Abstract
Aged animals show functional alterations in hippocampal neurons that lead to deficits in synaptic plasticity and changes in cognitive function. Transcription of immediate-early genes (IEGs), including Egr1, is necessary for processes such as long-term potentiation and memory consolidation. Here we show an age-related reduction in the transcription of Egr1 in the dentate gyrus following spatial behavior, whereas in the area CA1, Egr1 is reduced at rest, but its transcription can be effectively driven by spatial behavior to levels equivalent to those observed in adult animals. One mechanism possibly contributing to these aging-related changes is an age-associated, CpG site-specific change in methylation in DNA associated with the promoter region of the Egr1 gene. Our results add to a growing body of work demonstrating that complex transcriptional and epigenetic changes in the hippocampus significantly contribute to brain and cognitive aging.
Keywords: aging, transcriptional regulation, memory, epigenetic modulation
Introduction
Activity-regulated gene transcription is a molecular requisite for long-term synaptic plasticity and memory formation. Interfering with gene transcription mechanisms, either pharmacologically or via the use of knock out technology, results in deficits in memory consolidation and long-term potentiation (e.g., Alberini, 2009). In models of normal aging (i.e., in the absence of significant neurological changes), several large scale studies have observed significant and widespread changes in gene expression (e.g., Blalock et al., 2003; Verbitsky et al., 2004; Burger et al., 2007, 2008; Rowe et al., 2007; Kadish et al., 2009; Pawlowski et al., 2009; Haberman et al., 2011). These kinds of changes are consistently observed in the hippocampus, a brain structure critical for memory and vulnerable to the effects of advancing age.
Among the genes that are impacted by the aging process is Egr1, (Yau et al., 1996; Desjardins et al., 1997; Blalock et al., 2003; Marrone et al., 2011) a transcription factor and immediate-early gene belonging to the Egr family (Egr1 is also known as Zif268). This gene is rapidly induced in association with long-term potentiation (LTP) and in behaviorally relevant brain structures and circuits after specific learning experiences (e.g., Cole et al., 1989; Hall et al., 2001; Jones et al., 2001; Pinaud et al., 2002; Poirier et al., 2008; Renaudineau et al., 2009). Mice with targeted inactivation of the Egr1 gene can exhibit short-lasting hippocampal LTP, but durable LTP is impaired (Jones et al., 2001). Furthermore, on both spatial and non-spatial learning tasks, short-term memory is intact in mice with targeted disruption of Egr1, but performance is impaired on tests requiring long-term memory. In addition, work by Renaudineau et al. (2009) indicates that Egr1 is necessary for the long-term stability of spatial representations within the hippocampus, a process that is known to be deficient in the aged rat (for review, see Rosenzweig and Barnes, 2003).
The mechanisms that underlie age-related changes in gene transcription are not fully understood, although a number of possibilities exist, including changes in receptor function, and Ca++ dysregulation (see Foster and Norris, 1997; Toescu et al., 2004; Burke and Barnes, 2010; Toescu and Vreugdenhil, 2010). Furthermore, it has been demonstrated that DNA methylation and histone modifications can dynamically regulate the gene transcription necessary for synaptic plasticity and memory processes (e.g., Miller and Sweatt, 2007; Lubin et al., 2008; Gupta et al., 2010; Miller et al., 2010; for review see Day and Sweatt, 2011). In the aged brain, there is now a growing body of evidence demonstrating epigenetic changes that are likely to promote age-associated alterations in cognitive function (e.g., Jiang et al., 2008; Peleg et al., 2010; Penner et al., 2010, 2011; Castellano et al., 2012; Haberman et al., 2012; Kosik et al., 2012; Oliveira et al., 2012; Fletcher et al., 2014; Morse et al., 2015). For example, it has been shown that age-associated changes in the transcription of the memory-associated gene Arc (activity-regulated cytoskeletal gene) is modulated by aberrant methylation of the Arc gene, (Penner et al., 2011), and that degraded cognitive status of aged rats predicts an altered relationship between Arc mRNA transcription and Arc protein translation via a ubiquitin-mediated degradation process (Fletcher et al., 2014). Haberman et al. (2012) have also observed significant age-related increases in methylation of three genes known to influence the cognitive status of aged rats (Gabra5, Hspa5, and Syn1), and it has recently been shown that histone lysine methylation levels around the BDNF and Egr1 gene regions are abnormally regulated in the aged hippocampus (Morse et al., 2015). Here, we examine Egr1 transcription and the DNA methylation status of the Egr1 gene in CA1 and dentate gyrus regions of the hippocampus of normal adult and memory-impaired aged animals. The results indicate subregion specific age-related changes in Egr1 transcription, and in methylation of DNA associated with the Egr1 promoter.
Materials and Methods
Behavioral procedures
Experiments were performed in accordance with NIH guidelines for the care and use of animals and the Institutional Animal Care and Use Committee at the University of Arizona. Adult (9–12 months, n =44) and aged (24–32 months, n = 44) male Fischer-344 rats (National Institute on Aging at Charles River, Wilmington, MA) were used in these experiments. Prior to the beginning of experiments, rats were handled daily for 7 days, then the spatial and visual discrimination abilities of each rat were assessed using the Morris swim task as described in detail by Barnes et al. (1996). Briefly, the rats were tested on six spatial trials (hidden platform) per day for four consecutive days, followed by 12 visual discrimination trials over 2 days in which the platform was visible. The visual discrimination trials were used to assess the visual and motor functions of each rat, to rule out significant visual or physical deficits. Performance on the Morris swim task was analyzed using the corrected integrated path length (Gallagher et al., 1993) (CIPL), which is the sum of the rat’s distance from the platform at each point in time (sampled at 10 Hz), corrected for the rat’s initial distance from the platform.
Experiments were conducted 7–10 days following testing in the Morris swim task, and took place at least 2 hours after the house lights in the colony room were extinguished. Animals were divided into 2 groups for the FISH and PCR studies: (1) those that were sacrificed directly from their home cages to measure resting levels of Egr1 mRNA (resting); (2) animals that explored an environment twice for 5 minutes, with an intervening 20 minute rest period (behavior). Animals used for DNA methylation analysis, explored the identical environment as the behavior group described above for 5 minutes, after which they returned to their home cages for 25 minutes prior to tissue collection. The procedure for environmental exploration is described elsewhere (see Guzowski et al., 1999). Briefly, rats were transported to a room containing distinct visual cues. Once in the room, rats were placed within a square box measuring 61 × 61 cm with cardboard walls 20 cm high. The environment was divided into 9 equal squares and during the 5 minute exploration sessions, rats were gently placed within a different square every 15 seconds during the 5 minute exploration treatment. This partially passive exploration protocol ensures that both the adult rats and the less active aged rats sample all aspects of the environment equally. This was confirmed in a separate study by counting the line crossings made by the adult (n=6) and aged (n=6) rats during the 15 second periods between passive relocation to another grid square in the exploration arena (Supplemental Figure 1A). Immediate-early gene expression has been shown to be the same in the hippocampus of rats given this type of “assisted exploration”, compared with rats who freely explore the same size environment (Vazdarjanova and Guzowski, 2004). In support of this, we did not observe a significant difference in the number of line crossings made by adult and aged rats (Supplemental Figure 1B). This is an important demonstration, as differences in transcription might arise if the behavior is not carefully matched between age groups (Hartzell et al., 2013). It should also be noted that although the environment that the rats explored was novel to them on the first exploration, it has been demonstrated that novelty is not a requirement for immediate-early gene transcription induction under these conditions (Guzowski et al., 2006).
Tissue harvesting and fluorescence in situ hybridization
After decapitation, brains were removed and hemisected, and half of the brain (used for in situ hybridization) was quick-frozen in isopentane. The remaining half (used for RT-PCR or DNA methylation analysis) was dissected into the CA region and the DG. Although care was taken to ensure that these dissections were performed as accurately as possible, it is likely that a small proportion of area CA3 was incorporated into these samples. Nevertheless, both CA samples and dentate gyrus samples are rich in CA1 pyramidal neurons and dentate granule neurons, respectively, and thus will be referred to as CA1 and dentate gyrus samples throughout. To confirm that hemispheric differences in Egr1 transcription do not account for differences in cell count and PCR data, we analyzed a separate group of hemisected brains. Neither aged nor adult rats showed hemispheric differences in Egr1 mRNA levels in either the dentate gyrus (Supplemental Figure 2A) or in CA1 (Supplemental Figure 2B).
Hemisections containing the dorsal hippocampus from 8 rats were molded in a block with Tissue-Tek OCT compound, so that all experimental conditions were represented in each block for each time point. Use of tissue blocks helps control for slide-to-slide variation in signal detection. Brains were coronally sectioned at 20 µm through the dorsal hippocampus (−3.2 to −3.8 mm from bregma; Paxinos and Watson, 1998), thaw-mounted on slides, and stored at −70°C. Fluorescence in situ hybridization (FISH) was performed as described in detail elsewhere (Guzowski et al., 1999; Vazdarjanova and Guzowski, 2004).
Image acquisition and analysis
Images were collected using a Zeiss 510 Metaseries laser scanning confocal microscope. Photomultiplier tube assignment, pinhole size and contrast values were held constant for each brain region on a slide. The areas of analysis were z-sectioned in 1.0 µm optical sections. For area CA1, stacks were taken at 40× magnification in 3 non-overlapping areas of CA1. To determine the location of these 3 points for analysis, we used anatomical landmarks provided by the DG so that CA1 images were taken in reference to: (1) the crest of the DG, (2) the midpoint point between the crest and the lateral tip of the suprapyramidal blade, and (3) the lateral tip of suprapyramidal blade. Similar methods were used to acquire images of the DG, except that the whole structure was imaged (Penner et al., 2011).
