Abstract
Bifunctional chelators play an important role in developing metallic radionuclide-based radiopharmaceuticals. In this study, a new bifunctional ligand, p-SCN-PhPr-NE3TA, was synthesized and conjugated to a very late antigen-4 targeting peptidomimetic, LLP2A, for evaluating its application in 64Cu-based positron emission tomography (PET) imaging. The new ligand exhibited strong selective coordination of Cu(II), leading to a robust Cu complex, even in the presence of 10-fold Fe(III). The LLP2A conjugate of p-SCN-PhPr-NE3TA was prepared and successfully labeled with 64Cu under mild conditions. The conjugate 64Cu-NE3TA-PEG4-LLP2A showed significantly higher specific activity, compared with 64Cu-NOTA-PEG4-LLP2A, while maintaining comparable serum stability. Subsequent biodistribution studies and PET imaging in mice bearing B16F10 xenografts confirmed its favorable in vivo performance and high tumor uptake with low background, rendering p-SCN-PhPr-NE3TA a promising bifunctional chelator for 64Cu-based radiopharmaceuticals.
Graphical Abstract

INTRODUCTION
Positron emission tomography (PET) is a noninvasive and sensitive nuclear imaging method that provides high resolution and quantitative information on the diagnosis and prognosis of cancer and various other diseases.1 Fluorine-18 (18F)-based PET radiotacers have been successfully applied in clinical PET imaging. However, the short half-life of 18F (t1/2 = 110 min) has limited applications, where radiolabeled biomolecules serving as probes for disease diagnosis or drug development have a relatively long circulation time and slower tumor accumulation.2 Copper-64 (64Cu), which possesses a longer half-life of 12.7 h, enables delayed time point imaging, and is an attractive alternative of 18F for PET imaging with biomolecules having a longer circulation time in vivo.3
Considerable efforts have been devoted to develop novel bifunctional chelators (BFC) for conjugation to peptides or antibodies that form stable complexes with 64Cu.3 For the ideal BFC, mild labeling conditions, rapid chelation kinetics with good stability and high specific activity are desirable.4 Various BFCs have been investigated during past decades for copper radionuclides. Most of these BFCs are based on polyazamacrocyclic chelators, because of their enhanced kinetic inertness and thermodynamic stability in comparison to acyclic chelators such as diethylenetriamine pentaacetic acid (DTPA). Examples of polyazamacrocyclic chelators include DOTA (1,4,7,10-tetraazacyclododecane-N,N′,N″,N‴-tetraacetic acid), TE2A ((1,8-N,N′-bis(carboxymethyl)-1,4,8,11-tetraazacyclotetradecane), NOTA (1,4,7-triazacyclododecane-N,N′,N″-triacetic acid), or their derivatives.5–7 Besides carboxylic acid as the chelation group, DOTHA2 and NOTHA2 bearing hydroxamate arms have been developed and demonstrated fast labeling kinetics with 64Cu and good stability of the resulting labeled complexes.8 Other backbone-based BFCs, such as Sar-cage and Pycup derivatives, have also been developed and have shown promising results; however, the high positive charge is a concern, as it may cause high liver uptake.9,10
To further optimize the properties of BFCs, a series of cross-bridged TE2A (CB-TE2A)-based chelators were developed and demonstrated significantly enhanced in vivo stability, in comparison to DOTA chelators.11,12 Harsh labeling conditions such as high temperature and long labeling time were usually required for CB-TE2A chelators to get decent labeling yields, limiting their applications in labeling temperature-sensitive biomolecules such as antibodies, although the phosphonate-based cross-bridged chelators such as CB-TE1A1P can be labeled at lower temperatures and have demonstrated success in both antibody and peptide labeling.13–16 There remains a need for a diverse array of BFCs, particularly those that can be readily synthesized, labeled with radiometals at low temperatures and show specficity for a particular radiometal. Derivatives of the polyaminocarboxylate NOTA were revisited, because of their convenient radiolabeling of 64Cu and good in vivo stability.17,18 Recently, newly developed NOTA derivaitves, such as NETA, NE3TA as well as their analogues N-NE3TA and C-NE3TA,19–23 have been reported for radiolabeling of 64Cu.19,22 These chelators integrated advantages of both the macrocyclic and acyclic framework for the thermodynamic stability and a favorable chelating kinetics. Furthermore, 64Cu-N-NE3TA exhibited low uptake in normal organs, fast blood clearance, and great in vivo stability.24 Recently, a bifunctional version of N-NE3TA and its biomolecule conjugate were reported.25 In particular, the resulting NE3TA-Tf conjugate could be efficiently radiolabeled with 64Cu at room temperature. 64Cu-N-NE3TA–Tf displayed a rapid blood clearance and increased tumor uptake in the subsequent in vivo studies, suggesting that N-NE3TA-based BFCs can be applied in generating 64Cu-labeled tracers for PET imaging of tumors.
In the current study, a new bifunctional version of N-NE3TA, denoted as p-SCN-PhPr-NE3TA (Figure 1) was designed and synthesized. In particular, the BFC contains a p-SCN benzyl group for conjugation to targeting molecules via thiourea bond formation, and the long propyl chain in the structure was designed to reduce potential steric hindrance during the chelating process. The Cu(II) selectivity over Fe(III) of our newly developed BFC was evaluated using a mass-based analysis method and DFT calculations. The specific activity and serum stability of 64Cu-NE3TA conjugate of LLP2A-PEG4, a peptidomimetic with high affinity and specificity for very late antigen 4, was performed and compared to 64Cu-NOTA-PEG4-LLP2A conjugate in which the SCN-Bn-NOTA was used as a BFC. To investigate the in vivo performance of the conjugate, the biodistribution and PET/CT imaging of 64Cu-NE3TA-PEG4-LLP2A was performed in mice bearing B16F10 melanoma xenografts. In addition, the utility of p-SCN-PhPr-NE3TA in modifying antibodies was also investigated using cetuximab as a model agent, and the specific activity of the resulting 64Cu-NE3TA-cetuximab was determined and compared with 64Cu-NOTA-cetuximab.
Figure 1.
Chelators for copper radionuclides.
RESULTS AND DISCUSSION
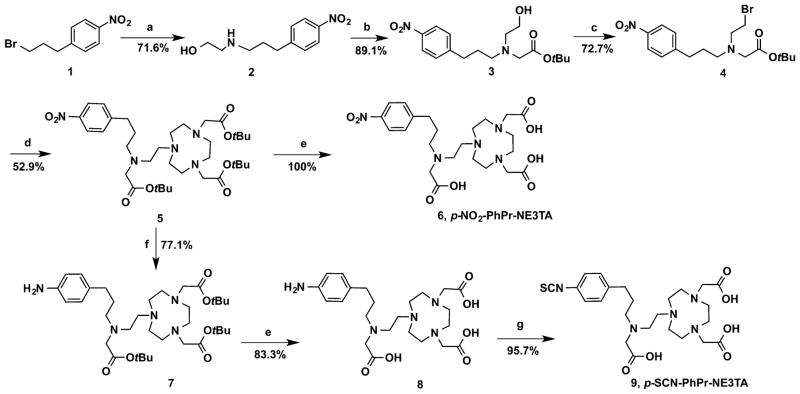
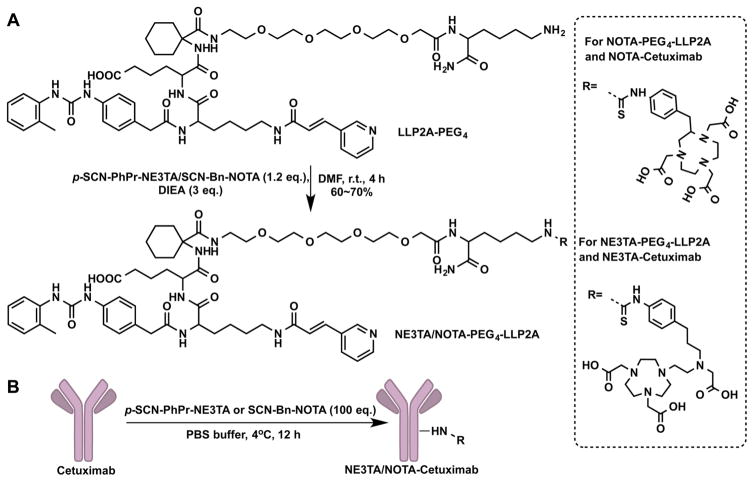
As illustrated in Scheme 1, p-SCN-PhPr-NE3TA (9) was readily synthesized in an overall yield of 15%. Starting material 1-(3-bromopropyl)-4-nitrobenzene (1) reacted with 2-amino-ethanol in MeCN in the presence of TEA to give 2-((3-(4-nitrophenyl)propyl)amino)ethan-1-ol (2). In this reaction, a mild base and excess amount of 2-aminoethanol were applied to prevent the formation of dialkylated byproduct, and the workup of the reaction mixture was carried out using a pH-controlled extraction. The purified compound 2 was further alkylated with tert-butyl bromoacetate to obtain tert-butyl (2-hydroxyethyl)(3-(4-nitrophenyl)propyl)carbamate (3). The alcohol group in 3 was brominated with N-bromosuccinimide and triphenylphosphine to its corresponding bromide 4 as a yellow oil. The introduction of the third carboxylate arm with an additional nitro-functional unit to the chelator NOTA backbone was carried out by the coupling reaction between 4 and di-tert-butyl 2,2′-(1,4,7-triazonane-1,4-diyl)diacetate (NO2A-t-Bu)26 in the presence of anhydrous K2CO3 in acetonitrile to yield the crucial intermediate 5, which was purified by column chromatography to obtain a white waxy solid. Deprotection of the tert-butyl group was achieved by treating compound 5 with trifluoroacetic acid (TFA) in methylene dichloride (DCM) at room temperature to afford the TFA salt of p-NO2-PhPr-NE3TA (6) as an oil, and the trace amount of water in the oil was removed by coevaporating the product with 20 mL CHCl3 three times. To prepare bifunctionalized version, the nitro group in the intermediate 5 was initially reduced by 10% Pd/C in methanol under H2 gas at room temperature to the aniline 7. The tert-butyl group in compound 7 was subsequently removed by treating with a 1:1 (v/v) mixture of TFA and DCM at room temperature to afford 8 as a yellow solid. The isothiocyanate (NCS), which is an amino-activated functional group for further bioconjugation, was then generated with thiophosgene in CHCl3. The reaction was carried out at room temperature for 4 h, and the desired compound p-SCN-PhPr-NE3TA (9) was obtained as a yellow solid by layer separation and lyophilization. A peptidomimetic ligand LLP2A-PEG4, which has a high affinity for very late antigen-4 (VLA-4, also called integrin α4β1),16,27 was chosen as a model of biomolecules. NE3TA-PEG4-LLP2A was conveniently obtained by attaching p-SCN-PhPr-NE3TA to LLP2A-PEG4 via thiourea bond formation and purification using HPLC (Scheme 2A). As a control, the LLP2A conjugate, NOTA-PEG4-LLP2A was also prepared using commercially available BFC SCN-Bn-NOTA.
