Grenfell et al. show that transcription and RNA processing occur in metaphase-arrested egg extracts and that noncoding RNA biogenesis is important for centromere, kinetochore, and mitotic spindle assembly.
Abstract
Transcription at the centromere of chromosomes plays an important role in kinetochore assembly in many eukaryotes, and noncoding RNAs contribute to activation of the mitotic kinase Aurora B. However, little is known about how mitotic RNA processing contributes to spindle assembly. We found that inhibition of transcription initiation or RNA splicing, but not translation, leads to spindle defects in Xenopus egg extracts. Spliceosome inhibition resulted in the accumulation of high molecular weight centromeric transcripts, concomitant with decreased recruitment of the centromere and kinetochore proteins CENP-A, CENP-C, and NDC80 to mitotic chromosomes. In addition, blocking transcript synthesis or processing during mitosis caused accumulation of MCAK, a microtubule depolymerase, on the spindle, indicating misregulation of Aurora B. These findings suggest that co-transcriptional recruitment of the RNA processing machinery to nascent mitotic transcripts is an important step in kinetochore and spindle assembly and challenge the idea that RNA processing is globally repressed during mitosis.
Introduction
Chromosome segregation requires that chromosomes attach to the spindle through kinetochores, complexes that assemble on specialized centromeric chromatin. Multiple cellular mechanisms contribute to this process, and mounting evidence suggests that components of the spliceosome, the dynamic RNP complex that removes introns from RNA polymerase II (Pol II) transcripts (Wahl et al., 2009), are also involved. Genome-wide screens in cultured cells identified splicing factors as important for cell division (Goshima et al., 2007; Kittler et al., 2007; Somma et al., 2008; Neumann et al., 2010), and microtubule- and mitotic chromatin-interacting proteins biochemically copurified with the catalytically active spliceosome isolated from HeLa cell nuclear extracts (Makarov et al., 2002). Recently, a role for the Prp19 splicing complex in Xenopus laevis egg extract spindle assembly was demonstrated (Hofmann et al., 2013); however, its specific function is unclear. Overall, mitotic functions for the RNA processing machinery have been largely unexplored.
Work in a variety of systems including plants, fission yeast, and cultured mammalian cells showed that Pol II transcription at the centromere during mitosis is important for kinetochore assembly (Chan and Wong, 2012; Gent and Dawe, 2012). Because splicing factors are co-transcriptionally recruited to Pol II transcripts (Listerman et al., 2006; David et al., 2011), involvement of the RNA processing machinery in centromeric noncoding RNA (ncRNA) biogenesis could explain its mitotic relevance. Support for this idea comes from the observation that the splicing factor Prp4 is kinetochore-localized during mitosis (Montembault et al., 2007) and that splicing factors interact with centromeric transcripts in mouse cells (Maison et al., 2011). Although little is known about the RNA biogenesis pathway at centromeres, centromeric transcription is important for centromere protein A (CENP-A) loading across eukaryotic species (Saffery et al., 2003; Nakano et al., 2008; Cardinale et al., 2009; Chueh et al., 2009; Bergmann et al., 2012; Quénet and Dalal, 2014; Chen et al., 2015). Furthermore, RNA binding by the inner kinetochore protein CENP-C promotes its association with centromeric DNA in plants and animals (Wong et al., 2007; Du et al., 2010; Rošić et al., 2014), which is a prerequisite for kinetochore assembly (Gascoigne et al., 2011; Rago et al., 2015). Studies in mouse cells and in X. laevis egg extracts have revealed that centromeric ncRNAs are also important for Aurora B kinase activation (Ferri et al., 2009; Blower, 2016), and RNA binding may be a general mechanism for regulating Aurora B activity (Jambhekar et al., 2014). Among its many roles (Carmena et al., 2012), centromere-associated Aurora B regulates the localization and activity of MCAK, a microtubule depolymerase that controls kinetochore fiber attachment to chromosomes as well as overall microtubule distribution (Andrews et al., 2004; Lan et al., 2004; Sampath et al., 2004; Zhang et al., 2007; Tanenbaum and Medema, 2011; Ems-McClung et al., 2013).
To investigate the mitotic functions of the RNA processing machinery, we used metaphase-arrested Xenopus egg extracts, which do not require global transcription and translation for spindle assembly (Newport and Kirschner, 1982a,b; Maresca and Heald, 2006; Blower et al., 2007), to identify two mitotic roles for splicing factors. First, we found that recruitment of the RNA processing machinery to nascent centromere-derived ncRNAs during mitosis enhances localization of CENP-A, CENP-C, and NDC80 to chromosomes. Second, ncRNA elongation and processing are required to properly regulate Aurora B kinase and maintain spindle integrity, at least in part by modulating MCAK localization. Our results indicate that multiple RNA biogenesis-dependent events contribute to kinetochore and spindle assembly, and that splicing is not globally inhibited during mitosis (Shin and Manley, 2002).
Results and discussion
The RNA processing machinery is required for spindle assembly
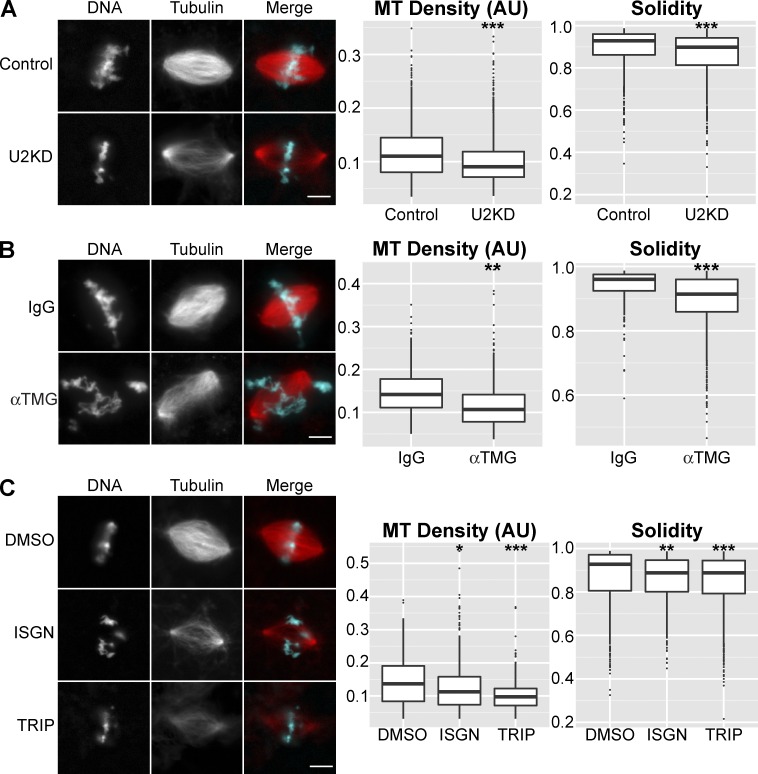
To investigate mitotic roles of the RNA processing machinery, we interfered with its function in both X. laevis and Xenopus tropicalis egg extracts. Abnormal spindle morphology was observed (Fig. 1 and Fig. S1), with defects including loss of spindle microtubule density, measured as median rhodamine tubulin fluorescence intensity, as well as disorganized structures with microtubules that projected off of the spindle, which manifested as changes in spindle solidity (Fig. S1 A). Multiple inhibition approaches were used, including RNase H–based knockdown of the U2 snRNA (Pan and Prives, 1988; Figs. 1 A and S1 B), immunodepletion of core spliceosomal snRNPs (Fig. 1 B and Fig. S1, C–E), and treatment with the splicing inhibitor isoginkgetin (ISGN; O’Brien et al., 2008) during mitosis (Fig. 1 C) or throughout the entire cell cycle (Fig. S1, F and G). Blocking Pol II transcription initiation with triptolide (Titov et al., 2011) resulted in similar spindle defects (Figs. 1 C and S1 H). These effects were not caused by aberrant expression of protein-coding RNAs, as blocking translation with cycloheximide did not result in detectable spindle defects (Fig. S1 I). These data suggest that the RNA processing machinery is involved in the mitotic regulation of one or more RNAs that are not acting in a protein coding capacity, hereafter referred to as ncRNAs, and that this regulation is important for spindle assembly.