Image analysis was conducted as described earlier (Guzowski et al., 1999; Vazdarjanova et al., 2002) using MetaMorph imaging software (Universal Imaging). Only whole neuron-like cells found in the middle 20% of each stack were included in the analyses. Neurons were classified as: 1) positive, having one or two intense intranuclear foci present in at least three planes, cytoplasmic staining surrounding at least 60% of the cell and visible in at least three plains, or both; 2) negative which did not have any detectable staining above background levels. Image analysis was performed by an experimenter blind to the experimental conditions.
Real-time RT-PCR
Samples used for RT-PCR were prepared using the RNAqueous-4PCR kit (Ambion) according to the manufacturer’s instructions. The RNA was DNase-treated and reverse transcribed using SuperScript II (Invitrogen). A negative control was included in which no reverse transcriptase was added. Primers for Egr1 and for glyceraldehydes-3-phosphatedehydrogenase (GapDH) were based on the rat sequences deposited in GenBank at the National Center for Biotechnology Information. GapDH was used to normalize data because its expression does not change with age or with various treatments (Tanic et al., 2007). The primer sequence for GapDH was as follows: forward, AATGGGAGTTGCTGTTGAAG; reverse, CTGGAGAAACCTGCCAAGTA. The primer sequence for Egr1 was as follows: forward, TCTAGTGCTGAAGGGAGCAA; reverse, ACTTTCAGCTGCCTGAAACAG. PCR amplification of cDNA was performed using the BioRad iCycler Real-Time Detection System (BioRad Laboratories). cDNA was added to a 1× reaction master mix (iQ SYBR Green Supermix, BioRad Laboratories) along with the gene specific primers (0.5mM each of forward and reverse 8 primer) and nuclease free water. For each sample, duplicate reactions were conducted in 96-well plates. A melt curve analysis was also conducted to determine the uniformity of product information, primer–dimer formation, and amplification of non-specific products. The presence of specific amplification products was confirmed by a single peak on the melt curve and plotted as the negative derivative of fluorescence as a function of temperature (−d(RFU)/dT). For all assays, the cycle threshold (Ct) values were chosen in the linear range of amplification. For all real-time PCR assays, the efficiencies of the target and control gene amplifications were tested in order to ensure high (90–100%) and similar efficiency.
Bisulfite sequencing
DNA was isolated and purified from hippocampal tissue, then subjected to bisulfite modification (for a detailed description of this method see Parrish et al., 2012). The bisulfite treated DNA was then amplified with a primer that targeted a CpG rich region within the Egr1 promoter (Forward: TTGTGAAGGAAGTGTTATTTTG; Reverse: CCAATCTAATAACCCCAAACTT). The resulting PCR products were run on an agarose gel and purified with a gel extraction kit (Qiagen). The purified PCR products were then sequenced with the reverse primer at the University of Alabama at Birmingham Genomics Core Facility of the Heflin Center for Human Genetics. The electropherogram was read on Chromas software, where the percent methylation of each CpG site was determined by the ratio between peak values of G and A (G/[G +A]). For a control, unmethylated and methylated standards (0–100% methylation; EpigenDx) were run to determine the accuracy of the sample results (Table 1). The TRANSFAC resources database was used to identify binding sites within the Egr1 sequence analyzed here (Heinemeyer et al., 1998).
Table 1.
Summary of results from standardized control samples
| Expected | Actual (Mean ± SEM) |
|---|---|
| Standard, 0% Methylation | 4.08% ± 0.78 |
| Standard, 5% Methylation | 6.14% ± 0.32 |
| Standard, 10% Methylation | 10.55% ± 0.34 |
| Standard, 25% Methylation | 18.07% ± 2.47 |
| Standard, 50% Methylation | 46.41% ± 0.0 |
| Standard, 75% Methylation | 75.06% ± 0.0 |
| Standard, 100% Methylation | 88.40% ± 1.26 |
Data analysis
Performance on the Morris swim task was analyzed using repeated measures ANOVA, with training day as the repeated factor, and age as the between-subjects factor. Performance on probe trials was assessed using a one-way ANOVA and paired t-tests to compare the time spent in the target quadrant of the pool (i.e., where the escape platform was located on acquisition trials). Relative gene expression was determined using the 2−ΔΔCT method (Livak and Schmittgen, 2001). To determine differences in the relative levels of Egr1 mRNA between adult and aged rats at baseline and following the behavioral treatment, the ΔCT was determined for each sample (CT Egr1 − CT GapDH), with the adult samples used as the calibrator samples in place of the caged control samples. Unpaired t-tests were used to assess mRNA levels measured by RT-PCR, and group differences for the cell count data were analyzed by ANOVA with age and treatment as dependent variables, followed by Bonferroni’s post hoc tests. The BSP data were analyzed by two-way ANOVA followed by Bonferroni’s post hoc tests. Data are shown as the mean and ± SEM. For all tests, the null hypotheses were rejected at the 0.05 level of significance.
Results
Aged rats show impaired spatial memory
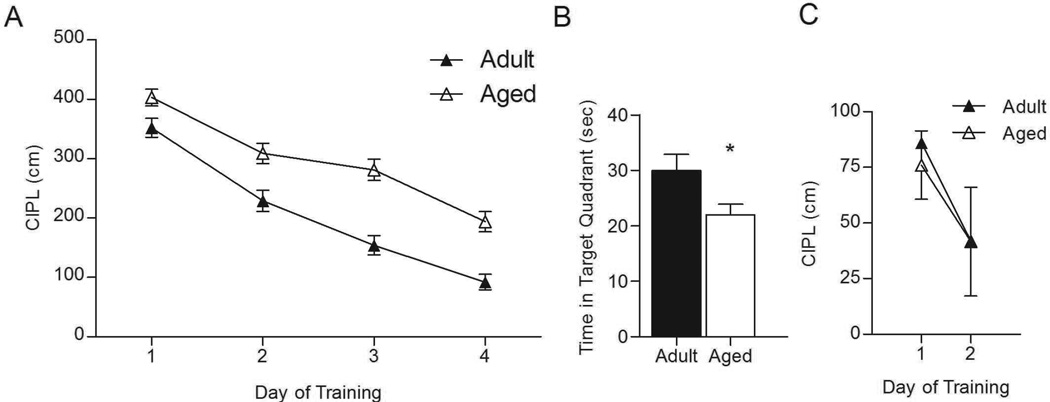
As shown in Figure 1, and consistent with previous reports (e.g., Gage et al., 1984; Gallagher et al., 1993; Barnes et al., 1996), aged animals showed a deficit in spatial, but not cued trials on the Morris swim task. There was a significant effect of age on the corrected integrated path length (CIPL) over the 4-day training period, with the aged rats taking significantly longer path lengths to the platform than adult rats (F(16, 134) = 16.887, p < 0.0001). Post-hoc analysis showed that this effect appeared on the first day of testing and persisted through the 4th day of training (day 1: p = 0.018; day 2: p = 0.001; day 3: p < 0.0001; day 4: p < 0.0001). As shown in Figure 1A, adult rats showed significant improvement across days, while the aged rats showed less robust learning. Moreover, on the probe trial with the platform removed from the pool (Figure 1B), adult rats spent significantly more time in the target quadrant of the pool than did the aged rats (F(1, 33) = 4.821, p = 0.035). On visible trials (Figure 1C), there was no effect of age on CIPL for day 1 (p = 0.755) or day 2 (p = 0.989) of testing. Thus, both age groups showed significant learning across days, taking a shorter path to the platform on day 2 of the visible trials compared to day 1 (Figure 1C; aged, p = 0.001; adult, p = 0.029). These data suggest that it is unlikely that the observed age-related differences on the spatial trials were due to deficits in visual acuity or swimming ability.
Figure 1.
Aged rats are impaired in the spatial version of the Morris swim task. A. Over 4 days of training, adult rats took a significantly shorter path length to reach the hidden platform than did the aged rats. B. On the probe trial, aged rats spent significantly less time in the target quadrant than did adult rats (*p = 0.035). C. During cued trials, in which the escape platform was visible, both adult and aged rats showed improvement in performance over 2 days of training. There was no significant effect of age on either day.