Scheme 1.
Synthesis of p-SCN-PhPr-NE3TAa
aConditions and reagents: (a) 2-aminoethanol (10 equiv (eq.)), TEA (5 equiv), MeCN, room temperature (rt), 24 h; (b) tert-butyl bromoacetate (1.2 equiv), K2CO3 (2.2 equiv), MeCN, rt, 24 h; (c) PPh3 (1.1 equiv), NBS (1.1 equiv), MeCN, 0 °C to rt, 3 h; (d) NO2AtBu (1 equiv), K2CO3 (2 equiv), MeCN, rt, overnight; (e); TFA, DCM, overnight; (f) Pd/C, H2, MeOH, rt, 3 h; and (g) CSCl2, CHCl3, H2O, rt, 4 h.
Scheme 2.
Bioconjugations of p-SCN-PhPr-NE3TA and SCN-Bn-NOTA with LLP2A-PEG4 and Cetuximab: (A) NE3TA-PEG4-LLP2A and NOTA-PEG4-LLP2A, and (B) NE3TA-Cetuximab and NOTA-Cetuximab
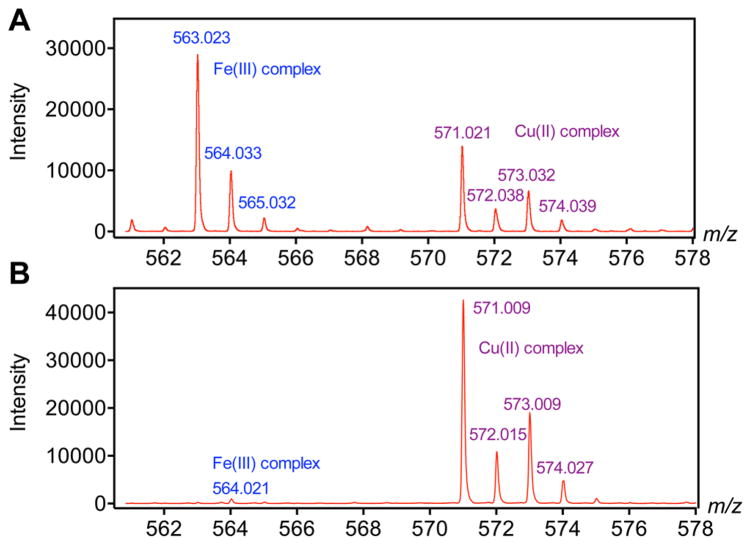
Although studies have already proven that NE3TA-based chelators form stable Cu(II) complexes, the adaptation of this ligand for 64Cu radiolabeling required additional characterization stringency in order to determine the possibility of preferential labeling impurities. Commercial radiocopper typically contains significant amounts of cold metals, especially Fe3+, which will heavily interfere the radiolabeling efficiency and decrease the specific activity (SA) of the corresponding radiopharmaceuticals.28,29 Thus, before labeling the p-SCN-PhPr-NE3TA with 64Cu, a rapid and sensitive LC-MS-based analysis method was developed to evaluate its Cu(II) chelating selectivity over Fe(III). As a model compound for the study, the nitro derivative 6 was used instead of p-SCN-PhPr-NE3TA (9), because the NCS group of 9 was unstable during complexation reaction, as confirmed by mass analysis of reaction mixture. To a solution of the model chelator 6, a metal ion mixture containing 5 mM CuCl2 and 5 mM FeCl3 was added at a molar ratio of 6 (compound 6):1 (metal ion mixture) in 0.1 M NH4OAc buffer (pH 4.0), and the reaction mixture was incubated at 70 °C for 30 min. Since the chelator was in excess, the metal ions were completely transformed into Fe(III)-6 and Cu(II)-6 complexes in the solution. The ionization capability of Fe(III)-6 and Cu(II)-6 complexes in the solution then was compared according to total ion counts determined from the same amount of these two complexes. The ratio of total ion counts was found to be ~2 (Fe(III)-6/Cu(II)-6) (see Figure 2A), suggesting that the Fe(III)-6 complex could more easily be ionized than Cu(II)-6 complex under the same conditions. Following the total ion counts ratio of the two complexes determined above, the chelating selectivity was then evaluated by incubating chelator 6 with 1 equiv of Cu(II) and 10 equiv of Fe(III) in the same buffer solution at room temperature for 30 min and subsequently comparing the ion counts of both complexes obtained from the MS spectra (Figure 2B). It was found that the ratio of Fe(III)-6/Cu(II)-6 was ~1:40 (back-calculated by the ionization capability correction), indicating a significant Cu(II) chelating selectivity over Fe(III) of 6. The study suggests that the chelator specifically chelates Cu(II) with minimal Fe(III) chelation even when the Fe(III) concentration was 10-fold to the Cu(II) concentration. This ensures improved specific activity for the 64Cu radiolabeling under the mild room-temperature labeling condition. A comparative study for chelator selectivity was carried out using commercially available p-NH2–Bn-NOTA (see Figure S4 in the Supporting Information). Compared with the chelator 6, the coordination of p-NH2–Bn-NOTA with Cu(II) and Fe(III) were much more complicated, because of the participation of the hydroxide functional group, making it difficult to quantify the intensity ratio of the cold metal complexes. However, given the fact of the obvious signal of Fe(III) complexes in Figure S4B in the Supporting Information, it is reasonable to conclude that the commercial chelator p-NH2–Bn-NOTA was less selective for Cu(II) versus Fe(III), compared to chelator 6.
Figure 2.
MS spectra of cold labeling solutions. Labeling conditions: (A) chelator 6, Cu(II) and Fe(III) were mixed at molar ratios 6:1:1 in 0.1 M NH4OAc buffer (pH 4) and incubated at 70 °C for 30 min; (B) chelator 6, Cu(II) and Fe(III) were mixed at molar ratios 1:1:10 in 0.1 M NH4OAc (pH 4).
