Meitinger et al. perform a genome-wide CRISPR/Cas9 screen for centrinone resistance and identify a 53BP1-USP28 module as critical for communicating mitotic challenges to the p53 circuit and TRIM37 as an enforcer of the singularity of centrosome assembly.
Abstract
In normal human cells, centrosome loss induced by centrinone—a specific centrosome duplication inhibitor—leads to irreversible, p53-dependent G1 arrest by an unknown mechanism. A genome-wide CRISPR/Cas9 screen for centrinone resistance identified genes encoding the p53-binding protein 53BP1, the deubiquitinase USP28, and the ubiquitin ligase TRIM37. Deletion of TP53BP1, USP28, or TRIM37 prevented p53 elevation in response to centrosome loss but did not affect cytokinesis failure–induced arrest or p53 elevation after doxorubicin-induced DNA damage. Deletion of TP53BP1 and USP28, but not TRIM37, prevented growth arrest in response to prolonged mitotic duration. TRIM37 knockout cells formed ectopic centrosomal-component foci that suppressed mitotic defects associated with centrosome loss. TP53BP1 and USP28 knockouts exhibited compromised proliferation after centrosome removal, suggesting that centrosome-independent proliferation is not conferred solely by the inability to sense centrosome loss. Thus, analysis of centrinone resistance identified a 53BP1-USP28 module as critical for communicating mitotic challenges to the p53 circuit and TRIM37 as an enforcer of the singularity of centrosome assembly.
Introduction
The tumor suppressor TP53, which encodes a transcription factor, is the most commonly mutated gene in cancer cells (Levine and Oren, 2009). The p53 circuit responds to a myriad of stress signals, including DNA damage, oncogene activation, metabolic fluctuations, and mitotic aberrations, and drives programs that can lead to temporary arrest, senescence, or cell death (Levine and Oren, 2009; Uetake and Sluder, 2010; Ganem et al., 2014; Gurpinar and Vousden, 2015; Kruiswijk et al., 2015). In the absence of stress, p53 is continuously targeted for proteasomal degradation by the ubiquitin ligase Mdm2 (Levav-Cohen et al., 2014; Karni-Schmidt et al., 2016). Layers of positive and negative regulation of p53 are imposed through an extensive array of posttranslational modifications (Gu and Zhu, 2012; Jenkins et al., 2012; Nguyen et al., 2014).
Recent work has shown that p53 monitors mitosis, although the underlying mechanisms remain poorly understood. Work in hTERT-RPE1 immortalized retinal pigment epithelial cells (RPE1 cells) has shown that if the duration of mitosis exceeds a relatively sharp temporal cutoff (∼1.5 h), the resulting daughter cells arrest in a p53-dependent fashion at the subsequent G1/S transition (Uetake and Sluder, 2010). Similarly, triggering cytokinesis failure by chemical disruption of the actin cytoskeleton leads to a p53-dependent arrest through activation of the Hippo tumor suppressor pathway (Ganem et al., 2014).
Centrosome loss has also been shown to activate p53 (Bazzi and Anderson, 2014; Lambrus et al., 2015; Wong et al., 2015). Centrioles organize pericentriolar material to form centrosomes that nucleate and anchor microtubules (Woodruff et al., 2014; Conduit et al., 2015). Centriole duplication is tightly controlled to ensure that mitotic cells have precisely two centrosomes. Centrioles duplicate in S-phase, when the polo family kinase Plk4 triggers formation of a single daughter adjacent to each mother centriole (Gönczy, 2012; Zitouni et al., 2014; Fu et al., 2015). To analyze the effect of centrosome removal in normal and cancer cells, we developed centrinone, a potent specific Plk4 inhibitor (Wong et al., 2015). Centrinone treatment blocks centriole duplication, leading to progressive loss of centrosomes as cells divide. Most cell lines with cancer-associated mutations (which frequently target the p53 circuit) continue to proliferate after centrinone-mediated centrosome removal, albeit at a reduced rate because of an increase in mitotic errors (Wong et al., 2015), which is consistent with prior work (Khodjakov and Rieder, 2001; Sir et al., 2013).
In contrast to cancer-derived cell lines, RPE1 and three primary cell cultures exhibited irreversible G1 arrest after centrinone-induced centrosome loss (Wong et al., 2015). Comparable results were also observed after auxin-induced Plk4 degradation (Lambrus et al., 2015). The centrosome loss–associated G1 arrest required p53 (Lambrus et al., 2015; Wong et al., 2015). Similarly, centriole removal from mouse embryos by genetic disruption of the centriole component Sas4 led to a wave of p53-dependent apoptosis at embryonic day 8.5, suggesting that a p53-based mechanism can detect centrosome loss beginning at midgestation (Bazzi and Anderson, 2014).