Egr1 transcription in the dentate gyrus
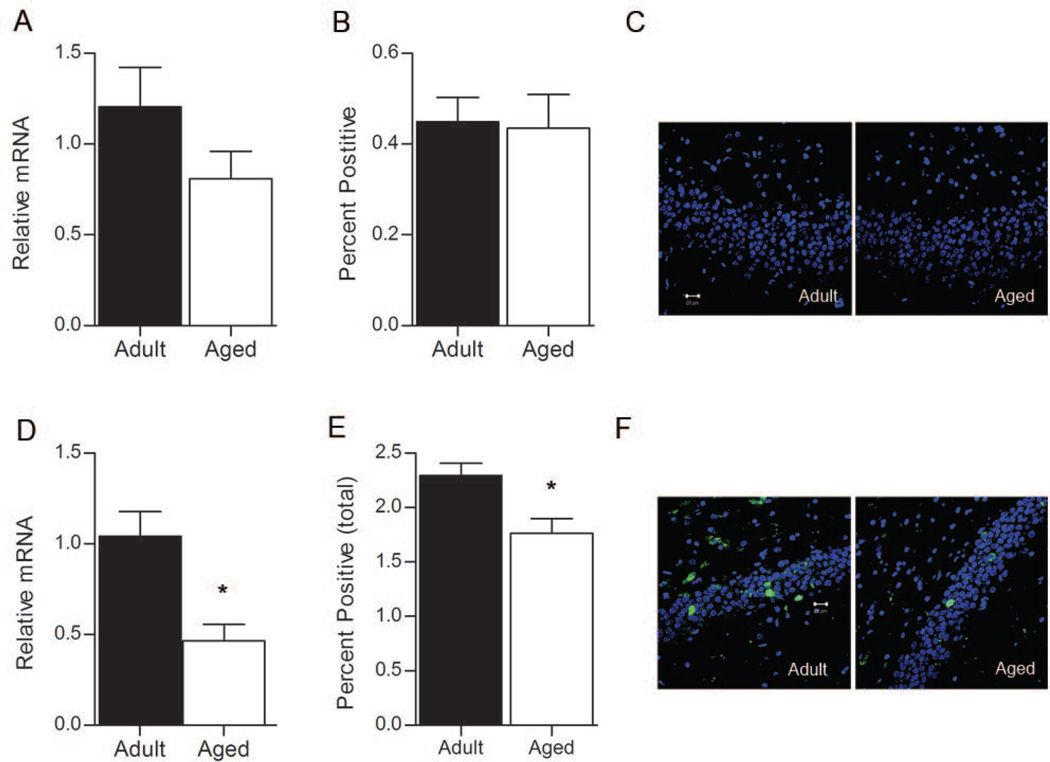
Resting levels of Egr1 mRNA were determined for a group of animals that remained undisturbed in their home cages prior to tissue collection. RT-PCR revealed that basal levels of Egr1 mRNA were not significantly different between adult and aged rats within the dentate gyrus (Figure 2A; p = 0.295). This effect is mimicked in the cell count data (Figure 2B), which indicates that similar proportions of dentate granule cells transcribed Egr1 under resting conditions in the adult and aged rat (t4 = 1.213, p = 0.292). Examples of the pattern of Egr1 activation for the caged control rats are shown in Figure 2C. These data indicate that under conditions in which rats are confined to familiar home cages, the same proportion of granule cells transcribe similar amounts of Egr1 mRNA in both the aged and adult dentate gyrus.
Figure 2.
Egr1 in the dentate gyrus. A. Basal levels of Egr1 mRNA are similar in adult rats (n=3) and aged rats (n=3), as measured by RT-PCR (p > 0.05). B. The proportion of dentate granule cells that transcribe Egr1 under resting conditions are similar in adult and aged rats (p > 0.05). C. Representative confocal images from the dentate gyrus in adult and aged rats under rest conditions. Green = Egr1, Blue = cell nuclei stained with TOPRO. Magnification = 40×. D. Behavior-induced relative levels of Egr1 mRNA are significantly lower in aged rats (n=6) compared to adult rats (n=6), as measured by RT-PCR (*p = 0.005). E. The proportion of dentate granule neurons that transcribe Egr1 after exploration is significantly lower in aged rats (*p = 0.030). Representative confocal images from the dentate gyrus of behavior-treated rats in both age groups are shown in F. Green = Egr1, Blue = cell nuclei stained with TOPRO. Magnification = 40×. Scale bar = 20 µm.
To determine if spatial behavior differentially affects the transcription of Egr1 in adult and aged rats, animals underwent 5 minutes of assisted exploration within a novel environment. Aged rats showed significantly lower levels of Egr1 transcription within the dentate gyrus (Figure 2D; t10 = −3.589, p = 0.005) after exploration. This reduction in Egr1 mRNA is likely the result of fewer neurons transcribing Egr1 following exploration, since FISH analysis revealed that the proportion of dentate granule neurons that transcribe Egr1 is also significantly reduced for aged rats relative to adult rats (Figure 2E and 2F; t10=2.529, p = 0.030).
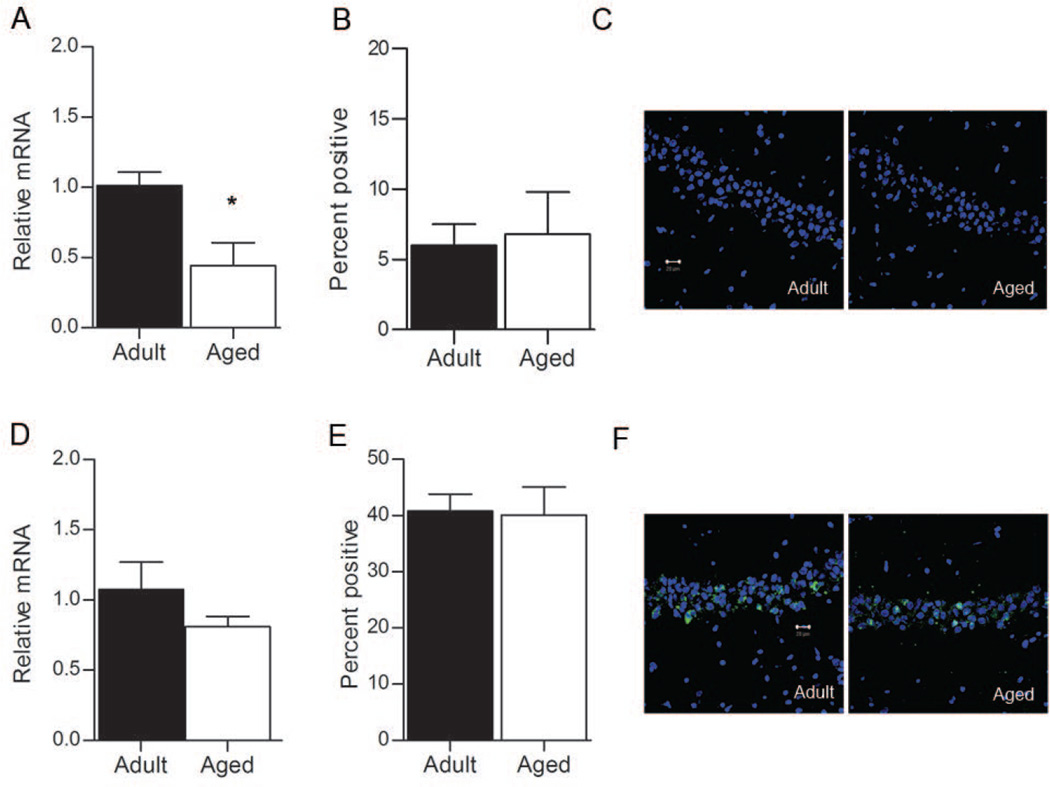
Egr1 transcription in area CA1
Resting levels of Egr1 transcription were also measured for area CA1. Here, RT-PCR revealed that Egr1 mRNA levels were significantly lower in aged rats relative to adult rats (Figure 3A; t6 = −3.033, p = 0.023). Because this difference may result from fewer pyramidal neurons transcribing Egr1 in aged rats, FISH was used to determine the proportion of CA1 pyramidal cells that transcribe Egr1 at rest, and indicated that similar proportions of CA1 neurons transcribed Egr1 in the aged and adult caged control groups (Figure 3B; t4 = 0.253, p = 0.813). Examples of the pattern of Egr1 activation for the caged control rats are shown in Figure 3C. Together, these data suggest that the basal rate of Egr1 transcription in area CA1 is reduced per cell in the aged rats, a finding that is similar to the one obtained for the effector gene Arc (Penner et al., 2011).
Figure 3.
Egr1 in area CA1. A. Basal levels of Egr1 mRNA are higher in adult rats (n=3) relative to aged rats (n=3), as measured by RT-PCR (*p = 0.023). B. The proportion of pyramidal neurons that transcribe Egr1 under resting conditions, are similar in adult and aged rats (p > 0.05). C. Representative confocal images from control rats in both age groups for area CA1. D. Behavior-induced levels of Egr1 mRNA are not different between adult (n=6) and aged (n=6) rats, as measured by RT-PCR (p > 0.05). E. The proportion of CA1 pyramidal neurons that transcribe Egr1 after exploration are similar in adult and aged rats (p > 0.05). F. Representative confocal images from CA1 of behavior-treated rats in both age groups. Green = Egr1, Blue = cell nuclei stained with TOPRO. Magnification = 40×. Scale bar = 20 µm.
Following exploration of a novel environment, the relative levels of Egr1 mRNA were not significantly different between adult and aged rats in area CA1 (Figure 3D; t10 = −1.268, p = 0.234), nor was there a significant age difference in the proportion of CA1 pyramidal neurons that transcribe Egr1 (Figure 3E and 3F; t10 = 0.039, p = 0.970).