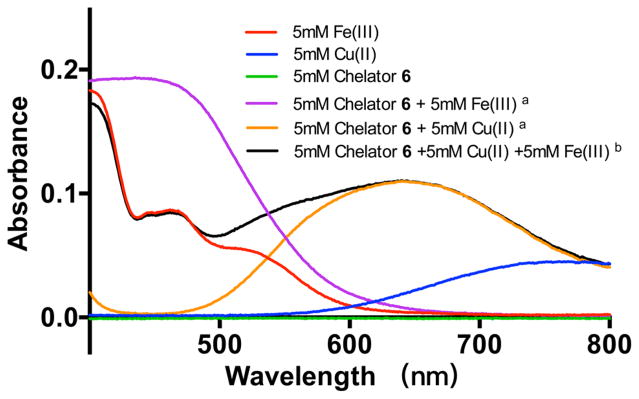
To further validate the selectivity of 6 for Cu(II) over Fe(III), a UV-vis-based analysis method was also developed. The spectra of Fe(III), Cu(II), chelator 6 itself, and the Cu(II)-6, Fe(III)-6 complexes are combined and shown in Figure 3. The maximum absorption values for the Fe(III) and Cu(II) solutions were 460 and 770 nm, respectively, while the absorption of the chelator 6 at the 400–800 nm is negligible. When the metal ions were complexed with chelator 6, the maximum absorption shifted to 435 nm for Fe(III)-6 and 640 nm for Cu(II)-6. In our study, chelator 6 (10 mM) was added to equal volume of the metal ion mixture containing CuCl2 (10 mM) and FeCl3 (10 mM) for a final concentration of 5 mM. The reaction mixture was incubated at room temperature for 30 min. The UV-vis spectra of the resulting mixture were measured (Figure 3). The wavelength-absorption curves showed that the absorbance at 640 nm matched with the absorbance of the pure Cu(II)-6 (5 mM), and there was no obviously absorbance increase at the wavelength of 435 nm, where Fe(III)-6 solution showed maximum absorption. It can be concluded that most of the chelator 6 selectively formed complexes with Cu(II) instead of Fe(III) under the above conditions, which confirms the excellent Cu(II) chelating selectivity over Fe(III) of 6.
Figure 3.
Ultraviolet–visible-light (UV-vis) spectra of cold labeling solutions. Labeling conditions: (a) chelator 6 and Cu(II) or Fe(III) (final concentration 5 mM for each component) were mixed at molar ratios of 1:1 in 0.1 M NH4OAc buffer (pH 4) and incubated at 70 °C for 30 min; (b) chelator 6, Cu(II) and Fe(III) (final concentration of 5 mM for each component) were mixed at molar ratios of 1:1:1 in 0.1 M NH4OAc buffer (pH 4) and incubated at room temperature for 30 min and incubated at room temperature for 30 min.
Because of the difficulties intrinsic to study the aqueous chelating chemistry of these e(III)-6/Cu(II)-6 complexes in the absence of crystal structures, the potentiometric titrations or extensive NMR studies were not used in the current study. In order to further investigate the coordination chemistry of the complexes, density functional theory (DFT) calculations were performed to determine their geometries and compare chelating selectivity of NE3TA with those of commercially available chelators (SCN-Bn-NOTA and NODAGA). All calculations were performed using the Gaussian 09 suite of computer programs.30 DFT was used employing uB3LYP hybrid functional. Geometry optimization was done in all internal degrees of freedom, while considering several different conformations of the ligands and complexes using a combined basis set 6-31G++(d,p). A frequency calculation was done at the same level of theory as geometry optimization to confirm that the stationary points were at the minima. We calculated the energies at 6-311++ G(d,p) level using water as solvent based on the geometry optimized structures of the Cu(II)-NE3TA and Fe(III)-NE3TA. We also considered SCN-Bn-NOTA and NODAGA with Cu(II) and Fe(III) for comparison, which are both commercially available chelators for good copper chelation capability. The optimized structures of the hypothetical models of Cu(II)-NE3TA and Fe(III)-NE3TA complexes are shown in Figure 4. In the Cu(II)-NE3TA complex, the bond distances of Cu–O1 (2.27) and Cu–N3 (2.83) are longer than the other coordinate bonds, consistent with the expected Jahn–Teller effects from the d9 electron configuration. However, in the Fe(III)-NE3TA complex, the bond distances of Fe and O, N atoms are almost the same (that of Fe–O is ~1.90 and that of Fe–N is ~2.05), which may result from the d2sp3 hybridization of Fe(III). While Fe(III)-NE3TA and Cu(II)-NE3TA complexes have similar configuration and interactions, we could not assume Cu(II)-NE3TA complex to be more stable under aqueous conditions. In order to identify their relative stability of the chelator-Fe(III) and chelator-Cu(II), the difference of free Gibbs energies (ΔG) for the reaction to form the complexes was calculated by eq 2 (see rxn 1). As shown in rxn 1, the aqua complexes for Cu(II) and Fe(III) were modeled as Cu2(OAc)4(H2O)2 and Fe3(μ-O) (OAc)6(H2O)3, since the NH4OAc buffer was used. The energies of the chelators and metal complexes were calculated at 6-31G++(d,p) level (with water as a solvent). The ΔG value of metal exchanging reaction of NE3TA chelator complexing with Cu(II) and Fe(III) ions was −510.6 kcal/mol, which is comparable to that of NOTA (−520.2 kcal/mol), but significant higher than that of NODAGA (−1358.6 kcal/mol). The data suggest that p-SCN-PhPr-NE3TA chelator has a comparable or better selectivity of Cu(II) over Fe(III) than two commercial chelators. However, because of the complexity of the model of Cu(II) and Fe(III) in buffer, the related ΔG value only could be used to evaluate and compare the complexing ability of these chelators.
Figure 4.
DFT-optimized structures of the hypothetical model of Cu(II)-NE3TA and Fe(III)-NE3TA complexes.
| (1) |
| (2) |
Promising results obtained from the chelating selectivity evaluation and DFT calculation drove us to further investigate the radiolabeling of NE3TA-PEG4-LLP2A with 64Cu. Preliminary radiolabeling experiments demonstrated quantitative radiolabeling of 50–200 pmol of NE3TA-PEG4-LLP2A with 64Cu within 10 min at room temperature under neutral or acidic conditions. Further experimentation revealed that complete labeling was dependent on ligand concentration and solution pH, with lower concentrations and higher pH requiring more labeling reaction time. Under the opitimal conditions, the purified NE3TA/NOTA-PEG4-LLP2A conjugates (50–200 pmol, μM level concentrations) were radiolabeled with 200 μCi 64Cu by incubation with 64CuCl2 in 0.1 M NH4OAc buffer (pH 4.0) at 37 °C for 30 min to ensure the best labeling, and the radiochemical yield and purity were confirmed by radio-HPLC. As shown in Figure 5, the specific activity of 64Cu-NE3TA-PEG4-LLP2A was determined to be 2–4 mCi/nmol (74–148 MBq/nmol), which was significantly higher than that of 64Cu-NOTA-PEG4-LLP2A (1–2 mCi/nmol, 37–74 MBq/nmol). In particular, when specific activity was 4 mCi/nmol, 90% labeling efficiency was achieved with NE3TA-PEG4-LLP2A, whereas the labeling yield of NOTA-PEG4-LLP2A was <50%. The lower specific activity of NOTA-PEG4-LLP2A could be due to the slower chelating kinetics, which required a longer incubation time and increased temperature to allow full complexation. Thus, slightly enhanced labeling efficiency (55%) was observed when the incubation time increased to 1 h with NOTA-PEG4-LLP2A, which was still significantly lower than that of NE3TA-PEG4-LLP2A. The remarkably enhanced labeling efficiency and SA of NE3TA-PEG4-LLP2A might be a result of the good Cu(II) selectivity of the chelator, and the conjugation did not affect the coordination properties of the chelating moiety in p-SCN-PhPr-NE3TA.
Figure 5.
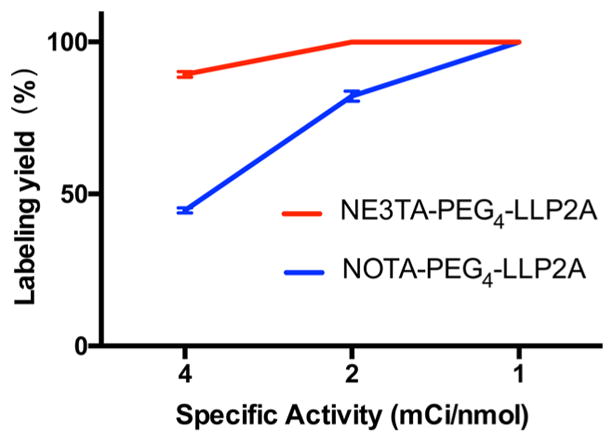
64Cu labeling yields of NE3TA-PEG4-LLP2A and NOTA-PEG4-LLP2A at 1, 2, and 4 mCi/nmol specific activities (p < 0.001).
In order to gain insight into the in vitro and in vivo kinetic inertness of 64Cu-NE3TA-PEG4-LLP2A, serum stability experiments were performed to determine if the conjugate radio-labeled with 64Cu remained stable under simulated biological conditions. The stability was assessed by measuring the disassociation of 64Cu from the complex in human serum using radio-HPLC. 64Cu-NE3TA-PEG4-LLP2A exhibited good in vitro stability with no sign of 64Cu disassociation after incubation in human serum at 37 °C for 1 d. Furthermore, <5% protein-bound 64Cu was observed in the serum samples containing the 64Cu-ligand complexes, confirming the complexes remained intact.
