Procko et al. blocked the transcriptional response to auxin within specific tissues to show that auxin signaling is required in many cell types for correct hypocotyl growth in shade, with a key role for the epidermis. Auxin acts in the epidermis in part by inducing activity of the locally acting, growth-promoting brassinosteroid pathway.
Keywords: auxin, brassinosteroid, epidermis, shade avoidance, stem growth
Abstract
Growth of a complex multicellular organism requires coordinated changes in diverse cell types. These cellular changes generate organs of the correct size, shape, and functionality. In plants, the growth hormone auxin induces stem elongation in response to shade; however, which cell types of the stem perceive the auxin signal and contribute to organ growth is poorly understood. Here, we blocked the transcriptional response to auxin within specific tissues to show that auxin signaling is required in many cell types for correct hypocotyl growth in shade, with a key role for the epidermis. Combining genetic manipulations in Arabidopsis thaliana with transcriptional profiling of the hypocotyl epidermis from Brassica rapa, we show that auxin acts in the epidermis in part by inducing activity of the locally acting, growth-promoting brassinosteroid pathway. Our findings clarify cell-specific auxin function in the hypocotyl and highlight the complexity of cell type interactions within a growing organ.
All complex multicellular organisms face a similar developmental challenge: how to coordinate the growth of diverse cell types. In plants and animals, this coordinated cellular growth is achieved in part by hormones and signaling molecules that instruct organs on whether to grow. In plants, this challenge of coordinating growth is further compounded due to their sessile nature, which necessitates the tuning of plant architecture to the local environment.
When in close proximity to other plants, shade-avoiding species sense changes in the local light environment. Neighboring plants absorb red wavelengths for photosynthesis and reflect far red, resulting in a decrease in the ratio of red to far-red light, referred to as shade. This battle for light causes changes in plant architecture to better position leaves for light acquisition (Casal 2012). In seedlings of Arabidopsis thaliana (Arabidopsis) and the crop species Brassica rapa (Brassica), shade perception by the cotyledons drives synthesis of the growth hormone auxin, which is transported to the hypocotyl to induce growth (Tao et al. 2008; Procko et al. 2014). The exact cell types in the hypocotyl that perceive the auxin signal and how the growth of these cells is coordinated remain poorly understood.
Observations first made during the 19th Century using stem segments have suggested that inner stem tissues have an inherent tendency to grow, while outer layers restrict this growth (Hofmeister 1859; Went 1934; Kutschera et al. 1987; Kutschera and Niklas 2007). While these findings and others have led to the hypothesis that auxin acts directly on the outer epidermis to relax constraints on inner tissue growth, the merits of this hypothesis have been actively debated (Kutschera et al. 1987; Kutschera and Briggs 1988; Peters and Tomos 1996). Indeed, transcriptional studies of soybean hypocotyls have shown that both inner and outer tissues respond to auxin (Gee et al. 1991). As such, it remains unclear which cell types respond to endogenous auxin signals in intact plants to help coordinate elongation growth in shade.
The ability of auxin to travel over long distances facilitates the communication between the cotyledons, where shade signals are perceived, and the hypocotyl, where cellular elongation occurs (Tao et al. 2008; Procko et al. 2014). However, not all plant hormones act over such distances. For example, brassinosteroids (BRs) are synthesized and act locally on nearby receptors (Savaldi-Goldstein et al. 2007). Furthermore, rescuing BR perception or biosynthesis specifically in the epidermis can restore shoot growth in BR receptor and biosynthesis mutants, respectively (Savaldi-Goldstein et al. 2007). Like auxin, BR is required for shade-induced hypocotyl growth, and interactions between auxin and BR in shade responses have been reported (Kozuka et al. 2010; Yu et al. 2015). However, the nature and location of the interaction between these pathways are unknown.
Here, we used shade-induced growth as a model to interrogate the cell types that are responsible for coordinating auxin responses in the hypocotyl. We present complementary evidence across Arabidopsis and Brassica to show that the epidermis is of particular importance and has a distinct auxin response compared with the inner tissues. We show that auxin functions in the epidermis in part by activating the BR signal transduction pathway upstream of the BR-activated transcription factor BZR1.
Results
Blocking the response to auxin in the epidermis affects hypocotyl growth
To determine in which tissues auxin responses are required, we blocked auxin transcriptional responses within specific cell types. Auxin acts through TIR1/AFB auxin receptor F-box proteins, which form part of the E3 ubiquitin protein ligase complex SCFTIR1/AFB that targets Auxin/indole-3-acetic acid (Aux/IAA) transcriptional repressors for degradation (Dharmasiri et al. 2005a,b; Kepinski and Leyser 2005). Gain-of-function mutations in Aux/IAA genes reduce the interaction of the Aux/IAA protein with SCFTIR1/AFB, resulting in constitutive repressor activity on auxin target genes. One of these mutations, axr3-1, produces a stabilized mutant protein of IAA17 (Leyser et al. 1996; Rouse et al. 1998; Gray et al. 2001). Here, we blocked the transcriptional response to auxin in specific cell types by expressing axr3-1 fused to the fluorescent protein mCitrine (mCit) from different promoters using a GAL4-based transactivation approach (F1 seedlings are referred to as cell-specific pro>>axr3-1::mCit) (Swarup et al. 2005).
Shade induces hypocotyl growth in an auxin-dependent manner (Tao et al. 2008). We found that this growth was greatly reduced in axr3-1 mutants compared with wild-type Arabidopsis seedlings. In contrast, in white light, the hypocotyl of axr3-1 mutants was marginally taller (Fig. 1A,B). To assess where axr3-1 is expressed, we cloned IAA17 (AXR3) promoter elements upstream of the coding sequence of β-glucuronidase (GUS) and mCit. GUS staining of IAA17pro::GUS::mCit seedlings revealed expression in roots and inner tissues of the hypocotyl–root junction. In the shoot, staining was observed in petioles and hypocotyls (Fig. 1C). Fluorescence imaging of cross-sections taken from the mid–upper hypocotyl showed reporter activity exclusively in the epidermis and cortex (Fig. 1D,E).
Figure 1.
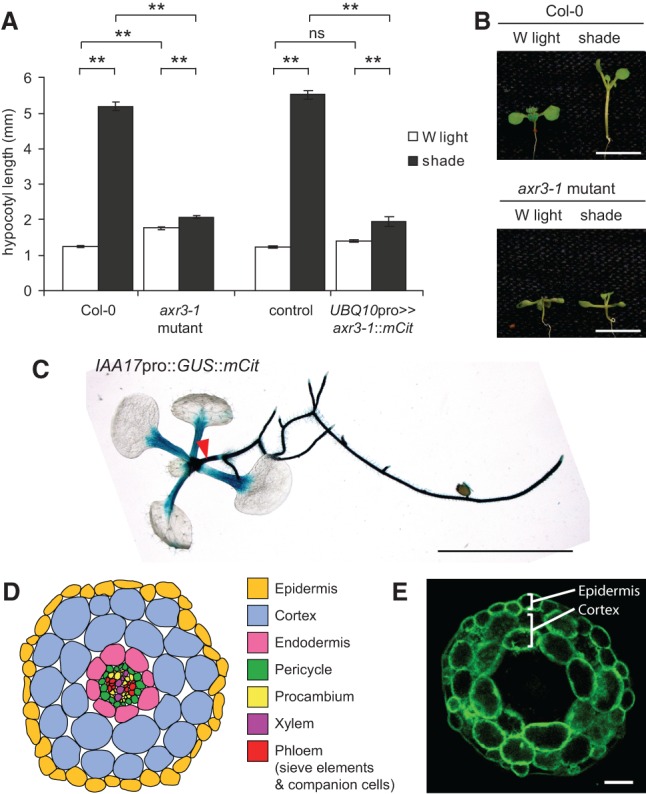
axr3-1 mutants of Arabidopsis have defects in shade-induced hypocotyl growth. (A) Hypocotyl length of 9-d-old seedlings grown in white (W) light or shade. Data show mean ± SEM. (**) P < 0.005; (ns) not significant, Tukey's HSD. The control genotype is plants hemizygous for UBQ10pro::GAL4::VP16. (B) Representative images of wild-type (Col-0) and axr3-1 mutants grown as in A. (C) Image of a GUS-stained 9-d-old white light-grown whole seedling carrying an IAA17pro::GUS::mCit transcriptional reporter. The red arrowhead marks the hypocotyl. (D) Schematic representation of cell types in a young Arabidopsis hypocotyl cross-section. (E) Fluorescence image showing GUS::mCit protein localization in a cross-section through the mid–upper portion of the hypocotyl from a 9-d-old IAA17pro::GUS::mCit plant. Note the GUS::mCit fluorescence in the epidermis and cortex. Bars: B,C, 5 mm; E, 50 µm.