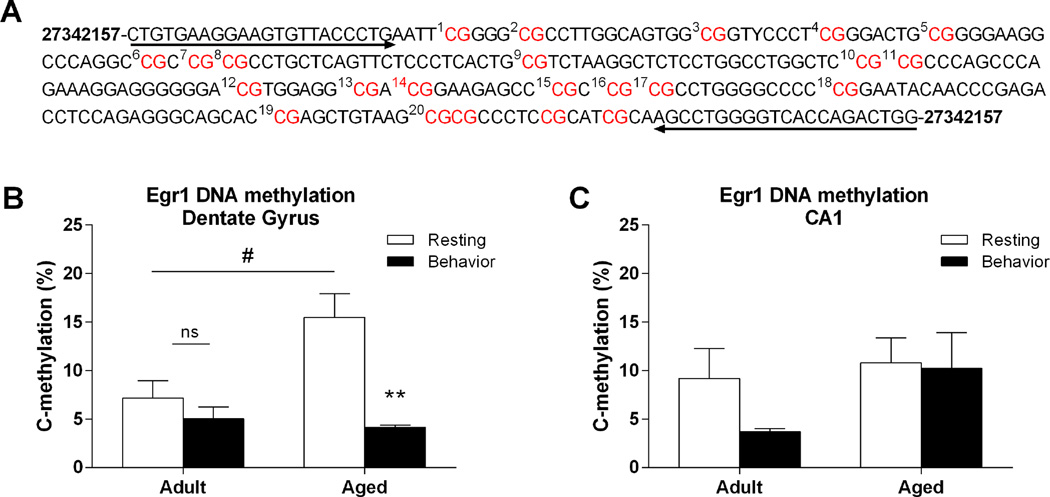
Methylation status of the Egr1 promoter
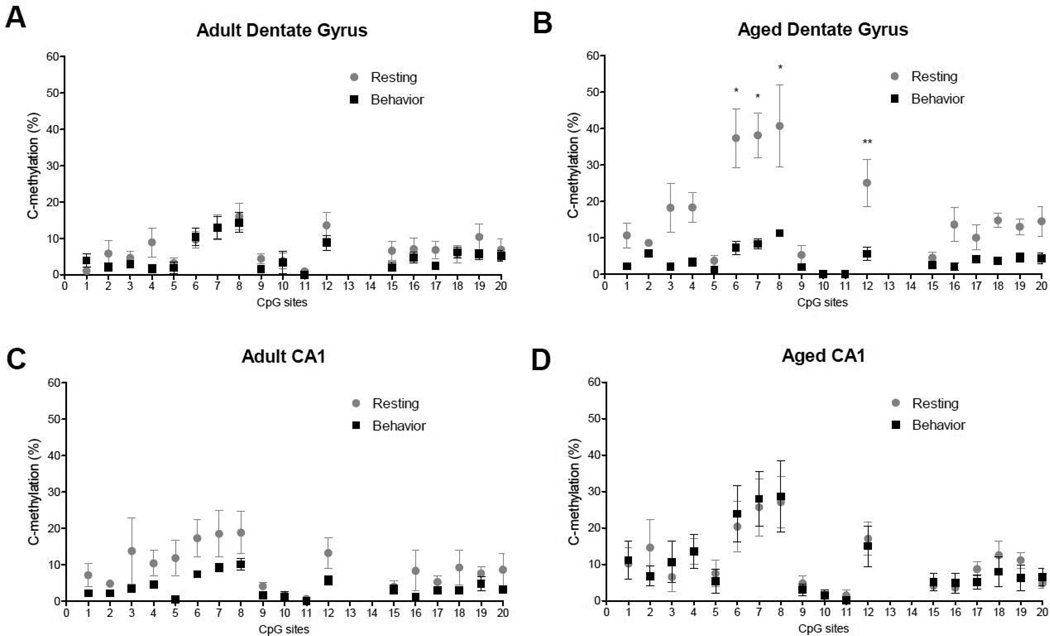
To determine if DNA methylation may play a role in regulating the transcription of Egr1 within the hippocampus, we analyzed the methylation status of a CpG island, consisting of 20 CpG sites, within the promoter region of the Egr1 gene (Figure 4A). To assess overall methylation levels of the CpG island, we averaged the percent methylation observed across the 20 CpG sites for both adult and aged rats under resting conditions and following exploration. In the dentate gyrus, this analysis revealed a significant effect of age (F(1, 17) = 4.607, p = 0.047), behavior (resting or exploration; F(1, 17) = 15.04, p = 0.001), as well as a significant interaction (F(1, 17) = 7.029, p = 0.017). Under resting conditions, aged rats showed significantly more methylation of Egr1 compared to adult rats (Figure 4B, p = 0.021). Following exploration, aged rats showed a significant reduction (compared to rest) in methylation of Egr1 within the dentate gyrus (p = 0.005), while adult rats did not show this dynamic change (p = 0.352), and no significant differences were observed between age groups following exploration (p = 0.585). Within CA1, there was no overall effect of either age (F(1, 18) = 2.224, p = 0.151) or behavior (F(1, 18) = 1.236, p = 0.280) on methylation of the Egr1 promoter (Figure 4C). Although adult rats did show a modest decrease in methylation of the Egr1 promoter after spatial behavior, this decrease was not statistically significant (p = 0.105)
Figure 4.
Methylation analysis of Egr1 dinucleotides from the hippocampus of adult and aged animals. A. Schematic of examined CpG sites in the promoter region of the Egr1 gene. Sequencing primer pair positions are indicated by the left and right arrows. B. In the DG, resting levels of methylation of the Egr1 promoter are significantly lower in adult rats (n=6) compared to aged rats (n=5; #p < 0.05). Following behavior, aged rats (n=4) show a significant decrease in methylation when compared to resting levels. Adult rats (n=6) do not show this dynamic change in methylation of the Egr1 promoter (**p < 0.01). C. Within CA1, there was no overall effect of age or behavior on methylation of Egr1.
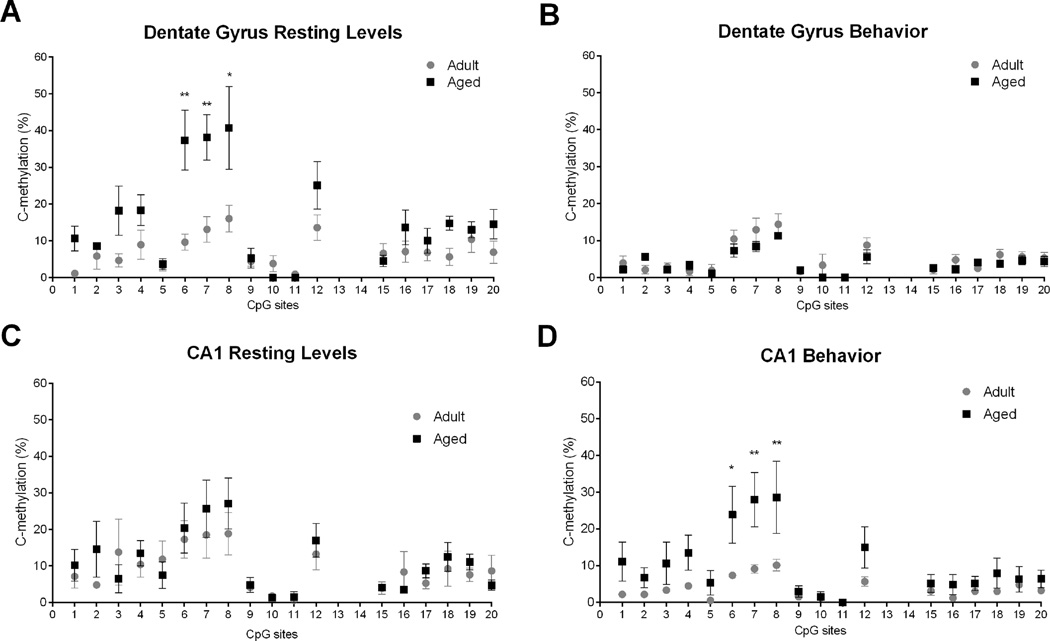
While examining methylation levels at each of the 20 CpG sites, it was apparent that the amount of methylation was variable among the CpG sites. Because average differences in methylation across a CpG island could mask potentially important differences at individual sites, we performed additional analysis to detect changes at each cytosine within (Figure 5) and between (Figure 6) age groups. Thus, the data in Figure 6 are a reorganization of those presented in Figure 5, specifically to illustrate more clearly the differences that emerged between age groups.
Figure 5.
Methylation analysis of individual CpG sites within age groups. A. In the dentate gyrus of adult rats, significant differences were not observed at individual CpG sites. B. In the dentate gyrus of aged rats, CpG6 (*p = 0.0001), CpG7 (*p = 0.0001), CpG8 (*p = 0.0001), and CpG12 (**p = 0.05) were significantly different between resting and behavioral conditions. C. In area CA1 of adult rats, there is no difference in methylation between individual CpG sites. D. In area CA1 of aged rats, there is no difference in methylation between individual CpG sites.
Figure 6.
Methylation analysis of individual CpG sites between age groups. A. In the dentate gyrus under resting conditions, a significant difference between adult and aged rats was observed at CpG6 (**p < 0.0001), CpG7 (**p < 0.0001), and Cpg8 (*p < 0.001). B. Following behavior, no significant differences in methylation were observed at individual CpG sites. C. In CA1, under resting conditions significant age differences were not observed at individual CpG sites. D. Following behavior, there was a significant difference in methylation between adult and aged rats at CpG6 (*p < 0.05), CpG7 (**p < 0.01) and CpG8 (**p < 0.01).
In adult rats, methylation of individual CpG sites in the dentate gyrus was not significantly different either under resting condition, or after exploration (Figure 5A). For the aged rat (Figure 5B), however, we observed a significant overall effect of behavior (resting versus exploration, F(1, 15) = 77.96, p = 0.0001), a significant effect of CpG site (F(1, 15) = 6.034, p = 0.0001) and a significant interaction (F(1, 15) = 2.757, p = 0.0012). Post-hoc analysis indicated that CpG6 (p = 0.0001), CpG7 (p = 0.0001), CpG8 (p = 0.0001), and CpG12 (p = 0.05) were significantly different between resting and exploration conditions in the dentate gyrus of aged rats, with all four CpG sites showing a significant reduction in methylation after exploration. In area CA1, methylation at individual CpG sites were not significantly different for adult (Figure 5C) or aged rats (Figure 5D).