Figure 1.
ncRNA biogenesis is important for mitotic spindle assembly. (A) Example images and quantification of spindle microtubule (MT) density and solidity after RNase H–based knockdown of the U2 snRNA (U2KD, n = 3,018) or treatment with a scrambled oligo (n = 1,454) in X. laevis egg extracts. Median microtubule density decreased 18.0% in U2 knockdown extract. (B) Example images and quantification of spindle microtubule density and solidity for X. tropicalis spindles formed after splicing factor immunodepletion with an antibody against the trimethylguanosine cap of snRNAs (α-TMG, n = 581) compared with mock depletion (IgG, n = 325). Median microtubule density decreased 24.5% in splicing factor-depleted extract. (C) Example images and quantification of spindle microtubule density and solidity in X. laevis extracts treated with splicing inhibitor (ISGN, n = 518), transcription inhibitor (TRIP, n = 417), or solvent control (DMSO, n = 483). Inhibitors were added immediately before spindle assembly. Median microtubule density decreased 14.9% in ISGN-treated extract and 24.1% in TRIP-treated extract. Bars, 10 µm. Box plot horizontal lines correspond to median values. Bottom and top of the boxes are first and third quartiles, respectively; whiskers show highest and lowest values within 1.5 times the interquartile range and outliers are plotted as single points. *, P < 10−5; **, P < 10−10; ***, P < 10−15 (Kolmogorov–Smirnov test). AU, arbitrary units.
Centromere-derived ncRNAs are targets of the RNA processing machinery
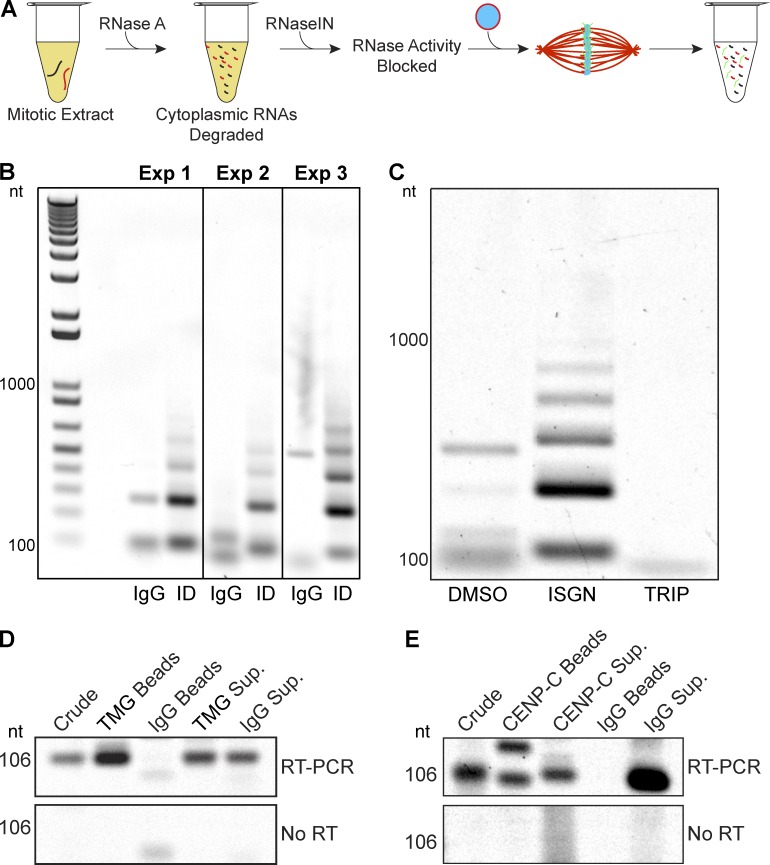
Given the growing number of studies showing a role for centromeric transcription (Chan and Wong, 2012; Gent and Dawe, 2012; Scott, 2013), we examined whether the RNA processing machinery was also involved in centromeric ncRNA biogenesis. Replicated, interphase nuclei were incubated in RNA-depleted, metaphase-arrested extract. After spindle assembly, RNAs were isolated and analyzed by RT-PCR with primers specific to the 174-bp fcr1 repeat (Edwards and Murray, 2005), the only known centromere sequence in X. laevis (Fig. 2 A). Whereas nuclei formed in mock-depleted extract produced amplification products that were 500 bp or less, nuclei formed in splicing factor–depleted extract produced a laddered pattern matching that observed when genomic DNA was amplified (Fig. 2 B and Fig. S2 C), indicating that tandem repeat–containing centromeric fcr1 RNAs were produced as large transcripts up to six repeat units in length in the absence of processing. Similar products were obtained in reactions treated with the splicing inhibitor ISGN, and no amplification was observed in extracts treated with triptolide, demonstrating that the Pol II products were not derived from RNA already present in the reaction (Fig. 2 C and Fig. S2, A–C). These products were derived from RNA, as no amplification was observed when reverse transcription was omitted (Fig. S2, A, D, and E), and products were synthesized from the added nuclei because no amplification was observed in their absence (Fig. S2 F).
Figure 2.
The RNA processing machinery contributes to centromeric ncRNA biogenesis during mitosis. (A) Schematic of the centromere RNA biogenesis assay in X. laevis egg extract. Nuclei containing replicated sperm chromosomes were added to RNA-depleted extract, and nascent centromeric ncRNAs were assessed by RT-PCR. (B) Centromeric RNAs appeared in a laddered pattern after splicing factor depletion with an antibody specific to the trimethylguanosine cap of core spliceosomal RNAs (ID), whereas mock depletion (IgG) resulted in one or a few centromeric RNA products. The fcr1 repeat is 174 bp. Primers used to amplify this sequence were 106 bp apart. (C) Centromeric RNAs appeared in a laddered pattern after splicing inhibition with ISGN and were not detected after transcription inhibition with triptolide. (D) RT-PCR analysis of centromeric fcr1 ncRNAs in immunoprecipitates using nonspecific IgG antibodies (IgG Beads) or antibodies specific to the trimethylguanosine cap of core spliceosomal RNAs (TMG Beads), compared with extract supernatants (Sup.). (E) RT-PCR analysis of centromeric fcr1 ncRNAs in CENP-C immunoprecipitates (Beads) or extract supernatants (Sup.). An amplification product from a potentially processed transcript appears at ∼150 bp. Note that PCR-amplified centromeric RNAs varied in size, likely because predicted splice sites present in the degenerate fcr1 RNA sequence could lead to a variety of splice products because of heterogeneity of centromeric sequences.
Supporting association of centromeric transcripts with the spliceosome, we found that fcr1 transcripts coimmunoprecipitated with the core splicing machinery (Fig. 2 D). Fcr1 transcripts, including potentially processed transcripts, also coimmunoprecipitated with the inner centromere protein CENP-C (Figs. 2 E and S2 G). Centromeric RNA amplicons varied in size, likely reflecting underlying repeat sequence heterogeneity (Miga et al., 2014) that could lead to variation in splice site selection. These results indicate that centromeric RNAs such as fcr1 are transcribed and processed during mitosis.
