Abstract
Epidemiological and experimental data implicate branched chain amino acids (BCAAs) in the development of insulin resistance, but the mechanisms underlying this link remain unclear.1–3 Insulin resistance in skeletal muscle stems from excess accumulation of lipid species4, a process that requires blood-borne lipids to first traverse the blood vessel wall. Little is known, however, of how this trans-endothelial transport occurs or is regulated. Here, we leverage PGC-1α, a transcriptional coactivator that regulates broad programs of FA consumption, to identify 3-hydroxy-isobutyrate (3-HIB), a catabolic intermediate of the BCAA valine, as a novel paracrine regulator of trans-endothelial fatty acids (FA) transport. 3-HIB is secreted from muscle cells, activates endothelial FA transport, stimulates muscle FA uptake in vivo, and promotes muscle lipid accumulation and insulin resistance in animals. Conversely, inhibiting the synthesis of 3-HIB in muscle cells blocks the promotion of endothelial FA uptake. 3-HIB levels are elevated in muscle from db/db mice and from subjects with diabetes. These data thus unveil a novel mechanism that regulates trans-endothelial flux of FAs, revealing 3-HIB as a new bioactive signaling metabolite that links the regulation of FA flux to BCAA catabolism and provides a mechanistic explanation for how increased BCAA catabolic flux can cause diabetes.
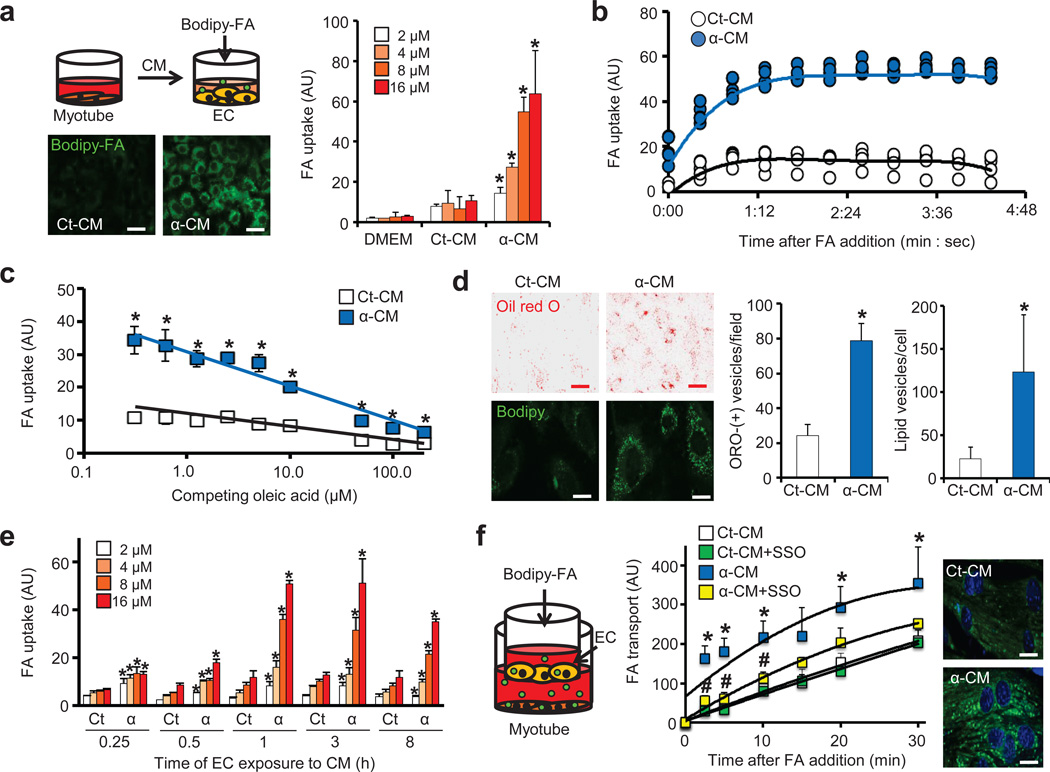
PGC-1α in skeletal muscle induces broad genetic programs, including mitochondrial biogenesis and FA β-oxidation.5,6 PGC-1α also induces the paracrine activation of angiogenesis, thus coordinating the consumption of FAs in mitochondria with their delivery via blood vessels.7 We reasoned here that PGC-1α, in order to maximize delivery of FAs to muscle, might also instruct the vessels to increase trans-endothelial FA transport from the vessel lumen to the extra-luminal myofibers, a process that remains poorly understood. To test this, we devised an assay in which conditioned medium (CM) from C2C12 skeletal myotubes was added to human umbilical vein endothelial cells (HUVECs), after which the ability of the HUVECs to take up FAs was measured with the fluorescent FA analog Bodipy-C12 (Supplementary Fig. 1a). Strikingly, CM from myotubes over-expressing PGC-1α (α-CM), either via adenovirus (Fig. 1a) or multiple retrovirus lines (Supplementary Fig. 1b–d), dramatically increased FA uptake in HUVECs, suggesting that a PGC-1α-regulated paracrine factor induces endothelial FA uptake. The uptake of FA was rapid (Fig. 1b), saturated by high FA concentrations (Supplementary Fig. 2a), competed by equimolar unlabeled oleic acid (Fig. 1c), limited to the uptake of long-chain FAs (Supplementary Fig. 2b), and persistent after dilution of α-CM (Supplementary Fig. 2c), observations that together strongly suggest that the uptake of FA is a specific, likely protein-mediated process. Prolonged exposure of ECs to α-CM increased intracellular lipids, measured by staining with oil red O or unconjugated Bodipy (Fig. 1d). The stimulation of FA uptake by α-CM was specific to ECs: only one of 10 non-EC types increased uptake of Bodipy-C12 in response to α-CM, while every EC type tested did (Supplementary Fig. 2d). The induction of FA uptake by ECs occurred within 15–60 minutes of exposure to α-CM (Fig. 1e) and was ATP-dependent (Supplementary Fig. 2e). α-CM strongly induced FA flux across a tight endothelial monolayer, which was blocked by Sulfo-N-succinimidyl oleate (SSO), an inhibitor of FA transport (Fig. 1f and Supplementary Fig. 2f–h). Conversely, an endothelial monolayer treated with CM from myotubes lacking both PGC-1α and PGC-1β, a related PGC-1 family member, showed reduced FA transport (Supplementary Fig. 2i–k). Together, these data demonstrate the existence of paracrine factor(s), induced in myotubes by PGC-1α, which stimulate(s) endothelial FA uptake and transport.
Figure 1. PGC-1α in muscle cells induces secretion of paracrine activity that stimulates endothelial FA transport.
(a) Experimental strategy (top), representative images (bottom) and quantification (right) of Bodipy-FA (2–16 µM) uptake by endothelial cells (ECs) after exposure to conditioned media (CM) from myotubes expressing control GFP (Ct-CM) or PGC-1α (α-CM). Scale bars, 50 µm (b,c) Endothelial FA uptake (8 µM) at different time points (b) and in the presence of the indicated concentrations of unlabeled oleic acid for 5 min (c). (d) Staining by oil red O (ORO) of intracellular neutral lipids in ECs after prolonged exposure (24 hrs) to α-CM. Representative images (left) and quantification (right). Red scale bars, 50 µm; white scale bars, 10 µm. (e) Endothelial FA uptake after exposure to the indicated CM for the indicated durations. (f) Experimental strategy (left), quantification of FA transport (8 µM) across a tight EC monolayer (middle), and representative images of myotubes taking up FA transported through the EC monolayer (right). Scale bars, 10 µm. Student’s t-test; *P < 0.05 vs. control; #P < 0.05 vs. α-CM. Two-way ANOVA was used for f. Data are mean ± standard deviation (s.d.) of at least three biological replicates.