Finally, we analyzed potential age differences between individual CpG sites (Figure 6). In the dentate gyrus under resting conditions. There was a significant main effect of age (F1, 17) = 43.630, p < 0.0001) and CpG site (F(1, 17) = 9.438, p < 0.001) as well as a significant interaction (F(1, 17) = 3.212, p = < 0.0001). Post hoc analysis of individual CpG sites revealed a significant difference between adult and aged rats at CpG6 (p < 0.0001), CpG7 (p < 0.0001), and Cpg8 (p < 0.001; Figure 6A). In all cases, aged rats showed significantly higher levels of CpG methylation compared to adult rats. Following behavior (Figure 6B) no significant differences were observed.
In CA1, under resting conditions significant age differences were not observed at individual CpG sites (Figure 6C). Following behavior, however, (Figure 6D) there was a significant main effect of age (F(1, 17) = 32.45, p < 0.0001), CpG site (F(1, 17) = 5.466, p < 0.0001), but only a trend toward a significant interaction (F(1, 17) = 1.549, p = 0.0825). Post hoc analysis revealed a significant difference in methylation between adult and aged rats at CpG6 (p < 0.05), CpG7 (p < 0.01) and CpG8 (p < 0.01). At these three CpG sites, aged rats showed significantly higher levels of methylation compared to adult rats.
Discussion
The present data indicate that aging is associated with distinct, hippocampal subregion-specific transcriptional alterations of the memory-associated gene, Egr1. While the role of Egr1 is well-established in transcriptional control of gene expression changes required for memory formation, very little is known about the impact of age on the regulation of this transcription factor. In aged, memory-impaired rats, the granule cells in the dentate gyrus show reduced Egr1 transcription following spatial behavior, while in area CA1, transcription is suppressed at rest, but can be driven to the levels observed in adult animals following behavior that engages the hippocampus. The data also suggest two levels of change in epigenetic mechanisms that may contribute to the aberrant transcriptional response observed in the aged hippocampus during aging: 1) differences between subregions CA1 and dentate gyrus in overall dynamic methylation of the Egr1 gene; and 2) CpG island site-specific alterations within those subregions. Analysis of a CpG island within the promoter region of the Egr1 gene revealed significant age-related differences in its dynamic regulation as well as methylation changes specific to individual CpG sites. Because it is known that gene transcription is necessary for processes such as LTP and memory consolidation, the effects described here and in other studies (Liu et al., 2011; Penner et al., 2011; Castellano et al., 2012; Oliveira et al., 2012) point to the potential role that age-related alterations in epigenetic regulation of gene transcription may play in altered synaptic plasticity and cognition during aging.
A role for Egr1 in learning and memory processes
It has been well-established that memory and synaptic plasticity processes in the cognitively healthy adult require transcription of IEGs, including Arc, Egr1, bdnf (Guzowski et al., 2000; French et al., 2001; Hall et al., 2001; Steward and Worley, 2001). When expression of these genes is blocked in otherwise normal and cognitively intact adult animals, the consolidation of memory is also blocked (e.g., Linnarsson et al., 1997; Guzowski et al., 2000; French et al., 2001). Moreover, changes in IEG expression are prevalent in many models of memory disorders (Dickey et al., 2003; Palop et al., 2005; Rosi et al., 2005) and as a result of the normal aging process (e.g., Blalock et al., 2003; Small et al., 2004; Rowe et al., 2007; Haberman et al., 2011; Marrone et al., 2012; Chawla et al., 2013).
Here, we report that Egr1 transcription shows distinct age-related changes that may contribute to the memory deficits that were observed in the aged rats on the Morris swim task. Previous studies investigating potential age-related changes in Egr1 transcription or expression have reported significant age-related changes in mRNA or protein expression under resting conditions (Yau et al., 1996; Desjardins et al., 1997; Blalock et al., 2003). For example, in agreement with our results, a large scale microarray study conducted by Blalock et al. (2003) found reduced resting levels of Egr1 mRNA in area CA1 of memory-impaired aged rats (they did not assess other subregions). Similarly, Yau et al. (1996) also reported reduced Egr1 in area CA1 of aged rats under resting condition. Our results are consistent with these studies, and expand our understanding of how behavior that engages the hippocampus, can drive Egr1 transcription in area CA1 of aged rats, to a level equivalent to that observed in adult rats. This suggests that, at least in area CA1, a compensatory mechanism may be recruited to drive low resting levels of Egr1, enabling aged pyramidal neurons to respond to novel, incoming information.
In the dentate gyrus, Egr1 mRNA levels were similar between adult and aged rats under resting conditions. When exploratory behavior was used to induce transcription, however, significantly fewer granule cells in the aged rats transcribed Egr1 and there were significantly lower levels of Egr1 mRNA compared to adult rats. There now exists an abundance of data obtained from humans, rodents and non-human primates pointing to the dentate gyrus as a site that is particularly vulnerable to advancing age (e.g., West, 1993; Gazzaley et al., 1996; Small et al., 2002, 2004; McHugh et al., 2007; Moreno et al., 2007; Marrone et al., 2011; Yassa and Stark, 2011; Yassa et al., 2011a; b). In line with the findings reported here, Marrone et al. (2011) has also demonstrated that fewer granule cells transcribe Egr1 in the aged dentate gyrus after exploration. Both here, and in the study by Marrone et al. (2011) resting levels of Egr1 were not different between adult and aged rats, indicating that behavior that engages the hippocampus is required to detect an age-related change in Egr1 transcription within this hippocampal subregion.
DNA Methylation at the Egr1 gene
DNA methylation is a powerful transcription mechanism that controls persistent gene expression in the central nervous system. The present study sought to determine whether decreased Egr1 gene expression in the aging hippocampus was associated with DNA methylation changes at the Egr1 gene locus. Our finding of a selective (granule cells not CA1 pyramidal cells), age-dependent change in Egr1 expression levels in the hippocampus suggests a long-lasting persistent effect on Egr1 transcription. However, the underlying transcriptional mechanisms that orchestrate abnormal steady-state gene expression changes in the aging brain are still uncertain. Investigation of these underlying mechanisms is important because targeting them may help to mitigate or improve memory retention with age.
Aging is associated with widespread changes in the methylome (e.g., Hannum et al., 2013), and in changes in the methylation of specific genes in the brain (e.g., Siegmund et al., 2007; Hernandez et al., 2011; Penner et al., 2011; Haberman et al., 2012; Morse et al., 2015), including Egr1 (Morse et al., 2015). In the present study, we used bisulfite sequencing to measure age-related changes in methylation of the CpG island within the Egr1 gene. In the dentate gyrus of adult animals, we observed consistent methylation levels across all 20 CpG sites of the CpG island. Furthermore, the methylation of the CpG island within the promoter region was low in adult animals, and the amount of methylation was not affected by spatial behavior. In aged rats, however, methylation of the CpG island within the promoter region of the Egr1 gene was significantly higher at rest when compared to adult rats. This suggests that, at baseline, the chromatin state at the Egr1 gene is resistant to transcriptional control in the dentate gyrus of aged rats. Interestingly, these higher rates of methylation were dynamically reduced as a result of behavior in old rats. Furthermore, we observed selective dynamic reductions in methylation at four individual CpG sites across the CpG island in the aged rats. Collectively, these findings suggest that Egr1 DNA methylation in the dentate gyrus of aged rats is indeed responsive to behavior, and that the elevated Egr1 DNA methylation levels observed during resting states can be effectively normalized by behavior in old animals. These active and highly specific changes in methylation may point to a unique compensatory mechanism at work within the aged dentate gyrus that can overcome transcriptional repression to result in at least some Egr1 transcription.
In contrast to the aged dentate gyrus, DNA methylation at the Egr1 promoter region was not significantly different in area CA1 between adult and aged rats at rest, nor was there a significant change in Egr1 DNA methylation after behavior, for either age group. Nonetheless, we found decreased Egr1 mRNA levels in area CA1 of aged rats compared to adults. Further analysis of individual CpG sites between groups revealed significantly higher levels of methylation at three CpG sites for aged rats after spatial behavior. How these changes in methylation relate to Egr1 mRNA transcription is not entirely clear, but an analysis of these individual CpGs reveals that they fall near (CpG 7 and CpG 8) or within (CpG 6) binding sites for nuclear factor-kappa B (NF-κB) and c-Rel. The NF-κB family of transcription factors (which included c-Rel) is known to play a key role in regulating the growth and elaboration of neural processes. For example, NF-κB signaling has been implicated in regulating dendritic spine density (Russo et al., 2009; Christoffel et al., 2011) and can also regulate neurogenesis (Widera et al., 2006; Denis-Donini et al., 2008; Koo et al., 2010) in the adult brain (see Gutierrez and Davies, 2011 for review). Increased methylation of CpGs near or at the NF-κB binding site, could inhibit activity at these specific sites, and have a significant impact on process that are known to promote learning and memory (i.e., dendritic spine density and neurogenesis). In fact, work by Haberman et al. (2012) and by Weaver et al. (2004, 2007) suggest that methylation at single CpG sites can have a significant impact on gene transcription. For example, Haberman et al. (2012) describe significant changes in methylation at a single CpG site within the promoter region for the Gabra5 gene. Because this site was found to be within a binding domain for an NFκB binding site, and its methylation status would have a significant effect on the transcription of the Gabra5 gene. Importantly, it was the methylation status of this CpG site that correlated with cognitive status in the aged rats.