The RNA processing machinery promotes kinetochore assembly
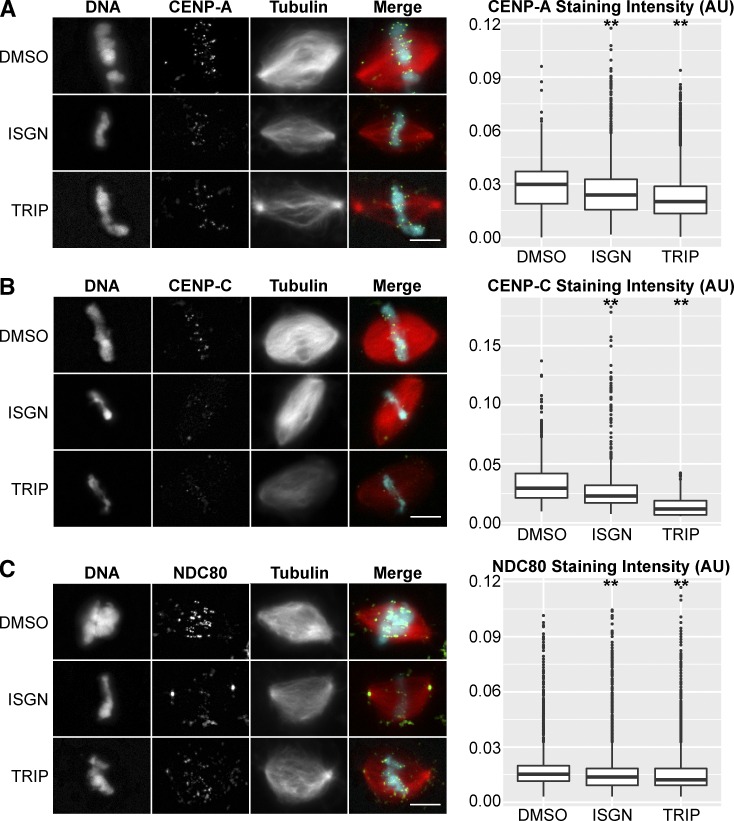
CENP-A and CENP-C depend in part on centromeric transcription for their localization (Chueh et al., 2009; Du et al., 2010). We therefore examined them by immunofluorescence after perturbation of RNA biogenesis and observed a decrease in staining density between 22.4% and 59.6% after inhibition of splicing or transcription initiation (Fig. 3, A and B). NDC80 staining intensity also decreased 10–19.9% under these conditions (Fig. 3 C), suggesting that centromere and inner kinetochore defects were propagated to the microtubule-binding interface of the outer kinetochore.
Figure 3.
Perturbation of RNA biogenesis leads to centromere and kinetochore defects. (A) Example images of CENP-A staining and quantification of median fluorescence intensity within antibody marked foci after inhibition of splicing (ISGN, n = 9,544) or transcription (TRIP, n = 9,613) compared with DMSO controls (n = 5,203). Median CENP-A staining density decreased 25.4% in ISGN-treated extract and 37.8% in TRIP-treated extract. (B) Example images of CENP-C staining and quantification of median fluorescence intensity within antibody marked foci after inhibition of splicing (ISGN, n = 1,214) or transcription (TRIP, n = 1,160) compared with DMSO controls (n = 2,411). Median CENP-C staining density decreased 22.4% in ISGN-treated extract and 59.6% in TRIP-treated extract. (C) Example images of NDC80 staining and quantification of median fluorescence intensity within antibody marked foci after inhibition of spicing (ISGN, n = 1,499) or transcription (TRIP, n = 1,623) compared with DMSO controls (n = 1,515). Median NDC80 staining density decreased 10.0% in ISGN-treated extract and 19.9% in TRIP-treated extract. Bars, 10 µm. **, P < 10−10 (Kolmogorov–Smirnov test). In the merged image, microtubules are red, centromere proteins are green, and DNA is cyan. AU, arbitrary units.
In contrast to results obtained with triptolide, we did not observe a decrease in CENP-A or NDC80 staining when transcription elongation was blocked with α-amanitin, which does not interfere with splicing catalysis (Bird et al., 2004) and leaves the nascent transcript associated with its chromosomal locus (Rudd and Luse, 1996). Interestingly, centromeric CENP-C staining increased significantly in α-amanitin–treated extract (Fig. S3 A), suggesting that increasing the residence time of nascent centromeric RNAs leads to greater CENP-C accumulation. Thus, although CENP-C RNA binding reinforces its centromeric localization (Du et al., 2010), our results suggest that splicing factor recruitment, but not transcript elongation or persistence of an RNA, plays a role in kinetochore assembly. Because recent measurements suggest that CENP-A is distributed throughout the genome (Bodor et al., 2014), centromere RNA biogenesis during mitosis could provide a second signal that helps mark the site of kinetochore assembly.
Our results are in contrast to previous studies reporting that α-amanitin treatment decreased CENP-C centromere localization (Chan et al., 2012; Quénet and Dalal, 2014). However, the long time course required in cell culture could result in α-amanitin dependent degradation of the Pol II large subunit (Nguyen et al., 1996), which would prevent splicing factor recruitment. In addition, our data are at odds with experiments in which RNAi depletion of centromeric transcripts led to centromere and kinetochore defects (Chueh et al., 2009; Quénet and Dalal, 2014). However, the RNAi machinery has been shown to trigger transcriptional gene silencing in vertebrates through siRNA-guided H3K9 di- and trimethylation, leading to the recruitment of HP1 proteins (Alló et al., 2009; Ameyar-Zazoua et al., 2012), which abolished the function of a human artificial chromosome centromere (Nakano et al., 2003). Thus, using RNAi to interrogate centromere RNA function could induce epigenetic centromere inactivation.
ncRNA processing contributes to spindle integrity by regulating Aurora B and MCAK localization
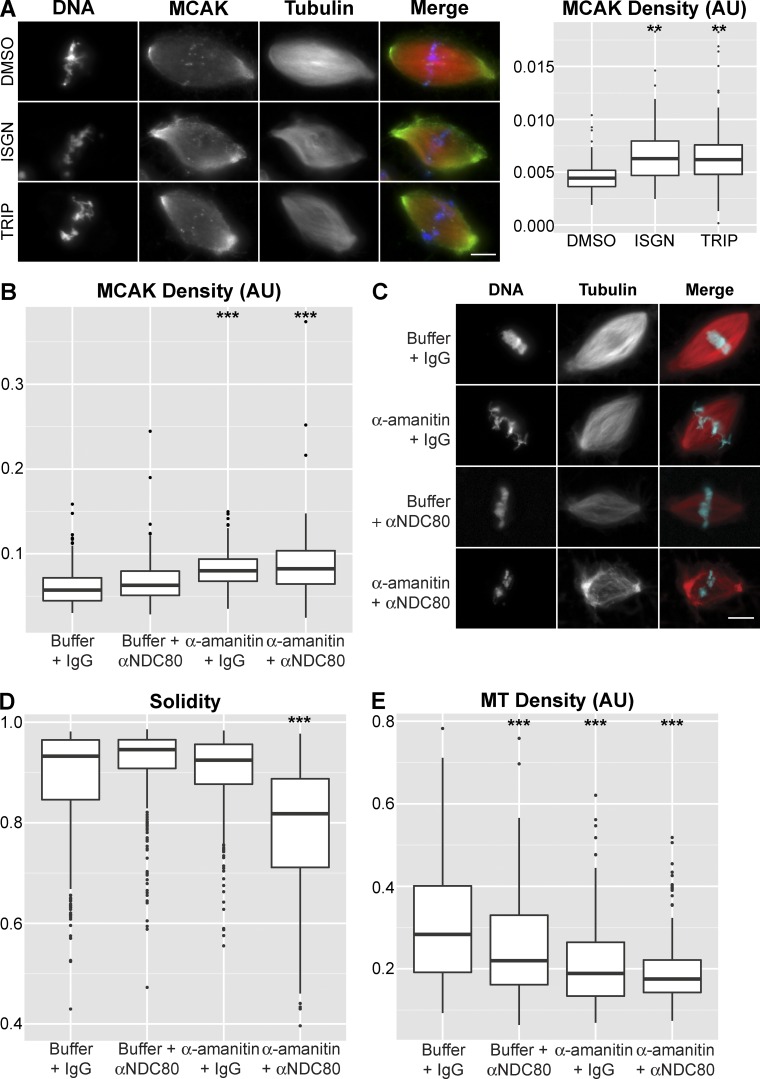
Because spindle defects observed upon inhibition of transcription or RNA processing were significant, we investigated whether ncRNA-dependent pathways in addition to kinetochore assembly contributed to spindle integrity. Aurora B kinase activity is stimulated by RNA binding (Ferri et al., 2009; Jambhekar et al., 2014), and centromere-localized Aurora B produces a phosphorylation gradient (Wang et al., 2011) that regulates the activity, localization, and microtubule binding of the microtubule depolymerase MCAK (Andrews et al., 2004; Lan et al., 2004; Zhang et al., 2007; Tanenbaum and Medema, 2011; Ems-McClung et al., 2013). RNase A treatment of egg extract reduced Aurora B activity and caused spindle assembly defects that could be partially rescued by inhibiting MCAK (Jambhekar et al., 2014). Furthermore, a reduction in Aurora B staining at the inner centromere upon inhibition of transcription was recently observed (Blower, 2016). We therefore tested whether interfering with ncRNA biogenesis affected MCAK, and we observed increased staining density within the spindle upon inhibition of transcription initiation or RNA processing (Figs. 4 A and S3 C). Although we have not assayed Aurora B activity directly, these results indicate that RNA biogenesis-dependent regulation of Aurora B and MCAK promotes spindle microtubule stability.