Hagberg et al. recently showed that VEGFB affects endothelial FA uptake via its cognate receptor VEGFR1 (Flt1)8, suggesting that VEGFB may be the paracrine factor in α-CM. Consistent with this notion, PGC-1α induces the expression of VEGFB (Supplementary Fig. 3a). However, neutralization of VEGFB by a neutralizing soluble receptor (sFlt1) or by receptor inhibition (SU11248) failed to block the activity of α-CM, despite efficient inhibition of VEGFB signaling (Fig. 2a and Supplementary Fig. 3b,c). Knockdown of Vegfb in myotubes (Fig. 2b and Supplementary Fig. 3d) and genetic deletion of either or both Vegfr1 (Flt) and Vegfr2 (Flk) in ECs also failed to inhibit α-CM activity (Fig. 2c and Supplementary Fig. 3e–g). The paracrine factor in α-CM is thus not VEGFB.
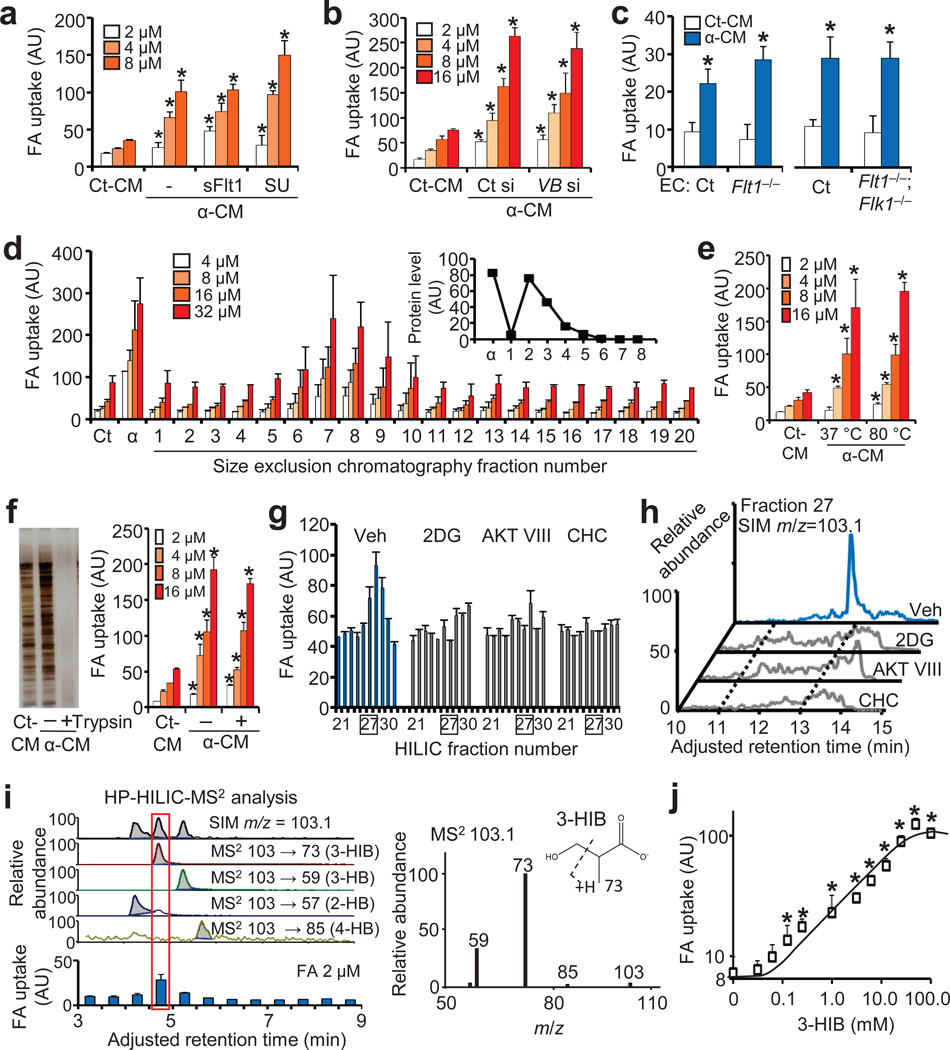
Figure 2. Identification of 3-hydroxyisobutyrate (3-HIB) as the paracrine factor.
(a) Endothelial FA uptake after exposure to conditioned media (CM) from myotubes expressing control GFP (Ct-CM) or PGC-1α (α-CM) after treatment of ECs with sFlt1 or SU11248 (SU). (b) Endothelial FA uptake after exposure to CM from PGC-1α-expressing myotubes treated with control siRNA (Ct si) or Vegfb siRNA (VB si). (c) FA uptake (2 µM) by ECs isolated from Flt1 KO mice (Flt1−/−) or Flt1 and Flk1 double KO mice (Flt1−/−; Flk1−/−). (d–f) Endothelial FA uptake after exposure to size-exclusion chromatography fractions of α-CM (d), to α-CM heat-inactivated at the indicated temperature (e), or to α-CM treated with trypsin (f). (g) Endothelial FA uptake (8 µM) after exposure to HILIC fractions of CM from PGC-1α-expressing myotubes treated with the indicated inhibitors. Veh, vehicle. (h) Identification by mass spectrometry of a molecule with m/z = 103.1 specific to HILIC fraction 27. (i) Selective ion monitoring (SIM) of HP-HILIC-MS2 identified the paracrine activity overlaid with MS2 signature of 103->73 (left), of which MS2 spectra matched with 3-HIB (right). (j) Endothelial FA uptake (2 µM) after incubation with the indicated concentrations of 3-HIB for 1 hr. Student’s t-test; *P < 0.05 vs. control. Data are mean ± s.d. of at least three biological replicates.