A role for epigenetic mechanisms in learning and memory processes
A number of observations suggest that DNA methylation is a key epigenetic mechanism that serves to regulate gene transcription within the hippocampus in response to environmental experiences. For example, factors such as maternal care and maltreatment (Weaver et al., 2004, 2007; Roth et al., 2009), fear conditioning (Miller and Sweatt, 2007; Lubin et al., 2008; Miller et al., 2010), stress (Roth et al., 2011), long-term potentiation (Levenson et al., 2006) and drug use (Anier et al., 2010; LaPlant et al., 2010) can have significant effects on DNA methylation levels and gene transcription (for review see Day and Sweatt, 2011). In addition, there is increasing support for the idea that DNA methylation patterns change as a result of the normal aging process (Jiang et al., 2008; Peleg et al., 2010; Penner et al., 2010, 2011; Castellano et al., 2012; Haberman et al., 2012; Kosik et al., 2012; Oliveira et al., 2012). Interestingly, environmental enrichment of old rats modified hippocampal histone methylation levels, an epigenetic process tightly linked to DNA methylation mechanisms, resulting in both object recognition memory performance and bdnf mRNA levels increasing in aged animals (Morse et al., 2015). It remains to be determined if environmental enrichment can also restore proper DNA methylation levels in old animals.
Experiential and environmental factors can lead to the accumulation of changes in patterns of DNA methylation across the lifespan, suggesting that these changes could contribute to the course of the normal aging process (Penner et al., 2010; Kosik et al., 2012). While it remains unclear what mechanism or mechanisms contribute to the altered patterns of methylation we report here, one possibility is that the aging process impacts activity of DNA methyltransferases (DNMTs). In fact, work by Liu et al. (2011) suggests that the activity of DNMT1 is necessary for normal healthy aging, because DNMT1 deficiency impairs learning and memory performance in an age-dependent manner. In addition, work by Oliveira et al. (2012) indicates that DNMT3a activity is reduced in the hippocampus and cortex of aged, memory-impaired mice. Importantly, when DNMT3a activity is restored in the aged brain (specifically in area CA1), cognitive function is restored. These authors also reported that reducing DNMT3a levels in the hippocampus impairs the regulation of well-known plasticity-related genes, including Arc and BDNF (Oliveira et al., 2012). In normal adult animals, previous work reported that inhibition of de novo DNMT activity (such as DNMT3a) not only blocks memory consolidation, but also results in the aberrant transcription of several genes important for normal memory function (Miller and Sweatt, 2007; Miller et al., 2010).
Although the activity of DNMTs may contribute to the changes in DNA methylation we observed here, it is unlikely to be the only mechanism at work. The complexity of epigenetic regulation of gene transcription is now well appreciated, and it is apparent that it is the coordinated regulation of epigenetic mechanisms that results in normal gene regulation and cognitive function (see Castellano et al., 2012). Overall, a unifying hypothesis is emerging that suggests that age-associated changes in epigenetic marks, including histone methylation and DNA methylation and demethylation, may be a driver of aging-related alterations in gene transcription and other cellular and physiologic changes that occur within the brain. These changes are complex, and interactive. A more complete understanding of the mechanisms that regulate these processes will not only lead to more effective prevention strategies for improved quality of life, but will also contribute to a better general understanding of the mechanisms underlying memory formation.
Supplementary Material
Acknowledgments
Grant sponsor: NIA; Grant number: R01 AG009219. Grant sponsor: NIH; Grant number: AG031722. Grant sponsor: NIH; Grant number: MH57014. Grant sponsor: the state of Arizona and ADHS. Grant sponsor: McKnight Brain Research Foundation. We thank Michelle Carroll and Luann Snyder for their administrative support, and Dr. David Sweatt for critical discussions and advice on these experiments.
References
- Alberini CM. Transcription factors in long-term memory and synaptic plasticity. Physiol Rev. 2009;89:121–145. doi: 10.1152/physrev.00017.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anier K, Malinovskaja K, Aonurm-Helm A, Zharkovsky A, Kalda A. DNA methylation regulates cocaine-induced behavioral sensitization in mice. Neuropsychopharmacology. 2010;35:2450–2461. doi: 10.1038/npp.2010.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes CA, Rao G, McNaughton BL. Functional integrity of NMDA-dependent LTP induction mechanisms across the lifespan of F-344 rats. Learn Mem. 1996;3:124–137. doi: 10.1101/lm.3.2-3.124. [DOI] [PubMed] [Google Scholar]
- Blalock EM, Chen K-C, Sharrow K, Herman JP, Porter NM, Foster TC, Landfield PW. Gene microarrays in hippocampal aging: statistical profiling identifies novel processes correlated with cognitive impairment. J Neurosci. 2003;23:3807–3819. doi: 10.1523/JNEUROSCI.23-09-03807.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger C, Lopez MC, Baker HV, Mandel RJ, Muzyczka N. Genome-wide analysis of aging and learning-related genes in the hippocampal dentate gyrus. Neurobiol Learn Mem. 2008;89:379–396. doi: 10.1016/j.nlm.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger C, López MC, Feller JA, Baker HV, Muzyczka N, Mandel RJ. Changes in transcription within the CA1 field of the hippocampus are associated with age-related spatial learning impairments. Neurobiol Learn Mem. 2007;87:21–41. doi: 10.1016/j.nlm.2006.05.003. [DOI] [PubMed] [Google Scholar]
- Burke SN, Barnes CA. Senescent synapses and hippocampal circuit dynamics. Trends Neurosci. 2010;33:153–161. doi: 10.1016/j.tins.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano JF, Fletcher BR, Kelley-Bell B, Kim DH, Gallagher M, Rapp PR. Age-related memory impairment is associated with disrupted multivariate epigenetic coordination in the hippocampus. PloS One. 2012;7:e33249. doi: 10.1371/journal.pone.0033249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla MK, Penner MR, Olson KM, Sutherland VL, Mittelman-Smith MA, Barnes CA. Spatial behavior and seizure-induced changes in c-fos mRNA expression in young and old rats. Neurobiol Aging. 2013;34:1184–1198. doi: 10.1016/j.neurobiolaging.2012.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoffel DJ, Golden SA, Dumitriu D, Robison AJ, Janssen WG, Ahn HF, Krishnan V, Reyes CM, Han M-H, Ables JL, Eisch AJ, Dietz DM, Ferguson D, Neve RL, Greengard P, Kim Y, Morrison JH, Russo SJ. IκB kinase regulates social defeat stress-induced synaptic and behavioral plasticity. J Neurosci. 2011;31:314–321. doi: 10.1523/JNEUROSCI.4763-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole AJ, Saffen DW, Baraban JM, Worley PF. Rapid increase of an immediate early gene messenger RNA in hippocampal neurons by synaptic NMDA receptor activation. Nature. 1989;340:474–476. doi: 10.1038/340474a0. [DOI] [PubMed] [Google Scholar]
- Day JJ, Sweatt JD. Epigenetic mechanisms in cognition. Neuron. 2011;70:813–829. doi: 10.1016/j.neuron.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denis-Donini S, Dellarole A, Crociara P, Francese MT, Bortolotto V, Quadrato G, Canonico PL, Orsetti M, Ghi P, Memo M, Bonini SA, Ferrari-Toninelli G, Grilli M. Impaired adult neurogenesis associated with short-term memory defects in NF-κB p50-deficient mice. J Neurosci. 2008;28:3911–3919. doi: 10.1523/JNEUROSCI.0148-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desjardins S, Mayo W, Vallée M, Hancock D, Le Moal M, Simon H, Abrous DN. Effect of aging on the basal expression of c-Fos, c-Jun, and Egr-1 proteins in the hippocampus. Neurobiol Aging. 1997;18:37–44. doi: 10.1016/s0197-4580(96)00206-0. [DOI] [PubMed] [Google Scholar]
- Dickey CA, Loring JF, Montgomery J, Gordon MN, Eastman PS, Morgan D. Selectively reduced expression of synaptic plasticity-related genes in amyloid precursor protein + presenilin-1 transgenic mice. J Neurosci. 2003;23:5219–5226. doi: 10.1523/JNEUROSCI.23-12-05219.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher BR, Hill GS, Long JM, Gallagher M, Shapiro ML, Rapp PR. A fine balance: Regulation of hippocampal Arc/Arg3.1 transcription, translation and degradation in a rat model of normal cognitive aging. Neurobiol Learn Mem. 2014;115:58–67. doi: 10.1016/j.nlm.2014.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster TC, Norris CM. Age-associated changes in Ca(2+)-dependent processes: relation to hippocampal synaptic plasticity. Hippocampus. 1997;7:602–612. doi: 10.1002/(SICI)1098-1063(1997)7:6<602::AID-HIPO3>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- French PJ, O’Connor V, Jones MW, Davis S, Errington ML, Voss K, Truchet B, Wotjak C, Stean T, Doyère V, Maroun M, Laroche S, Bliss TVP. Subfield-specific immediate early gene expression associated with hippocampal long-term potentiation in vivo. Eur J Neurosci. 2001;13:968–976. doi: 10.1046/j.0953-816x.2001.01467.x. [DOI] [PubMed] [Google Scholar]
