Esposito et al. employed live-imaging methods to visualize the Snail repressor. Snail target enhancers were attached to an MS2 reporter gene, permitting detection of nascent transcripts in living embryos. The transgenes exhibit initially broad patterns of transcription but are refined by repression in the mesoderm following mitosis.
Keywords: repression, mitosis, Drosophila embryo, transcription, live imaging
Abstract
Transcriptional repression is a pervasive feature of animal development. Here, we employ live-imaging methods to visualize the Snail repressor, which establishes the boundary between the presumptive mesoderm and neurogenic ectoderm of early Drosophila embryos. Snail target enhancers were attached to an MS2 reporter gene, permitting detection of nascent transcripts in living embryos. The transgenes exhibit initially broad patterns of transcription but are refined by repression in the mesoderm following mitosis. These observations reveal a correlation between mitotic silencing and Snail repression. We propose that mitosis and other inherent discontinuities in transcription boost the activities of sequence-specific repressors, such as Snail.
Transcriptional repression is essential for the patterning of the Drosophila embryo. Anterior–posterior patterning is initiated by the maternal Bicoid gradient, which produces sequential patterns of gap gene expression across the length of the early embryo (Driever and Nusslein-Volhard 1988, 1989; Ochoa-Espinosa et al. 2005). The encoded gap proteins function as sequence-specific transcriptional repressors that subdivide the embryo into head, thoracic, and abdominal territories (Nusslein-Volhard and Wieschaus 1980; Fowlkes et al. 2008; Surkova et al. 2008). They also delineate the borders of pair-rule stripes of gene expression underlying segmentation (Hiromi and Gehring 1987; Small et al. 1991; Tsai and Gergen 1994). Similarly, the maternal Dorsal gradient leads to localized expression of different transcriptional repressors across the dorsal–ventral axis of the early embryo (Ray et al. 1991; Casal and Leptin 1996; Stathopoulos et al. 2002; Stathopoulos and Levine 2005), including snail (sna) (Ashraf and Ip 2001; Hemavathy et al. 2004) and brinker (brk) (Jazwinska et al. 1999; Zhang et al. 2001), which establish the limits of the mesoderm and ventral ectoderm, respectively (Kosman et al. 2004; Markstein et al. 2004).
The mechanisms underlying transcriptional repression and hence the delineation of developmental boundaries have been studied extensively. Different mechanisms have been documented, including competition between DNA-binding proteins, “quenching” of activators bound to adjacent sites within target enhancers, or direct repression of the promoter (Levine and Manley 1989; Gray et al. 1994; Gray and Levine 1996a,b). Newly developed live-imaging methods permit the first opportunity to explore the temporal dynamics of these repression mechanisms in living embryos (Garcia et al. 2013; Lucas et al. 2013).
We examined the repression of two different tissue-specific enhancers: the 5′ brk enhancer and the intronic short-gastrulation (sog) enhancer. Both enhancers are activated by the maternal Dorsal gradient in ventral (presumptive mesoderm) and lateral (presumptive neurogenic ectoderm) regions of precellular embryos (Markstein et al. 2002; Stathopoulos et al. 2002). The Snail repressor binds to specific sites within each enhancer to exclude their activities within the mesoderm. We visualized the formation of this developmental boundary between mesoderm and neurogenic ectoderm in living embryos using the MS2/MCP detection method (e.g., see Bothma et al. 2014).
brk and sog transgenes mediate broad patterns of expression in the presumptive mesoderm and lateral ectoderm of precellular embryos. Nascent transcripts are lost during the general transcriptional silencing that occurs at mitosis. Strikingly, brk expression is virtually excluded from the mesoderm, while sog expression is significantly reduced upon reactivation of the transgenes during the final interphase prior to gastrulation. Mutant embryos expressing increased levels of the Snail repressor or undergoing protracted periods of mitosis exhibit more complete repression of sog upon reactivation in the final cell cycle. These observations suggest that the cell cycle is coupled to developmental patterning and raise the possibility that mitotic silencing somehow facilities the activities of Snail and other sequence-specific repressors.
Results and Discussion
The enhancers tested in this study were derived from two different dorsal–ventral patterning genes: brk and sog; both encode inhibitors of BMP signaling (Ashe and Levine 1999; Bray 1999; Campbell and Tomlinson 1999; Jazwinska et al. 1999). The brk enhancer is located ∼10 kb upstream of the transcription start site, while the sog enhancer is located within the first intron of the transcription unit, ∼1.5 kb downstream from the start site (Supplemental Fig. S1A). Each enhancer was placed immediately upstream of its cognate promoter and attached to a yellow reporter gene containing 24 MS2 stem–loops within the 5′ untranslated region (UTR). Nascent transcripts were visualized in living embryos using a maternally expressed MCP-GFP fusion protein (Supplemental Fig. S1B; see Garcia et al. 2013).