Although several potential mechanisms have been excluded (Lambrus et al., 2015; Wong et al., 2015), the molecular details of centrosome loss–mediated p53 stabilization remain unclear. To address this, we performed an unbiased genome-wide CRISPR/Cas9-based screen to identify genes whose loss enables RPE1 cells to proliferate in centrinone. This approach identified a two-protein module that can communicate centrosome loss or extended mitotic duration to the p53 circuit and revealed a ubiquitin ligase whose loss perturbs the singularity of centrosome assembly, resulting in ectopic assemblies that bypass the detrimental consequences of acentrosomal mitosis.
Results and discussion
A genome-wide CRISPR/Cas9 screen identifies genes required to activate p53 upon centrosome loss
To understand how centrosome loss elevates p53, we performed a genome-wide CRISPR/Cas9-based screen to identify genes whose mutation allows RPE1 cells to grow in the presence of centrinone (Fig. 1, A and B). We reasoned that genes required for p53-dependent arrest of centrosomeless cells would fall into two categories: genes specifically required to elevate p53 levels in response to centrosome loss (type 1) and p53 itself and its downstream effectors, such as p21, required to convert p53 activation into a durable G1 arrest (type 2; Levine and Oren, 2009; Kruiswijk et al., 2015). To identify type 1 genes, we used a secondary screen challenging colonies that grew in the presence of centrinone with an Mdm2 inhibitor (Mdm2i) that stabilizes p53 by preventing it from binding its negative regulator Mdm2 (Ding et al., 2009; Khoo et al., 2014). Type 1 colonies would grow in centrinone but not in Mdm2i; in contrast, type 2 colonies would grow in both centrinone and Mdm2i (Fig. 1 B).
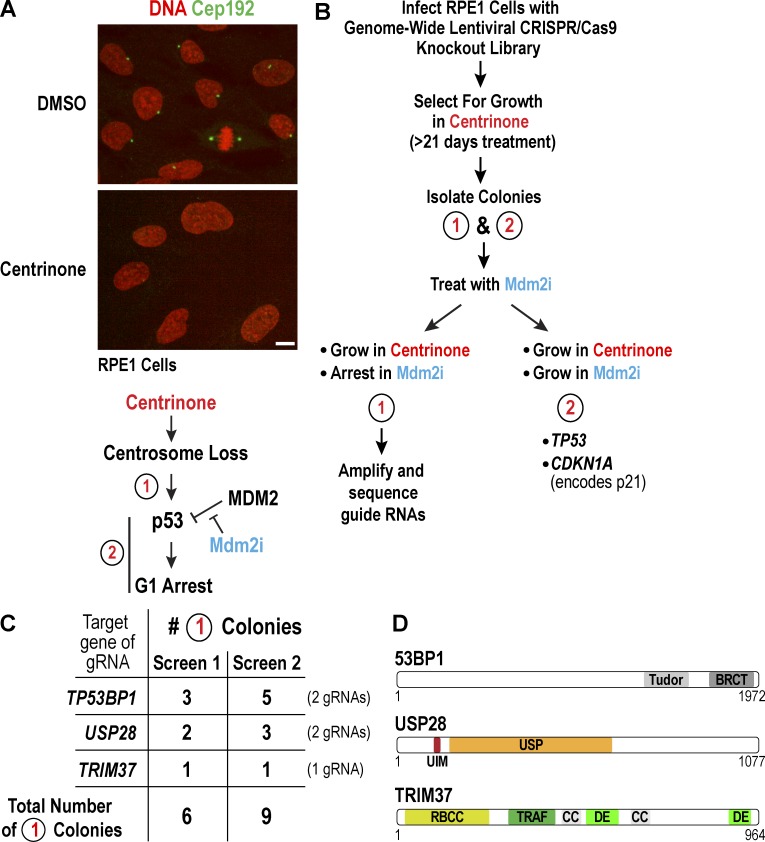
Figure 1.
Genome-wide CRISPR/Cas9 screen for genes involved in activating p53 upon centrosome loss. (A, top) Immunofluorescence images of RPE1 cells, stained for DNA (red) and the centrosomal protein Cep192 (green), after treatment with DMSO or centrinone for 5 d. Bar, 10 µm. (bottom) Schematic highlighting the two classes of genes that would be identified in a centrinone-resistance screen. (B) Summary of the screen designed to identify genes that activate p53 in response to centrosome loss. (C) Table summarizing the results of two independent screens. All 15 colonies had one of the three listed genes deleted; no colony had more than one. (D) Schematic of the three proteins 53BP1, USP28, and TRIM37, identified by the screen.