- Gage FH, Dunnett SB, Björklund A. Spatial learning and motor deficits in aged rats. Neurobiol Aging. 1984;5:43–48. doi: 10.1016/0197-4580(84)90084-8. [DOI] [PubMed] [Google Scholar]
- Gallagher M, Burwell R, Burchinal M. Severity of spatial learning impairment in aging: development of a learning index for performance in the Morris water maze. Behav Neurosci. 1993;107:618–626. doi: 10.1037//0735-7044.107.4.618. [DOI] [PubMed] [Google Scholar]
- Gazzaley AH, Siegel SJ, Kordower JH, Mufson EJ, Morrison JH. Circuit-specific alterations of N-methyl-D-aspartate receptor subunit 1 in the dentate gyrus of aged monkeys. Proc Natl Acad Sci U S A. 1996;93:3121–3125. doi: 10.1073/pnas.93.7.3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Kim SY, Artis S, Molfese DL, Schumacher A, Sweatt JD, Paylor RE, Lubin FD. Histone methylation regulates memory formation. J Neurosci. 2010;30:3589–3599. doi: 10.1523/JNEUROSCI.3732-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez H, Davies AM. Regulation of neural process growth, elaboration and structural plasticity by NF-κB. Trends Neurosci. 2011;34:316–325. doi: 10.1016/j.tins.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzowski JF, Lyford GL, Stevenson GD, Houston FP, McGaugh JL, Worley PF, Barnes CA. Inhibition of activity-dependent arc protein expression in the rat hippocampus impairs the maintenance of long-term potentiation and the consolidation of long-term memory. J Neurosci. 2000;20:3993–4001. doi: 10.1523/JNEUROSCI.20-11-03993.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzowski JF, McNaughton BL, Barnes CA, Worley PF. Environment-specific expression of the immediate-early gene Arc in hippocampal neuronal ensembles. Nat Neurosci. 1999;2:1120–1124. doi: 10.1038/16046. [DOI] [PubMed] [Google Scholar]
- Guzowski JF, Miyashita T, Chawla MK, Sanderson J, Maes LI, Houston FP, Lipa P, McNaughton BL, Worley PF, Barnes CA. Recent behavioral history modifies coupling between cell activity and Arc gene transcription in hippocampal CA1 neurons. Proc Natl Acad Sci U S A. 2006;103:1077–1082. doi: 10.1073/pnas.0505519103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberman RP, Colantuoni C, Stocker AM, Schmidt AC, Pedersen JT, Gallagher M. Prominent hippocampal CA3 gene expression profile in neurocognitive aging. Neurobiol Aging. 2011;32:1678–1692. doi: 10.1016/j.neurobiolaging.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberman RP, Quigley CK, Gallagher M. Characterization of CpG island DNA methylation of impairment-related genes in a rat model of cognitive aging. Epigenetics. 2012;7:1008–1019. doi: 10.4161/epi.21291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall J, Thomas KL, Everitt BJ. Cellular imaging of zif268 expression in the hippocampus and amygdala during contextual and cued fear memory retrieval: selective activation of hippocampal CA1 neurons during the recall of contextual memories. J Neurosci. 2001;21:2186–2193. doi: 10.1523/JNEUROSCI.21-06-02186.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannum G, Guinney J, Zhao L, Zhang L, Hughes G, Sadda S, Klotzle B, Bibikova M, Fan J-B, Gao Y, Deconde R, Chen M, Rajapakse I, Friend S, Ideker T, Zhang K. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 2013;49:359–367. doi: 10.1016/j.molcel.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartzell AL, Burke SN, Hoang LT, Lister JP, Rodriguez CN, Barnes CA. Transcription of the immediate-early gene Arc in CA1 of the hippocampus reveals activity differences along the proximodistal axis that are attenuated by advanced age. J Neurosci. 2013;33:3424–3433. doi: 10.1523/JNEUROSCI.4727-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemeyer T, Wingender E, Reuter I, Hermjakob H, Kel AE, Kel OV, Ignatieva EV, Ananko EA, Podkolodnaya OA, Kolpakov FA, Podkolodny NL, Kolchanov NA. Databases on transcriptional regulation: TRANSFAC, TRRD and COMPEL. Nucleic Acids Res. 1998;26:362–367. doi: 10.1093/nar/26.1.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez DG, Nalls MA, Gibbs JR, Arepalli S, van der Brug M, Chong S, Moore M, Longo DL, Cookson MR, Traynor BJ, Singleton AB. Distinct DNA methylation changes highly correlated with chronological age in the human brain. Hum Mol Genet. 2011;20:1164–1172. doi: 10.1093/hmg/ddq561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Langley B, Lubin FD, Renthal W, Wood MA, Yasui DH, Kumar A, Nestler EJ, Akbarian S, Beckel-Mitchener AC. Epigenetics in the nervous system. J Neurosci. 2008;28:11753–11759. doi: 10.1523/JNEUROSCI.3797-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MW, Errington ML, French PJ, Fine A, Bliss TVP, Garel S, Charnay P, Bozon B, Laroche S, Davis S. A requirement for the immediate early gene Zif268 in the expression of late LTP and long-term memories. Nat Neurosci. 2001;4:289–296. doi: 10.1038/85138. [DOI] [PubMed] [Google Scholar]
- Kadish I, Thibault O, Blalock EM, Chen K-C, Gant JC, Porter NM, Landfield PW. Hippocampal and cognitive aging across the lifespan: a bioenergetic shift precedes and increased cholesterol trafficking parallels memory impairment. J Neurosci. 2009;29:1805–1816. doi: 10.1523/JNEUROSCI.4599-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo JW, Russo SJ, Ferguson D, Nestler EJ, Duman RS. Nuclear factor-κB is a critical mediator of stress-impaired neurogenesis and depressive behavior. Proc Natl Acad Sci U S A. 2010;107:2669–2674. doi: 10.1073/pnas.0910658107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosik KS, Rapp PR, Raz N, Small SA, Sweatt JD, Tsai L-H. Mechanisms of age-related cognitive change and targets for intervention: epigenetics. J Gerontol A Biol Sci Med Sci. 2012;67:741–746. doi: 10.1093/gerona/gls110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaPlant Q, Vialou V, Covington HE, 3rd, Dumitriu D, Feng J, Warren BL, Maze I, Dietz DM, Watts EL, Iñiguez SD, Koo JW, Mouzon E, Renthal W, Hollis F, Wang H, Noonan MA, Ren Y, Eisch AJ, Bolaños CA, Kabbaj M, Xiao G, Neve RL, Hurd YL, Oosting RS, Fan G, Morrison JH, Nestler EJ. Dnmt3a regulates emotional behavior and spine plasticity in the nucleus accumbens. Nat Neurosci. 2010;13:1137–1143. doi: 10.1038/nn.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levenson JM, Roth TL, Lubin FD, Miller CA, Huang I-C, Desai P, Malone LM, Sweatt JD. Evidence that DNA (cytosine-5) methyltransferase regulates synaptic plasticity in the hippocampus. J Biol Chem. 2006;281:15763–15773. doi: 10.1074/jbc.M511767200. [DOI] [PubMed] [Google Scholar]
- Linnarsson S, Björklund A, Ernfors P. Learning deficit in BDNF mutant mice. Eur J Neurosci. 1997;9:2581–2587. doi: 10.1111/j.1460-9568.1997.tb01687.x. [DOI] [PubMed] [Google Scholar]
- Liu L, van Groen T, Kadish I, Li Y, Wang D, James SR, Karpf AR, Tollefsbol TO. Insufficient DNA methylation affects healthy aging and promotes age-related health problems. Clin Epigenetics. 2011;2:349–360. doi: 10.1007/s13148-011-0042-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lubin FD, Roth TL, Sweatt JD. Epigenetic regulation of BDNF gene transcription in the consolidation of fear memory. J Neurosci. 2008;28:10576–10586. doi: 10.1523/JNEUROSCI.1786-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrone DF, Adams AA, Satvat E. Increased pattern separation in the aged fascia dentata. Neurobiol Aging. 2011;32:2317.e23–2317.e32. doi: 10.1016/j.neurobiolaging.2010.03.021. [DOI] [PubMed] [Google Scholar]
- Marrone DF, Satvat E, Shaner MJ, Worley PF, Barnes CA. Attenuated long-term Arc expression in the aged fascia dentata. Neurobiol Aging. 2012;33:979–990. doi: 10.1016/j.neurobiolaging.2010.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHugh TJ, Jones MW, Quinn JJ, Balthasar N, Coppari R, Elmquist JK, Lowell BB, Fanselow MS, Wilson MA, Tonegawa S. Dentate gyrus NMDA receptors mediate rapid pattern separation in the hippocampal network. Science. 2007;317:94–99. doi: 10.1126/science.1140263. [DOI] [PubMed] [Google Scholar]
- Miller CA, Gavin CF, White JA, Parrish RR, Honasoge A, Yancey CR, Rivera IM, Rubio MD, Rumbaugh G, Sweatt JD. Cortical DNA methylation maintains remote memory. Nat Neurosci. 2010;13:664–666. doi: 10.1038/nn.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CA, Sweatt JD. Covalent modification of DNA regulates memory formation. Neuron. 2007;53:857–869. doi: 10.1016/j.neuron.2007.02.022. [DOI] [PubMed] [Google Scholar]
- Moreno H, Wu WE, Lee T, Brickman A, Mayeux R, Brown TR, Small SA. Imaging the Aβ-related neurotoxicity of Alzheimer disease. Arch Neurol. 2007;64:1467–1477. doi: 10.1001/archneur.64.10.1467. [DOI] [PubMed] [Google Scholar]
- Morse SJ, Butler AA, Davis RL, Soller IJ, Lubin FD. Environmental enrichment reverses histone methylation changes in the aged hippocampus and restores age-related memory deficits. Biology. 2015;4:298–313. doi: 10.3390/biology4020298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira AMM, Hemstedt TJ, Bading H. Rescue of aging-associated decline in Dnmt3a2 expression restores cognitive abilities. Nat Neurosci. 2012;15:1111–1113. doi: 10.1038/nn.3151. [DOI] [PubMed] [Google Scholar]