To clarify where axr3-1 might be functioning, we first tested the effect of expressing axr3-1 under the broad UBQ10 promoter (Marques-Bueno et al. 2016). All UBQ10pro>>axr3-1::mCit lines tested produced some seedlings with defects in embryonic patterning—including fused, missing, or additional cotyledons—and a lack of basal structures (Supplemental Fig. S1A). These phenotypes mimic those of higher-order auxin receptor mutants as well as the bodenlos mutant, which codes for a stabilized form of IAA12 (Hamann et al. 2002; Dharmasiri et al. 2005b). For those seedlings that developed a hypocotyl, we observed UBQ10pro>>axr3-1::mCit expression in the hypocotyl endodermis, cortex, and epidermis and defects in shade-induced growth similar to the axr3-1 mutant (Fig. 1A; Supplemental Fig. S1).
To further refine the cell types in which auxin responses are required, we expressed axr3-1 under cell-specific promoters. The Arabidopsis hypocotyl is composed of a central cylinder of vascular cells surrounded by radial layers of endodermis, cortex, and epidermis (Fig. 1D). Different promoters were used to drive axr3-1 in each of these tissue layers (Susek et al. 1993; Truernit and Sauer 1995; Helariutta et al. 2000; Wysocka-Diller et al. 2000; Hooker et al. 2002; Hirakawa et al. 2008). Surprisingly, blocking the response to auxin in a tissue-specific manner had a significant effect on hypocotyl growth in either white light or shade in all cell types tested (Fig. 2). However, the promoter that best recapitulated the hypocotyl growth phenotype of axr3-1 mutants or UBQ10pro>>axr3-1::mCit plants was CER6pro, which drove expression in the epidermis (Fig. 2A,G; Supplemental Fig. S4). To validate this finding, we reasoned that if blocking auxin transcriptional responses in the epidermis resulted in shorter plants in shade, then directly increasing or decreasing auxin levels in the epidermis should also alter hypocotyl length. Consistent with this, when we expressed the bacterial auxin biosynthesis gene iaaM (Thomashow et al. 1986) or the auxin-inactivating enzyme VAS2 (Zheng et al. 2016) in the epidermis using the CER6 promoter, we observed longer and shorter hypocotyls in shade, respectively (Supplemental Fig. S5); however, it is possible that auxin generated in the epidermis in this experiment moves to other cell types. Together, our results support the hypothesis that the epidermis is of particular importance in mediating auxin-induced growth in the hypocotyl.
Figure 2.
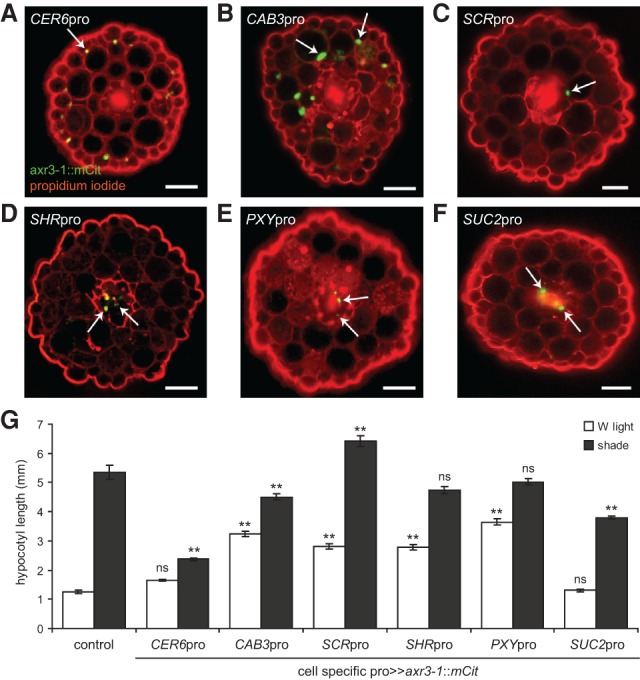
axr3-1 expression in different cell types alters hypocotyl growth in shade. (A–F) Representative fluorescence images of hypocotyl cross-sections of 9-d-old seedlings expressing axr3-1::mCit protein (green) under the control of different cell-specific promoters and counterstained with propidium iodide (red). CER6pro was used to drive axr3-1 expression in the epidermis (A), CAB3pro in green tissue (including strong cortical and weak epidermal expression) (B), SCRpro in the endodermis (C), SHRpro throughout the stele (D), PXYpro in procambium cells (E), and SUC2pro in the phloem companion cells (F). Arrows mark examples of nuclear-localized axr3-1::mCit. Not all nuclei of a given cell layer are in the plane of focus. In our hands, PXYpro had an expression pattern similar to that of SHRpro, including expression in the meristematic pericycle and procambium and perhaps other unidentified cells of the vasculature. SUC2pro also had occasional weak expression in the epidermis. For each promoter, the cell-specific pattern of axr3-1::mCit expression was unaffected by shade, and multiple independent lines had similar phenotypes that correlated with expression level (Supplemental Figs. S2, S3). In addition to hypocotyl expression, these promoters drove axr3-1::mCit in equivalent cell types of the cotyledons and, in some cases, various zones of the root. Bars, 50 µm. (G) Hypocotyl length in white (W) light and shade of 9-d-old seedlings expressing axr3-1::mCit under the control of a cell-specific promoter. Control is hemizygous for UAS::axr3-1::mCit. Data show mean ± SEM. Means are compared with the control in the same light condition using Tukey's HSD. (**) P < 0.005; (ns) not significant.
Auxin signaling in the epidermis regulates auxin production
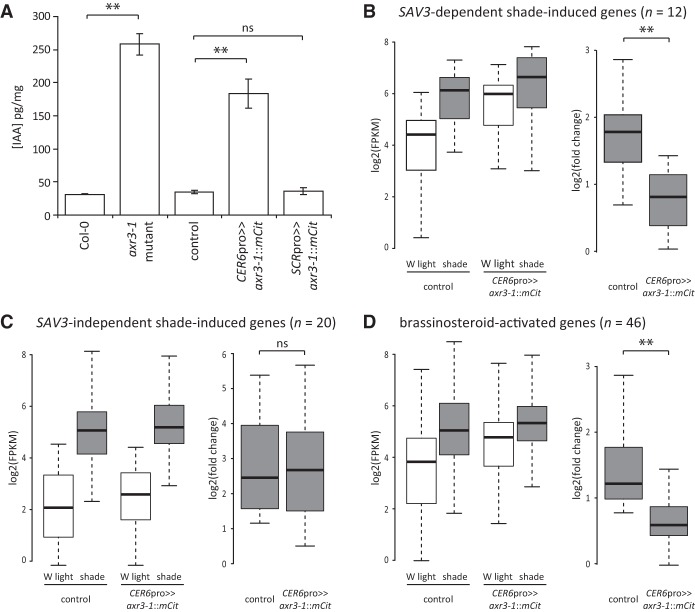
Previous studies in the root have shown that axr3-1 mutants have high levels of free IAA, the main auxin found in plants, as did axr1 loss-of-function mutants, which have defects in SCFTIR1/AFB activity (Jones et al. 2010). This suggests that auxin signaling may negatively feed back onto auxin production. Based on these observations, we hypothesized that blocking the transcriptional response to auxin in the epidermis might alter global auxin levels. To test this, we directly measured IAA levels in whole seedlings and found that both axr3-1 genomic mutants and CER6pro>>axr3-1::mCit plants had elevated levels compared with controls (Fig. 3A). This result could not be replicated by blocking auxin responses in just any cell type, as expressing axr3-1 in the endodermis using SCRpro had no significant effect.
Figure 3.
Blocking auxin responses in the epidermis feeds back onto auxin production and gene expression. (A) Levels of free IAA in 5-d-old white light-grown Arabidopsis seedlings. Data show mean ± SEM. (**) P < 0.005; (ns) not significant, Student's t-test. (B) Box plots showing gene expression values (left) and fold change in shade (right) of 5-d-old white (W) light-grown seedlings treated for 4 h of white light or shade measured by RNA sequencing (RNA-seq). Seedlings were either hemizygous for the UAS::axr3-1::mCit transgene only (control) or expressed axr3-1::mCit under the CER6pro (hemizygous for both UAS::axr3-1::mCit and CER6pro::GAL4::VP16 transgenes). Of 104 previously identified shade-induced, SAV3-dependent genes (Li et al. 2012), 12 were significantly up-regulated by shade in our control. P < 0.005, one-sided Fisher's exact test. Expression values for these 12 genes are shown. (**) P < 0.005; (ns) not significant, Student's t-test. (C) Same as in B, except showing SAV3-independent shade-induced genes (20 of 40 such genes were significantly induced by shade in our control). P < 0.005, one-sided Fisher's exact test. (D) Same as in B, except showing BR genes. Of 1005 genes previously identified as BR-activated (Sun et al. 2010), 46 were significantly induced by shade in our control and are shown here. P < 0.005, one-sided Fisher's exact test. Of these, three were also classified as SAV3-dependent genes, and two were classified as SAV3-independent genes.