We performed two independent screens using a lentiviral human CRISPR/Cas9 library (see Methods). After growth in centrinone for 20–25 d, colonies were picked and grown without centrinone for 5–10 d; 32 colonies were obtained in the first screen and 75 in the second. After expansion, a representative fraction of each colony was treated with Mdm2i and scored 7 d later. This approach identified 15 type 1 colonies (Fig. 1 C). Direct amplification of the guide RNAs (gRNAs) in each colony (Fig. S1 A) identified gRNAs targeting one of three genes: TP53BP1, encoding the DNA damage regulator 53BP1; USP28, encoding a deubiquitinase; and TRIM37, encoding a ubiquitin ligase (Fig. 1, C and D). For TP53BP1 and USP28, two different gRNAs were recovered (Fig. 1 C). In parallel, the 32 colonies that grew in centrinone in the first screen (type 1 and type 2) were pooled, and the amplified gRNAs were subjected to deep sequencing. In addition to identifying two gRNAs targeting TP53BP1 and USP28, this approach identified five gRNAs targeting TP53 and three targeting CDKN1A, both expected type 2 hits (Fig. S1 B). The one gRNA targeting TRIM37 was also on this list, but would not have stood out as a hit worth pursuing without the parallel colony sequencing approach.
Loss of 53BP1, USP28, or TRIM37 suppresses p53 elevation and proliferation arrest triggered by centrosome loss
To confirm the three type 1 hits from the screens and rule out possible contributions from other gRNAs, we used single gRNAs with centrinone selection to generate new RPE1 knockout cell lines for TP53BP1, TRIM37, and USP28 (Fig. 2 A and Fig. S2 A). All three cell lines were validated by immunoblotting (Fig. 2 A) and proliferated indefinitely in the absence of centrinone, indicating that these genes are not essential. By immunofluorescence, 53BP1 and USP28 exhibited a nuclear signal that was absent in the knockout lines (Fig. S2 B) and did not appear to change in intensity after centrosome loss (not depicted). For TRIM37, despite testing 10 different commercial antibodies, we were unable to observe any immunostaining that differed between control and knockout cells. Centrinone treatment depleted centrosomes (identified as foci costaining for the markers γ-tubulin and Cep192) in all knockout cell lines (Fig. S2 C).
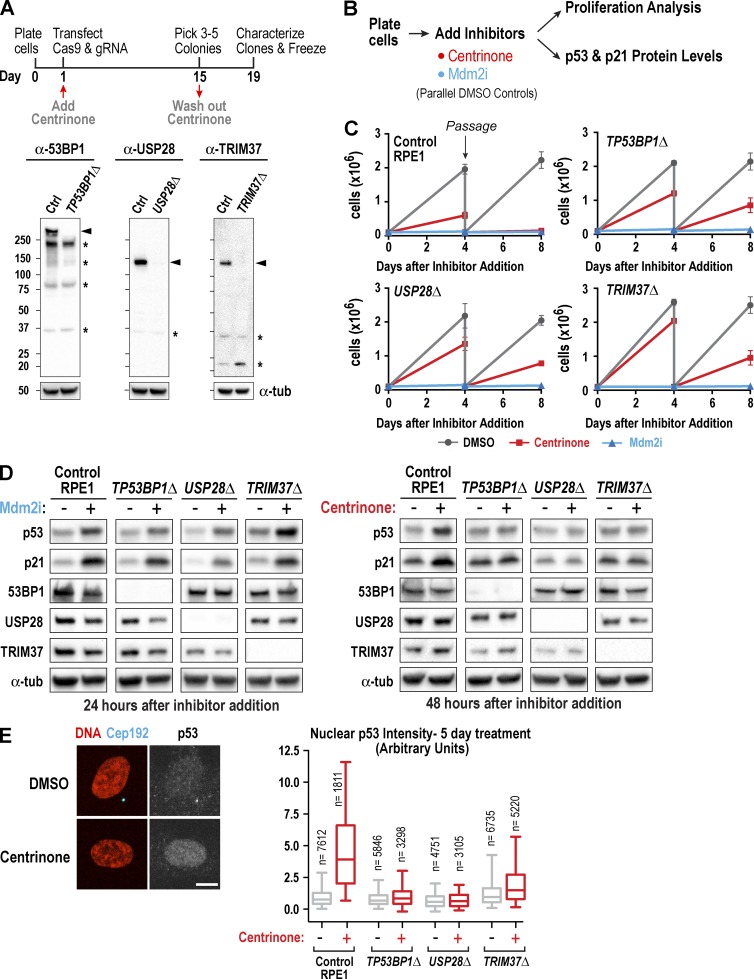
Figure 2.