Figure 4.
ncRNA-dependent MCAK regulation contributes to spindle integrity. (A) Example images and quantification of MCAK staining density after inhibition of splicing (ISGN, n = 100) or transcription (TRIP, n = 127) compared with the control (DMSO, n = 89). In the merged image, microtubules are red, MCAK is green, and DNA is blue. Median MCAK staining density increased 41.9% in ISGN-treated extract and 39.7% in TRIP-treated extract. (B) Quantification of MCAK staining density after treatment with buffer control (n = 466), αNDC80 antibodies (n = 477), α-amanitin (n = 350), or αNDC80 antibodies + α-amanitin (n = 262). Median MCAK staining density increased 9.8% in αNDC80-treated extract, 39.6% in α-amanitin–treated extract, and 43.8% in αNDC80 + α-amanitin–treated extract. (C) Example images of spindles formed under each of the conditions in B. Bar, 10 µm. (D) Quantification of spindle solidity under each of the conditions in C. (E) Quantification of spindle microtubule (MT) density under each of the conditions in C. Median microtubule density decreased 22.5% in αNDC80-treated extract, 33.4% in α-amanitin–treated extract, and 38.2% in αNDC80 + α-amanitin–treated extract. Bars, 10 µm. **, P < 10−10; ***, P < 10−15 (Kolmogorov–Smirnov test). AU, arbitrary units.
To determine whether the ncRNA-dependent Aurora B and centromere/kinetochore regulation we observed were separable processes, and together sufficient to explain the RNA biogenesis spindle phenotype, we developed an approach to inhibit them individually and in combination. We found that blocking transcription elongation with α-amanitin resulted in accumulation of MCAK on the spindle (Fig. 4 B), consistent with misregulation of Aurora B (Tanenbaum and Medema, 2011; Ems-McClung et al., 2013), but that centromere or kinetochore assembly was unaffected (Fig. S3 A). We combined this treatment with direct inhibition of kinetochore fiber formation with an inhibitory NDC80 antibody (McCleland et al., 2003) and found that inhibition of both processes simultaneously caused spindle defects similar to those produced by interfering with RNA biogenesis, whereas individual inhibitions did not (Fig. 4, B–E). Directly blocking Aurora B activity with the small molecule ZM447439 (Gadea and Ruderman, 2005) resulted in MCAK localization defects similar to α-amanitin treatment and gave additive effects with NDC80 inhibition, but it did not increase the severity of ISGN-induced spindle defects (Fig. S3, B and D). Thus, the mitotic function of the RNA processing machinery is largely accounted for by its involvement in Aurora B kinase activation and assembly of the centromere and kinetochore. However, there is likely cross talk between these two pathways in egg extracts because Aurora B inhibition interferes with outer kinetochore assembly (Emanuele et al., 2008), and centromeric transcription is required for Aurora B enrichment at centromeres and normal bipolar attachment of kinetochores (Blower, 2016).
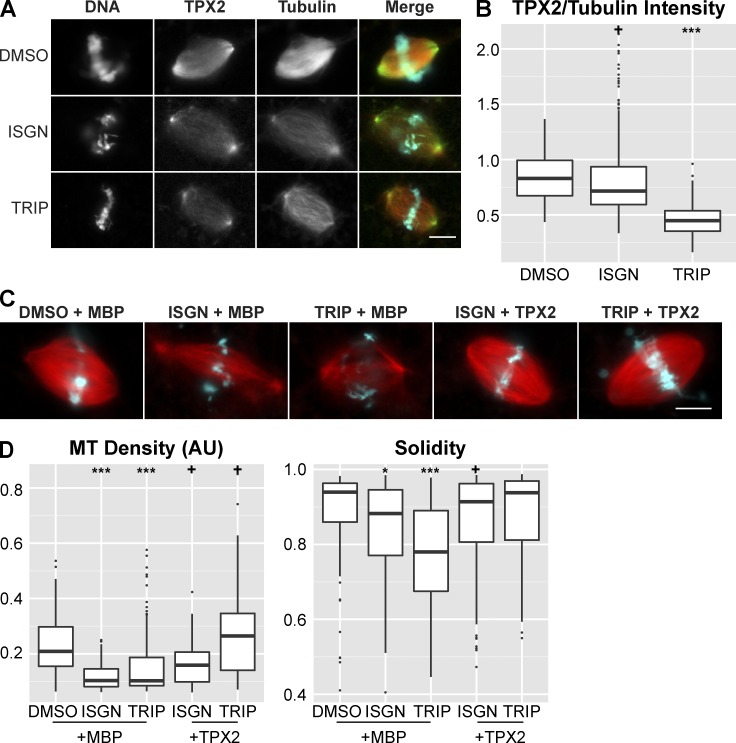
To begin to investigate the downstream function of RNA biogenesis in spindle assembly, we examined TPX2, a spindle assembly factor involved in kinetochore fiber formation (Ma et al., 2011) and microtubule branching nucleation (Petry et al., 2013), which has been shown to interact with the spliceosome in human cells (Makarov et al., 2002). We observed decreased levels of TPX2 on spindle microtubules upon treatment with transcription initiation or splicing inhibitors (Fig. 5, A and B) and found that addition of excess TPX2, but not the microtubule-stabilizing agent DMSO, rescued the spindle defects (Fig. 5, C and D; and Fig. S3 E). These results suggest that specific pathways affecting microtubule dynamics and organization are downstream of mitotic ncRNA biogenesis and act to stabilize the spindle. Given the known functions of TPX2, we propose that inhibition of RNA processing reduces amplification of spindle microtubules by interfering with nucleation from kinetochore fibers. However, it is also possible that excess TPX2 rescues spindle defects at least in part through the direct activation of Aurora B (Iyer and Tsai, 2012).
Figure 5.
Addition of TPX2 partially rescues spindle defects caused by perturbing ncRNA biogenesis. (A) Example images of TPX2 staining in control and inhibitor-treated reactions. In the merged image, microtubules are red, TPX2 is green, and DNA is cyan. Bar, 10 µm. (B) Plot showing the ratio of TPX2/tubulin intensity in spindles in control (n = 242) and inhibitor-treated reactions (ISGN n = 234, TRIP n = 120). The TPX2/tubulin ratio decreased 13.9% in ISGN-treated extract and 45.9% in TRIP-treated extract. (C) Example images of spindles assembled in extract after treatment with DMSO + 200 nM MBP (n = 193), ISGN + 200 nM MBP (n = 235), TRIP + 200 nM MBP (n = 199), ISGN + 200 nM TPX2 (n = 265), or TRIP + 200 nM TPX2 (n = 126). Bar, 10 µm. (D) Quantification of spindle microtubule density and spindle solidity under each of the conditions in C. Median microtubule density decreased 50.6% in MBP + ISGN–treated extract, 51.0% in MBP + TRIP–treated extract, 24.0% in TPX2 + ISGN–treated extract, and increased 26.9% in TPX2 + TRIP–treated extract. +, P < 0.05; *, P < 10−5; **, P < 10−10; ***, P < 10−15 (Kolmogorov–Smirnov test).