Given these high auxin levels, we reasoned that auxin target genes might be misexpressed. As such, we performed transcriptional profiling by RNA sequencing (RNA-seq) of whole seedlings. In control plants hemizygous for the UAS::axr3-1::mCit transgene, 251 genes were significantly up-regulated following 4 h of shade. These genes were enriched for gene ontology (GO) terms related to auxin (Supplemental Tables S1, S2). When we narrowed our analysis to only those shade-induced genes previously identified to be dependent on the SAV3 auxin biosynthesis gene (Li et al. 2012), we saw a lower fold induction in CER6pro>>axr3-1::mCit plants due to greatly elevated gene expression in white light (Fig. 3B; Supplemental Table S3). This is consistent with high auxin target gene expression in whole seedlings due to elevated auxin levels. In contrast, we found little effect of CER6pro>>axr3-1::mCit on shade-induced genes whose induction was identified as SAV3-independent (Fig. 3C). This suggests that CER6pro>>axr3-1::mCit specifically affects the induction of auxin target genes in shade but not auxin-independent genes or shade perception.
The increase in global auxin levels and auxin target gene expression in CER6pro>>axr3-1::mCit plants could be due to feedback on auxin production, reduced auxin degradation, or both. We found that three YUC flavin monooxygenases involved in auxin biosynthesis were up-regulated in CER6pro>>axr3-1::mCit seedlings (Supplemental Fig. S6; Won et al. 2011). Interestingly, these same YUCs were induced by shade, suggesting that they have an important role to play in regulating auxin levels in the shoot. These findings show that one explanation for our whole-seedling IAA measurements is that inhibiting auxin responses in the epidermis lead to positive feedback on auxin production.
The hypocotyl growth defect of CER6pro>>axr3-1::mCit plants is due to auxin insensitivity in the epidermis
Our observation that overexpressing iaaM did not result in a phenotype similar to that of axr3-1 mutants suggested that the axr3-1 hypocotyl growth defect is not a result of simply too much auxin but is rather due to auxin insensitivity. To confirm this, we reasoned that applying exogenous auxin to axr3-1 mutants would circumvent many of the effects of misregulated auxin production. As seen by others (Chapman et al. 2012), we observed a bell-shaped dose response curve of hypocotyl growth to the synthetic auxin picloram in wild-type plants. In contrast, axr3-1 mutants failed to respond (Fig. 4A). Furthermore, picloram caused less inhibition of root and cotyledon growth in axr3-1 mutants (Fig. 4B; see also Leyser et al. 1996). This suggests that axr3-1 reduces auxin sensitivity, and, as a result, YUC genes are up-regulated, leading to compensatory auxin overproduction. In addition, CER6pro>>axr3-1::mCit seedlings had reduced hypocotyl growth in the presence of picloram (Fig. 4C). Our observation that these plants retained some growth response demonstrates that endogenous auxin levels are not so high in CER6pro>>axr3-1::mCit plants as to repress hypocotyl growth. The different phenotypes of axr3-1 mutants and CER6pro>>axr3-1::mCit plants when treated with picloram might also indicate that nonepidermal cells have additional contributions to hypocotyl growth in this experiment. As such, we conclude that auxin perception is likely required tissue-autonomously within the epidermis of the hypocotyl for correct growth responses to shade, with minor contributions from other tissues. Our findings also offer an explanation for the seeming paradox that axr3-1 mutants have phenotypes reminiscent of both auxin insensitivity and constitutively high auxin levels (Leyser et al. 1996). For example, ectopic expression of an auxin-dependent transcriptional reporter for the SAUR-AC1/SAUR15 gene was previously observed in the root vasculature of axr3-1 mutants. This could be replicated in wild-type plants by exogenous auxin treatment, suggesting that axr3-1 might cause a loss of auxin sensitivity in some cell types and increased auxin target gene expression in others due to auxin overproduction (Leyser et al. 1996). Our conclusions are also applicable to another environmental condition that alters hypocotyl growth via auxin, as we found that hypocotyls of CER6pro>>axr3-1::mCit plants are shorter than controls at high temperature (Fig. 4D; Gray et al. 1998).
Figure 4.
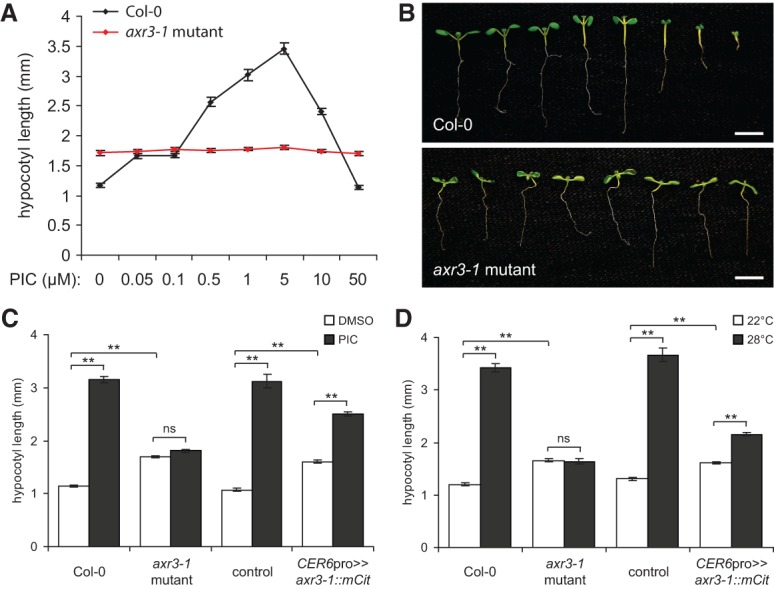
Auxin signaling is required in the epidermis for hypocotyl growth. (A) Hypocotyl length of 7-d-old white light-grown wild-type (Col-0) and axr3-1 mutant Arabidopsis seedlings grown on picloram (PIC). Data points show mean ± SEM. One-way ANOVA, Col-0, P < 0.005; axr3-1, not significant. (B) Representative images of Col-0 and axr3-1 seedlings grown as in A. Picloram concentrations were (from left to right) 0, 0.05, 0.1, 0.5, 1, 5, 10, and 50 µM. axr3-1 seedlings were agravitropic but are arranged vertically here for comparison with the Col-0 strain. Bars, 5 mm. (C) Hypocotyl length of 7-d-old white light-grown seedlings grown in the presence of 5 µM picloram or without picloram (DMSO vehicle). (D) Hypocotyl length of white light-grown seedlings grown for 5 d at 22°C followed by 4 d at either 22°C or 28°C. In C and D, the control genotype was plants hemizygous for the UAS::axr3-1::mCit transgene. Data show mean ± SEM. Tukey's HSD was calculated within a group. (**) P < 0.005; (ns) not significant.
Auxin regulates auxin and BR target genes in the hypocotyl epidermis
The BR hormone pathway similarly functions in the epidermis to promote shoot growth, and auxin and BR signaling interact to regulate shade-induced petiole elongation (Savaldi-Goldstein et al. 2007; Kozuka et al. 2010). As such, we sought to test whether shade alters BR target gene expression in our experiments. When we looked at genes previously shown to be up-regulated by BRs (Sun et al. 2010), we found that many were induced by shade in our RNA-seq data from Arabidopsis whole seedlings. Those that were significantly induced behaved similarly to the SAV3-dependent genes, with a reduced fold change in shade when auxin responses were blocked in the epidermis (Fig. 3D). We conclude that shade affects BR target genes in an auxin-dependent manner.
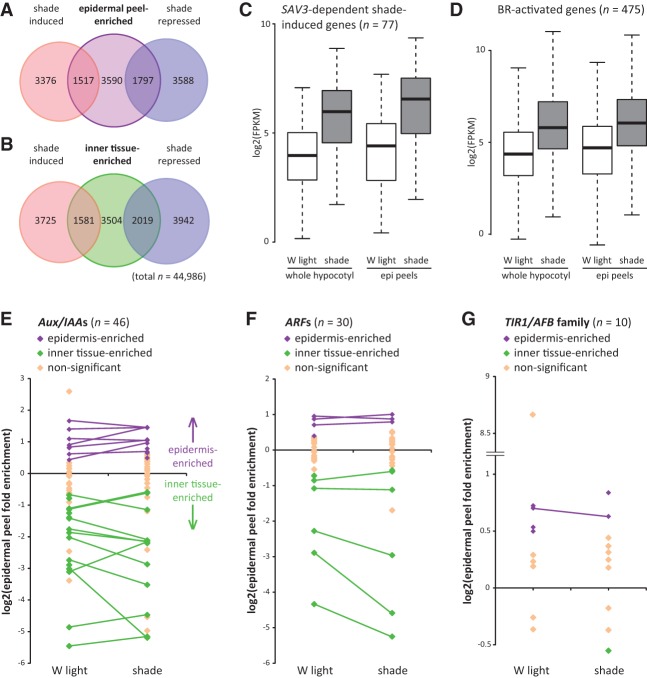
To test whether auxin and BR target genes are coregulated in the hypocotyl epidermis, we took advantage of the large size of Brassica hypocotyls, which allows for removal of the epidermis (with some cortical cells) for transcript analysis (Supplemental Fig. S7A–F). We collected hypocotyl epidermal peels from 4-d-old seedlings treated for 9 h with white light or shade and compared these with whole hypocotyl tissue by RNA-seq. Previously identified Brassica shade marker genes were induced by shade as expected (Supplemental Table S4; Procko et al. 2014). In addition, epidermal peels were enriched for expression of genes and GO terms associated with epidermal function, while whole hypocotyl tissue had greater expression and enrichment of genes and GO terms associated with inner cell types. We refer to these gene classes as “epidermis-enriched” and “inner tissue-enriched,” respectively. Some wounding response genes were also induced as an artifact of the peeling process (Supplemental Tables S5–S8; Supplemental Figs. S7–S8).