To identify the paracrine factor in α-CM, we fractionated α-CM with size exclusion chromatography. The FA uptake-inducing activity was found in fractions containing small molecules, and few proteins (Fig. 2d, Supplementary Fig. 3h). Treatment of α-CM at 80°C (Fig. 2e), trypsinization (Fig. 2f), or with charcoal (Supplementary Fig. 3i), failed to abolish the activity, suggesting that the paracrine factor is a hydrophilic non-protein metabolite(s). Initial efforts, using mass spectrometry (MS), to identify differences between α-CM and control CM revealed thousands of peaks, too many to identify individually. Thus, to find tools with that would help reduce the differences in metabolome between α-CM and control CM, we searched for compounds that blocked the production of FA uptake-inducing activity from PGC-1α-expressing C2C12 myotubes. We identified a number of such compounds, including inhibitors of the PI3K pathway (targeting PI3K, AKT, and mTORC2), of glycolysis (2-deoxy-glucose or 2DG), and of monocarboxylate transport (CHC) (Supplementary Fig. 4a–c). None of treatments directly affected FA uptake by ECs (Supplementary Fig. 4d), demonstrating that these treatments act on muscle cells to block production and/or secretion of the paracrine factor. Pyruvate supplementation partially restored the inhibition by AKT inhibitor or 2DG, but not by CHC (Supplementary Fig. 4e). The identification of these inhibitors thus allowed further purification with discerning negative controls that minimized differences between conditioned media (Supplementary Fig. 4f). The CMs were first treated with charcoal to remove hydrophobic compounds, and then fractionated by open column silica-gel chromatography (Supplementary Fig. 5a). The active fractions from this fractionation still contained too many MS peaks to allow identification of the active factor; the fractions were therefore further subjected to orthogonal fractionation by high-pressure hydrophilic interaction liquid chromatography (HP-HILIC) (Supplementary Fig. 5b). Evaluations by MS of active fractions revealed a peak with molecular weight of 104.1 ([M-H]=103.1), which was absent in parallel, inactive fractions from CM of cells pre-treated with 2DG, AKT inhibitor (AKT VIII), or CHC (Fig. 2g,h). Among multiple candidates based on the retention time in HILIC column, four isobaric hydroxybutyrates potentially corresponded to this molecular weight (Supplementary Fig. 5c). Among them, bioactivity was correlated with a molecule with MS2 fingerprint of 103→73, which is consistent with 3-hydroxyisobutyrate (3-HIB) (Fig. 2i). Further matching of chromatographic properties and MS2 spectra to synthetic standards unambiguously confirmed this conclusion. Synthetic 3-HIB alone was sufficient to increase Bodipy-FA uptake by HUVECs (Fig. 2j), Bodipy-FA transport across a tight monolayer of ECs (Supplementary Fig. 6a–c), and transport of 13C-labeled palmitate across a tight monolayer of ECs and consequent uptake and incorporation of the labeled palmitate into the TCA cycle of myotubes on the other side of the EC monolayer (Supplementary Fig. 6d). These data thus unequivocally identify 3-HIB as the active component in the fractions.
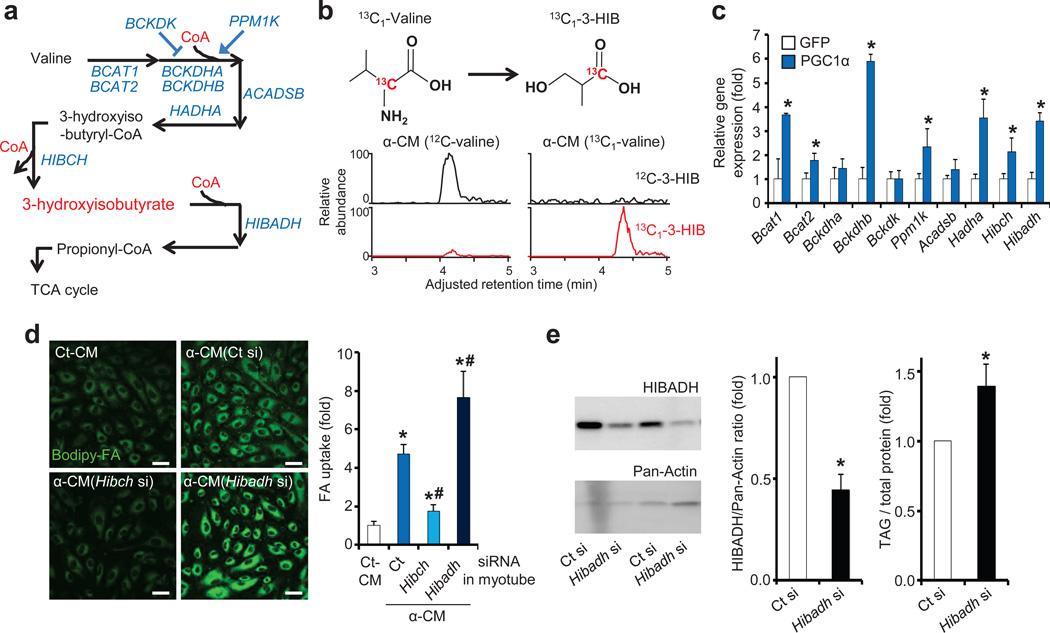
3-HIB is an intermediate of valine catabolism, derived from 3-hydroxyisobutyryl-CoA (HIBC) by HIBC hydrolase (HIBCH), and subsequently catabolized by 3-HIB dehydrogenase (HIBADH), eventually to form propionyl-CoA (Fig. 3a). 13C-labeling of valine in PGC-1α-expressing myotubes led to nearly complete 13C labeling of 3-HIB (Fig. 3b and Supplementary Fig. 6e,f), and the FA uptake-inducing activity of α-CM required valine (Supplementary Fig. 6g), demonstrating that 3-HIB in α-CM is indeed derived from valine. PGC-1α induced in myotubes the expression of nearly every enzyme of BCAA catabolism (Fig. 3c and Supplementary Fig. 7a,b).9 The increased levels of 3-HIB in α-CM (Supplementary Fig. 7c) thus reflect PGC-1α-induced catabolism of valine. PGC-1β induced the same genes, while deletion of both PGC-1α and PGC-1β repressed the valine catabolic genes (Supplementary Fig. 7d–f). Knockdown of Hibch in myotubes (Supplementary Fig. 7g) nearly abolished the induction of uptake of FAs in ECs (Fig. 3d). Conversely, knockdown of Hibadh (the downstream enzyme) in myotubes enhanced the uptake of FAs in ECs (Fig. 3d). Similarly, knockdown of Hibadh in mouse skeletal muscle in vivo increased triglyceride levels (Fig. 3e). ECs themselves express the valine catabolic enzymes at much lower levels than skeletal muscle (Supplementary Fig. 7h), and knockdown of HIBADH in ECs did not affect FA uptake (Supplementary Fig. 7i,j). Together, these data demonstrate that PGC-1α induces in myotubes the catabolism of valine to generate 3-HIB, which then acts as a paracrine factor to stimulate endothelial FA uptake.
Figure 3. 3-HIB is generated from valine catabolism induced by PGC-1α and stimulates endothelial FA uptake.
(a) Schematic of valine catabolism. (b) Detection of 13C-labeled 3-HIB in CM from PGC-1α-expressing myotubes incubated with 13C-labeled valine. (c) qPCR analysis of valine metabolic enzymes in myotubes expressing control GFP or PGC-1α. (d) Representative images of ECs taking up Bodipy-FA (left) and quantification (right) after exposure to CM from PGC-1α-expressing myotubes (α-CM) treated with Hibch or Hibadh siRNA. Scale bars, 50 µm. (e) Western blot of mouse skeletal muscle after injection in intact animals with Hibadh siRNA. Representative gel images (left), quantification (middle) and muscle triacylglyceride (TAG) levels (right, n = 8). Student’s t-test; *P < 0.05 vs. control; #P < 0.05 vs. α-CM. Two-way ANOVA for d. Data are mean ± s.d. of at least three biological replicates.
Fatty Acid Transport Proteins (FATP) 3 and 4, and CD36, have been proposed as putative transporters of FAs in ECs8. Knockdown of FATP3 or FATP4 in HUVECs, or primary ECs isolated from Fatp4 KO mice, revealed reduced FA uptake by α-CM or 3-HIB (Supplementary Fig. 8a–c). Knockdown of CD36 in HUVECs or primary ECs isolated from Cd36 KO mice had a lesser effect (Supplementary Fig. 8d–g). The uptake of FA by ECs in response to 3-HIB is thus largely mediated by FATP3 and 4. Interestingly, neither mRNA nor protein levels of FATP3 or FATP4 were affected by α-CM or 3-HIB treatment (Supplementary Fig. 8h–k), suggesting that post-translational mechanisms are likely involved, consistent with the rapid response of ECs to α-CM (Fig. 1e).