In summary, our results show that splicing factors are required for proper spindle assembly in Xenopus egg extracts in the absence of global gene expression caused by at least two RNA-dependent processes. The first is the involvement of RNA biogenesis at the centromere in kinetochore assembly, and the second is RNA-dependent activation of Aurora B kinase. Given the conservation of centromeric transcription across organisms (Chan and Wong, 2012), a role for the splicing machinery is likely to be conserved and could explain mitotic phenotypes observed upon splicing factor knockdown in cultured cells (Goshima et al., 2007; Kittler et al., 2007; Somma et al., 2008; Neumann et al., 2010). Although our analysis focused on a known centromeric transcript, other transcripts may be produced and function in mitosis. Importantly, our results show that RNA processing is not completely repressed during mitosis as previously thought (Shin and Manley, 2002). Further characterization of the RNAs and their downstream impact on cell division promises to be an interesting avenue for future research.
Materials and methods
Xenopus egg extracts, spindle assembly reactions, and drug treatments
X. laevis and X. tropicalis egg extracts were prepared and spindle assembly reactions performed according to established protocols (Maresca and Heald, 2006; Brown et al., 2007). In brief, metaphase-arrested extract was induced to enter interphase by the addition of 0.5 mM CaCl2, and subsequently induced to enter mitosis by the addition of one volume of metaphase-arrested extract after nuclei had completed DNA replication. Interphase nuclei for mitotic inhibition experiments were flash frozen in 25 µl aliquots of extract containing 8% (vol/vol) glycerol and stored at −80°C. Nuclei were thawed by the addition of 1 ml egg lysis buffer (250 mM sucrose, 50 mM KCl, 2.5 mM MgCl2, and 10 mM Hepes, pH 7.8) and pelleted at 1,600 g for 5 min at room temperature, then resuspended in fresh metaphase-arrested extract as described (Helmke and Heald, 2014). For mitotic inhibition experiments, fresh extract containing resuspended, cycled nuclei was split into separate reactions to which a final concentration of 100 µM ISGN (gift from D. Stanek, Institute of Molecular Genetics, Prague, Czech Republic; EMD Millipore) or 25 µM triptolide (TRIP; Sigma-Aldrich) was added. α-Amanitin (Sigma Aldrich) was used at a concentration of 50 µg/ml, ZM447439 at 5 µM, and cycloheximide (Sigma-Aldrich) at 100 and 1,000 µg/ml. Inhibitors were added and resuspended rapidly in mitotic extract, as lag times >60 s led to a significant decrease in phenotype severity and penetrance. Additionally, all egg lysis buffer was aspirated from pelleted nuclei as residual sucrose from the buffer masked the observed phenotypes. Great care was also taken during fixation of spindle reactions, as physical manipulation affected the recovery of fragile spindles resulting from inhibitor treatment. Centromere and kinetochore immunostaining experiments were performed in extracts treated throughout the entire cell cycle.
RNA-depleted extracts for centromere biogenesis assays were generated by treating cytostatic factor-arrested extract with 20 ng/µl RNase A for 30 min, then 1.5 U/µl RNasin (Promega) for 15 min before addition of nuclei and inhibitor.
Immunofluorescence, microscopy, and image analysis
All experiments were performed in at least biological triplicate. Immunofluorescence was performed as described (Maresca and Heald, 2006), except that all primary antibodies were used at 1:1000 dilution and allowed to reach equilibrium for at least 12 h at 4°C. Secondary antibody staining (Alexa Fluor 488–labeled anti-rabbit; Invitrogen) was performed concomitant with Hoechst staining at final concentrations of 1 µg/ml and 2 µg/ml, respectively, for 60 min at 21°C. Xenopus CENP-A and CENP-C antibodies were a gift from the Straight Lab (Stanford University, Palo Alto, CA), Xenopus NDC80 antibody was a gift from the Stukenberg Lab (University of Virginia, Charlottesville, VA), and Xenopus MCAK antibody was a gift from C. Walczak (Indiana University, Bloomington, IN). Images were obtained on an epifluorescence microscope (BX51; Olympus) under 40× magnification (0.75 NA; UPlanFl N; Olympus) with TRITC, DAPI, and FITC filters (Chroma Technology Corp.) by an Orca-ER cooled CCD camera (Hamamatsu Photonics).
Images were analyzed using a custom CellProfiler pipeline (Grenfell et al., 2016), which scales image intensities between 0 and 1 based on image bit depth. Spindle phenotypes were evaluated using microtubule density (median rhodamine tubulin fluorescence value within the segmented spindle structure) and solidity (area of spindle/area of the convex hull of the spindle; Fig. S1), and kinetochore phenotypes quantified by measuring the median FITC fluorescence intensity per antibody labeled focus. Analysis of centromeric (CENP-A, CENP-C, and NDC80) foci was performed on maximum intensity z-projected images assembled using FIJI from seven successive focal planes at 2-µm spacing. The fractional change in fluorescence intensity was calculated by subtracting the treatment median from the control median and dividing by the control median.
snRNA knockdown and immunodepletion
For U2 snRNA knockdown experiments, U2b DNA or scrambled control DNA oligonucleotides (Integrated DNA Technology; Black et al., 1985; Pan and Prives, 1988, 1989; Pan et al., 1989) were heated at 80°C for 8 min, cooled on ice, and added to metaphase-arrested extract to a final concentration of 2 mM and incubated at 19°C for 30 min to allow for antisense oligo–targeted degradation by endogenous RNase H. After incubation, 10 µl of extract was reserved for RNA isolation and the remaining extract was split and half was driven through interphase by the addition of 0.5 mM CaCl2 (final concentration) after addition of sperm nuclei. The other half was reserved on ice to drive the extract back into mitosis after DNA replication was complete.
For immunodepletion, 72.5 µl Protein G beads (Invitrogen) was used to couple 16 µg anti-TMG (trimethylguonosine; EMD Chemicals; Krämer et al., 1984) or nonimmune IgG (Sigma-Aldrich) antibodies in a PBS solution containing 0.2 µg/µl yeast tRNA (Sigma-Aldrich), 0.05% normal goat serum (Jackson ImmunoResearch), and 0.4 U/µl RNasin (Promega). Coupling was performed at room temperature for 1 h. 40 µl of extract was subjected to two rounds of immunodepletion (2 × 45 min on ice for X. laevis and 2 × 20 min at room temperature for X. tropicalis). After each round, 2 µl of extract was set aside to evaluate depletion efficiency by quantitative PCR (qPCR). After immunodepletion, extract was driven through interphase and analyzed for spindle assembly. RNPs associated with bead-coupled antibodies were analyzed as described previously (Stanĕk and Neugebauer, 2004), with minor modifications. In brief, beads containing antibody-bound RNP complexes were washed six times with NET-2 buffer (50 mM Tris-Cl, pH 7.5, 150 mM NaCl, and 0.05% Nonidet P-40) and kept on ice. After washing, beads were resuspended in NET-2 buffer containing 0.5% SDS and mixed in 1:1 (vol/vol) ratio with 5:1 acidic phenol chloroform (Sigma-Aldrich). After 1 h incubation at 37°C, they were centrifuged at 10,000 g and RNA was precipitated from the aqueous phase by acidic ethanol precipitation (300 mM sodium acetate, pH 5.5, and 70% ethanol). Isolated RNA was resolved on a 10% polyacrylamide urea gel and detected by silver staining or analyzed by RT-PCR (described in the next section). CENP-C immunoprecipitations were performed using the same protocol.