To determine the specific binding of 64Cu-NE3TA-PEG4-LLP2A to VLA-4 receptor, a cell internalization assay was performed using VLA-4-overexpressing B16F10 mouse melanoma cells. As shown in Figure 6, 64Cu-NE3TA-PEG4-LLP2A was rapidly internalized by B16F10 cells within the first 15 min, and the internalization became saturated at ~2 h, maintaining a high level up to 4 h. In a parallel blocking group that received an additional 10 μg unlabeled LLP2A-PEG4, the internalization was significantly blocked at all time-points (p < 0.001). The specific internalization was calculated by subtracting nonspecific internalization (blocked) from total internalization (non-blocked). High specific internalization was observed up to 4 h, indicating good VLA-4 receptor-binding affinity of 64Cu-NE3TA-PEG4-LLP2A.
Figure 6.
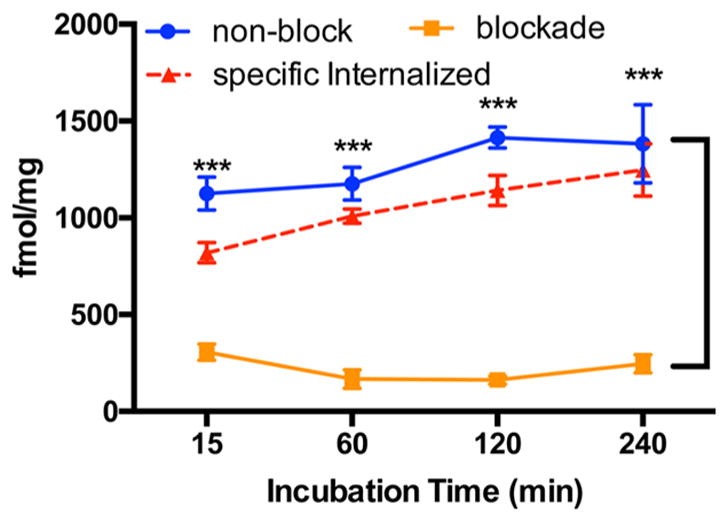
Cell internalization of 64Cu-NE3TA-PEG4-LLP2A (10 pmol per well) in B16F10 cells at 15 min, 1 h, 2 h, and 4 h. For blocking study, unlabeled LLP2A-PEG4 (10 μmol per well) was applied 30 min before the experiment (p < 0.001).
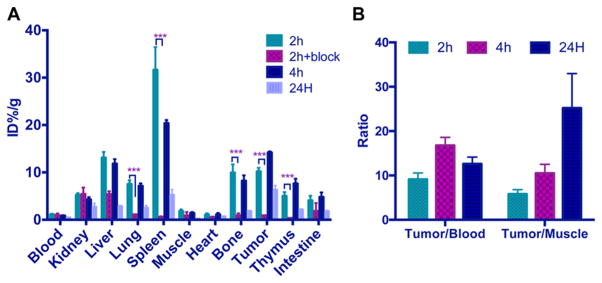
An ex vivo biodistribution study was performed on C57BL/6 mice bearing B16F10 xenografts to investigate the tumor and normal tissue uptakes of 64Cu-NE3TA-PEG4-LLP2A. The tumor uptake was high, with 10.2% ± 0.78%, 14.2% ± 0.25%, and 6.01% ± 1.32% ID/g at 2, 4, and 24 h, respectively (see Figure 7). Impressive tumor/blood ratios were observed at all examined time-points, with the highest value obtained at 4 h post-injection (16.8 ± 1.8). The tumor/muscle ratios increased over time, and the highest value was obtained at 24 h post-injection (25.2 ± 7.8). Uptake of 64Cu-NE3TA-PEG4-LLP2A in tumor, spleen, bone, lung, and thymus were significantly reduced after coinjecting an excess of unlabeled LLP2A-PEG4 with 64Cu-NE3TA-PEG4-LLP2A, indicating the VLA-4-mediated uptake in these tissues. The high uptake in VLA-4 receptor abundant organs such as the spleen, bone marrow, and thymus has been previously reported,31 and our results are consistent with the previous study using LLP2A as the high-affinity peptidomimetic ligand for VLA-4.16,32 In the presence of the blocking agent (excess LLP2A-PEG4), the radiotracer uptake was significantly reduced in the VLA-4–positive tissues, demonstrating the targeting specificity of 64Cu-NE3TA-PEG4-LLP2A. A similar biodistribution study was carried out with 64Cu-NODAGA-PEG4-LLP2A, which is a very close analogue to 64Cu-NOTA-PEG4-LLP2A.16 Compared with those results, both 64Cu-NODAGA-PEG4-LLP2A and 64Cu-NE3TA-PEG4-LLP2A showed relatively low liver and kidney uptake, and the tumor uptake of 64Cu-NE3TA-PEG4-LLP2A at 4 h (14.2% ± 0.25% ID/g) is slightly higher than that of 64Cu-NODAGA-PEG4-LLP2A (13.4% ± 1.7% ID/g). Taking into account both tumor uptake and the background contrast, it is concluded that time-points of 2 h and 4 h should be suitable for in vivo PET imaging.
Figure 7.
Biodistribution of 64Cu-NE3TA-PEG4-LLP2A (150–200 μCi) with or without a blocking agent (100 μg unlabeled LLP2A-PEG4) in B16F10 tumor bearing mice. (n = 3 or 4; p < 0.001).
Small animal PET/CT imaging with 64Cu-NE3TA-PEG4-LLP2A was also performed in the same mouse model (see Figure 8). All tumor xenografts were clearly visualized with a high signal to background ratio at both 2 and 4 h post-injection of 64Cu-NE3TA-PEG4-LLP2A. Good blocking was observed after co-injecting unlabeled LLP2A, together with 64Cu-NE3TA-PEG4-LLP2A, similar to the biodistribution study. Uptake values of tumor and background tissue were determined by quantitative analysis of the PET images. The tumor uptakes at 2 and 4 h post-injection of 64Cu-NE3TA-PEG4-LLP2A were 16.7% ± 1.0% and 19.2% ± 5.6% ID/g, respectively. The tumor/muscle ratios of 64Cu-NE3TA-PEG4-LLP2A were 13.1 ± 2.4 and 30.9 ± 0.7 at 2 and 4 h, respectively. In the blockade group, the tumor uptakes and tumor-to-muscle ratios decreased significantly to 1.4% ± 0.6% ID/g and 4.4 ± 1.8 (p < 0.001), respectively. Tumor/nontumor ratios of PET quantitative analysis were consistent with the biodistribution data, validating the in vivo specific binding of the tracer to its targeting receptor.
Figure 8.
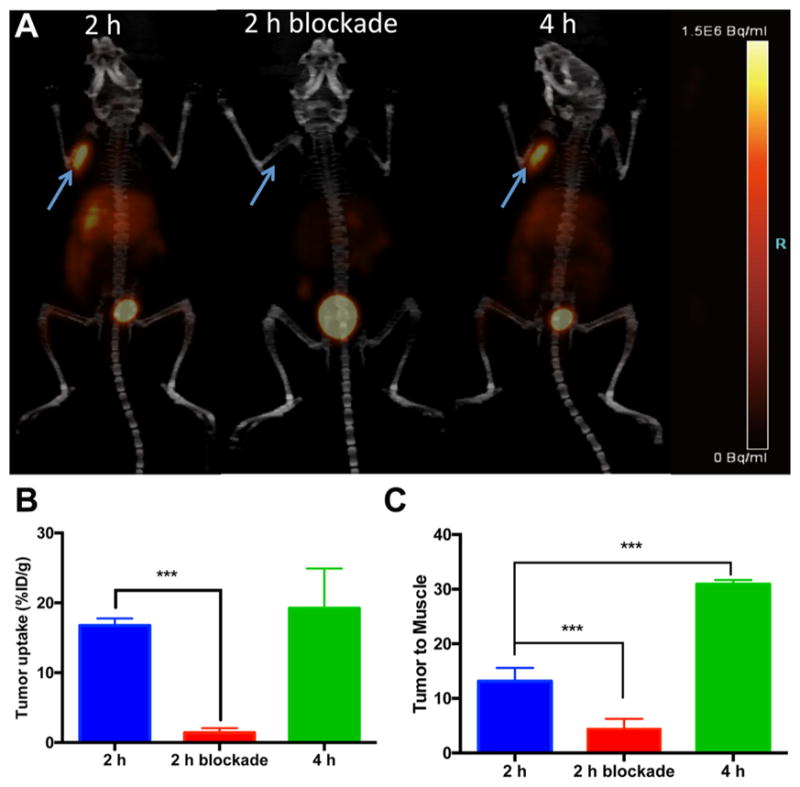
Maximum intensity projection (MIP) PET/CT images of mice bearing B16F10 tumors at 2 and 4 h post-injection of 64Cu-NE3TA-PEG4-LLP2A and blockade at 2 h (co-injected with unlabeled LLP2A-PEG4 (100 μg)): (A) tumor uptakes, (B) tumor-to-muscle ratios, (C) as determined by quantitative analysis of PET/CT images. Arrows indicate tumors (n = 3 or 4; p < 0.001).