We found that 6904 genes have epidermal peel-enriched expression in either white light or shade. Of these genes, we found that more than expected by chance alone were shade-regulated (Fig. 5A; Supplemental Tables S5, S9). A similar observation was made for genes with inner tissue-enriched expression (Fig. 5B; Supplemental Tables S6, S10). To test whether auxin and BR target genes are coregulated in the epidermis, we specifically looked at putative orthologs of SAV3-dependent shade-induced genes and BR-activated genes (Sun et al. 2010; Li et al. 2012). We found that 77 of 174 putative orthologs of SAV3-dependent genes and 475 of 1702 BR-activated genes were up-regulated by shade in both epidermal peels and whole hypocotyl tissue (Fig. 5C,D). This was more than expected by chance alone (one-sided Fisher's exact tests, P < 0.005). This confirms that auxin and BR target genes are induced in the epidermis and other cell types following shade perception.
Figure 5.
Shade increases the expression of auxin and BR target genes in the hypocotyl epidermis of Brassica plants. (A) Overlap of genes with epidermal peel-enriched expression (in either white [W] light or shade) with shade-induced and shade-repressed genes in epidermal peel samples. Tissue was collected from 4-d-old white light-grown Brassica seedlings treated with 9 h of white light or shade. Gene expression was measured by RNA-seq. (B) Overlap of genes expressed at higher levels in the whole hypocotyl tissue relative to epidermal peels (referred to here as “inner tissue-enriched genes”) with shade-induced and shade-repressed genes in whole hypocotyl tissue samples. In A and B, all overlapping regions are significant. P < 0.005, one-sided Fisher's exact tests. (C) Of 174 putative orthologs of SAV3-dependent shade-induced genes (Li et al. 2012), 77 were significantly induced by shade in both epidermal peel and whole hypocotyl tissue. Box plot shows expression values for these genes. (D) Of 1702 putative orthologs of BR-activated genes (Sun et al. 2010), 475 were significantly induced by shade in both epidermal peels and whole hypocotyl tissue. The box plot shows the expression values for these genes. Twenty-nine of these genes are also represented in C. (E–G) Fold enrichment of gene expression in epidermal peels compared with whole hypocotyl tissue for white light and shade treatments for putative orthologs of Aux/IAA (E), ARF (F), and TIR1/AFB (G) family members. Only genes with fragments per kilobase per million mapped fragments (FPKM) > 0 in all sample types are shown. Purple and green diamonds indicate expression values in epidermal peels that are significantly different from whole hypocotyl tissue (false discovery rate, q < 0.05). Purple indicates epidermis-enriched, and green indicates inner tissue-enriched. Lines connect genes that have significantly enriched expression in either the epidermis or inner tissues in both light conditions.
It has been hypothesized that auxin achieves diverse cellular outcomes through differences in the TIR1/AFB receptor and Aux/IAA interaction partners expressed in a given cell (Salehin et al. 2015). Consistent with this hypothesis, we found that many components of the auxin response machinery were expressed at different levels within the epidermis compared with hypocotyl inner tissues. Specifically, we noted differential expression of putative orthologs of TIR1/AFB and Aux/IAA genes as well as for ARF transcriptional regulators, which interact with Aux/IAAs and bind to auxin target gene promoters to affect transcription (Fig. 5E–G; Ulmasov et al. 1997). These results are consistent with the epidermis having a distinct auxin response.
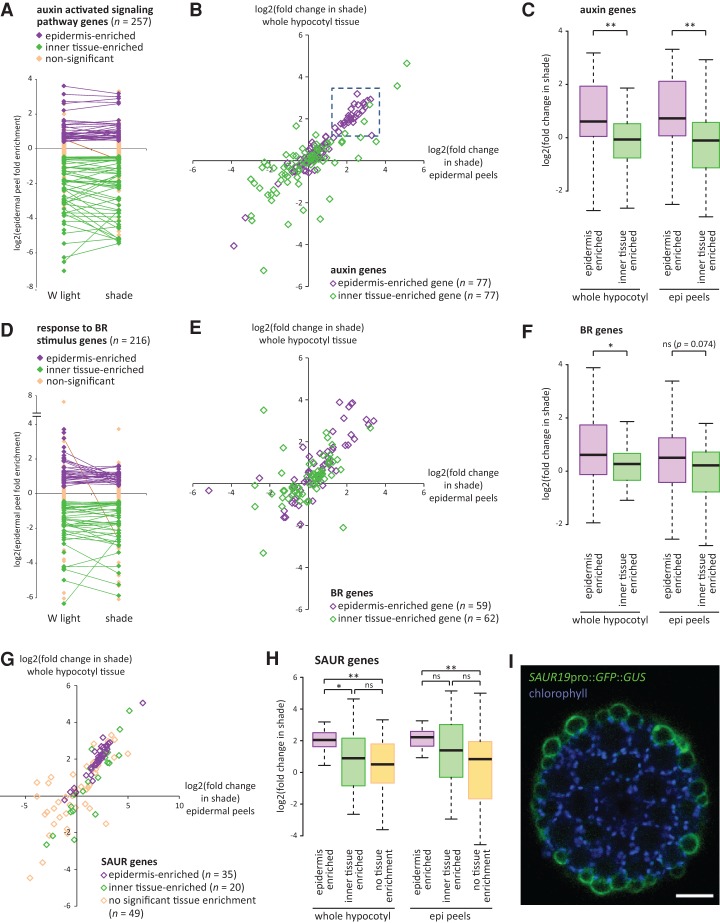
If the epidermis expresses distinct levels of auxin response components, we reasoned that there might also exist a differential response of auxin and BR target genes in this tissue. We therefore examined all of the epidermal peel-enriched and inner tissue-enriched genes that were associated with GO terms “auxin-activated signaling pathway” and “response to BR stimulus.” These GO terms were enriched in both tissue types, indicative of a high level of tissue specificity for these gene classes (Fig. 6A,D). Surprisingly, when we looked at auxin pathway genes, we noted that a large number of the epidermis-enriched genes had a higher fold change in shade compared with the inner tissue-enriched auxin genes (Fig. 6B,C). BR-associated genes followed a similar trend, although less pronounced (Fig. 6E,F). We conclude that the Brassica hypocotyl epidermis has a unique response to shade compared with the inner tissues and tends to be enriched for expression of auxin and BR target genes that have a high sensitivity to the 9-h shade treatment. Consistent with these conclusions, we found that a DR5 auxin reporter (Ulmasov et al. 1997) had a redistribution of relative activity from the vasculature to the outer layers of the Arabidopsis hypocotyl following shade treatment (Supplemental Fig. S9). Similar observations were previously made with DR5::GUS and Arabidopsis plants expressing an auxin-dependent IAA19pro::GUS reporter (Keuskamp et al. 2010; Zheng et al. 2016).
Figure 6.
The Brassica epidermis is enriched for expression of auxin and BR target genes with high sensitivity to 9 h of shade treatment. (A) Fold enrichment of gene expression in Brassica epidermal peels compared with whole hypocotyl tissue in white (W) light-treated and shade-treated seedlings for genes associated with the GO term “auxin-activated signaling pathway.” Purple and green diamonds indicate expression values in epidermal peels that are significantly different from whole hypocotyl tissue (false discovery rate, q < 0.05). Purple indicates epidermis-enriched, and green indicates inner tissue-enriched. Lines connect genes that have significantly enriched expression in either the epidermis or inner tissues in both light conditions. Note that most genes do not change the tissue type of enrichment. Only those genes with FPKM > 0 in all sample types are shown. (B) Fold change in shade of gene expression in whole hypocotyl tissue (Y-axis) plotted against fold change in epidermal peel samples (X-axis). “Auxin-activated signaling pathway” genes that are significantly enriched in either the epidermis or inner tissues (in either white light or shade) are shown. The boxed region represents a cluster of 29 epidermis-enriched genes highly induced by shade. (C) Box plot of genes shown in B, showing fold change in shade. Note that the epidermis-enriched genes are more sensitive to shade. (**) P < 0.005; (*) P < 0.05; (ns) not significant, Student's t-test. (D–F) Same as in A–C, except showing genes associated with the GO term “response to BR stimulus.” (G,H) Same as in B and C except showing SAUR family genes. Genes with FPKM > 0 in all samples but that are not enriched in any particular tissue type are also shown. In H, comparisons are Tukey's HSD. (I) Hypocotyl cross-section of a 6-d-old Arabidopsis seedling showing GFP localization (green) in a SAUR19pro::GFP::GUS plant. Chlorophyll autofluorescence (blue) shows the position of the cortical cells. Note the strong GFP localization to the epidermis. Bar, 50 µm.