RNA isolation, RT-PCR, and qPCR analysis
RNA was isolated using TRIzol Reagent (Thermo Fisher Scientific) and 5:1 acidic phenol chloroform (Sigma-Aldrich) using the manufacturer’s protocol with minor modification. In brief, sample was homogenized in TRIzol and the protocol was followed through isopropanol precipitation. After precipitation, RNA was resuspended in Diethylpyrocarbonate-treated water and treated with RNase-free DNase I (Roche) for 30 min at 37°C or DNA Removal kit (Ambion) then extracted twice with acidic phenol chloroform before reverse transcription with Superscript III (Thermo Fisher Scientific) according to the manufacturer’s protocols using random nonamers (qPCR), oligo dT18 (qPCR), or fcr1 gene-specific primers (centromeric ncRNA analysis). RNA integrity was assessed by running 1 µg of extracted RNA on an agarose gel and staining with ethidium bromide.
qPCR was performed on cDNA generated with random nonamers (U2 snRNA knockdown experiments) or oligo-dT18 (immunodepletion experiments) and SuperScript III (Invitrogen), using SYBR Green qPCR SuperMix on an ABI Thermalcycler. Analysis was performed using the ΔΔCT method with H3, 5S, or 18S as the reference transcripts (Table S1).
RT-PCR analysis of centromeric ncRNAs was performed with Phusion High Fidelity DNA Polymerase (New England Biolabs, Inc.) on cDNA generated using gene specific primers. Primers (Integrated DNA Technologies) were used at 0.5 µM final concentration and cDNA at 2 ng/µl in standard reaction buffer. Touchdown PCR (Don et al., 1991) was performed using 10 touchdown cycles with annealing temperatures starting at 69°C and ending at 59°C followed by 30 cycles with an annealing temperature of 59°C. Extension times were constant throughout at 120 s. Negative controls were processed in parallel using the same protocol except that reverse transcription was omitted.
Online supplemental material
Fig. S1 shows the validation of our knockdown and immunodepletion experiments as well as phenotypes associated with splicing and transcription inhibition in Xenopus egg extract. Fig. S2 shows replicates of the experiments shown in Fig. 2 as well as isolated RNA used for our PCR assay. Fig. S3 shows centromere and kinetochore staining after transcription elongation inhibition; spindle phenotypes associated with Aurora B, NDC80, and Aurora B plus splicing inhibition; MCAK linescan after transcription or splicing inhibition; and partial rescue of the splicing spindle phenotype after microtubule stabilization. Table S1 contains primer sequences used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201604029/DC1.
Supplementary Material
Acknowledgments
We thank Andy Lane and the rest of the Heald laboratory for helpful discussions and comments on the manuscript. Special thanks to Lauren Slevin for TPX2 antibodies and protein and Xiao Zhou for the linescan plugin. We thank David Stanek, Aaron Straight, Todd Stukenberg, Claire Walczak, and Minoru Yoshida for reagents.
R. Heald is supported by National Institutes of Health grant R35 GM118183. A.W. Grenfell is supported by the National Science Foundation Graduate Research Fellowship Program and National Institutes of Health training grant 2T32GM007232-36. M. Strzelecka was supported by a European Molecular Biology Organization long-term fellowship.
The authors declare no competing financial interests.
Author contributions: M. Strzelecka originally conceived the project. M. Strzelecka and A.W. Grenfell carried out the experiments. A.W. Grenfell, R. Heald, and M. Strzelecka wrote the paper. M. Strzelecka and A.W. Grenfell prepared figures.
Footnotes
Abbreviations used in this paper:
- CENP
- centromere protein
- ISGN
- isoginkgetin
- MBP
- maltose binding protein
- MCAK
- mitotic centromere associated kinesin
- ncRNA
- noncoding RNA
- NDC80
- nuclear division cycle 80
- Pol II
- RNA polymerase II
- qPCR
- quantitative PCR
- snRNA
- small nuclear RNA
- TMG
- trimethylguanosine cap
- TPX2
- targeting protein for Xklp2
- TRIP
- triptolide
References
- Alló M., Buggiano V., Fededa J.P., Petrillo E., Schor I., de la Mata M., Agirre E., Plass M., Eyras E., Elela S.A., et al. 2009. Control of alternative splicing through siRNA-mediated transcriptional gene silencing. Nat. Struct. Mol. Biol. 16:717–724. 10.1038/nsmb.1620 [DOI] [PubMed] [Google Scholar]
- Ameyar-Zazoua M., Rachez C., Souidi M., Robin P., Fritsch L., Young R., Morozova N., Fenouil R., Descostes N., Andrau J.-C., et al. 2012. Argonaute proteins couple chromatin silencing to alternative splicing. Nat. Struct. Mol. Biol. 19:998–1004. 10.1038/nsmb.2373 [DOI] [PubMed] [Google Scholar]
- Andrews P.D., Ovechkina Y., Morrice N., Wagenbach M., Duncan K., Wordeman L., and Swedlow J.R.. 2004. Aurora B regulates MCAK at the mitotic centromere. Dev. Cell. 6:253–268. 10.1016/S1534-5807(04)00025-5 [DOI] [PubMed] [Google Scholar]
- Bergmann J.H., Jakubsche J.N., Martins N.M., Kagansky A., Nakano M., Kimura H., Kelly D.A., Turner B.M., Masumoto H., Larionov V., and Earnshaw W.C.. 2012. Epigenetic engineering: histone H3K9 acetylation is compatible with kinetochore structure and function. J. Cell Sci. 125:411–421. 10.1242/jcs.090639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird G., Zorio D.A.R., and Bentley D.L.. 2004. RNA polymerase II carboxy-terminal domain phosphorylation is required for cotranscriptional pre-mRNA splicing and 3′-end formation. Mol. Cell. Biol. 24:8963–8969. 10.1128/MCB.24.20.8963-8969.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black D.L., Chabot B., and Steitz J.A.. 1985. U2 as well as U1 small nuclear ribonucleoproteins are involved in premessenger RNA splicing. Cell. 42:737–750. 10.1016/0092-8674(85)90270-3 [DOI] [PubMed] [Google Scholar]
- Blower M.D. 2016. Centromeric transcription regulates Aurora-B localization and activation. Cell Reports. 15:1624–1633. 10.1016/j.celrep.2016.04.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blower M.D., Feric E., Weis K., and Heald R.. 2007. Genome-wide analysis demonstrates conserved localization of messenger RNAs to mitotic microtubules. J. Cell Biol. 179:1365–1373. 10.1083/jcb.200705163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodor D.L., Mata J.F., Sergeev M., David A.F., Salimian K.J., Panchenko T., Cleveland D.W., Black B.E., Shah J.V., and Jansen L.E.. 2014. The quantitative architecture of centromeric chromatin. eLife. 3:e02137 10.7554/eLife.02137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown K.S., Blower M.D., Maresca T.J., Grammer T.C., Harland R.M., and Heald R.. 2007. Xenopus tropicalis egg extracts provide insight into scaling of the mitotic spindle. J. Cell Biol. 176:765–770. 10.1083/jcb.200610043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardinale S., Bergmann J.H., Kelly D., Nakano M., Valdivia M.M., Kimura H., Masumoto H., Larionov V., and Earnshaw W.C.. 2009. Hierarchical inactivation of a synthetic human kinetochore by a chromatin modifier. Mol. Biol. Cell. 20:4194–4204. 10.1091/mbc.E09-06-0489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmena M., Wheelock M., Funabiki H., and Earnshaw W.C.. 2012. The chromosomal passenger complex (CPC): from easy rider to the godfather of mitosis. Nat. Rev. Mol. Cell Biol. 13:789–803. 10.1038/nrm3474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan F.L., and Wong L.H.. 2012. Transcription in the maintenance of centromere chromatin identity. Nucleic Acids Res. 40:11178–11188. 