Both transgenes recapitulate the expression profiles of the endogenous genes; namely, they are activated throughout the presumptive mesoderm and neurogenic ectoderm and then repressed in the mesoderm. Prior studies with fixed embryos suggest that these enhancers respond to different levels of the Snail repressor (e.g., see Bothma et al. 2011). The brk 5′ enhancer appears to be more efficiently repressed by Snail as compared with the sog intronic enhancer. We examined both transgenes in living embryos to determine whether they exhibit distinctive repression dynamics.
brk repression dynamics is correlated with mitotic silencing
The brk>MS2 transgene exhibits an expression profile that is similar to that seen for the endogenous locus based on conventional in situ hybridization methods (Supplemental. Fig. S1C). The main difference is that the transgene produces a slightly narrower pattern due to the absence of the 3′ “shadow” enhancer (Perry et al. 2010; Dunipace et al. 2013).
There is broad activation of the brk>MS2 transgene in both ventral and lateral regions during nuclear cleavage cycles 10–13 (nc10–nc13) (see Supplemental Movie S1). brk>MS2 nascent transcripts are lost during the general silencing of transcription at each mitosis. Interestingly, upon reactivation of the transgene at the onset of nc14, we observed a sudden loss of de novo transcription in the mesoderm (Fig. 1A,B). Transcripts are restricted to the neurogenic ectoderm, suggesting that the mature brk expression pattern is established immediately following mitosis.
Figure 1.
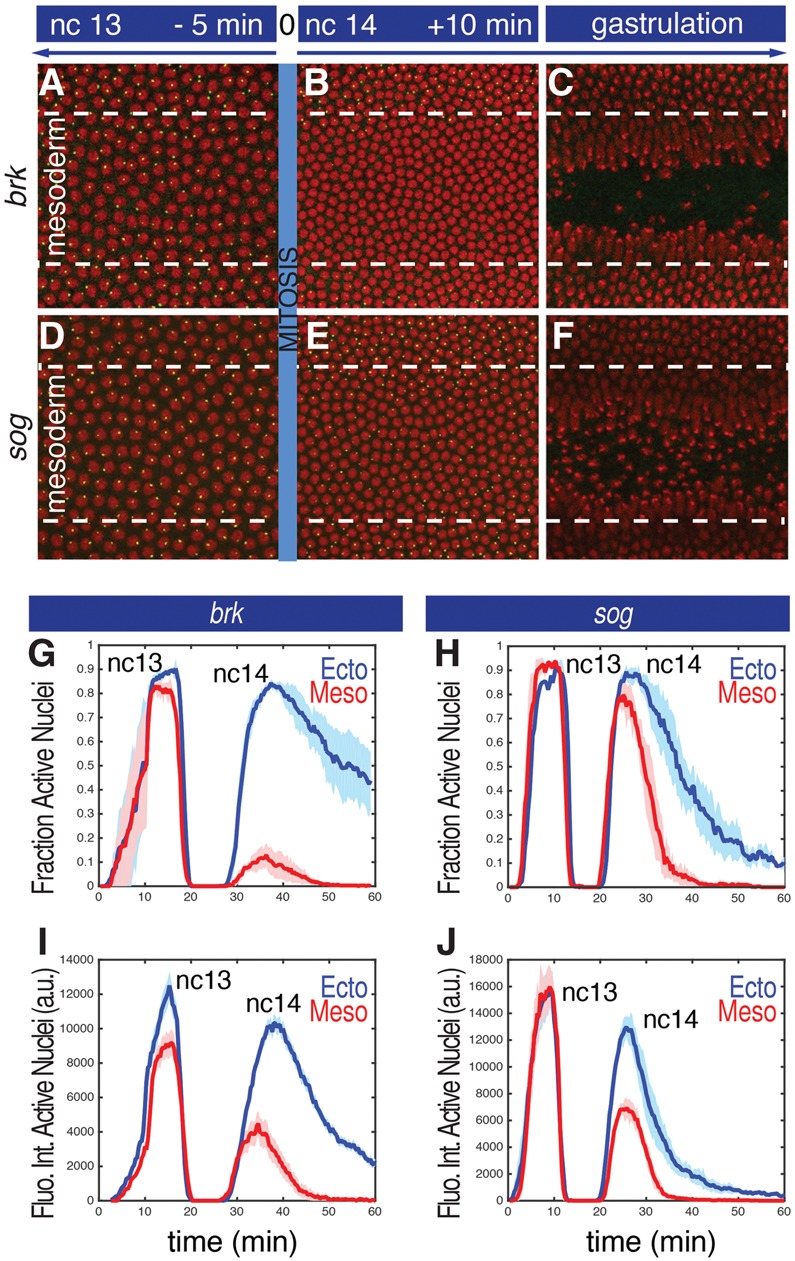
Visualization of transcriptional repression in the mesoderm. (A–F) Projected confocal stacks of a live embryo at three different time points during nc13 and nc14 expressing brk>MS2 (A–C) or sog>MS2 (D–F). Green dots correspond to nascent transcripts visualized with an MCP::GFP fusion protein. Histone-RFP was used to visualize the nuclei (red). The presumptive mesoderm encompasses the nuclei included between dashed lines and corresponds to the region that invaginates at the onset of gastrulation. (G,H) The fraction of nuclei that display nascent RNA signals during a 1-h interval encompassing nc13, mitosis, and the first half of nc14. (Blue) Ectoderm nuclei; (red) mesoderm nuclei. Mean values from three biological replicate embryos are represented by the continuous line. The shaded area corresponds to the standard error of the mean (SEM) of three biological replicates. (I–J) Mean fluorescence intensities in active nuclei. (Blue) Ectoderm; (red) mesoderm. The shaded areas correspond to the SEM of three biological replicate embryos. The times above each panel in A–C are scaled to the general silencing that occurs at mitosis (time = 0).