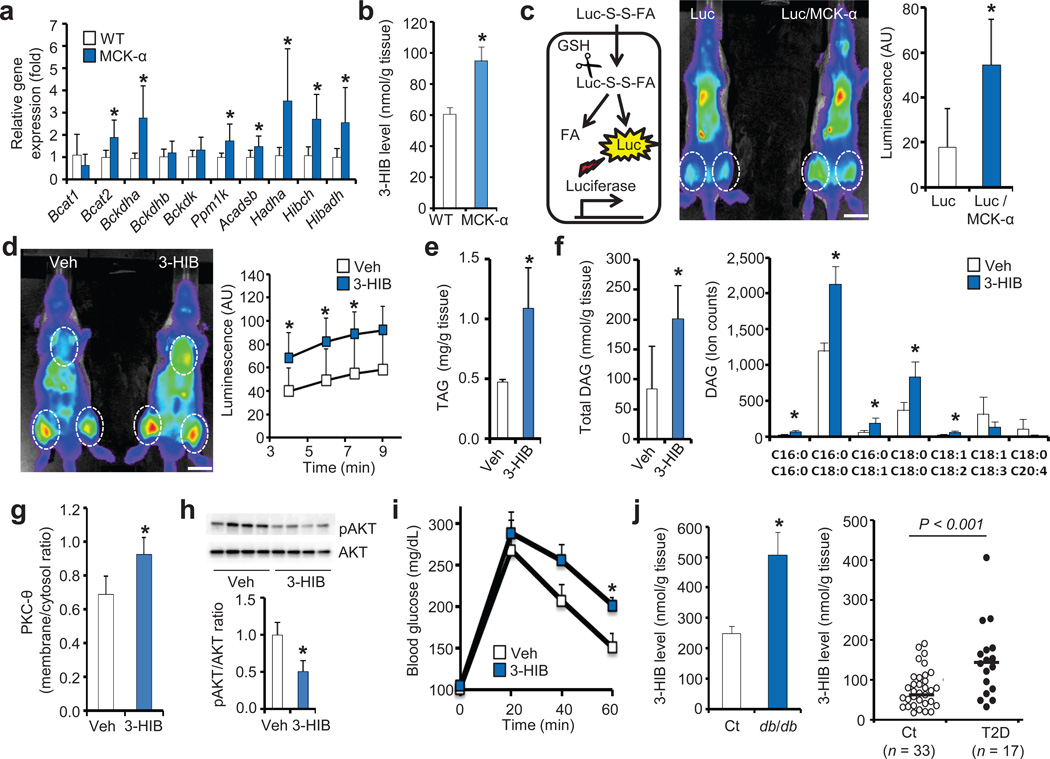
The majority of genes that mediate valine catabolism are induced in the muscle of mice that transgenically overexpress PGC-1α or β in skeletal muscle (MCK-α and MCK-β mice10,11) (Fig. 4a, Supplementary Fig. 9a), while conversely repressed in mice lacking both PGC-1α and β in skeletal muscle (Supplementary Fig. 9b). Consistent with increased catabolism of valine, 3-HIB is increased in skeletal muscle of MCK-α mice (Fig. 4b), while BCAAs are reduced.12 FA uptake in skeletal muscle of MCK-α mice is dramatically increased in vivo, as shown noninvasively with the use of FAs linked to luciferin that is released upon entry into cells, thus allowing for quantification via luminometry of FA uptake in vivo in luciferase-expressing animals (Fig. 4c).13 These findings are thus consistent with the seemingly paradoxical observation that MCK-α mice accumulate excess lipid in muscle, leading to lipotoxicity and insulin resistance.14 Similar glucose intolerance is observed in MCK-β mice (Supplementary Fig. 9c), while conversely glucose tolerance is improved and lipid levels are decreased in mice lacking both PGC-1α and β in skeletal muscle (Supplementary Fig. 9d,e). Altered 3-HIB secretion thus likely contributes to altered lipid accumulation in these mice, probably as part of a broader comprehensive program controlled by these powerful metabolic transcription factors.
Figure 4. 3-HIB induces FA uptake in vivo and causes glucose intolerance.
(a,b) qPCR analysis of valine metabolic enzymes (a, n = 6) and measurement of 3-HIB levels (b, n = 3) in muscle from wild type (WT) or PGC-1α-muscle specific transgenic mice (MCK-α). (c) Schematic of in vivo FA uptake assay (left). GSH, glutathione. Representative image (middle) and quantification (right, n = 4) of FA uptake in thigh of Luciferase transgenic (Luc) or Luc/MCK-α double transgenic mice. Scale bars, 1 cm. (d) Representative image (left) and quantification (right, n = 4) of FA uptake in thigh of Luc mice fed with vehicle (Veh) or 3-HIB for 1.5 hr. Scale bars, 1 cm. (e–i) Measurements of triacylglyceride (e, n = 3) and diglyceride (DAG, f, n = 4), PKC-θ membrane translocation (g, n = 4), AKT activation (h, n = 4) in muscle, and systemic glucose tolerance (i, n = 8) of mice provided with vehicle (Veh) or 3-HIB in the drinking water for 2 weeks. (j) Measurements of 3-HIB levels in muscle of db/db mice (left, n = 10) and muscle biopsies of type II diabetic (T2D) subjects (right). Student’s t-test; *P < 0.05 vs. control. In j, indicated P was with Student’s t-test; with Mann-Whitney u-test, P < 0.005 vs. control. Data are mean ± s.d. (a,c–h) or ± s.e.m. (b,i,j).
Provision of 3-HIB in the drinking water of wild type animals led to a transient 8-fold induction of 3-HIB in serum and skeletal muscle (Supplementary Fig. 9f–h), and dramatically increased uptake of FAs into the heart and thigh without affecting vessel leakiness (Fig. 4d and Supplementary Fig. 9i–k) demonstrating that 3-HIB alone is sufficient to cause FA accumulation acutely in vivo. To test for a long-term effect, mice were provided with 3-HIB in the drinking water for two weeks, leading to modest 1.6-fold induction of 3-HIB in muscle (Supplementary Fig. 10a), similar to that found in MCK-α mice (Fig. 4b) and diabetic muscles (see below). The mice receiving 3-HIB accumulated triacylglycerides (TAG) and diglycerides (DAG) in skeletal muscle (Fig. 4e,f), despite the absence of any increase in plasma FA or TAG levels (Supplementary Fig. 10b–e) or any apparent tissue damage (Supplementary Fig. 10f). The plasma membrane localization of PKC-θ was increased (Fig. 4g and Supplementary Fig. 10g) and phosphorylation of AKT was blunted (Fig. 4h), consistent with the known inhibition of insulin signaling by DAG and activated PKC-θ.4 Mice provided with 3-HIB for two weeks revealed systemic intolerance to a glucose load (Fig. 4i), and insulin resistance in hyperinsulinemic euglycemic clamp studies (Supplementary Fig. 10h–j). Neither hepatic gluconeogenesis gene expression nor glucose output was significantly different (Supplementary Fig. 10k,l), suggesting that 3-HIB has little impact on hepatic function. Together, these data demonstrate that paracrine secretion of 3-HIB, an intermediate of BCAA catabolism, induces FA uptake in vivo, and causes excess accumulation of incompletely esterified lipids in skeletal muscle, blunted AKT signaling, and glucose intolerance.