10.1093/nar/gks921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan F.L., Marshall O.J., Saffery R., Kim B.W., Earle E., Choo K.H.A., and Wong L.H.. 2012. Active transcription and essential role of RNA polymerase II at the centromere during mitosis. Proc. Natl. Acad. Sci. U S A. 109:1979–1984. 10.1073/pnas.1108705109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.-C., Bowers S., Lipinszki Z., Palladino J., Trusiak S., Bettini E., Rosin L., Przewloka M.R., Glover D.M., O’Neill R.J., and Mellone B.G.. 2015. Establishment of centromeric chromatin by the CENP-A assembly factor CAL1 requires FACT-mediated transcription. Dev. Cell. 34:73–84. 10.1016/j.devcel.2015.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chueh A.C., Northrop E.L., Brettingham-Moore K.H., Choo K.H.A., and Wong L.H.. 2009. LINE retrotransposon RNA is an essential structural and functional epigenetic component of a core neocentromeric chromatin. PLoS Genet. 5:e1000354 (published erratum appears in PLoS Genet. 2009. 5) 10.1371/journal.pgen.1000354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David C.J., Boyne A.R., Millhouse S.R., and Manley J.L.. 2011. The RNA polymerase II C-terminal domain promotes splicing activation through recruitment of a U2AF65-Prp19 complex. Genes Dev. 25:972–983. 10.1101/gad.2038011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Don R.H., Cox P.T., Wainwright B.J., Baker K., and Mattick J.S.. 1991. ‘Touchdown’ PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 19:4008 10.1093/nar/19.14.4008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y., Topp C.N., and Dawe R.K.. 2010. DNA binding of centromere protein C (CENPC) is stabilized by single-stranded RNA. PLoS Genet. 6:e1000835 10.1371/journal.pgen.1000835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards N.S., and Murray A.W.. 2005. Identification of Xenopus CENP-A and an associated centromeric DNA repeat. Mol. Biol. Cell. 16:1800–1810. 10.1091/mbc.E04-09-0788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuele M.J., Lan W., Jwa M., Miller S.A., Chan C.S.M., and Stukenberg P.T.. 2008. Aurora B kinase and protein phosphatase 1 have opposing roles in modulating kinetochore assembly. J. Cell Biol. 181:241–254. 10.1083/jcb.200710019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ems-McClung S.C., Hainline S.G., Devare J., Zong H., Cai S., Carnes S.K., Shaw S.L., and Walczak C.E.. 2013. Aurora B inhibits MCAK activity through a phosphoconformational switch that reduces microtubule association. Curr. Biol. 23:2491–2499. 10.1016/j.cub.2013.10.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferri F., Bouzinba-Segard H., Velasco G., Hubé F., and Francastel C.. 2009. Non-coding murine centromeric transcripts associate with and potentiate Aurora B kinase. Nucleic Acids Res. 37:5071–5080. 10.1093/nar/gkp529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadea B.B., and Ruderman J.V.. 2005. Aurora kinase inhibitor ZM447439 blocks chromosome-induced spindle assembly, the completion of chromosome condensation, and the establishment of the spindle integrity checkpoint in Xenopus egg extracts. Mol. Biol. Cell. 16:1305–1318. 10.1091/mbc.E04-10-0891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascoigne K.E., Takeuchi K., Suzuki A., Hori T., Fukagawa T., and Cheeseman I.M.. 2011. Induced ectopic kinetochore assembly bypasses the requirement for CENP-A nucleosomes. Cell. 145:410–422. 10.1016/j.cell.2011.03.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gent J.I., and Dawe R.K.. 2012. RNA as a structural and regulatory component of the centromere. Annu. Rev. Genet. 46:443–453. 10.1146/annurev-genet-110711-155419 [DOI] [PubMed] [Google Scholar]
- Goshima G., Wollman R., Goodwin S.S., Zhang N., Scholey J.M., Vale R.D., and Stuurman N.. 2007. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science. 316:417–421. 10.1126/science.1141314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenfell A.W., Strzelecka M., Crowder M.E., Helmke K.J., Schlaitz A.-L., and Heald R.. 2016. A versatile multivariate image analysis pipeline reveals features of Xenopus extract spindles. J. Cell Biol. 213:127–136. 10.1083/jcb.201509079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmke K.J., and Heald R.. 2014. TPX2 levels modulate meiotic spindle size and architecture in Xenopus egg extracts. J. Cell Biol. 206:385–393. 10.1083/jcb.201401014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann J.C., Tegha-Dunghu J., Dräger S., Will C.L., Lührmann R., and Gruss O.J.. 2013. The Prp19 complex directly functions in mitotic spindle assembly. PLoS One. 8:e74851 10.1371/journal.pone.0074851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer J., and Tsai M.-Y.. 2012. A novel role for TPX2 as a scaffold and co-activator protein of the chromosomal passenger complex. Cell. Signal. 24:1677–1689. 10.1016/j.cellsig.2012.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jambhekar A., Emerman A.B., Schweidenback C.T.H., and Blower M.D.. 2014. RNA stimulates Aurora B kinase activity during mitosis. PLoS One. 9:e100748 10.1371/journal.pone.0100748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittler R., Pelletier L., Heninger A.-K., Slabicki M., Theis M., Miroslaw L., Poser I., Lawo S., Grabner H., Kozak K., et al. 2007. Genome-scale RNAi profiling of cell division in human tissue culture cells. Nat. Cell Biol. 9:1401–1412. 10.1038/ncb1659 [DOI] [PubMed] [Google Scholar]
- Krämer A., Keller W., Appel B., and Lührmann R.. 1984. The 5′ terminus of the RNA moiety of U1 small nuclear ribonucleoprotein particles is required for the splicing of messenger RNA precursors. Cell. 38:299–307. 10.1016/0092-8674(84)90551-8 [DOI] [PubMed] [Google Scholar]
- Lan W., Zhang X., Kline-Smith S.L., Rosasco S.E., Barrett-Wilt G.A., Shabanowitz J., Hunt D.F., Walczak C.E., and Stukenberg P.T.. 2004. Aurora B phosphorylates centromeric MCAK and regulates its localization and microtubule depolymerization activity. Curr. Biol. 14:273–286. 10.1016/j.cub.2004.01.055 [DOI] [PubMed] [Google Scholar]
- Listerman I., Sapra A.K., and Neugebauer K.M.. 2006. Cotranscriptional coupling of splicing factor recruitment and precursor messenger RNA splicing in mammalian cells. Nat. Struct. Mol. Biol. 13:815–822. 10.1038/nsmb1135 [DOI] [PubMed] [Google Scholar]
- Ma N., Titus J., Gable A., Ross J.L., and Wadsworth P.. 2011. TPX2 regulates the localization and activity of Eg5 in the mammalian mitotic spindle. J. Cell Biol. 195:87–98. 10.1083/jcb.201106149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maison C., Bailly D., Roche D., Montes de Oca R., Probst A.V., Vassias I., Dingli F., Lombard B., Loew D., Quivy J.-P., and Almouzni G.. 2011. SUMOylation promotes de novo targeting of HP1α to pericentric heterochromatin. Nat. Genet. 43:220–227. 10.1038/ng.765 [DOI] [PubMed] [Google Scholar]
- Makarov E.M., Makarova O.V., Urlaub H., Gentzel M., Will C.L., Wilm M., and Lührmann R.. 2002. Small nuclear ribonucleoprotein remodeling during catalytic activation of the spliceosome. Science. 298:2205–2208. 10.1126/science.1077783 [DOI] [PubMed] [Google Scholar]
- Maresca T.J., and Heald R.. 2006. Methods for studying spindle assembly and chromosome condensation in Xenopus egg extracts. Methods Mol. Biol. 322:459–474. 10.1007/978-1-59745-000-3_33 [DOI] [PubMed] [Google Scholar]