In an effort to quantify the dynamics of this repression, we partitioned individual nuclei within the presumptive mesoderm and ectoderm (see the Materials and Methods; Supplemental Fig. S2) and calculated the fraction of active nuclei in these regions throughout nc13–nc14. Active nuclei are defined as those exhibiting nascent RNA signals in at least one z-series (20 sec). We did not observe any significant variation in the fraction of active nuclei between the mesoderm and lateral ectoderm during nc13 (Fig. 1G). However, at the onset of nc14, ∼90% of the nuclei in the mesoderm are silenced, while expression persists in the lateral ectoderm (Fig. 1G).
To obtain more detailed information on the dynamics of repression in the ventral mesoderm, we quantified the fluorescence of individual transcription foci, since previous studies have shown that it scales with the number of RNA polymerase II (Pol II) complexes engaged in active transcription (Garcia et al. 2013; Bothma et al. 2014). Quantitative analysis of the brk>MS2 transgene reveals that the ventral-most nuclei in the mesoderm exhibit ∼25% reduction in signal intensity during nc13 as compared with active nuclei in the lateral ectoderm (Fig. 1I, nc13). The majority of nuclei that display nascent transcripts at the onset of nc14 is located in the ectoderm, and the few active nuclei in the mesoderm show a substantial reduction in signal intensity (∼60% reduction) (Fig. 1I, nc14). Thus, Snail begins to attenuate the brk 5′ enhancer during nc13, and this repression appears to be strongly reinforced during mitosis. We suggest that mitotic silencing augments the activity of the localized Snail repressor (see below).
sog expression is attenuated following mitosis
sog is regulated by two enhancers with overlapping activities: a distal enhancer located ∼20 kb upstream of the sog promoter and an intronic enhancer located ∼1.5 kb downstream from the transcription start site (Hong et al. 2008; Perry et al. 2010). The distal 5′ enhancer contains high-affinity Snail-binding sites and exhibits repression dynamics similar to that of the brk>MS2 transgene following mitosis (Supplemental. Fig. S3; Supplemental Movie S2). The intronic enhancer contains weak Snail-binding sites and shows modest repression in the ventral mesoderm of wild-type embryos (Fig. 1D–F; Supplemental Movie S3). Most nuclei are reactivated following mitotic silencing (Fig. 1D–F,H) but display a significant reduction (more than twofold) in expression (Fig. 1J).
To explore the dynamics of sog repression, we examined embryos carrying three copies of the snail locus. They exhibit significantly more complete repression of the sog>MS2 transgene during the onset of nc14 as compared with wild-type embryos (cf. Figs. 2A–C and 1D–F; Supplemental Movie S4). Fewer than half the nuclei in the mesoderm reactivate sog>MS2 expression at the onset of nc14 (Fig. 2J).
Figure 2.
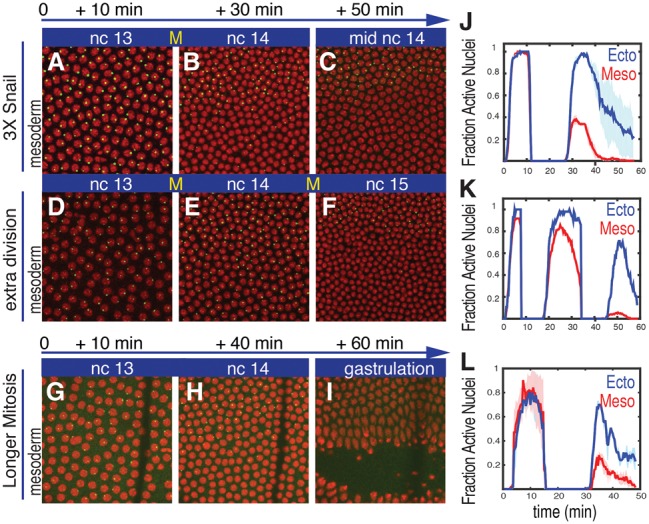
Increasing doses of Snail augments repression of sog>MS2. Projected confocal stacks of live embryos at three different time points during nc13 and nc14 expressing sog>MS2 and visualized with a MCP::GFP fusion protein (green). (A–C) Normal embryo containing three copies of snail due to an ∼25-kb snail BAC transgene. This embryo exhibits more immediate repression of sog>MS2 at the onset of nc14 as compared with the wild-type control (cf. B and Fig. 1E). (D–F) An embryo exhibiting an additional nuclear cycle. There is a substantial loss of sog>MS2 expression in the mesoderm at the onset of the final cell cycle (nc15). (G–I) A wild-type embryo where mitosis occurs at 17°C rather than room temperature. The line near the right of each panel corresponds to a glue line used to immobilize the embryo within the microfluidic chamber. Histone-RFP was used to visualize the nuclei (red). The times indicated above each panel are scaled to the general silencing that occurs at mitosis. (J–L) The fraction of nuclei that displays nascent transcripts in embryos carrying three copies of snail (J), undergoing an extra nuclear division (K), or undergoing extended mitosis (L). The shaded area in J and L corresponds to the SEM of two biological replicate embryos.