In addition to peptidomimetic ligand, the developed p-SCN-PhPr-NE3TA has also been used to modify cetuximab (anti-EGFR monoclonal antibody), and each antibody contains ~2 NE3TA (Scheme 2B). Because of the excellent 64Cu chelating selectivity (over Fe(III)), the specific activity (9–10 μCi/μg) of resulting 64Cu-NE3TA-cetuximab was significantly higher than that (3–4 μCi/μg) of 64Cu-NOTA-cetuximab prepared from cetuximab and SCN-Bn-NOTA using the same procedures. Although the specific activity of 64Cu-NOTA-cetuximab could be improved by increasing the number of NOTA on each antibody, increasing of the number of NOTA on cetuximab may decrease the immunoactivity of resulting NOTA-cetuximab conjugate. Therefore, the developed highly 64Cu-selected p-SCN-PhPr-NE3TA BFC will also have a big impact on the radiolabeling of the modification-sensitive antibodies. Currently, comprehensive evaluation of the resulting 64Cu-NE3TA-cetuximab is in progress, and the results will be compared to those obtained from 64Cu-NOTA-cetuximab as part of a future study.
CONCLUSIONS
p-SCN-PhPr-NE3TA, which is a new bifunctional chelator (BFC) that contains an extra isothiocyanate (NCS) pendant group for the bioconjugation with tumor targeting vector was developed for positron emission tomography (PET) imaging. The chelator exhibited the ability to form stable Cu(II) complexes with excellent Cu(II) chelating selectivity over Fe(III). p-SCN-PhPr-NE3TA was easily conjugated with peptidomimetic to obtain NE3TA-PEG4-LLP2A, which was radiolabeled with 64Cu under mild labeling conditions at high specific activity (SA). The radiolabeled conjugate 64Cu-NE3TA-PEG4-LLP2A demonstrated promising in vivo performances in both biodistribution and small animal-PET imaging studies, confirming the excellent VLA-4-overexpressed-tumor targeting efficacy of LLP2A-PEG4. Besides using the peptidomimetic ligand, LLP2A, p-SCN-PhPr-NE3TA was successfully used to modify cetuximab, and NE3TA-cetuximab was labeled with 64Cu at higher SA than NOTA-cetuximab, showing a great potential in radiolabeling modification-sensitive antibodies. In addition, the study also confirmed a proof of concept using NE3TA-PEG4-LLP2A and NE3TA-cetuximab as the example to render this BFC a promising tool for generating biomolecule-based tracers.
EXPERIMENTAL SECTION
All chemicals were purchased from Sigma–Aldrich Chemical Co. (St. Louis, MO), unless otherwise specified. Aqueous solutions were prepared using ultrapure water (resistivity, 18 M). Copper-64 was obtained from Washington University (St. Louis, MO) and the University of Wisconsin (Madison, WI). A Model Cobra II gamma counter (PerkinElmer (Packard)) and a Model 2470 Wizard automatic gamma counter (PerkinElmer, Waltham, MA, USA) were used to measure γ radiation. 1H NMR and 13C NMR spectra were recorded on a Bruker DRX 400 MHz spectrometer (Billerica, MA), and electron-spray ionization mass spectroscpy (ESI-MS) spectra were measured on a Model LCT-Premier XE LC-MS station (Waters Corp., Milford, MA, USA). High-performance liquid chromatography (HPLC) columns (Model Luna C-18, Phenomenex, Torrance, CA, USA) were obtained. HPLC analyses were performed on a binary HPLC pump (Model 1525, Waters Corp., Milford, MA, USA) with an ultraviolet–visible-light (UV-vis) detector (Model 2489, Waters Corp., Milford, MA, USA) and a radioactivity detector (Model 106, Bioscan) for purification of some peptide conjugates and analysis of their 64Cu-labeled conjugates using two elution buffers (0.1 vol % TFA in deionized water as elution buffer A and 0.1 vol % TFA in acetonitrile as elution buffer B). PET/CT data were acquired using an Inveon Preclinical Imaging Station (Siemens Medical Solutions).
Synthesis of 2-(3-(4-nitrophenyl)propylamino)ethanol (2)
To a solution of 2-aminoethanol (6.1 g, 100 mmol) and triethylamine (TEA) (5 g, 50 mmol) in 20 mL of MeCN at 0 °C was added 1 (4.88 g, 10 mmol) in 30 mL of MeCN over 2 h. The reaction mixture was allowed to warm to room temperature and stirred for 24 h. The solvent was evaporated and the residue was treated with 10 mL of water. The resulting solution was adjusted to pH 3 using 1 M HCl and extracted with ethyl acetate (EA) (50 mL × 2). The aqueous layer was then adjusted to pH 12 using 1 M NaOH and extracted with dichloromethane (DCM) (50 mL × 2). The organic layer was dried and evaporated to afford a yellow solid. The crude product is recrystallized from 50% EA in petroleum ether (PE) to give 2 (3.22 g, 71.6%) as a white solid, mp: 82.6–84.0 °C. 1H NMR (400 MHz, CDCl3) δ 8.09 (d, J = 7.2 Hz, 2H, NO2–Ar–H), 7.31 (d, J = 8.3 Hz, 2H, Ar–H), 3.70–3.56 (m, 2H, HO–CH2–), 2.74 (m, 4H, CH2–N), 2.64 (t, J = 7.2 Hz, 2H, Ar–CH2), 2.52 (br, 2H, –NH–, –OH), 1.89–1.71 (m, 2H, –CH2–).13C NMR (101 MHz, CDCl3) δ 149.96, 146.32, 129.14, 123.64, 60.74, 51.16, 48.74, 33.41, 31.17. MALDI-HRMS (matrix: HCCA) calculated for C11H16N2O3, [M + H]+ m/z 225.1234, found 225.1233.
Synthesis of tert-Butyl 2-((2-hydroxyethyl) (3-(4-nitrophenyl)propyl)amino)acetate (3)
To a solution of 2 (2.24 g, 10 mmol) and K2CO3 (2.78 g, 22 mmol) in 50 mL MeCN at 0 °C was dropwise added t-butyl-bromoacetate (2.34 g, 12 mmol) over 30 min. The reaction mixture was gradually warmed to room temperature and stirred for 24 h. The result reaction mixture was filtered, and the filtrate was concentrated. The residue was then passed through a short silica column eluted with 10% EA in PE to provide 3 (3.01 g, 89.1%) as a yellow oil. 1H NMR (400 MHz, CDCl3) δ 8.11 (d, J = 8.4 Hz, 2H, NO2–Ar–H), 7.33 (d, J = 8.5 Hz, 2H, Ar–H), 3.52 (t, J = 5.1 Hz, 2H, HO–CH2–), 3.23 (s, 2H, COCH2N), 2.82–2.69 (m, 4H, CH2–N), 2.64 (t, J = 7.0 Hz, 2H, Ar–CH2), 1.80 (m, 2H, –CH2–), 1.44 (s, 9H, –CH3).13C NMR (101 MHz, CDCl3) δ 171.51, 150.02, 146.34, 129.21, 123.63, 81.51, 59.12, 57.24, 56.24, 54.28, 33.07, 29.21, 28.07. MALDI-HRMS (matrix: HCCA) calculated for C17H26N2O5, [M + H]+ m/z 339.1914, found 339.1914.
Synthesis of tert-Butyl 2-((2-bromoethyl) (3-(4-nitrophenyl)-propyl)amino)acetate (4)
To a solution of 3 (3.42 g, 12.9 mmol) in 50 mL MeCN at 0 °C was added PPh3 (3.72 g, 14.2 mmol). NBS (2.53 g, 14.2 mmol) was then added, in portions, over 1 h. The reaction mixture was stirred at 0 °C for 30 min, after which the reaction mixture was allowed to warm to room temperature and stirred for 3 h. The resulting mixture was evaporated into a yellowish solid. The residue was passed through a short silica column eluted with 5% EA in PE to obtain 4 (3.75 g, 72.7%) as a yellow oil. 1H NMR (400 MHz, CDCl3) δ 8.12 (d, J = 8.7 Hz, 2H, NO2–Ar–H), 7.34 (d, J = 8.6 Hz, 2H, Ar–H), 3.41–3.34 (m, 2H, Br–CH2–), 3.32 (s, 2H, COCH2N), 3.05 (t, J = 7.1 Hz, 2H, –CH2CH2Br), 2.81–2.74 (m, 2H, NH–CH2CH2–), 2.70 (t, J = 7.0 Hz, 2H, Ar–CH2–), 1.87–1.73 (m, 2H, –CH2–), 1.51–1.38 (m, 9H, CH3).13C NMR (101 MHz, CDCl3) δ 170.48, 150.12, 146.33, 129.24, 123.63, 81.30, 56.13, 55.68, 53.38, 33.07, 30.55, 29.34, 28.16.