- McCleland M.L., Gardner R.D., Kallio M.J., Daum J.R., Gorbsky G.J., Burke D.J., and Stukenberg P.T.. 2003. The highly conserved Ndc80 complex is required for kinetochore assembly, chromosome congression, and spindle checkpoint activity. Genes Dev. 17:101–114. 10.1101/gad.1040903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miga K.H., Newton Y., Jain M., Altemose N., Willard H.F., and Kent W.J.. 2014. Centromere reference models for human chromosomes X and Y satellite arrays. Genome Res. 24:697–707. 10.1101/gr.159624.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montembault E., Dutertre S., Prigent C., and Giet R.. 2007. PRP4 is a spindle assembly checkpoint protein required for MPS1, MAD1, and MAD2 localization to the kinetochores. J. Cell Biol. 179:601–609. 10.1083/jcb.200703133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano M., Okamoto Y., Ohzeki J., and Masumoto H.. 2003. Epigenetic assembly of centromeric chromatin at ectopic alpha-satellite sites on human chromosomes. J. Cell Sci. 116:4021–4034. 10.1242/jcs.00697 [DOI] [PubMed] [Google Scholar]
- Nakano M., Cardinale S., Noskov V.N., Gassmann R., Vagnarelli P., Kandels-Lewis S., Larionov V., Earnshaw W.C., and Masumoto H.. 2008. Inactivation of a human kinetochore by specific targeting of chromatin modifiers. Dev. Cell. 14:507–522. 10.1016/j.devcel.2008.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann B., Walter T., Hériché J.-K., Bulkescher J., Erfle H., Conrad C., Rogers P., Poser I., Held M., Liebel U., et al. 2010. Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature. 464:721–727. 10.1038/nature08869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newport J., and Kirschner M.. 1982a A major developmental transition in early Xenopus embryos: I. characterization and timing of cellular changes at the midblastula stage. Cell. 30:675–686. 10.1016/0092-8674(82)90272-0 [DOI] [PubMed] [Google Scholar]
- Newport J., and Kirschner M.. 1982b A major developmental transition in early Xenopus embryos: II. Control of the onset of transcription. Cell. 30:687–696. 10.1016/0092-8674(82)90273-2 [DOI] [PubMed] [Google Scholar]
- Nguyen V.T., Giannoni F., Dubois M.F., Seo S.J., Vigneron M., Kédinger C., and Bensaude O.. 1996. In vivo degradation of RNA polymerase II largest subunit triggered by alpha-amanitin. Nucleic Acids Res. 24:2924–2929. 10.1093/nar/24.15.2924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien K., Matlin A.J., Lowell A.M., and Moore M.J.. 2008. The biflavonoid isoginkgetin is a general inhibitor of Pre-mRNA splicing. J. Biol. Chem. 283:33147–33154. 10.1074/jbc.M805556200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Z.Q., and Prives C.. 1988. Assembly of functional U1 and U2 human-amphibian hybrid snRNPs in Xenopus laevis oocytes. Science. 241:1328–1331. 10.1126/science.2970672 [DOI] [PubMed] [Google Scholar]
- Pan Z.Q., and Prives C.. 1989. U2 snRNA sequences that bind U2-specific proteins are dispensable for the function of U2 snRNP in splicing. Genes Dev. 3(12a, 12A):1887–1898. 10.1101/gad.3.12a.1887 [DOI] [PubMed] [Google Scholar]
- Pan Z.Q., Ge H., Fu X.Y., Manley J.L., and Prives C.. 1989. Oligonucleotide-targeted degradation of U1 and U2 snRNAs reveals differential interactions of simian virus 40 pre-mRNAs with snRNPs. Nucleic Acids Res. 17:6553–6568. 10.1093/nar/17.16.6553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petry S., Groen A.C., Ishihara K., Mitchison T.J., and Vale R.D.. 2013. Branching microtubule nucleation in Xenopus egg extracts mediated by augmin and TPX2. Cell. 152:768–777. 10.1016/j.cell.2012.12.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quénet D., and Dalal Y.. 2014. A long non-coding RNA is required for targeting centromeric protein A to the human centromere. eLife. 3:e03254 10.7554/eLife.03254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rago F., Gascoigne K.E., and Cheeseman I.M.. 2015. Distinct organization and regulation of the outer kinetochore KMN network downstream of CENP-C and CENP-T. Curr. Biol. 25:671–677. 10.1016/j.cub.2015.01.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rošić S., Köhler F., and Erhardt S.. 2014. Repetitive centromeric satellite RNA is essential for kinetochore formation and cell division. J. Cell Biol. 207:335–349. (published erratum appears in J. Cell Biol. 2014. 207:673) 10.1083/jcb.201404097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd M.D., and Luse D.S.. 1996. Amanitin greatly reduces the rate of transcription by RNA polymerase II ternary complexes but fails to inhibit some transcript cleavage modes. J. Biol. Chem. 271:21549–21558. 10.1074/jbc.271.35.21549 [DOI] [PubMed] [Google Scholar]
- Saffery R., Sumer H., Hassan S., Wong L.H., Craig J.M., Todokoro K., Anderson M., Stafford A., and Choo K.H.A.. 2003. Transcription within a functional human centromere. Mol. Cell. 12:509–516. 10.1016/S1097-2765(03)00279-X [DOI] [PubMed] [Google Scholar]
- Sampath S.C., Ohi R., Leismann O., Salic A., Pozniakovski A., and Funabiki H.. 2004. The chromosomal passenger complex is required for chromatin-induced microtubule stabilization and spindle assembly. Cell. 118:187–202. 10.1016/j.cell.2004.06.026 [DOI] [PubMed] [Google Scholar]
- Scott K.C. 2013. Transcription and ncRNAs: at the cent(rome)re of kinetochore assembly and maintenance. Chromosome Res. 21:643–651. 10.1007/s10577-013-9387-3 [DOI] [PubMed] [Google Scholar]
- Shin C., and Manley J.L.. 2002. The SR protein SRp38 represses splicing in M phase cells. Cell. 111:407–417. 10.1016/S0092-8674(02)01038-3 [DOI] [PubMed] [Google Scholar]
- Somma M.P., Ceprani F., Bucciarelli E., Naim V., De Arcangelis V., Piergentili R., Palena A., Ciapponi L., Giansanti M.G., Pellacani C., et al. 2008. Identification of Drosophila mitotic genes by combining co-expression analysis and RNA interference. PLoS Genet. 4:e1000126 10.1371/journal.pgen.1000126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanĕk D., and Neugebauer K.M.. 2004. Detection of snRNP assembly intermediates in Cajal bodies by fluorescence resonance energy transfer. J. Cell Biol. 166:1015–1025. 10.1083/jcb.200405160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanenbaum M.E., and Medema R.H.. 2011. Localized Aurora B activity spatially controls non-kinetochore microtubules during spindle assembly. Chromosoma. 120:599–607. 10.1007/s00412-011-0334-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titov D.V., Gilman B., He Q.-L., Bhat S., Low W.-K., Dang Y., Smeaton M., Demain A.L., Miller P.S., Kugel J.F., et al. 2011. XPB, a subunit of TFIIH, is a target of the natural product triptolide. Nat. Chem. Biol. 7:182–188. 10.1038/nchembio.522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl M.C., Will C.L., and Lührmann R.. 2009. The spliceosome: design principles of a dynamic RNP machine. Cell. 136:701–718. 10.1016/j.cell.2009.02.009 [DOI] [PubMed] [Google Scholar]
- Wang E., Ballister E.R., and Lampson M.A.. 2011. Aurora B dynamics at centromeres create a diffusion-based phosphorylation gradient. J. Cell Biol. 194:539–549. 10.1083/jcb.201103044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong L.H., Brettingham-Moore K.H., Chan L., Quach J.M., Anderson M.A., Northrop E.L., Hannan R., Saffery R., Shaw M.L., Williams E., and Choo K.H.A.. 2007. Centromere RNA is a key component for the assembly of nucleoproteins at the nucleolus and centromere. Genome Res. 17:1146–1160. 10.1101/gr.6022807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Lan W., Ems-McClung S.C., Stukenberg P.T., and Walczak C.E.. 2007. Aurora B phosphorylates multiple sites on mitotic centromere-associated kinesin to spatially and temporally regulate its function. Mol. Biol. Cell. 18:3264–3276. 10.1091/mbc.E07-01-0086 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.