The preceding results suggest a clear correlation between mitotic silencing and repression of the brk>MS2 transgene and attenuation of sog>MS2. There is an approximately twofold reduction in the levels of sog>MS2 expression in the mesoderm following mitotic silencing in wild-type embryos and a substantial reduction in the number of nuclei that reactivate the transgene at the onset of nc14 in embryos containing three copies of snail. Nonetheless, it is not clear whether the ∼8-min interval during mitosis is a more effective period of Snail-mediated repression than a comparable interphase period.
Additional support stems from the analysis of a rare haploid embryo (Fig. 2D–F). Following fertilization, the paternal and maternal haploid pronuclei sometimes fail to fuse, and development proceeds with successive divisions of the maternal pronucleus. Due to the reduced amount of DNA, these haploid nuclei undergo an additional, 14th mitotic division (Edgar et al. 1986). As expected, the resulting nc15 nuclei are half the volume of normal nc14 diploid nuclei due to the extra division cycle (Fig. 2, cf. F and C; Supplemental Movie S5). Strikingly, the sog>MS2 transgene exhibits a dramatic loss of expression in mesoderm nuclei at the onset of nc15 (Fig. 2, cf. F and E). There is only a modest loss in the number of nuclei that exhibit repression in the mesoderm during nc14 (∼10%). This is followed by an ∼10-fold reduction in the number of nuclei that reactivate the transgene at the onset of nc15 (Fig. 2K). This loss in expression following mitosis is similar to that seen for the brk>MS2 transgene in wild-type embryos (e.g., see Fig. 1B). Unfortunately, given the scarcity of spontaneous haploid embryos, we were able to perform these measurements on only one embryo.
These observations reinforce the correlation between mitosis and repression. Additional evidence was obtained by extending the normal period of mitosis by lowering the temperature of developing embryos. A temperature-controlled microfluidic chamber was used to produce a transient reduction in temperature—from 22°C to 17oC—during the 13th mitosis while maintaining the temperature at 22°C during interphases 13 and 14. This treatment diminishes the rate of embryonic development and extends the time of mitosis from ∼8 to ∼15 min (see the Materials and Methods). There is a more pervasive loss of expression in the mesoderm upon reactivation of the sog>MS2 transgene at the onset of nc14 (Fig. 2G–I; Supplemental Movie S6). Less than half the mesoderm nuclei exhibit expression of the sog>MS2 transgene upon reactivation at the onset of nc14 (Fig. 2L).
We do not believe that the correlation between mitosis and repression is a peculiarity of the Snail repressor. Mitotic silencing might influence gap repressors such as Krüppel, since there is a substantial refinement in the eve stripe-2 expression pattern at the onset of nc14 (see Bothma et al. 2014).
Allele-by-allele repression
The preceding analyses employed heterozygous embryos carrying a single copy of the brk or sog transgenes. We next examined homozygous embryos to determine whether the two alleles of a locus display coordinated or uncoupled patterns of repression. The brk>MS2 transgene exhibits a sharp transition from active to inactive mesoderm nuclei following mitosis (see Supplemental Fig. S4A–C,F–G; Supplemental Fig. S5; Supplemental Movie S7). We therefore focused our analysis on the regulation of sog>MS2 homozygotes (Fig. 3).
Figure 3.
