the “tube only” state may be a partially assembled MAC, whereas the “extended” MAC is in a ready-to-fire configuration (12, 22). “Contracted with fired tube” and “contracted without tube” are likely MACs that have fired. “Contracted with jammed tube” could represent fired MACs that failed to propel the tube. In this conformation, the sheath is contracted as indicated by the helical ridges [assembling T6SS sheaths are in the extended conformation (12)], and the sheath length of 5 of 7 structures matches the length of a fired MAC (table S2).
We averaged 20 and 25 subtomograms of MACs in the “extended” (Fig. 4P, movie S6) and “contracted without tube” (Fig. 4Q, movie S7) states, respectively. Averages of subtomograms show the different sheath diameters, helical surface ridges on contracted sheath (ridges indicated in Fig. 4K), baseplate symmetry and tail fibers in longitudinal (Fig. 4, J and O) and cross-sectional views (Fig. 4, F to I and K to N). In both the extended and contracted forms twelve fibers emerge from the baseplate, cross paths, and separate to meet at the ring-shaped vertices of the hexagonal net surrounding individual MACs (Fig. 4R, movie S8). We speculate that six of the tail fibers originate from a single MAC, with the remaining six fibers stemming from neighboring MACs to connect the array (Fig. 4, P to S, orange). This six-tail fiber per MAC model (Fig. 4S) is supported by the fact that the two arms of a phage tail fiber have a length ratio of 1:1 (23) and that the length is similar to the tail fiber connections in MACs (fig. S10). The model also predicts the presence of an as-yet-unidentified protein that forms the hexagonal net. A set of six tail pins (Fig. 4P, red) face outwards. Because the tail pins are the most distal structure in the arrays, they are likely the first structure to engage and sense MAC targets.
We have shown that an ordered array of contractile phage tail–like structures produced by an environmentally occurring bacterium induces metamorphosis of a marine invertebrate larva. This discovery begins to explain how marine biofilms can trigger metamorphosis of benthic animals. Our data suggest that MAC arrays are synthesized intracellularly by P. luteoviolacea, released by cell lysis, and expand extracellularly into an ordered multi-MAC array. How these arrays engage with larvae of H. elegans is an open question. In the arrays imaged, all contracted MACs were clustered together, which suggests that their linkages might support cooperative firing. Array formation might also multiply the total payload delivered per interaction or favor specific engagement sites and/or geometries with MAC targets. The evolutionary pressure to produce MACs is probably strong, given that MAC production leads to the lysis and death of a subpopulation of cells. Whether this represents an instance of altruistic behavior that facilitates group selection, or a neutral lytic event with a set frequency remains to be determined. Although MAC production is beneficial for tubeworm larvae by inducing metamorphosis, it is currently unclear how larval settlement and metamorphosis might benefit the bacterium. It is equally possible that MACs evolved for a completely different purpose. Note that P. luteoviolacea has been found to induce the metamorphosis of coral and sea-urchin larvae (24, 25). Other bacterial species also induce metamorphosis of H. elegans larvae (8, 26, 27), and mac-like gene clusters have been identified in the genomes of other marine bacteria (28). Future research into how MACs interact with larvae might yield new insights into the mechanisms underpinning marine animal development and ecology, with potentially important practical applications for aquaculture and biofouling.
Supplementary Material
Acknowledgments
We thank B. Pernet for help with locating and identifying tubeworms and for giving us the algal strain used in this work; A. McDowall for help with EM; Y. Huang, who created the StrR-strain (7); A. Asahina and S. Wilbur for laboratory assistance; J. Levine for help with time-lapse microscopy; J. Ricci for help with phylogenetic analyses; and members of the Newman group for discussions and comments on the manuscript. The Howard Hughes Medical Institute, Z. Yu, and J. de la Cruz are acknowledged for providing access to the FEI Titan Krios at Janelia Farm and support in data collection. N.J.S. was supported by a California Institute of Technology (Caltech) Division of Biology Postdoctoral Fellowship. This collaboration was supported by the Caltech Center for Environmental Microbiology Interactions, the Howard Hughes Medical Institute (D.K.N. and G.J.J.), Office of Naval Research grants N00014-08-1-0413 and N00014-05-1-0579 (M.G.H.), NIH grant GM094800B (G.J.J.), and a gift from the Gordon and Betty Moore Foundation (Caltech). D.K.N. and G.J.J. are Investigators of the Howard Hughes Medical Institute. Strains obtained from the American Type Culture Collection listed in table S2 (ATCC 33492, ATCC 14393, ATCC 15057). DNA sequences encoding for mac, T6SS, and bacteriocin-2 genes are deposited under GenBank accession numbers KF724687, KF724688, and KF724689, respectively. Subtomogram averages were deposited in the Electron Microscopy Data Bank (accession numbers EMD-2543, EMD-2544, and EMD-2545).
Footnotes
Author contributions: All authors designed research. N.J.S., M.P. and G.L.W. performed research. All authors wrote the paper.