Valine is one of three BCAAs, all three of which are essential dietary components. BCAAs are abundant, comprising up to 30% of muscle protein. Catabolic flux of the BCAAs is tightly regulated in multiple organs, including skeletal muscle. Interestingly, all catabolic products of the three branched-chain α-keto acids are trapped inside the cell by covalent linkage to Coenzyme A, with the single exception of 3-HIB (Fig. 3a). 3-HIB is thus ideally suited to act as a secreted reporter of BCAA catabolic flux in muscle. Excess BCAAs have recently been implicated in the progression to diabetes both in rodents and human.1–3 Data from the Framingham Heart Study, for example, show that elevated blood BCAAs precede the onset of diabetes by decades.1 A mechanistic explanation for these observations has been lacking. Our data indicate that increased catabolic flux of BCAAs can cause the secretion of 3-HIB from muscle, leading to excess trans-endothelial FA import into muscle, accumulation of lipotoxic incompletely esterified intermediates like DAG, and blunted insulin signaling. Consistent with this notion, we found highly increased levels of 3-HIB in skeletal muscle from db/db mice and in muscle biopsies from diabetic subjects (Fig. 4j), and elevated 3-HIB in serum of diabetic subjects has been reported.15,16. Interestingly, elevations in 3-HIB occur in the absence of changes in gene expression of BCAA catabolic enzymes in either db/db mice or human diabetic muscle biopsies (Supplementary Fig. 10m,o); in contrast, proteins levels of HIBADH, the enzyme that degrades 3-HIB, are repressed in muscle from db/db mice (Supplementary Fig. 10n), as has been reported in diabetic rats17, suggesting that post-transcriptional regulation may contribute to aberrant valine catabolism in insulin resistance. Individuals with severe 3-HIB aciduria and significant neurological sequelae have also been described18, but their glucose homeostasis have not been reported.
In summary, our data unveil 3-HIB as a novel bioactive paracrine metabolite that regulates trans-endothelial flux of FAs. The data highlight the importance of the vasculature in whole-body metabolic homeostasis. The data also uncover a novel cross-regulatory link between the catabolism of BCAAs and FAs. Finally, the data provide a mechanistic explanation for how excess catabolic flux of BCAAs can lead to insulin resistance, suggesting this pathway as a new target to treat diabetes.
Online Methods
FA uptake assay
Confluent HUVECs (passage 2–8) were transferred from a 10 cm dish to a gelatin-coated 96 well black clear bottom plate (Corning #3603) with empty corner wells for no-cell controls. After overnight incubation, the cells were serum-starved with EBM2 containing EGM supplements (Lonza #CC-3162) for at least 8 hours. The cells were then treated with CMs or 3-HIB (2.5 mM) for an hour and briefly washed with PBS. Then, Bodipy-FA (Molecular Probes #D3823) pre-incubated with FA-free BSA (2:1 molar ratio) in PBS for 10 min in a 37 °C water bath was added to the cells for 5 min at 37 °C. The Bodipy-FA/BSA solution was then completely aspirated and the cells were washed with 0.5% BSA in PBS for 1.5 min twice (50 µL/well). To quench extracellular fluorescence, 0.4% trypan blue (MP Biomedicals #1691049) was added (50 µL/well) and intracellular fluorescence was measured immediately (bottom-read) with a microplate reader (excitation 488 nm, emission 515 nm, cut-off 495 nm, SpectraMax M5, Molecular Probes). Readings from wells without Bodipy addition were used to subtract background signals. The cells were then quickly washed twice with PBS (50 µL/well) and incubated with 44 µM resazurin (#R7017, Sigma) in DMEM (50 µL/well) containing 10% FBS for 2 hours. The resazurin fluorescence was then measured with a microplate reader (excitation 530 nm, emission 590 nm, cut-off 550 nm) and used to normalize Bodipy signals to cell number. Bodipy FL C16 (#D3821) and Bodipy FL C5 (#D3834) were purchased from Molecular Probes. Oleic acid-BSA was purchased from Sigma (#O3008). To block VEGFB signaling in ECs, cells were pre-treated with 1 µM SU11248 (sc-220178, Santa Cruz) for 10 min before CM treatment, or CMs were pre-incubated with 1 µg/mL sFlt1 (#14-923, Calbiochem) for 6 hours at 4 °C with shaking before being treated to ECs. For the trypsin treatment, CMs were pre-incubated with 30 µg/ml trypsin (Gibco) for 3 hours at 37 °C, then incubated with 500 µg/mL soy bean trypsin inhibitor (17075-029, Gibco) before being treated to the cells.
FA transport assay
Isolation of primary rat brain ECs was performed as described previously.19 Cells were counted and seeded (5 × 104 cells/well) on a 0.4 µm transwell (Corning #07-200-147), then grown for 4 days until they formed compact monolayers. Meanwhile, C2C12 was differentiated into myotubes in 24 well plate for 4 days (see below). The brain ECs in a transwell were then incubated with CMs or 3-HIB (2.5 mM) for an hour and the transwell was inserted into the 24 well plate in which C2C12 myotubes were grown. Bodipy-FA/BSA or [U-13C]-palmitate/BSA in CMs (80 µL) with or without Dextran Texas-Red (70 kDa, Molecular Probes #D1830) was then added to the top chamber of the transwell. CMs containing only BSA (520 µL) were added to the bottom chamber of transwell. After each time point, 10 µL of media was taken from the bottom transwell to measure the Bodipy-FA and Dextran Texas-Red fluorescences. To measure [U-13C]-palmitate metabolism in C2C12 myotubes, media from the bottom transwell were collected after 30 min of the transport assay and incubated with C2C12 myotubes for an hour. Metabolites were then isolated from the C2C12 myotubes and quantified (UPenn Metabolomics Core, Princeton). Trans-endothelial electrical resistance (TEER) was measured as described previously.20
Preparation of CMs
C2C12 cells were grown in 10 cm dishes until 90% confluency and differentiated into myotubes with 5 µg/mL insulin plus 5 µg/mL transferrin (Sigma) in DMEM for 2 days, followed by 2% horse serum in DMEM (C2C12 differentiation media) for additional 2 days. The cells were then infected with an adenovirus expressing GFP, PGC-1α or PGC-1β. Two days after infection, the cells were washed twice with PBS to remove the adenovirus and incubated with 12 mL DMEM (with or without 2% horse serum) for 2 days. 2DG (5 mM), AKT VIII (5 µM) or CHC (5 mM) was added if necessary. CMs were then collected, centrifuged at 13,000 g for 10 min at 4 °C, aliquoted and stored at –80 °C for future use. To generate PGC-1α-expressing C2C12 cell lines, cells were infected with retrovirus expressing PGC-1α or an empty-vector control. Two days after infection, the infected cells were selected with 2.5 µg/mL puromycin for 2 days. To generate VEGFB CM, a VEGFB construct was obtained from the hORFeome Database and cloned into the Gateway pcDNA-DEST40 vector (Invitrogen) and transfected into HEK293T cells. Fifteen hours after transfection, the cells were washed once with PBS and incubated with DMEM for 2 days before collection. The control CM was generated with GFP construct.