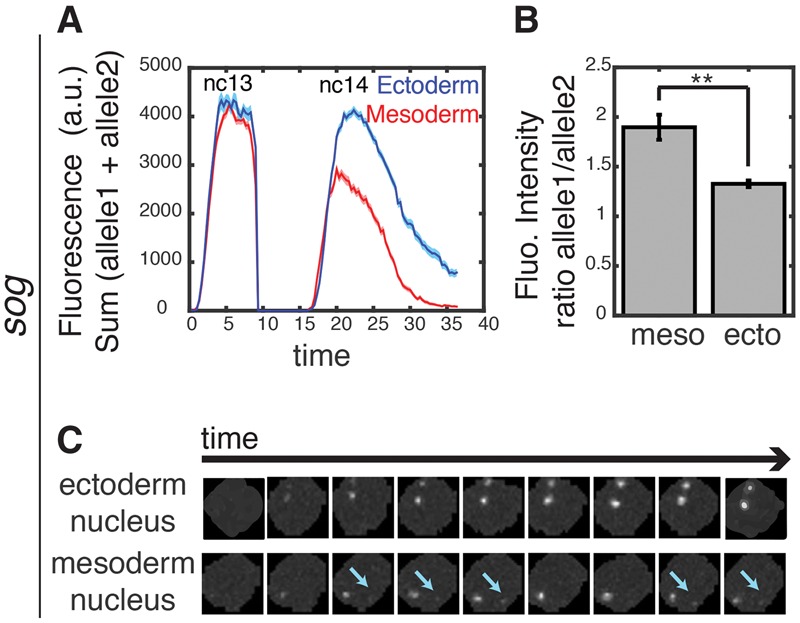
Asymmetric repression of sog>MS2. Mean fluorescence intensities in active nuclei (sum of allele 1 and allele 2 intensity, where allele 1 is defined as the one with the higher expression level). (Blue) Ectoderm; (red) mesoderm. The shaded areas correspond to the SEM of two biological replicates. (B) The sog>MS2 transgene exhibits the asymmetric activities of homologous chromosomes. The ratio of transcriptional activity between the two alleles in ∼30 nuclei in the mesoderm and ∼30 nuclei in the ectoderm. (**) P-value < 0.02. Allele 1 is defined as the one with the higher expression level, and allele 2 is an allele with a lower expression level. (C) Raw snapshots of a nucleus with two active alleles in the ectoderm and mesoderm for the sog>MS2 transgene. Cyan arrows indicate the second (weak) allele in the nucleus.
There is no detectable repression of either sog>MS2 allele during nc13 (Supplemental Figs. S4D,H,S6A; Supplemental Movie S8). At the onset of nc14, most of the daughter nuclei reactivate expression following mitotic silencing (Supplemental Figs. S4E,I,S6B) but produce fewer transcripts than the nuclei in the ectoderm (Fig. 3A). Moreover, mesoderm nuclei exhibit asymmetric reactivation of the two alleles (Fig. 3B,C). One of the alleles exhibits expression that is comparable with each allele in the ectoderm, whereas the other allele is either silent or exhibits very weak and transient expression (Fig. 3C). These observations suggest that the diminished levels of expression that are observed for sog>MS2 at the onset of nc14 are mainly due to the repression of one of the alleles. Thus, Snail represses sog expression in the mesoderm one allele at a time. Instead of diminishing the levels of both alleles, there is a clear trend to silence one of the alleles (Supplemental Fig. S4I). The basis for this “digital” mode of repression is uncertain, but it is possible that it reflects the exact time when each homolog is silenced and then decondensed following mitosis.
Snail exploits intrinsic discontinuities in transcription
Here we present evidence that transcriptional repression is intimately linked to the cell cycle and propose that Snail exploits the general silencing of transcription that occurs during mitosis. Mitotic silencing offers an opportunity to reset the balance between transcriptional activators and repressors. It is possible that Snail outcompetes the Dorsal activator during mitosis. Indeed, immunohistochemical localization assays suggest that the Snail repressor remains associated with the apical cytoplasm during mitosis (Supplemental Fig. S7A–C), whereas Dorsal becomes distributed throughout the cytoplasm (Supplemental Fig. S7D–F). This might give Snail “the jump” on Dorsal after the completion of mitosis. It is also possible that the balance between the Dorsal activator and the Snail repressor is influenced by “titration” of Dorsal due to increases in chromosomal templates arising from replication.
We propose that Snail and other developmental repressors exploit natural discontinuities in transcription (summarized in Fig. 4). In addition to mitotic silencing, many genes exhibit transcriptional bursts (e.g., see Bothma et al. 2014; Fukaya et al. 2016). It is possible that repressors like Snail get the upper hand during the refractory periods between bursts. Indeed, we observed inhibition between successive bursts of sog>MS2 expression in the mesoderm following mitotic silencing. Approximately 80% of mesoderm nuclei exhibit a single burst of expression before falling silent (Fig. 4D), and we observed a similar trend in the repression of the brk>MS2 transgene (Supplemental. Fig. S8). The Snail repressor may be more effective in maintaining the off state following a burst (or mitotic silencing) than inhibiting a gene at the peak of its activity. Mitotic silencing and transcriptional bursting might represent intrinsic mechanisms that foster dynamic repression of gene expression during development.
Figure 4.