Synthesis of Di-tert-butyl-2,2′-(7-(2-((2-(tert-butoxy)-2-oxoethyl)(3-(4-nitrophenyl)propyl)amino) ethyl)-1,4,7-triazonane-1,4-diyl)diacetate (5)
To a solution of NO2AtBu26 (1 g, 2.8 mmol) in 10 mL of MeCN was added ethyldiisopropylamine (DIEA) (0.72 g, 5.6 mmol) and 4 (1.12 g, 2.8 mmol). The reaction mixture was left at room temperature overnight. The resulting solution was filtered and concentrated. The residue was purified using silica column eluted with 10% MeOH in DCM to obtain 5 (1 g, 52.9%) as a waxy solid. 1H NMR (400 MHz, CDCl3) δ 8.09 (d, J = 8.6 Hz, 2H, NO2–Ar–H), 7.31 (d, J = 8.6 Hz, 2H, Ar–H), 3.27 (s, 4H, COCH2N), 3.22 (s, 2H, COCH2N), 3.01–2.63 (m, 18H, N–CH2), 2.63–2.56 (m, 2H, Ar–CH2–), 1.81–1.70 (m, 2H, –CH2–), 1.40 (s, 18H, CH3), 1.40 (s, 9H, CH3).13C NMR (101 MHz, CDCl3) δ 171.40, 170.79, 150.33, 146.29, 129.22, 123.56, 80.84, 80.71, 59.78, 56.14, 55.48, 54.04, 33.32, 29.06, 28.19, 28.16. MALDI-HRMS (matrix: HCCA) calculated for C35H59N5O8, [M + H]+ m/z 678.4436, found 678.4435.
Synthesis of 2,2′-(7-(2-((carboxymethyl)(3-(4-nitrophenyl)-propyl)amino)ethyl)-1,4,7-triazonane-1,4-diyl)diacetic acid (6)
Compound 5 (500 mg, 0.74 mmol) in 2 mL of DCM on an ice bath was treated with 10 mL TFA/DCM (1:1). The resulting mixture was gradually warmed to ambient temperature and stirred overnight. The solvents were evaporated and further coevaporated with 20 mL of CHCl3 three times to obtain 6 (400 mg, 100%) as a yellow oil as its TFA salts. 1H NMR (400 MHz, D2O) δ 7.98 (d, J = 8.8 Hz, 2H, NO2–Ar–H), 7.33 (d, J = 8.7 Hz, 2H, Ar–H), 3.99 (s, 2H, COCH2N), 3.86 (s, 4H, COCH2N), 3.47–3.37 (m, 2H, N–CH2), 3.24 (m, 4H, N–CH2), 3.21–3.04 (m, 8H, N–CH2), 2.92 (m, 4H, N–CH2), 2.71 (t, J = 7.3 Hz, 2H, Ar–CH2–), 2.11–1.91 (m, 2H, –CH2–).13C NMR (101 MHz, D2O) δ 171.19, 168.53, 148.59, 146.21, 129.48, 123.85, 56.17, 54.42, 54.33, 51.10, 50.92, 49.86, 49.81, 48.55, 31.43, 23.94. MALDI-HRMS (matrix: HCCA) calculated for C23H35N5O8, [M + H]+ m/z 510.2558, found 510.2588.
Synthesis of Di-tert-butyl-2,2′-(7-(2-((3-(4-aminophenyl)-propyl)(2-(tert-butoxy)-2-oxoethyl)amino) ethyl)-1,4,7-triazonane-1,4-diyl)diacetate (7)
To a solution of 6 (136 mg, 0.2 mmol) in 5 mL MeOH was added 10% Pd/C. The resulting mixture was bubbled with H2 at room temperature for 3 h. The reaction mixture was filtered and the filtrate was concentrated in vacuo to provide pure 7 (100 mg, 77.1%). 1H NMR (400 MHz, CDCl3) δ 6.88 (d, J = 8.3 Hz, 2H, NO2–Ar–H), 6.56 (d, J = 8.3 Hz, 2H, NO2–Ar–H), 3.61 (brs, 2H, –NH2–), 3.33 (s, 4H, COCH2N), 3.20 (s, 2H, COCH2N), 3.29–2.47 (m, 18H, –NCH2–), 2.42 (t, J = 7.5 Hz, 2H, Ar–CH2–), 1.69–1.59 (m, 2H, –CH2–), 1.39 (s, 18H, –CH3), 1.37 (s, 9H, –CH3). 13C NMR (101 MHz, CDCl3) δ 170.66, 170.56, 144.50, 131.35, 129.07, 115.28, 81.52, 81.39, 58.05, 55.58, 53.90, 53.45, 53.23, 52.66, 49.93, 49.87, 32.35, 28.68, 28.14, 28.11. MALDI-HRMS (matrix: HCCA) calculated for C35H61N5O6, [M + H]+ m/z 648.4695, found 648.4701.
Synthesis of 2,2′-(7-(2-((3-(4-aminophenyl)propyl) (carboxymethyl)amino)ethyl)-1,4,7-triazonane-1,4-diyl)-diacetic acid (8)
Compound 7 (65 mg, 0.2 mmol) in 0.5 mL of DCM in an ice bath was treated with 1 mL TFA/DCM (1:1). The reaction mixture was warmed to room temperature and stirred overnight. The solvent was evaporated in vacuo. The residue was taken up in deionized (DI) water and passed through a 0.45 μm nylon syringe filter. The aqueous solution was concentrated in vacuo to provide 8 (50 mg, 83.3%) as a yellow solid. 1H NMR (400 MHz, D2O) δ 7.35–7.17 (m, 4H, Ar–H), 3.96 (s, 2H, N–CH2CO–), 3.88 (s, 4H, N–CH2CO–), 3.48–3.35 (m, 2H, N–CH2–), 3.27 (s, 4H, N–CH2–), 3.23–3.05 (m, 8H, N–CH2–), 2.96–2.88 (m, 4H, N–CH2–), 2.65 (t, J = 7.3 Hz, 2H, Ar–CH2–), 2.07–1.85 (m, 2H, –CH2CH2CH2–). 13C NMR (101 MHz, D2O) δ 171.33, 168.68, 141.49, 130.07, 128.02, 123.20, 56.24, 54.48, 51.12, 50.02, 49.80, 48.59, 31.04, 24.25. MALDI-HRMS (matrix: HCCA) calculated for C23H37N5O6, [M + H]+ m/z 480.2817, found 480.2822.
Synthesi s of 2,2′-(7 -(2-((carboxymethyl)(3-(4-isothiocyanatophenyl)propyl)amino)ethyl)-1,4,7-triazonane-1,4-diyl)diacetic acid (9, p-SCN-PhPr-NE3TA)
To a solution of 8 (4.8 mg, 0.01 mmol) in DI water was added a 1 M solution of thiophosgene in CHCl3. The resulting mixture was stirred for 4 h at room temperature. The aqueous layer was separated and washed with CHCl3. The aqueous layer was evaporated to dryness to provide 9 (5 mg, 95.7%) as a yellowish solid. 1H NMR (400 MHz, D2O) δ 7.26–7.14 (m, 4H, Ar–H), 3.85 (s, 2H, –NCH2CO–), 3.72–3.57 (m, 4H, N–CH2–), 3.40–3.29 (m, 2H, N–CH2–), 3.12 (s, 4H, –NCH2CO–), 3.09–2.58 (m, 14H, N–CH2–, Ar–CH2–), 2.08–1.92 (m, 2H, –CH2CH2CH2–). MALDI-HRMS (matrix: HCCA) calculated for C24H35N5O6S, [M + H]+ m/z 522.2381, found 522.2387.
Synthesis of NE3TA-PEG4-LLP2A
To a solution of LLP2A-PEG4 and DIEA (3 equiv) in DMF was added p-SCN-PhPr-NE3TA (1.2 equiv). The mixture was reacted at room temperature for 4 h. The solvent was lyophilized, and the residue was purified using HPLC with a stepwise gradient method at a flow rate of 1.5 mL/min. The elution method started with 18% B for 3 min, followed by 18% to 35% B over 30 min (retention time of ~22 min). The purity was determined to be >95%, while the isolation yield was 65%. ESI-MS: observed, m/z (M + 2H)2+ = 846.544, calculated, (M + 2H)2+ = 847.437.