Loss of TP53BP1, USP28, or TRIM37 suppresses p53 elevation and proliferation arrest triggered by centrosome loss. (A, top) Outline of the procedure used to generate RPE1 knockouts. (A, bottom) Immunoblots of extracts from control (Ctrl) and knockout RPE1 lines. Bands corresponding to each protein (arrowheads) and nonspecific bands (asterisks) are indicated. α-Tubulin serves as a loading control. (B) Outline of cell proliferation analysis and assessment of p53 and p21 levels after acute treatment with centrinone or Mdm2i. (C) Graphs plotting the results of passaging assays monitoring the growth of control and knockout RPE1 cell lines after addition at day 0 of DMSO (vehicle), centrinone, or Mdm2i. (D) Immunoblots probed with the indicated antibodies after addition of Mdm2i (left) or centrinone (right). α-Tubulin (α-tub) serves as a loading control. (E) Immunofluorescence analysis of Cep192 and p53 after 5-d centrinone treatment. Representative images (left) and graph (right) plotting the distributions of nuclear p53 fluorescence for one of three experiments (for quantification of the other two experiments, see Fig. S2 E). Graph shows 5–95% box-and-whiskers plots. Bar, 10 µm.
Next, we used a passage-based cell counting assay to measure the proliferative capacity of the three knockout lines in the presence of centrinone and Mdm2i. Consistent with prior work (Wong et al., 2015), control RPE1 cells failed to proliferate when they were passaged 4 d after centrinone or Mdm2i addition (Fig. 2, B and C). The TP53BP1Δ, USP28Δ, and TRIM37Δ cell lines all ceased proliferation after Mdm2i addition, with kinetics identical to that of controls. In contrast, the three knockouts continued to proliferate, albeit at slower rates, when they were passaged 4 d after centrinone addition (Fig. 2, B and C). The reduced proliferation rates were comparable to those observed previously for centrinone-treated cancer cell lines (Wong et al., 2015), suggesting that they result from error-prone mitosis after centrosome removal. Consistent with these results, immunoblotting revealed that elevation of p53 and p21 was still observed after Mdm2i treatment but was greatly suppressed in the three knockout cell lines after 2-d centrinone treatment (Fig. 2 D). We also quantified nuclear p53 signal by immunofluorescence 5 d after centrinone addition. The centrinone-dependent approximately fivefold increase in nuclear p53 observed in control RPE1 cells was absent in the TP53BP1Δ and USP28Δ cell lines and was largely, but not completely, suppressed in the TRIM37Δ cell line (Figs. 2 E and S2 D).
53BP1 and USP28, but not TRIM37, are essential for p53 activation in response to prolonged mitotic duration
Beyond centrosome loss, p53 is activated by stresses such as DNA damage, cytokinesis failure, and extended mitotic duration (Jackson and Bartek, 2009; Uetake and Sluder, 2010; Ganem et al., 2014; Williams and Schumacher, 2016). To understand their roles, we assessed the impact of 53BP1, USP28, and TRIM37 loss on each of these distinct means of p53 activation. We found that all three knockouts exhibited a normal p53 response after DNA damage caused by doxorubicin (Fig. 3 A), a DNA-intercalating topoisomerase poison (Jackson and Bartek, 2009). To analyze cytokinesis failure, wild-type and mutant cell lines expressing RFP::histone H2b were treated for 24 h with the actin polymerization inhibitor cytochalasin D, and the fate of the resulting tetraploid cells was followed by live imaging for 3 d (Fig. 3 B). Approximately 97% of tetraploid cells expressing a stable shRNA targeting p53 entered mitosis. In contrast, the TP53BP1Δ, USP28Δ, and TRIM37Δ cell lines all exhibited low percentages of tetraploid cells entering mitosis, comparable to control RPE1 cells (Fig. 3 B), indicating that these genes are not required for the cell cycle arrest induced by cytokinesis failure.