We noted a cluster of auxin genes with epidermis-enriched expression that have high induction in shade (Fig. 6B). Interestingly, 23 of these 29 genes were SAURs (in contrast, only one of the remaining 48 auxin genes was a SAUR), which represent the largest family of early auxin response genes. Some SAURs are also regulated by BR signaling (Ren and Gray 2015). When we looked at all Brassica SAUR genes, we again observed that those with enriched expression in the epidermis tended to have a greater collective response to shade (Fig. 6G,H). Similar to other auxin- and BR-related genes (Fig. 6B,E), some specific SAURs with inner tissue-enriched expression were also highly induced by shade, indicating that inner tissues do respond to auxin but that this response is distinct (Supplemental Fig. S10). Our observations made with the SAUR gene family were not simply an artifact of our RNA-seq analysis methods assigning sequencing reads to incorrect but highly related SAUR genes, as we observed a similar trend with a more stringent multimapped read correction analysis (Supplemental Fig. S11A). Previous studies have shown that some SAURs are expressed in the epidermis and cortex of soybean hypocotyls, while another SAUR was expressed in the vasculature of Arabidopsis, suggesting tissue-specific expression patterns within the family (McClure and Guilfoyle 1989; Gee et al. 1991; Kong et al. 2013). Here, we found that most epidermis-enriched SAUR genes in Brassica were related to the SAUR19 subfamily from Arabidopsis (Supplemental Fig. S11B,C). Previous studies have shown that this subfamily is induced by shade and that its knockdown results in shorter hypocotyls, suggesting that it codes for positive regulators of growth (Spartz et al. 2012). We generated SAUR19pro::GFP::GUS Arabidopsis plants and found that GFP was almost exclusively localized to the epidermis in hypocotyls (Fig. 6I). This finding highlights the conservation between Brassica and Arabidopsis species and our ability to identify new epidermis-enriched factors.
Together, our Brassica RNA-seq results show that the hypocotyl epidermis expresses distinct components of the auxin sensory and transcriptional response machinery and that the epidermis is enriched for distinct auxin and BR target genes with high sensitivity to shade. This includes a subfamily of SAUR genes implicated in hypocotyl elongation.
Auxin mediates hypocotyl growth in part through the BR pathway upstream of BZR1
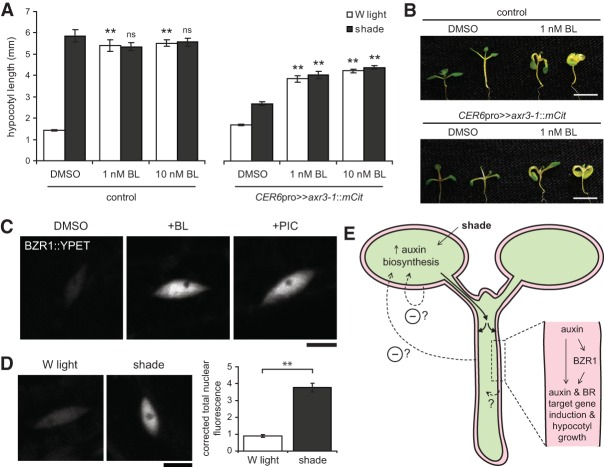
Our results show that auxin signaling in the epidermis induces both auxin and BR target genes. Because BRs signal through the epidermis to regulate shoot size (Savaldi-Goldstein et al. 2007), we reasoned that auxin might affect hypocotyl growth by activating the BR pathway in this tissue. Importantly, we were able to partially rescue the growth defect of CER6pro>>axr3-1::mCit Arabidopsis plants by exogenously applying the BR brassinolide (BL) (Fig. 7A,B). BL treatment increased hypocotyl length of CER6pro>>axr3-1::mCit plants in shade. However, this rescue was incomplete, as hypocotyl length was consistently lower than that seen in the control genotype. As such, we conclude that auxin signaling in the epidermis has both BR-dependent and BR-independent effects on growth.
Figure 7.
Auxin functions to regulate hypocotyl growth in part through the BR pathway, upstream of BZR1. (A) Hypocotyl length of 9-d-old Arabidopsis seedlings treated with white (W) light or shade in the presence of BL or without BL (DMSO vehicle). Data show mean ± SEM. Statistical comparisons are shown for each mean compared with the control genotype grown in the same light condition. (**) P < 0.005; (ns) not significant, Tukey's HSD. The CER6pro>>axr3-1::mCit line shown is number 4. Control plants were CER6pro::GAL4::VP16 hemizygotes. (B) Representative images of plants from A. For each genotype and pharmacological treatment, white light-grown is at the left, and shade-treated is at the right. Bars, 5 mm. (C,D) Representative fluorescence images of nuclear BZR1::YPET protein in hypocotyl epidermal cells of 5-d-old white light-grown plants treated for 24 h with DMSO, 1 µM BL, or 5 µM picloram (PIC) (C), or, in a separate experiment, 6-d-old white light-grown plants treated with 24 h of white light or shade (D). Images were taken near the center of the hypocotyl and with the same exposure in a given panel. Bars, 10 µm. The graph shows the corrected total nuclear fluorescence for plants treated as in D. Nuclear fluorescence was measured from 54 and 59 nuclei from 12 white light-treated and 12 shade-treated plants, respectively. Data show mean ± SEM. (**) P < 0.005, Student's t-test. (E) A model for auxin-induced hypocotyl growth in shade. Shade stimulates auxin production in the cotyledons, which is transported to the hypocotyl (Tao et al. 2008). As auxin travels down the hypocotyl via the vasculature, it is transported laterally and is perceived by the different cell layers (Keuskamp et al. 2010). Auxin perception by nonepidermal cell types has minor contributions to hypocotyl elongation but may have greater roles to play in other auxin-dependent phenotypes not assessed here. In the epidermis (red), auxin induces auxin and BR target genes and cellular growth through both BR-dependent (upstream of BZR1) and BR-independent mechanisms. How the epidermis signals back to instruct the inner cell layers on how to grow is unknown but may involve a chemical or mechanical signal. In addition, auxin signaling in the epidermis negatively feeds back onto auxin production. We were not able to resolve whether this feedback occurs in the cotyledons, from the hypocotyl, or from another organ.
Auxin signaling may interact with the BR signal transduction pathway (1) by directly activating target genes that are common between the two pathways, (2) by inducing BR biosynthesis, or (3) through cross-talk with one or more BR pathway signaling components. From our gene expression analysis of Brassica hypocotyls, we noted that expression of a putative ortholog of BZR1 (Brara.G02244) was induced by shade. BZR1 codes for a terminal transcriptional regulator of the BR signal transduction pathway, and expression of a stabilized mutant form specifically in the epidermis can rescue the short hypocotyl defect of Arabidopsis BR receptor mutants (Wang et al. 2002; Savaldi-Goldstein et al. 2007). To more closely examine BZR1 expression, we generated Arabidopsis plants carrying a BZR1::2xYPET::3xHA (referred to here as BZR1::YPET) translational fusion gene in the context of a large bacterial artificial chromosome (BAC) clone to better recapitulate endogenous BZR1 expression. BZR1::YPET nuclear expression was observed along the hypocotyl, with the exception of the first few cells from the base, and was present in most if not all cell layers (Supplemental Fig. S12). As reported for other BZR1 reporter lines, expression was stronger following exogenous BR treatment (Fig. 7C), indicating that BZR1 both regulates and is regulated by BR signaling (Wang et al. 2002). We further show that hypocotyl expression increased with picloram or shade treatment, suggesting that auxin feeds into the BR signal transduction pathway upstream of BZR1 (Fig. 7C,D). Because exogenous BR has a similar effect, this may be through increased BR biosynthesis. For example, we found that 68 of 203 genes related to the GO term “BR biosynthetic process” were induced by shade in the Brassica epidermis (Supplemental Table S11). This includes a putative ortholog of DWF7 (Brara.A03878), which is required for sterol biosynthesis, and putative orthologs of CYP85A1 and CYP85A2 (Brara.D01180 and Brara.F03069, respectively), which code for enzymes catalyzing the final oxidation reactions necessary to generate BL (Choe et al. 1999; Nomura et al. 2005). These latter reactions are likely rate-limiting steps in BL biosynthesis, and cyp85a1 cyp85a2 double mutants have a severe dwarf phenotype (Nomura et al. 2005).
Together, our results show that auxin has both BR-dependent and BR-independent effects on plant growth and interacts with the BR pathway in the epidermis and other tissues upstream of BZR1, perhaps through increased BL biosynthesis. In addition, these findings demonstrate the power of our Brassica transcriptional profiling data set to make biologically accurate predictions across species.