Cell culture
HUVECs (Lonza), human endothelial colony forming cells (ECFCs, a gift from J. Bischoff, Children’s Hospital, Boston), primary rat brain ECs, primary mouse heart and muscle ECs were grown in EBM2 containing EGM supplements with 20 % FBS. C2C12 and all other cell lines (ATCC) were grown in DMEM with 10 % FBS. Isolation of primary ECs and myoblasts was performed as described previously.21,22 Pericyte-like cells were differentiated from 10T1/2 with TGF-β for 4 days as described previously.23 For the siRNA transfection, control siRNA (SIC001), FATP3 siRNA (SASI_Hs01_00100092), FATP4 siRNA (SASI_Hs01_00047530 ), FLK1 siRNA (SASI_Hs01_00073461), CD36 esiRNA (EHU089321), HIBADH siRNA (SASI_Hs01_00061462), Hibch siRNA (SASI_Mm02_00341755), Hibadh siRNA (SASI_Mm01_00120340) or Vegfb siRNA (SASI_Mm01_00114251, all from Sigma) was mixed with Lipofectamine RNAi Max (Invitrogen) in Opti-MEM (Sigma) for 20 min before being treated to cells.
Bodipy and immunofluorescence staining
ECs were grown on fibronectin-coated cover glass. The cells were then washed with PBS once and fixed with 4% formaldehyde in PBS for 20 min at room temperature. The cells were then washed with washing buffer (0.1 % Triton X-100 in PBS) three times for 30 min in total, incubated in blocking buffer (5 % BSA in washing buffer) for an hour, and incubated with 1 µg/ml Bodipy (Molecular Probes) in blocking buffer with or without a primary antibody (1:100 dilution) overnight at 4°C. The cells were then washed with washing buffer 3 times for 30 min in total, incubated with a secondary antibody (1:1,000 dilution in blocking buffer) for 3 h at room temperature. The cells were then washed with washing buffer 6 times for an hour in total, and mounted with DAPI mounting media (Molecular Probes #P36931). Images were then taken with a confocal microscopy.
Quantitative RT-PCR (qPCR)
mRNA was isolated using a TurboCapture mRNA kit (Qiagen) and reverse-transcribed with an RT kit (Applied Biosystems) according to the manufacturers’ protocols. Primers used for qPCR are indicated in Supplementary Table 1.
Subcellular fractionation
All procedures were performed on ice. Fresh or frozen muscle (~150 mg) was minsed with scissors and homogenized with a Dounce homogenizer (Corning, PYREX 2 ml, 30 strokes) in 1.3 mL homogenization buffer (0.1 M sucrose, 10 mM EDTA, 46 mM KCl, 5 mM NaN3, 100 mM Tris-HCl pH 7.4 and protease inhibitors). The samples were then centrifuged at 4,000 g for 10 min at 4 °C to precipitate nuclear fractions and cell debris. The supernatant (1 mL) was then collected and mixed with 0.33 mL Opti-Prep (Sigma D1556, 60% iodixanol) to make a 15% iodixanol solution. Then, 1.33 mL of 40% iodixanol gradient solution containing 0.1 M sucrose, 10 mM EDTA, 46 mM KCl, 5 mM NaN3, 10 mM Tris-HCl pH 7.4 and protease inhibitor, was added into a 4 mL ultracentrifuge tube and the sample was carefully added on top of the gradient solution. Then, 1.33 mL of 5% iodixanol gradient solution containing 0.1 M sucrose, 10 mM EDTA, 46 mM KCl, 5 mM NaN3, 10 mM Tris-HCl pH 7.4 and protease inhibitor, was carefully added on top of the sample. After ultracentifugation at 80,000 g for 16 hours at 4 °C, fractions (250 µL) were collected from the top. Then each fraction was mixed with 6X SDS sample loading buffer, boiled for 5 min at 95 °C, and loaded to 4–20% gradient SDS-PAGE gels for Western blot.
Chromatography
Silica was baked to remove moisture and mixed to n-BuOH:MeOH = 3:1 for packing. CMs were treated with charcoal, lyophilized and resuspended in the same solvent. The column was eluted with three 5 mL of n-BuOH:MeOH = 3:1, 1:1 mixture and 100% methanol. Each fraction was dried and resuspended in media for the activity test. For HILIC separation, active fractions were resuspended in-phase and injected to HPLC (Dionex LC20 with GP50 quad pump and AD25 single wavelength UV detector) equipped with an HILIC column (Agilent HILIC plus, 4.6 mm × 100 mm × 3.5 µm). Ammonium acetate buffer (10 mM adjusted to pH 4.0) in water/acetonitrile mix was used as eluent, initially at water:acetonitrile = 5:95 for 5 min, ramped to 55:45 over 8 min and held for 2 min. Thirty fractions were collected every 30 sec.
Tandem Mass spectrometry for structural identification
HP-HILIC (Waters BEH amide, 2.1 mm × 100 mm × 2.5 µm, equipped with HP1100 quad pump HPLC) coupled with tandem mass spectrometers (Thermo Scientific LTQ XL for structural identification and pathway analysis of 3-HIB, or Thermo Scientific Q Exactive orbitrap for high-sensitivity quantitation of in vitro and in vivo samples) was used. Ammonium formate buffer (2 mM adjusted at pH 9.0) in water/acetonitrile mix was used as eluent, initially at water:acetonitrile = 10:90 for 4 min, ramped to 40:60 over 3 min and held for 5 min for washing and reconditioned for 12 min. Selective ion monitoring (SIM)/parallel reaction monitoring (PRM) at 103.1 (hydroxybutyrate (HB) isomers), 106.1 (d3-2-HB standard) and 109.1 (d6-4-HB standard) were carried out for MS2 spectra acquisition. Quantification of each HB species was carried out with specific Q1→Q3 transition fingerprint (2-HB: 103→57, 3-HB:103→59, 3-HIB:103→73, 4-HB:103→85). 13C-2-Valine was purchased from Sigma (#604917). 13C2-1,2-Leucine was purchased from Cambridge Isotope Laboratories (#CLM-3524). d3-2HB was purchased from CDN Isotopes (#D-7002) and d6-4HB was purchased from Cerilliant (#G-006).