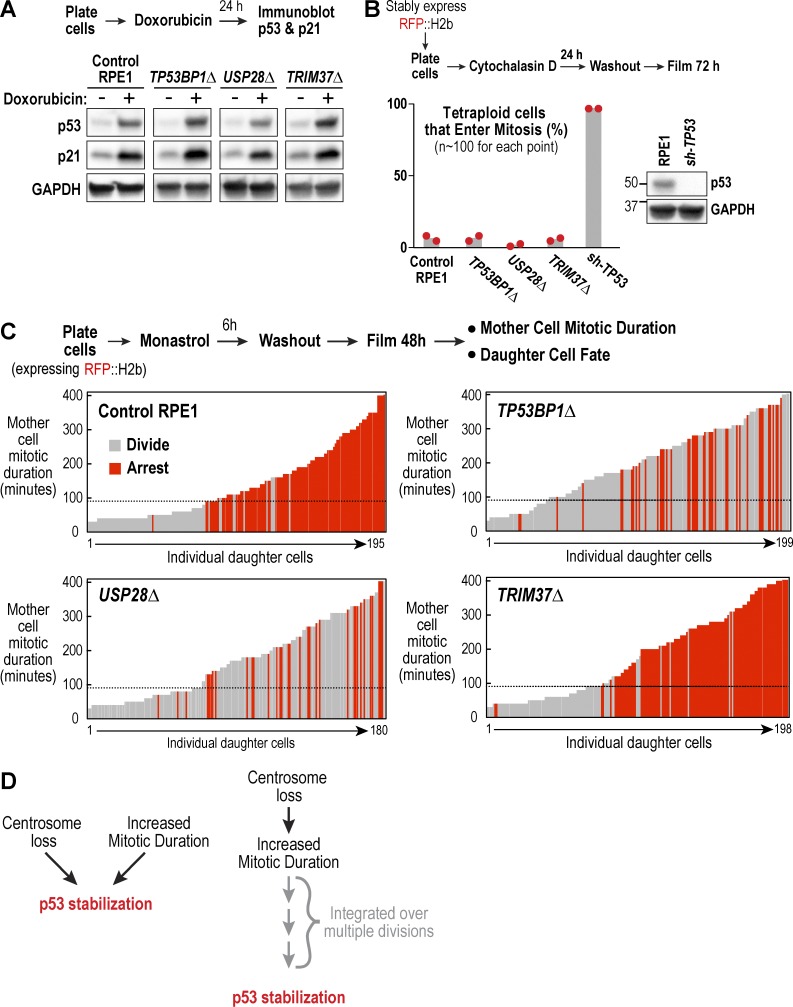
Figure 3.
53BP1 and USP28, but not TRIM37, are essential for activating p53 in response to prolonged mitotic duration. (A) Analysis of p53 and p21 levels after induction of DNA damage with doxorubicin; schematic describes experimental protocol and GAPDH serves as a loading control. (B) Analysis of cytokinesis failure–induced division arrest; schematic describes experimental protocol. Red dots show results from two independent experiments. Immunoblot confirms efficient p53 depletion. (C) Analysis of extended mitotic duration–induced division arrest; schematic describes experimental protocol. Vertical bars represent individual daughter cells. Bar height shows the time the mother cells spent in mitosis, and bar color indicates whether they arrested (red) or divided (gray). Black dashed line marks the mitotic duration cutoff in control RPE1 cells, after which resulting daughter cells arrest in G1. (D) Schematic shows two possible models for how centrosome loss might trigger p53 activation, either directly (left) or indirectly through successive prolonged mitoses (right).
To analyze p53-mediated arrest caused by extended mitotic duration, we used the procedure schematized in Fig. 3 C (based on Uetake and Sluder [2010]) in which treatment with the Eg5 inhibitor monastrol was used to prolong mitosis for varying amounts of time, and daughter cells were followed for 48 h after drug washout to determine whether they arrested or divided. In control RPE1 and TRIM37Δ cells, mitotic durations greater than 90 min led to penetrant arrest of the resulting daughter cells. In contrast, both TP53BP1Δ and USP28Δ cells failed to arrest in response to extended mitotic duration. Thus, 53BP1 and USP28 are required to arrest cells after either centrosome loss or extended mitotic duration.
Centrosome loss slows spindle assembly and increases mitotic duration (Wong et al., 2015; see Fig. 5, A and C), raising the question of whether centrosome loss–mediated p53 activation occurs via extending mitosis (Fig. 3 D). In an attempt to address this, we pretreated cells for 24 h with centrinone, at which point the majority of cells have one centrosome and do not yet have elevated p53 (Fig. S2 E). The cells were then incubated with centrinone plus an Mps1 inhibitor (NMS-P715, which suppresses the spindle checkpoint) for 24 h to determine whether shortening mitosis could block the p53 elevation that occurs as the first wave of cells go from having one to zero centrosomes. Mps1 inhibition suppressed centrinone-mediated p53 elevation but also resulted in penetrant cytokinesis failure that likely prevented centrosome removal (Fig. S2 F), thereby complicating interpretation of this result.