Discussion
Auxin has a central role in plant organ development, and biologists have long debated in which cell types it acts to affect growth. Here, we show an important role for the epidermis in auxin-mediated growth of the hypocotyl. While auxin responses were required in many cell types for correct hypocotyl growth in shade, blocking the response specifically in the epidermis best recapitulated the phenotype of plants broadly expressing the stabilized mutant protein axr3-1. Thus, our results support the epidermal growth control hypothesis (Kutschera and Niklas 2007). Interestingly, epidermal responses to auxin are also required for root gravitropism, suggesting some level of conservation of epidermal function across different organs (Swarup et al. 2005). We further found that defective auxin responses in the shoot epidermis cause compensatory feedback on auxin production and that auxin regulates hypocotyl growth through both BR-dependent and BR-independent means (Fig. 7E).
While other studies have noted interactions between auxin and BR hormone pathways, how auxin specifically affects the BR pathway has remained elusive (Goda et al. 2004; Nemhauser et al. 2004; Kozuka et al. 2010). Previously, the observation that the BR-regulated transcription factor BES1 does not change subcellular localization in the hypocotyl in response to exogenous IAA treatment has been cited as evidence that auxin does not alter BR biosynthesis (Yin et al. 2002; Nemhauser et al. 2004). However, unlike the artificial auxin picloram, IAA treatment in our hands and others often has either little or an inhibitory effect on hypocotyl growth, possibly because this natural auxin is inefficiently transported to the shoot when applied exogenously to the growth medium (Hanson and Trewavas 1982; Collett et al. 2000). Here, we show that auxin induces nuclear expression of the transcriptional regulator BZR1 in the hypocotyl. The observed increase in BZR1 expression may be due to either direct activity of the auxin pathway on the BZR1 promoter or indirect activity via increased BR biosynthesis. Although we were not able to distinguish between these two possibilities, we noted that multiple BR biosynthesis genes were affected by shade in Brassica. As such, we favor the hypothesis that auxin acts as a long-distance messenger that increases biosynthesis of the local effector BR to induce cellular elongation. Consistent with this idea, exogenous auxin application induces promoter activity of the BR biosynthesis enzyme DWF4 in the root, leading to an increase in BR intermediates (Chung et al. 2011). However, we cannot rule out further interactions between the two pathways; for example, ARFs may also bind to BR target genes directly and interact physically with BZR1 (Oh et al. 2014).
Perturbing the transcriptional response to auxin within the epidermis resulted in changes to the hypocotyl as a whole, with other cell types adjusting their growth accordingly. How, then, does the epidermis instruct another cell layer how to grow? Because inner tissues have an inherent tendency to grow when the outer layers are removed, it has been hypothesized that the epidermis restricts growth of the inner layers by mechanical means (Kutschera and Niklas 2007). Indeed, the epidermis has thick outer cell walls, and auxin and BR might relax cell wall rigidity by controlling the expression of cell wall-loosening enzymes (Kutschera and Niklas 2007; Kozuka et al. 2010). SAUR19 subfamily proteins might further facilitate the activation of such enzymes by lowering apoplastic pH (Spartz et al. 2014). Alternatively, these restraints could be chemical or a combination of the two mechanisms.
Converting hormonal signals into cell-specific transcriptional and phenotypic outcomes is a challenge faced by plants and animals alike. Indeed, an outstanding question in the auxin field is whether distinct auxin receptor–Aux/IAA–ARF modules can generate different outcomes in different cell types (Salehin et al. 2015). Here, we present data showing that the Brassica epidermis expresses distinct levels of auxin receptor and response components. It remains to be seen whether this is in part causative for the transcriptional differences observed in the epidermis, which we found is enriched for distinct auxin target genes, which collectively have a high sensitivity to shade and include a subfamily of SAURs. Together, these findings help clarify how hormones generate complex growth responses in multicellular organs.
Materials and methods
Plant materials, growth conditions, and hypocotyl measurements
The Arabidopsis strains used were Col-0 and axr3-1 (Leyser et al. 1996). The B. rapa strain was FPsc. Seeds were sterilized, stratified, and germinated in constant white light on 0.7% agar plates with 0.5× Linsmaier and Skoog salts and vitamins. The temperature used was 22°C. To generate cell-specific pro>>axr3-1::mCit plants, a GAL4-based transactivation expression approach was used, as some of the promoters driving axr3-1 used in this study caused severe defects in adult plants (Swarup et al. 2005; data not shown). Plants homozygous for UAS::axr3-1::mCit (T3 generation) were crossed to lines homozygous for cell-specific pro::GAL4::VP16 (T3), and F1 seedlings hemizygous for both transgenes were scored. Two of three randomly selected UAS::axr3-1::mCit lines gave similar results when crossed to a single cell-specific pro::GAL4::VP16 line, and one of these was chosen for all subsequent crosses. F1 seeds from a given silique were evenly distributed across white light and shade treatments. Siliques bearing seeds that failed to germinate or grow were omitted from downstream analyses. Hypocotyl growth measurements of shade-treated Arabidopsis plants were of 9-d-old seedlings grown for 5 d in white light followed by 4 d in white light or shade. The shade conditions used were as described in Tao et al. (2008) and Procko et al. (2014). For hypocotyl measurements at high temperature, plants were grown for 5 d at 22°C followed by 4 d at 22°C or 28°C in white light. CER6pro>>axr3-1::mCit line shown is number 22 unless otherwise indicated.
DNA constructs and plant transformation
The multisite Gateway system (Invitrogen) was used for all constructs except the BZR1::2xYPET::3xHA recombineered BAC clone. See the Supplemental Material for details. Col-0 plants were transformed with Agrobacterium GV3101 carrying the various constructs using standard techniques. Segregation analyses at T2 and T3 generations were used to isolate single-insertion homozygous lines.
Imaging
GUS staining was performed using standard protocols and imaged on a Leica MZ FLIII stereo microscope. Fluorescence images were taken with a Zeiss LSM 710 confocal microscope. For propidium iodide (PI) staining of hypocotyls, seedlings were incubated in 50 μg/mL PI with 0.4% Triton X-100 for 10–20 min, washed, and embedded in 2% low-melting-temperature agarose for sectioning. Corrected total nuclear fluorescence (CTNF) of BZR1::YPET-expressing plants was calculated from confocal images of epidermal cells taken using the same exposure settings. Images were taken from the center of the hypocotyl, and only the first five in-focus nuclei in a given microimage were measured. CTNF was calculated using ImageJ (McCloy et al. 2014).
Free IAA measurements
Samples containing 10–20 mg of plant material were purified and analyzed by gas chromatography-tandem mass spectrometry (GC-MS/MS) as previously described (Andersen et al. 2008) with minor modifications. Five-hundred picograms of 13C6-IAA internal standard was added to each sample before purification, and five biological replicates were analyzed per tissue type.
Collection of B. rapa and Arabidopsis tissues for RNA-seq
For RNA-seq of Brassica tissue, 4-d-old white light-grown plants were treated with 9 h of white light or shade. Tissue was then dissected and snap-frozen in liquid N2. Each whole-hypocotyl tissue sample was collected from four hypocotyls pooled (seedlings were dissected just above the root–hypocotyl junction and just below the junction with the petioles). Each epidermal peel sample was collected from ∼60 seedlings. Outer layers at either the tops or bottoms of hypocotyl segments were gripped with fine forceps and pulled, removing a strip of cuticle and attached epidermal cells. One to two peels were made from each hypocotyl segment. During the peeling process, peels were left for up to 15 min at incubator temperature before snap-freezing in liquid N2. Samples were collected in triplicate, with collection times staggered randomly across a 24-h period. One of the shade-treated epidermal peel samples was removed from downstream analysis at the Cuffdiff step due to contamination with an unacceptable number of cells from the inner tissues (Supplemental Fig. S7E). For RNA-seq of Arabidopsis seedlings, plants were grown for 5 d in white light and then treated for 4 h with white light or shade. Seeds from a given silique were divided equally between conditions. Four replicates for each genotype and condition were collected. Ten to 15 whole seedlings were collected per replicate, and replicates were collected over two different days. We noted possible batch effects on gene expression related to silique of origin and day of sample collection (data not shown).
High-throughput whole-transcriptome sequencing and analysis
Total RNA was extracted using Qiagen RNeasy microkit and digested with DNase I. Agilent tape station was used to determine RNA integrity numbers prior to library preparation. Stranded mRNA-seq libraries were prepared using Illumina TruSeq stranded mRNA library preparation kit according to the manufacturer's instructions. Libraries were then quantified, pooled, and sequenced at single-end 50-base-pair reads using the Illumina HiSeq 2500 platform at the Salk Next-Generation Sequencing Core. Raw sequencing data were demultiplexed and converted into FASTQ files using CASAVA (version 1.8.2). Libraries were sequenced at an average depth of 16.5 million and 14.7 million reads per library for Brassica and Arabidopsis samples, respectively.