Lipidomics
Liquid chromatography-mass spectrometry (LC-MS) grade acetonitrile (ACN), isopropanol (IPA), methanol, and chloroform were purchased from Fisher Scientific. Diglyceride standard mix (d5-DAG, LM-6004) was purchased from Avanti Polar Lipids and leucine-enkephalin from Sigma. To extract lipids, vehicle or 3-HIB-fed mouse muscle samples were pulverized in liquid nitrogen and lipids were extracted from muscle powders (50 mg) with a methanol-chloroform (2:1) mixture (300 µL), lysed by mechanical disruption (Tissue laser from QIAGEN) with 25 pulses/sec for 2 min and sonicated for an additional 15 min. Chloroform and water were added (100 µL each) and vortexed. Organic and aqueous layers were separated by centrifugation for 7 min at 13,300 rpm at 4 °C. The d5-DAG internal standard mix was added to the organic layer and samples were dried under N2 gas. The dried lipid samples were re-dissolved in a mixture of 60% solvent A (40% H2O, 60% ACN, 10 mM Ammonium formate) and 40% solvent B (90% IPA, 10% ACN, 10 mM Ammonium formate). The samples were centrifuged at 13,300rpm for 5 min to remove fine particulates. The supernatant was transferred to a glass vial for ultra-performance liquid chromatography combined with qTOF Xevo G2S (Waters) detector for high through put LC-MS lipidomics. For UPLC QTOF MS based data acquisition for untargeted lipidomic profiling, each 10 µL sample was injected on to a reverse-phase column (XSELECTTM CSHTM C18, 2.1 mm × 100 mm × 2.5 µm) using an Aquity H-class UPLC system (Waters Corporation). Each sample was chromatographed for 9 min at a flow rate of 0.5 mL/min. The UPLC gradient consisted of 75% A and 25% B for 0.5 min, a quick ramp of 50% A and 50% B for 0.5 min, 25% A and 75% B for 4 min followed by a ramp of 10% A and 90% B for 2 min, and finally a ramp to 1% A and 99% B for 2 min. The column eluent was introduced directly into the mass spectrometer. MS was performed with a positive ion sensitive mode with a Capillary voltage of 3,000 V and Sampling Cone voltage of 40 °C. The desolvation gas flow was set to 800 L/hr and the temperature was set to 450 °C. The source temperature was set at 80 °C. Accurate mass was maintained by introduction of a lock-spray interface of leucine-enkephalin (556.2771 m/z) at a concentration of 0.5 ng/µL in 50 % aqueous acetonitrile and a rate of 5 µL/min. Data was acquired in centroid MSe mode from 50–1,200 m/z mass ranges for both MS (low energy) and MSe (high energy) modes. Low energy or fragmented data were collected without collision energy, while high energy or fragmented data were collected by using collision energy ramp from 15–40 eV. The entire set of triplicate sample injections was bracketed with a test mix of standard metabolites at the beginning and at the end of the run to evaluate instrument performance with respect to sensitivity and mass accuracy. The sample queue was randomized to remove bias. Lipid analysis and identifications were performed using Progenesis software (Waters Corporation). Endogenous DG (16:0/16:0)-m/z 551.50 Da, DG (16:0/18:0)-596.54 Da, DG (16:0/18:1)-577.51 Da, DG (18:0/18:0)-607.56 Da, DG (18:1/18:2)-601.51 Da, DG (18:1/18:3)-1234.07, DG (18:0/20:4)-689.51 Da fragmentation was confirmed by DDA and TOF-MS MS methods along with MSe.
Animal Models
All animal experiments were performed according to procedures approved by the University of Pennsylvania or Beth Israel Deaconess Medical Center (BIDMC) Institutional Animal Care and Use Committees. For 3-HIB feeding, 12-week-old C57BL/6 male mice (Jackson Lab) were fed with 3-HIB (Sigma 36105) in drinking water (300 mg/kg/day) for 2 weeks. Free FAs (Cayman), TAG (Cayman) and DAG (Mybiosource) kits were used according to manufacturer’s protocols. For glucose tolerance test, 8-week-old male mice were fasted for 16 hours, weighed, and a baseline blood glucose level was measured using a glucometer. Each mouse was intraperitoneally injected with sterilized 10% w/v D-glucose (10 µL/g body weight), then blood glucose was measured at each time point. Flt1flox/flox and Flk1flox/flox mice were kindly shared by Genentech. Hyperinsulinemic euglycemic clamp studies were performed as described previously24. Luciferase-transgenic mice were purchased from Jackson Lab (#008450). Luc-S-S-FA was purchased from INTRACE MEDICAL SA. To measure in vivo FA uptake, 10-week-old female Luciferase-TG or Luciferase/MCK-α double transgenic mice were fasted for 16 hours, then ad lib fed with food paste mixed with 30 mM 3-HIB in water for 1.5 hours. The mice were then anesthetized and intravenously injected with 100 µL warm PBS containing 20 µM Luc-S-S-FA and 10 µM FA-free BSA. Luminescence was measured immediately after injection of Luc-S-S-FA with Xenogen IVIS-50 Bioluminescence Imaging System at the BIDMC Small Animal Imaging Core. Images and quantifications were performed by personnel blinded to the treatment or genotypes. To measure vessel leakiness in vivo, mice were anesthetized and intravenously injected with 50 µL of 2% Evans blue in PBS. After 5 min, blood was collected and intracardiac perfusion was performed with 100 mL PBS. Skeletal muscle was then isolated, minced with scissors in PBS, homogenized with metal beads, centrifuged at 13,000 g for 10 min, and Evans blue in supernatants was quantified (absorbance at 611 nm) and normalized to total protein concentrations measured by BCA assay (Promega). To perform plasmid and siRNA electroporation into the tibialis anterior muscle, 8-week-old mice were anesthetized with isofuran, hair was then removed from legs, and a small incision was made on the skin to expose the muscle. Then, 25 µg of CMV-EGFP plasmid (Addgene) with 5 µg of control or Hibadh siRNA in 25 µL sterilized saline was slowly injected into the muscle with an insulin syringe. The skin incision was then sutured and electropersation was applied by touching the two pin electrodes around the injection site (Voltage, 50 V/cm; pulse duration, 100 ms; frequency of pulses, 1 Hz). A train of total 8 pulses (4 pulses first, then additional 4 pulses after switching electrode position) was delivered. The left leg was injected with control siRNA and the right leg was injected with Hibadh siRNA. After 9 days of recovery, the injected muscle was isolated and GFP-positive muscle fibers (5–10 mg) were collected to measure TAG and protein concentrations.
Human samples
The cohort of skeletal muscle biopsy samples was described previously.25 Human studies were approved by the University of Pennsylvania institutional review board (IRB). Informed consent was obtained from all subjects.
Reagents
Anti-Pan-Actin (#4968), Anti-ERK1/2 (#9102), anti-pERK1/2 Thr 202/Tyr204 (#9101), anti-AKT (#4685), anti-pAKT Ser473 (#9271 all from Cell Signaling), anti-FATP3 (Proteintech 12943-1-AP), anti-FATP4 (Abnova H00010999-M01), anti-PKC-θ (BD Transduction lab 610089), anti-Na,K ATPase A1 (Novus Biologicals NB300-146SS) and anti-HIBADH (Proteintech 13466-1-AP) antibodies were used for Western blot (1:1000 dilution). Anti-Occludin-1 (Abcam ab31721) and anti-Calnexin (Thermo MA3-27) antibodies were used for immunostaining (1:200 dilution). Mitotracker® Red CMXRos (Cell Signaling 9082S) was used for mitochondria staining. Sulfo-N-succinimidyl Oleate (sc-208408) was purchased from Santa Cruz. Akt VIII (#124018) was purchased from Calbiochem. CHC (#5029) was purchased from Tocris. Recombinant VEGFA (#293-VE-010) and VEGFB (#767-VE-010) were purchased from R&D systems. Human insulin (Humulin R U-100) was purchased from Harvard Drug Group (#821501). Membrane filters (MWCO 3 kDa #Z677094), Calcimycin (#C9275), 2,4-Dinitrophenol (#D198501), 2-deoxyglucose (#D8375), activated charcoal (#C4386) and other chemicals were purchased from Sigma unless otherwise stated.
Statistics
P-values were calculated using the two-tailed Student’s t-test. For statistical comparisons between study groups, two-way ANOVA was used followed by Bonferroni post hoc testing. P < 0.05 was considered statistically significant. Data are displayed as mean ± standard deviation or standard error (as indicated). All cell culture experiments included at least 3 biological replicates. All animal cohorts included at least 3 animals in each study group (as indicated). Animals were randomized to treatment groups.