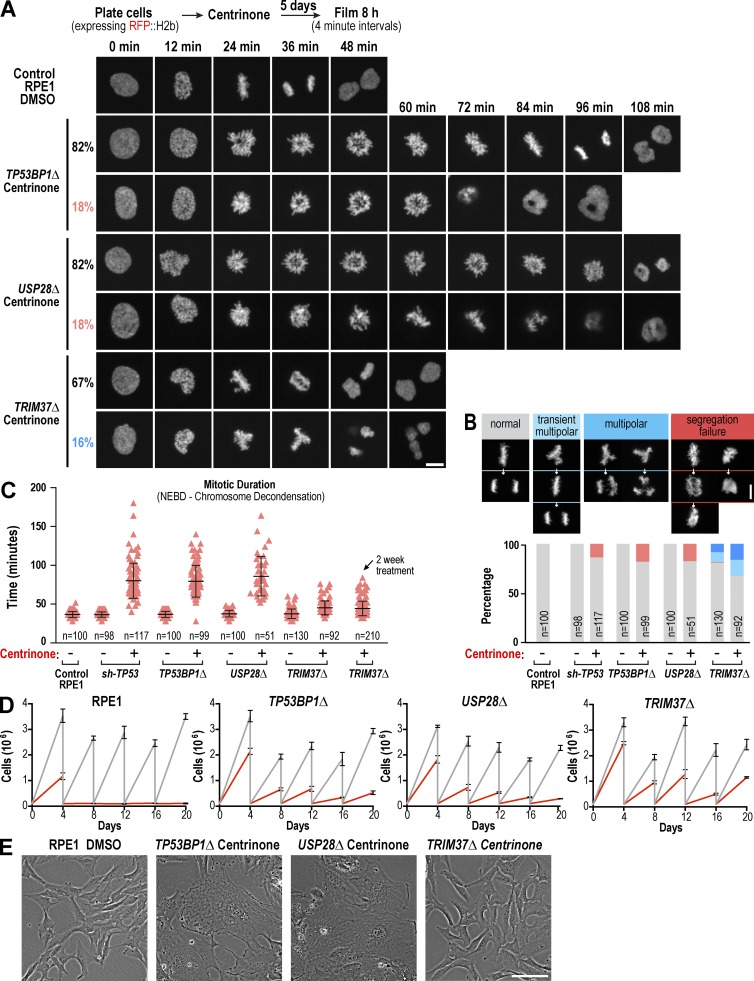
Figure 5.
Knockout of TRIM37, but not TP53BP1 or USP28, suppresses mitotic defects in centrosomeless cells. (A) Selected images from timelapse series of representative DMSO-treated control RPE1 cells and centrinone-treated TP53BP1Δ, USP28Δ, and TRIM37Δ mutant cells, acquired as outlined in the schematic. (B) Graph plotting the distribution of mitotic phenotypes observed for each condition along with representative images. (C) Graph plotting mitotic duration. Individual cell values (red triangles) are shown along with the mean and SD (black bars) for each condition. NEBD, nuclear envelope breakdown. (D) Graphs plotting the results of passaging assays monitoring the growth of control and knockout RPE1 cell lines after acute addition at day 0 of DMSO (vehicle) or centrinone. (E) Representative phase-contrast images of fields of DMSO-treated control RPE1 and knockout mutant cells after prolonged (>20 d) treatment with centrinone. Bars: (A and B) 10 µm; (E) 100 µm.
If centrosome loss is detected solely because of its effect on mitotic duration, prior work (Lambrus et al., 2015; Wong et al., 2015) suggests that the p53 circuit must be able to “integrate” mitotic duration over multiple generations. In our prior study, we found that only 13% of cells undergoing their first division in the absence of centrosomes spent longer in mitosis than the mitotic duration threshold, yet 70% of their daughters arrested; in addition, whether daughter cells arrested was not strongly correlated with mitotic duration of the mother cells (Wong et al., 2015). Because mitosis is also extended in one-centrosome cells, it is possible that two sequential moderately prolonged mitoses below the mitotic duration threshold could arrest daughter cells via the same mechanism that arrests daughter cells after a single mitosis above the threshold. Alternatively, centrosome loss and mitotic duration may independently contribute to p53 elevation via a module that requires 53BP1 and USP28 (Fig. 3 D). Resolving this question will likely require additional approaches focused on 53BP1 and USP28 as well as live-cell monitoring of p53 levels after centrosome removal.