Supplementary Materials
www.sciencemag.org/content/343/6170/529/suppl/DC1
Materials and Methods
Figs. S1 to S10
Tables S1 to S4
References (30–46)
Movies S1 to S8
References and Notes
- 1.Hadfield MG, Paul VJ. In: Marine Chemical Ecology. McClintock JB, Baker BJ, editors. Boca Raton, FL: CRC Press; 2001. p. 431. [Google Scholar]
- 2.Hadfield MG. Annu. Rev. Mar. Sci. 2011;3:453–470. doi: 10.1146/annurev-marine-120709-142753. [DOI] [PubMed] [Google Scholar]
- 3.Food and Agriculture Organization of the United Nations. The State of World Fisheries and Aquaculture 2012. Rome: FAO; 2012. [Google Scholar]
- 4.Schultz MP, Bendick JA, Holm ER, Hertel WM. Biofouling. 2011;27:87–98. doi: 10.1080/08927014.2010.542809. [DOI] [PubMed] [Google Scholar]
- 5.McFall-Ngai M, et al. Proc. Natl. Acad. Sci. U.S.A. 2013;110:3229–3236. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nedved BT, Hadfield MG. Marine Indust.l Biofoul. 2009;4:203–217. [Google Scholar]
- 7.Huang Y, Callahan S, Hadfield MG. Sci. Rep. 2012;2:228. doi: 10.1038/srep00228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang SY, Hadfield MG. Mar. Ecol. Prog. Ser. 2003;260:161–172. [Google Scholar]
- 9.Holmström C, Kjelleberg S. FEMS Microbiol. Ecol. 1999;30:285–293. doi: 10.1111/j.1574-6941.1999.tb00656.x. [DOI] [PubMed] [Google Scholar]
- 10.Michel-Briand Y, Baysse C. Biochimie. 2002;84:499–510. doi: 10.1016/s0300-9084(02)01422-0. [DOI] [PubMed] [Google Scholar]
- 11.Uratani Y, Hoshino T. J. Bacteriol. 1984;157:632–636. doi: 10.1128/jb.157.2.632-636.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Basler M, Pilhofer M, Henderson GP, Jensen GJ, Mekalanos JJ. Nature. 2012;483:182–186. doi: 10.1038/nature10846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hurst MR, Glare TR, Jackson TA. J. Bacteriol. 2004;186:5116–5128. doi: 10.1128/JB.186.15.5116-5128.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang G, Dowling AJ, Gerike U, ffrench-Constant RH, Waterfield NR. J. Bacteriol. 2006;188:2254–2261. doi: 10.1128/JB.188.6.2254-2261.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pukatzki S, et al. Proc. Natl. Acad. Sci. U.S.A. 2006;103:1528–1533. doi: 10.1073/pnas.0510322103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Penz T, et al. PLOS Genet. 2012;8:e1003012. doi: 10.1371/journal.pgen.1003012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Penz T, Horn M, Schmitz-Esser S. Virulence. 2010;1:541–545. doi: 10.4161/viru.1.6.13800. [DOI] [PubMed] [Google Scholar]
- 18.Pédelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS. Nat. Biotechnol. 2006;24:79–88. doi: 10.1038/nbt1172. [DOI] [PubMed] [Google Scholar]
- 19.Pilhofer M, Ladinsky MS, McDowall AW, Jensen GJ. Methods Cell Biol. 2010;96:21–45. doi: 10.1016/S0091-679X(10)96002-0. [DOI] [PubMed] [Google Scholar]
- 20.Köhler T, Donner V, van Delden C. J. Bacteriol. 2010;192:1921–1928. doi: 10.1128/JB.01459-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gebhart D, et al. J. Bacteriol. 2012;194:6240–6247. doi: 10.1128/JB.01272-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leiman PG, et al. Virol. J. 2010;7:355. doi: 10.1186/1743-422X-7-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cerritelli ME, Wall JS, Simon MN, Conway JF, Steven AC. J. Mol. Biol. 1996;260:767–780. doi: 10.1006/jmbi.1996.0436. [DOI] [PubMed] [Google Scholar]
- 24.Tran C, Hadfield MG. Mar. Ecol. Prog. Ser. 2011;433:85–96. [Google Scholar]
- 25.Huggett MJ, Williamson JE, de Nys R, Kjelleberg S, Steinberg PD. Oecologia. 2006;149:604–619. doi: 10.1007/s00442-006-0470-8. [DOI] [PubMed] [Google Scholar]
- 26.Unabia CRC, Hadfield MG. Mar. Biol. 1999;133:55–64. [Google Scholar]
- 27.Lau SC, Mak KK, Chen F, Qian P-Y. Mar. Ecol. Prog. Ser. 2002;226:301–310. [Google Scholar]
- 28.Persson OP, et al. Environ. Microbiol. 2009;11:1348–1357. doi: 10.1111/j.1462-2920.2008.01861.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anisimova M, Gascuel O. Syst. Biol. 2006;55:539–552. doi: 10.1080/10635150600755453. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.