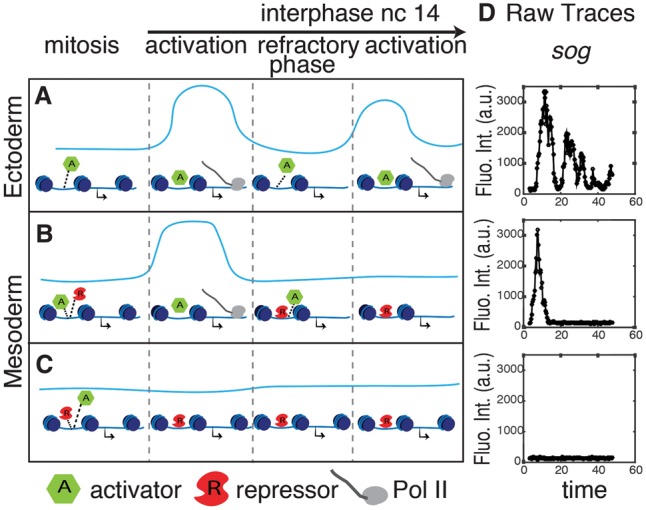
Snail exploits intrinsic pauses in transcription. Blue lines represent successive transcriptional bursts (levels, Y-axis; time, X-axis). Successive bursts are interrupted by mitotic silencing (“mitosis” above the diagram) or inherent refractory phases between bursts. (A) The Snail repressor is absent in the ectoderm, and there are successive bursts during nc14 following mitotic silencing. (B,C) In the mesoderm, the Snail repressor either prevents reactivation of transcription at the onset of nc14 (C) or inhibits successive bursts during nc14 (B). (D) Raw sog>MS2 fluorescence intensity traces for individual nuclei in the ectoderm (top panel), near the mesoderm/ectoderm boundary (middle panel), or within the mesoderm (bottom panel).
Materials and methods
Cloning and transgenesis
The plasmids containing MS2 transgenes were generated using pbPHi, which contains the yellow reporter gene (Venken et al. 2006; Perry et al. 2010). We amplified the enhancer (sog distal, sog intronic, and brk 5′) and promoter (sog and brk) regions from Drosophila genomic DNA using primers listed in Supplemental Table 1. The enhancer was inserted into the pbPHi multiple cloning site using the NotI–XhoI restriction enzyme, while the promoter was inserted using the XhoI–BamHI sites. Twenty-four copies of the MS2 stem–loops were enzymatically released from the 24XMS2SL stable vector (Addgene, 31865) by digestion with BamHI and BglII restriction enzymes. It was subsequently inserted downstream from the promoter in the pbPHi vector containing an enhancer, promoter, and yellow reporter gene after being linearized with BamHI. All MS2 transgenes were integrated into the VK33 site of chromosome 3.
FISH and immunostaining
Two-hour-old to 4-h-old yellow;white (yw) embryos were fixed as previously described (Kosman et al. 2004; Bothma et al. 2011). Hapten-tagged RNA probes were used for hybridization, including the first intron of yellow, brk full-length cDNA, and sog intronic sequence. Hybridized embryos were imaged with a Zeiss700 laser-scanning microscope in z stacks through the nuclear layer at 0.5-μm intervals using a Plan-Apochromat 20×/0.8 air lens. Immunostained embryos were imaged on a Zeiss700 laser-scanning microscope in z-stacks through the nuclear layer at 0.5-μm intervals using a Plan-Apochromat 63×/1.4 air lens. The following primary antibodies were used for detection: sheep anti-digoxigenin and mouse anti-biotin (Roche Applied Sciences, Invitrogen). Both were labeled with Alexa dyes using Alexa fluor 555 donkey anti-sheep and Alexa fluor 488 donkey anti-mouse secondary antibodies (Invitrogen). Nuclei were stained with DAPI (Invitrogen). We used a mouse anti-Dorsal (Developmental Studies Hybridoma Bank, 74A) antibody and a guinea-pig anti-Snail antibody (kindly provided by Dr. Wieschaus, Princeton University).
Live-imaging sample preparation and data acquisition
Virgin yw; Histone-RFP; MCP-NoNLS-GFP (Garcia et al. 2013) females were mated with homozygous males carrying brk > MS2, sogIntronic > MS2, or sogDistal > MS2 transgenes on chromosome 3. Embryos were collected and mounted as described in Bothma et al. (2014)
Embryos were imaged on a Zeiss LSM 780 confocal microscope using a Plan-Apochromat 40×/1.4 N.A oil immersion objective. At each time point, a stack of 21 images separated by 0.5 µm was acquired with a final time resolution of 20 sec. Images were taken at 512 × 512 pixels with a pinhole set to 115-µm diameter using bidirectional laser scanning.
Fly genetics
Increases in snail copy number were obtained using a fly strain homozygous for a BAC transgene (25 kb) located on chromosome 3 (Lagha et al. 2013). Males of this strain were mated with yw; Histone-RFP; MCP-NoNLS-GFP virgin females to obtain yw; Histone-RFP/+;MCP-NoNLS-GFP/snaBAC. Virgin females were collected and mated with transgenic males carrying sogIntronic Enhancer > MS2, and embryos were imaged as described above. The progeny with the additional copy of the snail gene (50%) were retrospectively identified after imaging by PCR.
Live-imaging data analysis
Analysis of live images (mean intensity fluorescence of active nuclei and fraction of active nuclei) was performed as described by Garcia et al. (2013) and Bothma et al. (2014). The mesodermal and ectodermal regions were calculated using a heuristic algorithm that calculates the relative distance between nuclei in nc14 during gastrulation. In this way, we were able to identify nuclei that are fated to undergo ventral furrow formation.
Lineage tracking
A custom-made Matlab code was implemented to trace a nucleus splitting into two daughter nuclei during mitosis and assign these daughter nuclei to the lineage of the mother nucleus. The lineage was manually corrected.