Synthesis of NOTA-PEG4-LLP2A
To a solution of LLP2A-PEG4 and DIEA (3 equiv) in DMF was added SCN-Bn-NOTA (1.2 equiv). The mixture was reacted at room temperature for 4 h. The solvent was lyophilized, and the residue was purified using HPLC with a stepwise gradient method at a flow rate of 1.5 mL/min. The elution method started with 18% B for 3 min, followed by 18% to 35% B over 30 min (retention time of ~26 min). The purity was determined to be >95%, while the isolation yield was 63%. MALDI-TOF: observed, m/z (M + H)+ = 1642.67, calculated, (M + H)+ = 1622.78.
Cu(II) Selectivity Evaluation (LC-MS)
Stock solution A, which contained 5 mM CuCl2 and 5 mM FeCl3, stock solution B, which contained 5 mM CuCl2 and 50 mM FeCl3, and chelator 6 (20 mM) were prepared using 0.1 M NH4OAc buffer (pH 4). All reagents used were trace metal grade. Fifteen microliters (15 μL) of chelator 6 (300 nmol) was mixed with 5 μL stock solution A (25 nmol Cu(II), 25 nmol Fe(III)) and 192 μL buffer. The mixture was incubated at 70 °C for 30 min to make all ions chelated. After cooling to room temperature, an ESI-MS analysis was applied to check the mass intensity of Fe(III)-6 complex and Cu(II)-6 complex. To further check the Cu(II) selectivity of chelator 6 over Fe(III), 2 μL of chelator 6 (40 nmol) stock solution was mixed with 8 μL stock solution B (40 nmol Cu(II), 400 nmol Fe(III)) and 190 μL buffer, and the resulting mixture was incubated at 37 °C for 30 min. ESI-MS was used to check the mass intensity of Fe(III)-6 complex and Cu(II)-6 complex. For comparison purposes, the same procedures were also performed using chelator p-NH2–Bn-NOTA instead of chelator 6.
Cu(II) Selectivity Evaluation (UV-vis)
All stock solutions were prepared using 0.1 M NH4OAc buffer (pH 4). All reagents used were trace metal grade. Chelator 6 (10 mM) was mixed with 10 mM Cu(II) or 10 mM Fe(III) at a volume ratio of 1:1, and the mixtures were incubated at 70 °C for 30 min to make all ions chelated. Chelator 6 (10 mM) was added to a metal ion mixture containing CuCl2 (10 mM) and FeCl3 (10 mM) at a volume ratio of 1:1, and the reaction mixture was incubated at room temperature for 30 min. The UV–vis absorption spectra of CuCl2 (5 mM), FeCl3 (5 mM), chelator 6 (5 mM), and the above reaction mixtures were recorded using a Cary 100 Bio UV-vis spectrophotometer.
Radiolabeling
The 64Cu-labeling of bioconjugates were performed in NH4OAc buffer as follows: 50–200 pmol of bioconjugates was added to 100 μL of 0.1 M NH4OAc (pH 4) in a 1.5 mL tube, and then 200 μCi 64Cu in 0.1 M NH4OAc solution (pH 4.0) was added. The mixture was vortexed for 10 s and incubated in a thermomixer at 37 °C for 0.5 h. The radiolabeling yield was determined by radio-HPLC.
Serum Stability
64Cu-labeled bioconjugates (~100 μCi) were mixed with 0.5 mL of human serum, and the resulting mixtures were incubated at 37 °C for 1 day. After incubation, plasma protein was precipitated with 0.5 mL of acetonitrile and centrifuged at 14 000 rpm for 5 min. The supernatants were collected and >90% activity was recovered, as determined by a dose calibrator. The radiopurity was determined by radio-HPLC. Free 64Cu2+ comes out at ~1.5 min and the radiolabeled conjugates come out at 7–8 min. More than 95% intact radiolabeled conjugates were observed.
Cell Studies
B16F10 cell line was purchased from the American Tissue Culture Collection (ATCC). All cell handling was aseptically performed in a laminar flow hood. The B16F10 cells were cultured in Dulbecco’s Modified Eagle Medium, supplemented with 10% FBS, penicillin (100 unit/mL), streptomycin (100 μg/mL) L-glutamine (300 μg/mL) and sodium pyruvate (100 mg/mL), glucose (4.5 g/L) and maintained at 37 °C, 5% CO2.
Internalization Assay
Internalization assays were performed to determine the internalization of 64Cu-NE3TA-PEG4-LLP2A in VLA-4 positive B16F10 mouse melanoma cells. Cells were seeded in 12-well plates (200 000 cells per well) 24 h prior to the experiment. Before the experiment, cells were washed with 1 mL of HBSS twice and 1 mL media (DMEM with 0.1% BSA and 1 mM Mn2+) was added to each well. Cells were then incubated with the 64Cu-NE3TA-PEG4-LLP2A (10 pmol per well) with and without the addition of excess LLP2A-PEG4 (10 μg per well) to determine nonspecific internalization. At each time point (15 min, 1 h, 2 h, and 4 h) radioactive media were aspirated, and the plate was washed twice with HBSS (pH 7.2). To collect the surface-bound fraction, each well was treated with 20 mM sodium acetate-HBSS (pH 4.0) and was incubated at 37 °C for 10 min. After removal of the surface-bound fraction, cell pellets were dissolved in 0.5% SDS. All of the fractions were counted for radioactivity on the gamma counter. The radioactivity in each fraction was measured in a well counter (Packard Cobra II gamma counter). The amount internalized was the amount of activity in the final cell pellet, corrected for activity in the blocked fractions and background activity. The protein content of each cell lysate sample was determined (BCA Protein Assay Kit, Pierce). The measured radioactivity associated with the cells was normalized to the amount of cell protein present (%AD/mg protein).
Animal Model
Four-week old female C57BL/6 mice purchased from Jackson Laboratories (Bar Harbor, ME, USA) were used in this study. All animal studies were performed under the Guide for the Care and Use of Laboratory Animals, under the auspices of Division of Laboratory Animal Resources (DLAR) of the University of Pittsburgh. This study was approved by the IACUC Committee. For xenograft B16F10 tumors, mice were injected subcutaneously at the right shoulder with one million cells in PBS and BD matrigel (1:1).
Small Animal PET/CT Imaging
Mice bearing B16F10 xenografts (n = 3 and 4 per group) were injected intravenously with 64Cu-NE3TA-PEG4-LLP2A (150–200 μCi per mouse) with and without 100 μg unlabeled LLP2A-PEG4. At 2 and 4 h, after injection mice were anesthetized with 2% isoflurane and small-animal PET/CT was performed. Static images were collected for 15 min using a small animal Inveon PET/CT scanner (Siemens Medical Solution), with a tangential and radial full width at half-maximum (fwhm) of 1.5 mm at the center of the field of view and 1.8 mm at the edge of the field of view. PET and CT images were coregistered using Inveon Research Workstation (IRW) software (Siemens Medical Solutions). PET images were reconstructed with the ordered-subsets expectation maximization three-dimensional/maximum a posteriori probability algorithm, and the analysis of images was done using the IRW software. Regions of interest were drawn using the CT scan, and the associated PET activities were calculated using the IRW software.
DFT Computational Method
All calculations were performed using the Gaussian 09 suite of programs.1 Density functional theory (DFT) method was used, employing the uB3LYP hybrid functional.2 Geometry optimization was done using a combined basis set 6-31+G(d,p). Frequency calculation was done at the same level of theory as geometry optimization, to confirm the stationary points to be minima. Single-point energy calculations were done using the B3LYP method at a larger basis set 6-311++G(d,p). Solvent effect was accounted for using self-consistent reaction field (SCRF) method, SMD model, and UAKS radii.3 Water was used as the solvent. (See the Supporting Information for detailed calculation data.)
Conjugation of BFCs to Cetuximab and Radiolabeling with 64Cu
BFC (SCN-Bn-NOTA or p-SCN-PhPr-NE3TA) was mixed with cetuximab (2 mg/mL) at a molar ratio of 100:1 (BFC: cetuximab) in PBS buffer, followed by incubation at 4 °C for 1 day with end-over-end rotation. The conjugates were then transferred to Centricon 100 centrifugal filter tubes, washed five times with 0.1 M ammonium acetate (pH 6.8, trace metal) to remove unreacted BFCs. Purity and concentration of the conjugated cetuximab were determined by size-exclusion HPLC. The number of chelators per antibody of NE3TA/NOTA–cetuximab was determined to be ~2, using a published method.33 For radiolabeling, 64CuCl2 was added to NE3TA/NOTA–cetuximab in 0.1 M ammonium acetate (pH 6.8), followed by incubation at 37 °C for 1 h. The radiochemical purity of 64Cu-NETA/NOTA-cetuximab was determined by radio-iTLC.