RNA-seq analysis was performed using the Tuxedo protocol (Trapnell et al. 2012) using iPlant/CyVerse (Goff et al. 2011). Briefly, raw reads were aligned to Arabidopsis TAIR10 or B. rapa FPsc version 1.3 (DOE-JGI, http://phytozome.jgi.doe.gov) using TopHat version 2.0.9 and Bowtie 2.1.0 (strand-specific). Cufflinks, Cuffmerge, and Cuffdiff versions 2.1.1 were used for transcriptome assembly and differential expression analysis using default parameters. For reanalysis of SAUR gene expression in the Brassica data set, the analysis was reperformed in the same manner except using multimapped read correction in Cufflinks and Cuffdiff. Test_ids (referred to here as genes) that had FPKM (fragments per kilobase per million mapped fragments) > 0 in all samples tested and a Benjamini and Hochberg-corrected P-value (false discovery rate, q) of <0.05 between two samples were designated as differentially expressed. Only nuclear genes in Arabidopsis were analyzed. Putative orthologs of Brassica genes were identified from the annotation file accompanying the B. rapa FPsc version 1.3 genome release (generally, the top result from a BLASTP alignment of the FPsc proteome to the Arabidopsis TAIR10 proteome; determined by Department of Energy Joint Genome Institute). BiNGO 3.03 was used for GO (biological process only) enrichment analysis with a Benjamini and Hochberg-corrected P-value of <0.05 (Maere et al. 2005). For Brassica, a custom annotation file was provided to BiNGO (see the Supplemental Material), where each Brassica gene was assigned the same GO terms as its putative ortholog from Arabidopsis (Arabidopsis TAIR10 annotation data downloaded September 2015) on the assumption that the two genes would have conserved function. For Figure 6, genes associated with GO terms 9734 (“auxin-activated signaling pathway;” synonym “auxin-mediated signaling pathway”) and 9741 (“response to BR stimulus”) are shown, including genes associated with child terms. Unless otherwise noted, genes are classified as epidermis-enriched or inner tissue-enriched if expression is higher in the corresponding tissue type in at least one of the two light conditions (white light or shade). Box plots show whiskers using the Tukey method; outliers are not shown. Log2 transformed data were used for statistical comparisons of fold change values.
Previously, 144 genes were identified by RNA-seq to be induced >1.7 log2 fold by 1 h of shade in wild-type Arabidopsis seedlings (Li et al. 2012). Only 40 of these genes were induced above this threshold in sav3 mutants (Li et al. 2012), and these genes are defined here as sav3-independent shade-induced genes. These include genes that are likely to be independent of auxin signals (e.g., PIL1, ATHB2, and HFR1) (Procko et al. 2014) as well as some auxin target genes whose induction may have decreased in the sav3 mutant background but not below 1.7 log2 fold. The remaining 104 shade-induced genes are classified here as sav3-dependent shade-induced genes.
Quantitative RT–PCR (qRT–PCR) of Brassica samples
qRT–PCR was performed using standard techniques. Previously, we reported qRT–PCR primers for shade marker genes in Brassica using a candidate approach (Procko et al. 2014). Here, we used our RNA-seq results to design improved primers to a reference gene as well as shade and wounding response genes (see the Supplemental Material). Because the B. rapa genome has undergone a recent genome triplication event (Wang et al. 2011), it is possible that our primers also report the expression of paralagous genes with high sequence identity. This limitation was even more apparent in our previously published primer sets (see the Supplemental Material).
Accession numbers
RNA-seq raw and processed data have been deposited into the Gene Expression Omnibus with accession number GSE79881.
Supplementary Material
Acknowledgments
We thank Scott Woody and Rick Amasino for providing B. rapa FPsc seeds and sharing information. The FPsc genome data were produced by the US Department of Energy Joint Genome Institute, with additional information provided by Stephen Fairclough. We also thank Roger Granbom for assistance with IAA measurements; Ullas Pedmale for the CER6pro plasmid; Javier Gallego-Bartolome and Björn Willige for reagents and discussion; Carol Huang for guidance on statistical analysis; Adam Seluzicki and J. Gallego-Bartolome for critical reading of the manuscript; and Leah Baum, Cory Chen, and Kira Bender for technical assistance. RNA-seq was performed with assistance from Manching Ku at the Next-Generation Sequencing Core of the Salk Institute with funding from NIH-National Cancer Institute Cancer Center Support Grants (P30 014195), the Chapman Foundation, and the Helmsley Charitable Trust. RNA-seq analysis was performed using iPlant/CyVerse (http://www.iplantcollaborative.org), supported by the NSF (DBI-0735191 and DBI-1265383). This work was supported by National Institutes of Health (NIH) fellowship 1F32GM101876-01 (C.P.); NIH grants GM52413 and GM94428 (to J.C.); National Science Foundation (NSF) EAGER grant IOS-1045256 (to J.C. and J.A.L.); and the Swedish Governmental Agency for Innovation Systems (Vinnova), the Swedish Research Council (VR) and the Marianne and Marcus Wallenberg Foundation (to K.L.).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.283234.116.
References
- Andersen SU, Buechel S, Zhao Z, Ljung K, Novak O, Busch W, Schuster C, Lohmann JU. 2008. Requirement of B2-type cyclin-dependent kinases for meristem integrity in Arabidopsis thaliana. Plant Cell 20: 88–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal JJ. 2012. Shade avoidance. Arabidopsis Book 10: e0157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman EJ, Greenham K, Castillejo C, Sartor R, Bialy A, Sun TP, Estelle M. 2012. Hypocotyl transcriptome reveals auxin regulation of growth-promoting genes through GA-dependent and -independent pathways. PLoS One 7: e36210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S, Noguchi T, Fujioka S, Takatsuto S, Tissier CP, Gregory BD, Ross AS, Tanaka A, Yoshida S, Tax FE, et al. 1999. The Arabidopsis dwf7/ste1 mutant is defective in the Δ7 sterol C-5 desaturation step leading to brassinosteroid biosynthesis. Plant Cell 11: 207–221. [PMC free article] [PubMed] [Google Scholar]
- Chung Y, Maharjan PM, Lee O, Fujioka S, Jang S, Kim B, Takatsuto S, Tsujimoto M, Kim H, Cho S, et al. 2011. Auxin stimulates DWARF4 expression and brassinosteroid biosynthesis in Arabidopsis. Plant J 66: 564–578. [DOI] [PubMed] [Google Scholar]
- Collett CE, Harberd NP, Leyser O. 2000. Hormonal interactions in the control of Arabidopsis hypocotyl elongation. Plant Physiol 124: 553–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Estelle M. 2005a. The F-box protein TIR1 is an auxin receptor. Nature 435: 441–445. [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Weijers D, Lechner E, Yamada M, Hobbie L, Ehrismann JS, Jurgens G, Estelle M. 2005b. Plant development is regulated by a family of auxin receptor F box proteins. Dev Cell 9: 109–119. [DOI] [PubMed] [Google Scholar]
- Gee MA, Hagen G, Guilfoyle TJ. 1991. Tissue-specific and organ-specific expression of soybean auxin-responsive transcripts GH3 and SAURs. Plant Cell 3: 419–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda H, Sawa S, Asami T, Fujioka S, Shimada Y, Yoshida S. 2004. Comprehensive comparison of auxin-regulated and brassinosteroid-regulated genes in Arabidopsis. Plant Physiol 134: 1555–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff SA, Vaughn M, McKay S, Lyons E, Stapleton AE, Gessler D, Matasci N, Wang L, Hanlon M, Lenards A, et al. 2011. The iPlant collaborative: cyberinfrastructure for plant biology. Front Plant Sci 2: 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, Ostin A, Sandberg G, Romano CP, Estelle M. 1998. High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. Proc Natl Acad Sci 95: 7197–7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M. 2001. Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature 414: 271–276. [DOI] [PubMed] [Google Scholar]
- Hamann T, Benkova E, Baurle I, Kientz M, Jurgens G. 2002. The Arabidopsis BODENLOS gene encodes an auxin response protein inhibiting MONOPTEROS-mediated embryo patterning. Genes Dev 16: 1610–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson JB, Trewavas AJ. 1982. Regulation of plant cell growth: the changing perspective. New Phytol 90: 1–18. [Google Scholar]
- Helariutta Y, Fukaki H, Wysocka-Diller J, Nakajima K, Jung J, Sena G, Hauser MT, Benfey PN. 2000. The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell 101: 555–567. [DOI] [PubMed] [Google Scholar]
- Hirakawa Y, Shinohara H, Kondo Y, Inoue A, Nakanomyo I, Ogawa M, Sawa S, Ohashi-Ito K, Matsubayashi Y, Fukuda H. 2008. Non-cell-autonomous control of vascular stem cell fate by a CLE peptide/receptor system. Proc Natl Acad Sci 105: 15208–15213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmeister W. 1859. Über die Beugungen saftreicher Pflanzenteile nach Erschütterung. Ber Verh Ges Wiss Leipzig 11: 175–204. [Google Scholar]