Analysis of homozygous embryos
A custom Matlab code was implemented to record the fluorescent intensity of the MS2 loci in a given nucleus. Each nucleus was segmented, and the maximum projected MS2 channel (488-nm laser excitation) was converted into a binary image in a threshold-dependent manner. Each MS2 locus from a single nucleus was traced over time, and the fluorescent intensity of each locus was recorded. Nuclei with ambiguous separation of the two signals were excluded in this analysis. To calculate the ratio between fluorescent intensity of two alleles, we defined “allele 1” as the one that shows a higher expression level and “allele 2” as the one showing a lower expression level from a given nucleus.
Microfluidics-mediated lengthening of mitosis
A microfluidics device was fabricated as described in Lucchetta et al. (2005) with slight modifications to allow for imaging with a 40× water objective. Due to the short working distance of this objective, the embryos were mounted on a 48 × 60-mm cover glass using heptane glue. The cover glass was coated with a thin layer of PDMS to reduce the heat transfer with the environment. The microfluidics channel was placed on top of this cover glass. Using this device, the embryo was exposed to a flow of 17°C during nc13/nc14 mitosis. The temperature of the flow was measured with a built-in thermometer placed 5 mm upstream of the embryo.
Supplementary Material
Acknowledgments
We thank Emma Farley, Yuji Yamazaki, Mounia Lagha, Sam Ng, Jacques Bothma, Avaneesh Narla, and members of the Levine laboratory for helpful comments and discussions. We also thank Thomas Gregor and Eric Wieschaus for helpful comments and reagents, and Evangelos Gatzogiannis for help at the microscope facility. This work was funded by a grant from the National Institutes of Health (GM46638).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.281188.116.
References
- Ashe HL, Levine M. 1999. Local inhibition and long-range enhancement of Dpp signal transduction by Sog. Nature 398: 427–431. [DOI] [PubMed] [Google Scholar]
- Ashraf SI, Ip YT. 2001. The Snail protein family regulates neuroblast expression of inscuteable and string, genes involved in asymmetry and cell division in Drosophila. Development 128: 4757–4767. [DOI] [PubMed] [Google Scholar]
- Bothma JP, Magliocco J, Levine M. 2011. The snail repressor inhibits release, not elongation, of paused Pol II in the Drosophila embryo. Curr Biol 21: 1571–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bothma JP, Garcia HG, Esposito E, Schlissel G, Gregor T, Levine M. 2014. Dynamic regulation of eve stripe 2 expression reveals transcriptional bursts in living Drosophila embryos. Proc Natl Acad Sci 111: 10598–10603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray S. 1999. DPP on the brinker. Trends Genet 15: 140. [DOI] [PubMed] [Google Scholar]
- Campbell G, Tomlinson A. 1999. Transducing the Dpp morphogen gradient in the wing of Drosophila: regulation of Dpp targets by brinker. Cell 96: 553–562. [DOI] [PubMed] [Google Scholar]
- Casal J, Leptin M. 1996. Identification of novel genes in Drosophila reveals the complex regulation of early gene activity in the mesoderm. Proc Natl Acad Sci 93: 10327–10332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driever W, Nusslein-Volhard C. 1988. A gradient of bicoid protein in Drosophila embryos. Cell 54: 83–93. [DOI] [PubMed] [Google Scholar]
- Driever W, Nusslein-Volhard C. 1989. The bicoid protein is a positive regulator of hunchback transcription in the early Drosophila embryo. Nature 337: 138–143. [DOI] [PubMed] [Google Scholar]
- Dunipace L, Saunders A, Ashe HL, Stathopoulos A. 2013. Autoregulatory feedback controls sequential action of cis-regulatory modules at the brinker locus. Dev Cell 26: 536–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar BA, Kiehle CP, Schubiger G. 1986. Cell cycle control by the nucleo-cytoplasmic ratio in early Drosophila development. Cell 44: 365–372. [DOI] [PubMed] [Google Scholar]
- Fowlkes CC, Hendriks CL, Keranen SV, Weber GH, Rubel O, Huang MY, Chatoor S, DePace AH, Simirenko L, Henriquez C, et al. 2008. A quantitative spatiotemporal atlas of gene expression in the Drosophila blastoderm. Cell 133: 364–374. [DOI] [PubMed] [Google Scholar]
- Fukaya T, Lim B, Levine M. 2016. Enhancer control of transcriptional bursting. Cell 166: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia HG, Tikhonov M, Lin A, Gregor T. 2013. Quantitative imaging of transcription in living Drosophila embryos links polymerase activity to patterning. Curr Biol 23: 2140–2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray S, Levine M. 1996a. Short-range transcriptional repressors mediate both quenching and direct repression within complex loci in Drosophila. Genes Dev 10: 700–710. [DOI] [PubMed] [Google Scholar]