Supplementary Material
Acknowledgments
Fatp4−/− and Cd36−/− mice were kindly provided by J. Miner (Washington University School of Medicine) and J. Lawler (Harvard Medical School), respectively. C.J. is supported by Lotte Scholarship and American Heart Association (AHA). S.F.O. is supported by Crohn’s and Colitis Foundation of America (Research Fellowship Award). S.W. is supported by Toyobo biotechnology foundation. G.C.R. is supported by US National Institute of Arthritis and Musculoskeletal and Skin Diseases (AR062128). J.R. is supported by US National Institute of Health 5 T32 GM7592-35. S.M.P. was supported by US National Heart, Lung, and Blood Institute (NHLBI) (HL093234, HL125275) and US National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) (DK095072). Q.C. and J.A.B. are supported by US NIDDK (DK098656, DK049210). Z.A. is supported by US NHLBI (HL094499), the AHA, and the Geis Realty Group Emerging Initiatives Fund and Dean and Ann Geis.
Footnotes
Author Contributions
C.J. led the studies and was directly involved in most experiments. S.F.O. assigned the structure of the paracrine factor as 3-HIB and performed mass spectrometric profiling. S.W., G.C.R., L.L., M.C.C, J.R., A.H., B.K., A.I., L.G.B., E.K., and A.J. assisted with experiments throughout, including qPCR, cell culture, and animal studies. Q.C. and J.A.B. performed the mouse clamps studies. S.K. and A.M.W. performed the lipidomics studies. D.E.F. and S.H.L. isolated the human muscle biopsies. C.C.G. and S.M.P. performed the TEER studies. J.D.R. performed the metabolic flux analysis. D.L.K. and Z.A. oversaw the studies. C.J. and Z.A. designed experiments, interpreted results and wrote the paper. All authors discussed the results and commented on the manuscript.
The authors have no competing interests as defined by Nature Publishing Group, or other interests that might be perceived to influence the results and/or discussion reported in this paper.
References
- 1.Wang TJ, et al. Metabolite profiles and the risk of developing diabetes. Nat. Med. 2011;17:448–453. doi: 10.1038/nm.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Newgard CB, et al. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab. 2009;9:311–326. doi: 10.1016/j.cmet.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Newgard CB. Interplay between lipids and branched-chain amino acids in development of insulin resistance. Cell Metab. 2012;15:606–614. doi: 10.1016/j.cmet.2012.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shulman GI. Ectopic fat in insulin resistance, dyslipidemia, and cardiometabolic disease. N. Engl. J. Med. 2014;371:1131–1141. doi: 10.1056/NEJMra1011035. [DOI] [PubMed] [Google Scholar]
- 5.Chan MC, Arany Z. The many roles of PGC-1alpha in muscle - recent developments. Metabolism. 2014;63:441–451. doi: 10.1016/j.metabol.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Handschin C, Spiegelman BM. Peroxisome proliferator-activated receptor gamma coactivator 1 coactivators, energy homeostasis, and metabolism. Endocr. Rev. 2006;27:728–735. doi: 10.1210/er.2006-0037. [DOI] [PubMed] [Google Scholar]
- 7.Arany Z, et al. HIF-independent regulation of VEGF and angiogenesis by the transcriptional coactivator PGC-1α. Nature. 2008;451:1008–1012. doi: 10.1038/nature06613. [DOI] [PubMed] [Google Scholar]
- 8.Hagberg CE, et al. Vascular endothelial growth factor B controls endothelial fatty acid uptake. Nature. 2010;464:917–921. doi: 10.1038/nature08945. [DOI] [PubMed] [Google Scholar]
- 9.Roberts LD, et al. beta-Aminoisobutyric acid induces browning of white fat and hepatic beta-oxidation and is inversely correlated with cardiometabolic risk factors. Cell Metab. 2014;19:96–108. doi: 10.1016/j.cmet.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin J, et al. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature. 2002;418:797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- 11.Arany Z, et al. The transcriptional coactivator PGC-1beta drives the formation of oxidative type IIX fibers in skeletal muscle. Cell Metab. 2007;5:35–46. doi: 10.1016/j.cmet.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 12.Hatazawa Y, et al. Metabolomic Analysis of the Skeletal Muscle of Mice Overexpressing PGC-1α. PLoS One. 2015;10:e0129084. doi: 10.1371/journal.pone.0129084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henkin AH, et al. Real-time noninvasive imaging of fatty acid uptake in vivo. ACS Chem. Biol. 2012;7:1884–1891. doi: 10.1021/cb300194b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choi CS, et al. Paradoxical effects of increased expression of PGC-1alpha on muscle mitochondrial function and insulin-stimulated muscle glucose metabolism. Proc. Natl. Acad. Sci. U S A. 2008;105:19926–19931. doi: 10.1073/pnas.0810339105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Avogaro A, Bier DM. Contribution of 3-hydroxyisobutyrate to the measurement of 3-hydroxybutyrate in human plasma: comparison of enzymatic and gas-liquid chromatography-mass spectrometry assays in normal and in diabetic subjects. J. Lipid Res. 1989;30:1811–1817. [PubMed] [Google Scholar]
- 16.Giesbertz P, et al. Metabolite profiling in plasma and tissues of ob/ob and db/db mice identifies novel markers of obesity and type 2 diabetes. Diabetologia. 2015;58:2133–2143. doi: 10.1007/s00125-015-3656-y. [DOI] [PubMed] [Google Scholar]
- 17.Mullen E, Ohlendieck K. Proteomic profiling of non-obese type 2 diabetic skeletal muscle. Int. J. Mol. Med. 2010;25:445–458. doi: 10.3892/ijmm_00000364. [DOI] [PubMed] [Google Scholar]
- 18.Sasaki M, et al. A severely brain-damaged case of 3-hydroxyisobutyric aciduria. Brain Dev. 2001;23:243–245. doi: 10.1016/s0387-7604(01)00196-6. [DOI] [PubMed] [Google Scholar]
References
- 19.Abbott NJ, Hughes CC, Revest PA, Greenwood J. Development and characterization of a rat brain capillary endothelial culture: towards an in vitro blood-brain barrier. J. Cell Sci. 1992;103:23–37. doi: 10.1242/jcs.103.1.23. [DOI] [PubMed] [Google Scholar]
- 20.Xie Z, et al. Vascular endothelial hyperpermeability induces the clinical symptoms of Clarkson disease (the systemic capillary leak syndrome) Blood. 2012;119:4321–4332. doi: 10.1182/blood-2011-08-375816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sawada N, Salomone S, Kim HH, Kwiatkowski DJ, Liao JK. Regulation of endothelial nitric oxide synthase and postnatal angiogenesis by Rac1. Circ. Res. 2008;103:360–368. doi: 10.1161/CIRCRESAHA.108.178897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rowe GC, et al. Disconnecting mitochondrial content from respiratory chain capacity in PGC-1-deficient skeletal muscle. Cell Rep. 2013;3:1449–1456. doi: 10.1016/j.celrep.2013.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Darland DC, D'Amore PA. TGF beta is required for the formation of capillary-like structures in three-dimensional cocultures of 10T1/2 and endothelial cells. Angiogenesis. 2001;4:11–20. doi: 10.1023/a:1016611824696. [DOI] [PubMed] [Google Scholar]
- 24.Titchenell PM, Chu Q, Monks BR, Birnbaum MJ. Hepatic insulin signalling is dispensable for suppression of glucose output by insulin in vivo. Nat Commun. 2015;6:7078. doi: 10.1038/ncomms8078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Forman DE, et al. Analysis of skeletal muscle gene expression patterns and the impact of functional capacity in patients with systolic heart failure. J. Card. Fail. 2014;20:422–430. doi: 10.1016/j.cardfail.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.