TRIM37Δ cells form foci containing centrosomal markers after centrinone-mediated centrosome loss
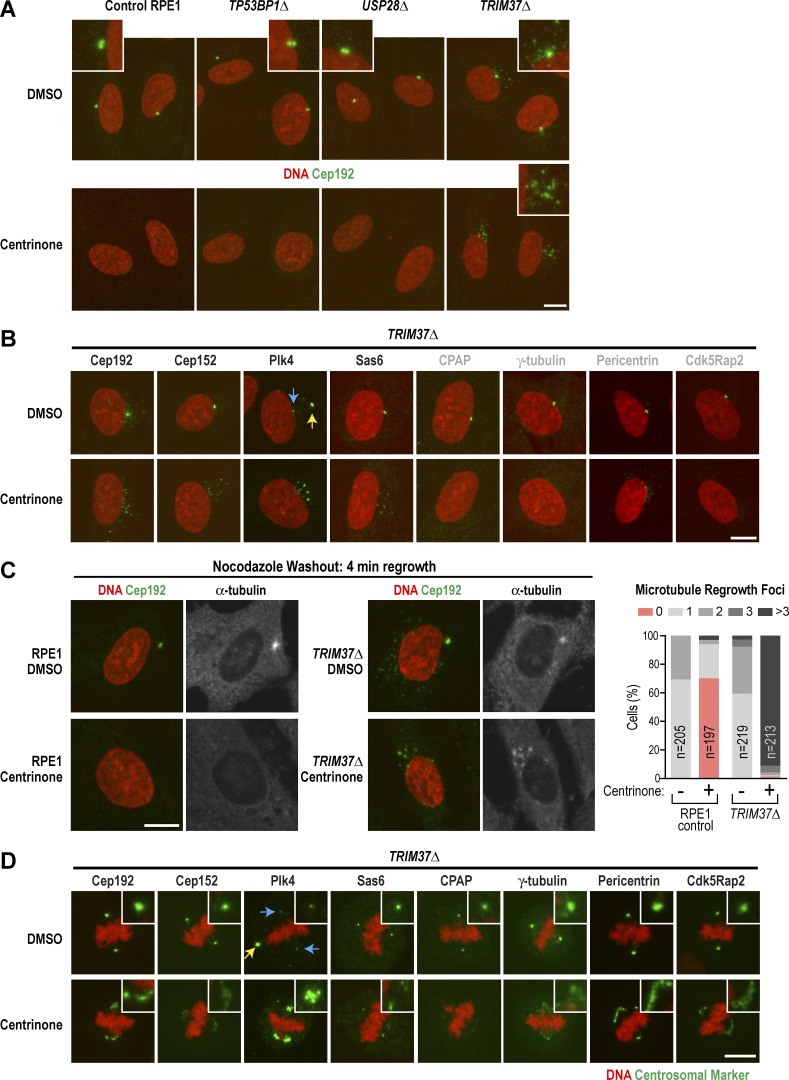
While analyzing DMSO-treated knockout lines, we noticed that Cep192 localized to small foci in TRIM37Δ cells, often in a halo surrounding the centrosome, a phenomenon not observed in control RPE1 cells or the other two knockout cell lines (Fig. 4 A, top row). Costaining with the centriolar satellite protein PCM-1 (Balczon et al., 1994; Kubo et al., 1999; Dammermann and Merdes, 2002) suggested that loss of TRIM37 causes Cep192 to accumulate at satellites (Fig. S3 A), whereas other centrosomal markers did not localize to satellites (Fig. S3 B), Plk4 exhibited a different behavior, concentrating in a single bright noncentriolar focus (Fig. 4 B, yellow arrow) in ∼40% of cells (Fig. S3 B). In centrinone-treated interphase TRIM37Δ cells, Cep192 localized to an array of small foci, as did Plk4 and two other components at the top of the centriole assembly hierarchy, Cep152 and Sas6 (Fig. 4, A and B, and Fig. S3 B). The centriole outer wall component CPAP and three pericentriolar material (PCM) components (γ-tubulin, pericentrin, and Cdk5rap2) did not localize to these interphase foci (Figs. 4 B and S3 B). The interphase foci in centrinone-treated TRIM37Δ cells were able to serve as sites for microtubule regrowth after nocodazole washout (Fig. 4 C). In mitotic TRIM37Δ cells treated with DMSO, the localization of centrosome markers was similar to that in controls except for the presence in ∼90% of cells (24/26) of a large bright noncentriolar focus of Plk4 (Figs. 4 D and S3 D, yellow arrow). Consistent with a prior study showing that TRIM37 inhibition can cause centriole reduplication (Balestra et al., 2013) ∼15–20% of mitotic DMSO-treated TRIM37Δ cells also appeared to contain one or occasionally two extra centrioles. In contrast, centrinone-treated mitotic TRIM37Δ cells contained an array of small foci that stained for the PCM markers γ-tubulin, pericentrin, and Cdk5rap2 in addition to Cep192, Cep152, Plk4, and Sas6 (Fig. 4 D and Fig. S3, C and D); however, CPAP was not detected. Similar foci were not detected for any of these markers in DMSO- or centrinone-treated control, TP53BP1Δ, or USP28Δ cells (Fig. S3, C and D).