- Gray S, Levine M. 1996b. Transcriptional repression in development. Curr Opin Cell Biol 8: 358–364. [DOI] [PubMed] [Google Scholar]
- Gray S, Szymanski P, Levine M. 1994. Short-range repression permits multiple enhancers to function autonomously within a complex promoter. Genes Dev 8: 1829–1838. [DOI] [PubMed] [Google Scholar]
- Hemavathy K, Hu X, Ashraf SI, Small SJ, Ip YT. 2004. The repressor function of snail is required for Drosophila gastrulation and is not replaceable by Escargot or Worniu. Dev Biol 269: 411–420. [DOI] [PubMed] [Google Scholar]
- Hiromi Y, Gehring WJ. 1987. Regulation and function of the Drosophila segmentation gene fushi tarazu. Cell 50: 963–974. [DOI] [PubMed] [Google Scholar]
- Hong JW, Hendrix DA, Levine MS. 2008. Shadow enhancers as a source of evolutionary novelty. Science 321: 1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazwinska A, Kirov N, Wieschaus E, Roth S, Rushlow C. 1999. The Drosophila gene brinker reveals a novel mechanism of Dpp target gene regulation. Cell 96: 563–573. [DOI] [PubMed] [Google Scholar]
- Kosman D, Mizutani CM, Lemons D, Cox WG, McGinnis W, Bier E. 2004. Multiplex detection of RNA expression in Drosophila embryos. Science 305: 846. [DOI] [PubMed] [Google Scholar]
- Lagha M, Bothma JP, Esposito E, Ng S, Stefanik L, Tsui C, Johnston J, Chen K, Gilmour DS, Zeitlinger J, et al. 2013. Paused Pol II coordinates tissue morphogenesis in the Drosophila embryo. Cell 153: 976–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M, Manley JL. 1989. Transcriptional repression of eukaryotic promoters. Cell 59: 405–408. [DOI] [PubMed] [Google Scholar]
- Lucas T, Ferraro T, Roelens B, De Las Heras Chanes J, Walczak AM, Coppey M, Dostatni N. 2013. Live imaging of bicoid-dependent transcription in Drosophila embryos. Curr Biol 23: 2135–2139. [DOI] [PubMed] [Google Scholar]
- Lucchetta EM, Lee JH, Fu LA, Patel NH, Ismagilov RF. 2005. Dynamics of Drosophila embryonic patterning network perturbed in space and time using microfluidics. Nature 434: 1134–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markstein M, Markstein P, Markstein V, Levine MS. 2002. Genome-wide analysis of clustered Dorsal binding sites identifies putative target genes in the Drosophila embryo. Proc Natl Acad Sci 99: 763–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markstein M, Zinzen R, Markstein P, Yee KP, Erives A, Stathopoulos A, Levine M. 2004. A regulatory code for neurogenic gene expression in the Drosophila embryo. Development 131: 2387–2394. [DOI] [PubMed] [Google Scholar]
- Nusslein-Volhard C, Wieschaus E. 1980. Mutations affecting segment number and polarity in Drosophila. Nature 287: 795–801. [DOI] [PubMed] [Google Scholar]
- Ochoa-Espinosa A, Yucel G, Kaplan L, Pare A, Pura N, Oberstein A, Papatsenko D, Small S. 2005. The role of binding site cluster strength in Bicoid-dependent patterning in Drosophila. Proc Natl Acad Sci 102: 4960–4965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry MW, Boettiger AN, Bothma JP, Levine M. 2010. Shadow enhancers foster robustness of Drosophila gastrulation. Curr Biol 20: 1562–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray RP, Arora K, Nusslein-Volhard C, Gelbart WM. 1991. The control of cell fate along the dorsal-ventral axis of the Drosophila embryo. Development 113: 35–54. [DOI] [PubMed] [Google Scholar]
- Small S, Kraut R, Hoey T, Warrior R, Levine M. 1991. Transcriptional regulation of a pair-rule stripe in Drosophila. Genes Dev 5: 827–839. [DOI] [PubMed] [Google Scholar]
- Stathopoulos A, Levine M. 2005. Genomic regulatory networks and animal development. Dev Cell 9: 449–462. [DOI] [PubMed] [Google Scholar]
- Stathopoulos A, Van Drenth M, Erives A, Markstein M, Levine M. 2002. Whole-genome analysis of dorsal-ventral patterning in the Drosophila embryo. Cell 111: 687–701. [DOI] [PubMed] [Google Scholar]
- Surkova S, Kosman D, Kozlov K, Manu, Myasnikova E, Samsonova AA, Spirov A, Vanario-Alonso CE, Samsonova M, Reinitz J. 2008. Characterization of the Drosophila segment determination morphome. Dev Biol 313: 844–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai C, Gergen JP. 1994. Gap gene properties of the pair-rule gene runt during Drosophila segmentation. Development 120: 1671–1683. [DOI] [PubMed] [Google Scholar]
- Venken KJ, He Y, Hoskins RA, Bellen HJ. 2006. P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science 314: 1747–1751. [DOI] [PubMed] [Google Scholar]
- Zhang H, Levine M, Ashe HL. 2001. Brinker is a sequence-specific transcriptional repressor in the Drosophila embryo. Genes Dev 15: 261–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