- Palop JJ, Chin J, Bien-Ly N, Massaro C, Yeung BZ, Yu G-Q, Mucke L. Vulnerability of dentate granule cells to disruption of Arc expression in human amyloid precursor protein transgenic mice. J Neurosci. 2005;25:9686–9693. doi: 10.1523/JNEUROSCI.2829-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish RR, Day JJ, Lubin FD. Direct bisulfite sequencing for examination of DNA methylation with gene and nucleotide resolution from brain tissues. Curr Protoc Neurosci. 2012;Chapter 7(Unit 7.24) doi: 10.1002/0471142301.ns0724s60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlowski TL, Bellush LL, Wright AW, Walker JP, Colvin RA, Huentelman MJ. Hippocampal gene expression changes during age-related cognitive decline. Brain Res. 2009;1256:101–110. doi: 10.1016/j.brainres.2008.12.039. [DOI] [PubMed] [Google Scholar]
- Paxinos G, Watson C. The Rat Brain in Stereotaxic Coordinates. 4th. San Diego: Academic Press; 1998. [Google Scholar]
- Peleg S, Sananbenesi F, Zovoilis A, Burkhardt S, Bahari-Javan S, Agis-Balboa RC, Cota P, Wittnam JL, Gogol-Doering A, Opitz L, Salinas-Riester G, Dettenhofer M, Kang H, Farinelli L, Chen W, Fischer A. Altered histone acetylation is associated with age-dependent memory impairment in mice. Science. 2010;328:753–756. doi: 10.1126/science.1186088. [DOI] [PubMed] [Google Scholar]
- Penner MR, Roth TL, Barnes CA, Sweatt JD. An epigenetic hypothesis of aging-related cognitive dysfunction. Front Aging Neurosci. 2010;2:9. doi: 10.3389/fnagi.2010.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penner MR, Roth TL, Chawla MK, Hoang LT, Roth ED, Lubin FD, Sweatt JD, Worley PF, Barnes CA. Age-related changes in Arc transcription and DNA methylation within the hippocampus. Neurobiol Aging. 2011;32:2198–2210. doi: 10.1016/j.neurobiolaging.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinaud R, Tremere LA, Penner MR, Hess FF, Robertson HA, Currie RW. Complexity of sensory environment drives the expression of candidate-plasticity gene, nerve growth factor induced-A. Neuroscience. 2002;112:573–582. doi: 10.1016/s0306-4522(02)00094-5. [DOI] [PubMed] [Google Scholar]
- Poirier GL, Amin E, Aggleton JP. Qualitatively different hippocampal subfield engagement emerges with mastery of a spatial memory task by rats. J Neurosci. 2008;28:1034–1045. doi: 10.1523/JNEUROSCI.4607-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renaudineau S, Poucet B, Laroche S, Davis S, Save E. Impaired long-term stability of CA1 place cell representation in mice lacking the transcription factor zif268/egr1. Proc Natl Acad Sci U S A. 2009;106:11771–11775. doi: 10.1073/pnas.0900484106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenzweig ES, Barnes CA. Impact of aging on hippocampal function: plasticity, network dynamics, and cognition. Prog Neurobiol. 2003;69:143–179. doi: 10.1016/s0301-0082(02)00126-0. [DOI] [PubMed] [Google Scholar]
- Rosi S, Ramirez-Amaya V, Vazdarjanova A, Worley PF, Barnes CA, Wenk GL. Neuroinflammation alters the hippocampal pattern of behaviorally induced Arc expression. J Neurosci. 2005;25:723–731. doi: 10.1523/JNEUROSCI.4469-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth TL, Lubin FD, Funk AJ, Sweatt JD. Lasting epigenetic influence of early-life adversity on the BDNF gene. Biol Psychiatry. 2009;65:760–769. doi: 10.1016/j.biopsych.2008.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth TL, Zoladz PR, Sweatt JD, Diamond DM. Epigenetic modification of hippocampal Bdnf DNA in adult rats in an animal model of post-traumatic stress disorder. J Psychiatr Res. 2011;45:919–926. doi: 10.1016/j.jpsychires.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe WB, Blalock EM, Chen K-C, Kadish I, Wang D, Barrett JE, Thibault O, Porter NM, Rose GM, Landfield PW. Hippocampal expression analyses reveal selective association of immediate-early, neuroenergetic, and myelinogenic pathways with cognitive impairment in aged rats. J Neurosci. 2007;27:3098–3110. doi: 10.1523/JNEUROSCI.4163-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo SJ, Wilkinson MB, Mazei-Robison MS, Dietz DM, Maze I, Krishnan V, Renthal W, Graham A, Birnbaum SG, Green TA, Robison B, Lesselyong A, Perrotti LI, Bolaños CA, Kumar A, Clark MS, Neumaier JF, Neve RL, Bhakar AL, Barker PA, Nestler EJ. Nuclear factor κB signaling regulates neuronal morphology and cocaine reward. J Neurosci. 2009;29:3529–3537. doi: 10.1523/JNEUROSCI.6173-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegmund KD, Connor CM, Campan M, Long TI, Weisenberger DJ, Biniszkiewicz D, Jaenisch R, Laird PW, Akbarian S. DNA methylation in the human cerebral cortex is dynamically regulated throughout the life span and involves differentiated neurons. PLoS ONE. 2007;2:e895. doi: 10.1371/journal.pone.0000895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small SA, Chawla MK, Buonocore M, Rapp PR, Barnes CA. Imaging correlates of brain function in monkeys and rats isolates a hippocampal subregion differentially vulnerable to aging. Proc Natl Acad Sci U S A. 2004;101:7181–7186. doi: 10.1073/pnas.0400285101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small SA, Tsai WY, DeLaPaz R, Mayeux R, Stern Y. Imaging hippocampal function across the human life span: is memory decline normal or not? Ann Neurol. 2002;51:290–295. doi: 10.1002/ana.10105. [DOI] [PubMed] [Google Scholar]
- Steward O, Worley PF. Selective targeting of newly synthesized Arc mRNA to active synapses requires NMDA receptor activation. Neuron. 2001;30:227–240. doi: 10.1016/s0896-6273(01)00275-6. [DOI] [PubMed] [Google Scholar]
- Tanic N, Perovic M, Mladenovic A, Ruzdijic S, Kanazir S. Effects of aging, dietary restriction and glucocorticoid treatment on housekeeping gene expression in rat cortex and hippocampus-evaluation by real time RT-PCR. J Mol Neurosci. 2007;32:38–46. doi: 10.1007/s12031-007-0006-7. [DOI] [PubMed] [Google Scholar]
- Toescu EC, Verkhratsky A, Landfield PW. Ca2+ regulation and gene expression in normal brain aging. Trends Neurosci. 2004;27:614–620. doi: 10.1016/j.tins.2004.07.010. [DOI] [PubMed] [Google Scholar]
- Toescu EC, Vreugdenhil M. Calcium and normal brain ageing. Cell Calcium. 2010;47:158–164. doi: 10.1016/j.ceca.2009.11.013. [DOI] [PubMed] [Google Scholar]
- Vazdarjanova A, Guzowski JF. Differences in hippocampal neuronal population responses to modifications of an environmental context: evidence for distinct, yet complementary, functions of CA3 and CA1 ensembles. J Neurosci. 2004;24:6489–6496. doi: 10.1523/JNEUROSCI.0350-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazdarjanova A, McNaughton BL, Barnes CA, Worley PF, Guzowski JF. Experience-dependent coincident expression of the effector immediate-early genes arc and Homer 1a in hippocampal and neocortical neuronal networks. J Neurosci. 2002;22:10067–10071. doi: 10.1523/JNEUROSCI.22-23-10067.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbitsky M, Yonan AL, Malleret G, Kandel ER, Gilliam TC, Pavlidis P. Altered hippocampal transcript profile accompanies an age-related spatial memory deficit in mice. Learn Mem. 2004;11:253–260. doi: 10.1101/lm.68204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver ICG, Cervoni N, Champagne FA, D’Alessio AC, Sharma S, Seckl JR, Dymov S, Szyf M, Meaney MJ. Epigenetic programming by maternal behavior. Nat Neurosci. 2004;7:847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- Weaver ICG, D’Alessio AC, Brown SE, Hellstrom IC, Dymov S, Sharma S, Szyf M, Meaney MJ. The transcription factor nerve growth factor-inducible protein A mediates epigenetic programming: altering epigenetic marks by immediate-early genes. J Neurosci. 2007;27:1756–1768. doi: 10.1523/JNEUROSCI.4164-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West MJ. Regionally specific loss of neurons in the aging human hippocampus. Neurobiol Aging. 1993;14:287–293. doi: 10.1016/0197-4580(93)90113-p. [DOI] [PubMed] [Google Scholar]
- Widera D, Mikenberg I, Elvers M, Kaltschmidt C, Kaltschmidt B. Tumor necrosis factor α triggers proliferation of adult neural stem cells via IKK/NF-κB signaling. BMC Neurosci. 2006;7:64. doi: 10.1186/1471-2202-7-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yassa MA, Lacy JW, Stark SM, Albert MS, Gallagher M, Stark CEL. Pattern separation deficits associated with increased hippocampal CA3 and dentate gyrus activity in nondemented older adults. Hippocampus. 2011a;21:968–979. doi: 10.1002/hipo.20808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yassa MA, Mattfeld AT, Stark SM, Stark CEL. Age-related memory deficits linked to circuit-specific disruptions in the hippocampus. Proc Natl Acad Sci U S A. 2011b;108:8873–8878. doi: 10.1073/pnas.1101567108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yassa MA, Stark CEL. Pattern separation in the hippocampus. Trends Neurosci. 2011;34:515–525. doi: 10.1016/j.tins.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yau JLW, Olsson T, Morris RGM, Noble J, Seckl JR. Decreased NGFI-A gene expression in the hippocampus of cognitively impaired aged rats. Brain Res Mol Brain Res. 1996;42:354–357. doi: 10.1016/s0169-328x(96)00220-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.