- Hooker TS, Millar AA, Kunst L. 2002. Significance of the expression of the CER6 condensing enzyme for cuticular wax production in Arabidopsis. Plant Physiol 129: 1568–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones B, Gunneras SA, Petersson SV, Tarkowski P, Graham N, May S, Dolezal K, Sandberg G, Ljung K. 2010. Cytokinin regulation of auxin synthesis in Arabidopsis involves a homeostatic feedback loop regulated via auxin and cytokinin signal transduction. Plant Cell 22: 2956–2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepinski S, Leyser O. 2005. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435: 446–451. [DOI] [PubMed] [Google Scholar]
- Keuskamp DH, Pollmann S, Voesenek LA, Peeters AJ, Pierik R. 2010. Auxin transport through PIN-FORMED 3 (PIN3) controls shade avoidance and fitness during competition. Proc Natl Acad Sci 107: 22740–22744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Y, Zhu Y, Gao C, She W, Lin W, Chen Y, Han N, Bian H, Zhu M, Wang J. 2013. Tissue-specific expression of SMALL AUXIN UP RNA41 differentially regulates cell expansion and root meristem patterning in Arabidopsis. Plant Cell Physiol 54: 609–621. [DOI] [PubMed] [Google Scholar]
- Kozuka T, Kobayashi J, Horiguchi G, Demura T, Sakakibara H, Tsukaya H, Nagatani A. 2010. Involvement of auxin and brassinosteroid in the regulation of petiole elongation under the shade. Plant Physiol 153: 1608–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutschera U, Briggs WR. 1988. Interaction between cortical cylinder and epidermis during auxin-mediated growth in pea internodes. Plant Sci 54: 23–28. [Google Scholar]
- Kutschera U, Niklas KJ. 2007. The epidermal-growth-control theory of stem elongation: an old and a new perspective. J Plant Physiol 164: 1395–1409. [DOI] [PubMed] [Google Scholar]
- Kutschera U, Bergfeld R, Schopfer P. 1987. Cooperation of epidermis and inner tissues in auxin-mediated growth of maize coleoptiles. Planta 170: 168–180. [DOI] [PubMed] [Google Scholar]
- Leyser HM, Pickett FB, Dharmasiri S, Estelle M. 1996. Mutations in the AXR3 gene of Arabidopsis result in altered auxin response including ectopic expression from the SAUR-AC1 promoter. Plant J 10: 403–413. [DOI] [PubMed] [Google Scholar]
- Li L, Ljung K, Breton G, Schmitz RJ, Pruneda-Paz J, Cowing-Zitron C, Cole BJ, Ivans LJ, Pedmale UV, Jung HS, et al. 2012. Linking photoreceptor excitation to changes in plant architecture. Genes Dev 26: 785–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maere S, Heymans K, Kuiper M. 2005. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21: 3448–3449. [DOI] [PubMed] [Google Scholar]
- Marques-Bueno MM, Morao AK, Cayrel A, Platre MP, Barberon M, Caillieux E, Colot V, Jaillais Y, Roudier F, Vert G. 2016. A versatile Multisite Gateway-compatible promoter and transgenic line collection for cell type-specific functional genomics in Arabidopsis. Plant J 85: 320–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCloy RA, Rogers S, Caldon CE, Lorca T, Castro A, Burgess A. 2014. Partial inhibition of Cdk1 in G2 phase overrides the SAC and decouples mitotic events. Cell Cycle 13: 1400–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure BA, Guilfoyle T. 1989. Rapid redistribution of auxin-regulated RNAs during gravitropism. Science 243: 91–93. [DOI] [PubMed] [Google Scholar]
- Nemhauser JL, Mockler TC, Chory J. 2004. Interdependency of brassinosteroid and auxin signaling in Arabidopsis. PLoS Biol 2: E258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura T, Kushiro T, Yokota T, Kamiya Y, Bishop GJ, Yamaguchi S. 2005. The last reaction producing brassinolide is catalyzed by cytochrome P-450s, CYP85A3 in tomato and CYP85A2 in Arabidopsis. J Biol Chem 280: 17873–17879. [DOI] [PubMed] [Google Scholar]
- Oh E, Zhu JY, Bai MY, Arenhart RA, Sun Y, Wang ZY. 2014. Cell elongation is regulated through a central circuit of interacting transcription factors in the Arabidopsis hypocotyl. Elife 3: e03031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters WS, Tomos D. 1996. The epidermis still in control? Bot Acta 109: 264–267. [Google Scholar]
- Procko C, Crenshaw CM, Ljung K, Noel JP, Chory J. 2014. Cotyledon-generated auxin is required for shade-induced hypocotyl growth in Brassica rapa. Plant Physiol 165: 1285–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren H, Gray WM. 2015. SAUR proteins as effectors of hormonal and environmental signals in plant growth. Mol Plant 8: 1153–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse D, Mackay P, Stirnberg P, Estelle M, Leyser O. 1998. Changes in auxin response from mutations in an AUX/IAA gene. Science 279: 1371–1373. [DOI] [PubMed] [Google Scholar]
- Salehin M, Bagchi R, Estelle M. 2015. SCFTIR1/AFB-based auxin perception: mechanism and role in plant growth and development. Plant Cell 27: 9–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savaldi-Goldstein S, Peto C, Chory J. 2007. The epidermis both drives and restricts plant shoot growth. Nature 446: 199–202. [DOI] [PubMed] [Google Scholar]
- Spartz AK, Lee SH, Wenger JP, Gonzalez N, Itoh H, Inze D, Peer WA, Murphy AS, Overvoorde PJ, Gray WM. 2012. The SAUR19 subfamily of SMALL AUXIN UP RNA genes promote cell expansion. Plant J 70: 978–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spartz AK, Ren H, Park MY, Grandt KN, Lee SH, Murphy AS, Sussman MR, Overvoorde PJ, Gray WM. 2014. SAUR inhibition of PP2C-D phosphatases activates plasma membrane H+-ATPases to promote cell expansion in Arabidopsis. Plant Cell 26: 2129–2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Fan XY, Cao DM, Tang W, He K, Zhu JY, He JX, Bai MY, Zhu S, Oh E, et al. 2010. Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev Cell 19: 765–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susek RE, Ausubel FM, Chory J. 1993. Signal transduction mutants of Arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell 74: 787–799. [DOI] [PubMed] [Google Scholar]
- Swarup R, Kramer EM, Perry P, Knox K, Leyser HM, Haseloff J, Beemster GT, Bhalerao R, Bennett MJ. 2005. Root gravitropism requires lateral root cap and epidermal cells for transport and response to a mobile auxin signal. Nat Cell Biol 7: 1057–1065. [DOI] [PubMed] [Google Scholar]
- Tao Y, Ferrer JL, Ljung K, Pojer F, Hong F, Long JA, Li L, Moreno JE, Bowman ME, Ivans LJ, et al. 2008. Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133: 164–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow MF, Hugly S, Buchholz WG, Thomashow LS. 1986. Molecular basis for the auxin-independent phenotype of crown gall tumor tissues. Science 231: 616–618. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7: 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truernit E, Sauer N. 1995. The promoter of the Arabidopsis thaliana SUC2 sucrose-H+ symporter gene directs expression of β-glucuronidase to the phloem: evidence for phloem loading and unloading by SUC2. Planta 196: 564–570. [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. 1997. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZY, Nakano T, Gendron J, He J, Chen M, Vafeados D, Yang Y, Fujioka S, Yoshida S, Asami T, et al. 2002. Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev Cell 2: 505–513. [DOI] [PubMed] [Google Scholar]
- Wang X, Wang H, Wang J, Sun R, Wu J, Liu S, Bai Y, Mun JH, Bancroft I, Cheng F, et al. 2011. The genome of the mesopolyploid crop species Brassica rapa. Nat Genet 43: 1035–1039. [DOI] [PubMed] [Google Scholar]
- Went FW. 1934. On the pea test method for auxin, the plant growth hormone. Proc Kon Akad Wet Amst 37: 547–555. [Google Scholar]
- Won C, Shen X, Mashiguchi K, Zheng Z, Dai X, Cheng Y, Kasahara H, Kamiya Y, Chory J, Zhao Y. 2011. Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAs in Arabidopsis. Proc Natl Acad Sci 108: 18518–18523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka-Diller JW, Helariutta Y, Fukaki H, Malamy JE, Benfey PN. 2000. Molecular analysis of SCARECROW function reveals a radial patterning mechanism common to root and shoot. Development 127: 595–603. [DOI] [PubMed] [Google Scholar]
- Yin Y, Wang ZY, Mora-Garcia S, Li J, Yoshida S, Asami T, Chory J. 2002. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109: 181–191. [DOI] [PubMed] [Google Scholar]
- Yu J, Qiu H, Liu X, Wang M, Gao Y, Chory J, Tao Y. 2015. Characterization of tub4(P287L), a β-tubulin mutant, revealed new aspects of microtubule regulation in shade. J Integr Plant Biol 57: 757–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z, Guo Y, Novak O, Chen W, Ljung K, Noel JP, Chory J. 2016. Local auxin metabolism regulates environment-induced hypocotyl elongation. Nat Plants 2: 16025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.