Data Analysis
Data are expressed as mean ± standard deviation (SD). Prism software (GraphPad, San Diego, CA, USA) was used to determine statistical significance. Statistical significance was calculated using a paired t-test. A P-value of <0.01 was considered significant.
Supplementary Material
Acknowledgments
This work was supported by the National Institute of Biomedical Imaging and Bioengineering (Grant No. R21-EB017317). It was also partially supported by Specialized Research Fund for the Doctoral Program of Higher Education of China (No. 20120142120095), the Fundamental Research Fund for the Central Universities (Nos. 2016YXMS140 and 2014TS090). Small animal PET/CT imaging at UPCI was supported in part by UPCI CCSG (No. P30CA047904).
Footnotes
Notes
The authors declare no competing financial interest.
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.inorg-chem.6b00395.
DFT calculation details and other supplemental data (PDF)
References
- 1.Ametamey SM, Honer M, Schubiger PA. Chem Rev. 2008;108:1501–1516. doi: 10.1021/cr0782426. [DOI] [PubMed] [Google Scholar]
- 2.Gambhir SS. Nat Rev Cancer. 2002;2:683–693. doi: 10.1038/nrc882. [DOI] [PubMed] [Google Scholar]
- 3.Wadas TJ, Wong EH, Weisman GR, Anderson CJ. Chem Rev. 2010;110:2858–2902. doi: 10.1021/cr900325h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartholomä MD. Inorg Chim Acta. 2012;389:36–51. [Google Scholar]
- 5.Ferreira CL, Yapp DT, Lamsa E, Gleave M, Bensimon C, Jurek P, Kiefer GE. Nucl Med Biol. 2008;35:875–882. doi: 10.1016/j.nucmedbio.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 6.Moi MK, Meares CF, McCall MJ, Cole WC, DeNardo SJ. Anal Biochem. 1985;148:249–253. doi: 10.1016/0003-2697(85)90653-0. [DOI] [PubMed] [Google Scholar]
- 7.Pandya DN, Kim JY, Park JC, Lee H, Phapale PB, Kwak W, Choi TH, Cheon GJ, Yoon YR, Yoo J. Chem Commun. 2010;46:3517–3519. doi: 10.1039/b925703a. [DOI] [PubMed] [Google Scholar]
- 8.Ait-Mohand S, Denis Cl, Tremblay Gv, Paquette M, Guérin B. Org Lett. 2014;16:4512–4515. doi: 10.1021/ol5020575. [DOI] [PubMed] [Google Scholar]
- 9.Ma MT, Karas JA, White JM, Scanlon D, Donnelly PS. Chem Commun. 2009:3237–3239. doi: 10.1039/b903426a. [DOI] [PubMed] [Google Scholar]
- 10.Boros E, Rybak-Akimova E, Holland JP, Rietz T, Rotile N, Blasi F, Day H, Latifi R, Caravan P. Mol Pharmaceutics. 2014;11:617–629. doi: 10.1021/mp400686z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wadas TJ, Anderson CJ. Nat Protoc. 2007;1:3062–3068. doi: 10.1038/nprot.2006.431. [DOI] [PubMed] [Google Scholar]
- 12.Dale AV, An GI, Pandya DN, Ha YS, Bhatt N, Soni N, Lee H, Ahn H, Sarkar S, Lee W, Huynh PT, Kim JY, Gwon MR, Kim SH, Park JG, Yoon YR, Yoo J. Inorg Chem. 2015;54:8177–8186. doi: 10.1021/acs.inorgchem.5b01386. [DOI] [PubMed] [Google Scholar]
- 13.Guo Y, Ferdani R, Anderson CJ. Bioconjugate Chem. 2012;23:1470–1477. doi: 10.1021/bc300092n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zeng D, Ouyang Q, Cai Z, Xie XQ, Anderson CJ. Chem Commun. 2014;50:43–45. doi: 10.1039/c3cc45928d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zeng D, Guo Y, White AG, Cai Z, Modi J, Ferdani R, Anderson CJ. Mol Pharmaceutics. 2014;11:3980–3987. doi: 10.1021/mp500004m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beaino W, Anderson CJ. J Nucl Med. 2014;55:1856–1863. doi: 10.2967/jnumed.114.144881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dumont RA, Deininger F, Haubner R, Maecke HR, Weber WA, Fani M. J Nucl Med. 2011;52:1276–1284. doi: 10.2967/jnumed.111.087700. [DOI] [PubMed] [Google Scholar]
- 18.Fani M, Del Pozzo L, Abiraj K, Mansi R, Tamma ML, Cescato R, Waser B, Weber WA, Reubi JC, Maecke HR. J Nucl Med. 2011;52:1110–1118. doi: 10.2967/jnumed.111.087999. [DOI] [PubMed] [Google Scholar]
- 19.Chong HS, Mhaske S, Lin M, Bhuniya S, Song HA, Brechbiel MW, Sun X. Bioorg Med Chem Lett. 2007;17:6107–6110. doi: 10.1016/j.bmcl.2007.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McBride WJ, D’Souza CA, Sharkey RM, Karacay H, Rossi EA, Chang CH, Goldenberg DM. Bioconjugate Chem. 2010;21:1331–1340. doi: 10.1021/bc100137x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kang CS, Song HA, Milenic DE, Baidoo KE, Brechbiel MW, Chong HS. Nucl Med Biol. 2013;40:600–605. doi: 10.1016/j.nucmedbio.2013.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chong HS, Sun X, Zhong Y, Bober K, Lewis MR, Liu D, Ruthengael VC, Sin I, Kang CS. Eur J Org Chem. 2014;2014:1305–1313. [Google Scholar]
- 23.Chong HS, Sun X, Chen Y, Sin I, Kang CS, Lewis MR, Liu D, Ruthengael VC, Zhong Y, Wu N, Song HA. Bioorg Med Chem. 2015;23:1169–1178. doi: 10.1016/j.bmc.2014.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Silva RA, Jain S, Lears KA, Chong HS, Kang CS, Sun X, Rogers BE. Nucl Med Biol. 2012;39:1099–1104. doi: 10.1016/j.nucmedbio.2012.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kang CS, Wu N, Chen Y, Sun X, Bandara N, Liu D, Lewis MR, Rogers BE, Chong HS. J Inorg Biochem. 2016;154:60–66. doi: 10.1016/j.jinorgbio.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gai Y, Hu Z, Rong Z, Ma X, Xiang G. Molecules. 2015;20:19393–19405. doi: 10.3390/molecules201019393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peng L, Liu R, Marik J, Wang X, Takada Y, Lam KS. Nat Chem Biol. 2006;2:381–389. doi: 10.1038/nchembio798. [DOI] [PubMed] [Google Scholar]
- 28.Ducry L, Stump B. Bioconjugate Chem. 2010;21:5–13. doi: 10.1021/bc9002019. [DOI] [PubMed] [Google Scholar]
- 29.Zeng D, Anderson CJ. Chem Commun. 2013;49:2697–2699. doi: 10.1039/c3cc39071c. [DOI] [PubMed] [Google Scholar]
- 30.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Scalmani G, Barone V, Mennucci B, Petersson GA, Nakatsuji H, Caricato M, Li X, Hratchian HP, Izmaylov AF, Bloino J, Zheng G, Sonnenberg JL, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Vreven T, Montgomery JA, Jr, Peralta JE, Ogliaro F, Bearpark MJ, Heyd J, Brothers EN, Kudin KN, Staroverov VN, Kobayashi R, Normand J, Raghavachari K, Rendell AP, Burant JC, Iyengar SS, Tomasi J, Cossi M, Rega N, Millam NJ, Klene M, Knox JE, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Martin RL, Morokuma K, Zakrzewski VG, Voth GA, Salvador P, Dannenberg JJ, Dapprich S, Daniels AD, Farkas Ö, Foresman JB, Ortiz JV, Cioslowski J, Fox DJ. Gaussian 09. Gaussian, Inc; Wallingford, CT, USA: 2009. [Google Scholar]
- 31.Hyduk SJ, Oh J, Xiao H, Chen M, Cybulsky MI. Blood. 2004;104:2818–2824. doi: 10.1182/blood-2003-12-4402. [DOI] [PubMed] [Google Scholar]
- 32.Jiang M, Ferdani R, Shokeen M, Anderson CJ. Nucl Med Biol. 2013;40:245–251. doi: 10.1016/j.nucmedbio.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun X, Rossin R, Turner JL, Becker ML, Joralemon MJ, Welch MJ, Wooley KL. Biomacromolecules. 2005;6:2541–2554. doi: 10.1021/bm050260e. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.