Figure 4.
TRIM37Δ cells form foci containing centrosomal markers. (A) Immunofluorescence images after 5-d treatment with DMSO (top row) or centrinone (bottom row). Cells were stained for DNA (red) and with antibodies to the centrosomal protein Cep192 (green; insets 2.5× magnified). (B) Immunofluorescence images of interphase TRIM37Δ cells stained for DNA (red) and with antibodies to the indicated centrosomal proteins (green) after 5-d treatment with DMSO (top) or centrinone (bottom). Images are representative, and each marker was equivalently scaled for the two conditions. Yellow arrow points to bright ectopic Plk4 focus; blue arrow points to centrioles. (C) Immunofluorescence images of a microtubule regrowth experiment. Control RPE1 (left) and TRIM37Δ (middle) cells were pretreated for 5 d with DMSO (top) or centrinone (bottom) followed by 2-h treatment with nocodazole to depolymerize microtubules. Microtubules were allowed to grow for 4 min after nocodazole washout before fixation. Graph shows quantification of microtubule regrowth foci. (D) Immunofluorescence images of mitotic TRIM37Δ cells stained for DNA (red) and with antibodies to the indicated centrosomal proteins (green) after 5-d treatment with DMSO (top) or centrinone (bottom). Each marker was equivalently scaled for the two conditions. Yellow arrow points to bright ectopic Plk4 focus; blue arrows point to centrioles. Bars, 10 µm.
Consistent with our finding that TRIM37 loss leads to the accumulation of Cep192 at satellites, TRIM37 was previously reported to associate with two centriolar satellite components and Cep192 (Firat-Karalar et al., 2014). Our results suggest that after centrosome removal, the Cep192-containing satellites in TRIM37Δ cells acquire additional components (including Plk4, Cep152, and Sas6) and the ability to nucleate microtubules. In mitotic TRIM37Δ cells, Cep192-containing foci acquire PCM components and are observed at spindle poles, suggesting that TRIM37 loss leads to the assembly of centrosome-like structures in centrinone-treated cells that attenuate the detrimental consequences of centrosome removal.
Deletion of TRIM37, but not TP53BP1 or USP28, suppresses mitotic defects in centrosomeless cells
To test whether, in contrast to TP53BP1Δ or USP28Δ, TRIM37Δ suppresses p53 elevation by improving mitotic fidelity after centrosome loss, we imaged mitosis in cells expressing RFP::histone H2b after 5-d centrinone or DMSO treatment. In addition to the three knockout cell lines, we imaged an RPE1 cell line stably expressing a p53 shRNA to allow continued proliferation after centrosome loss (sh-TP53; Fig. 3 B). Mitosis in DMSO-treated TP53BP1Δ, USP28Δ, and sh-TP53 cells was comparable to that in DMSO-treated control RPE1 cells in terms of duration and the percentage of cells undergoing a normal bipolar division (Video 1; Fig. 5, B and C; and Fig. S3 E). Although most (∼81%) DMSO-treated TRIM37Δ cells also underwent a normal bipolar division, some (∼18%) underwent a multipolar mitosis or had metaphase plates that transiently appeared multipolar before a normal bipolar division (Video 1 and Fig. 5 B), consistent with the presence of extra centrosomes as a result of reduplication events (Fig. S3 D; Balestra et al., 2013).
Centrinone-treated TP53BP1Δ, USP28Δ, and p53 shRNA–expressing cells took longer to align their chromosomes at the metaphase plate (Fig. 5 A and Video 1), as expected after centrosome removal (Bazzi and Anderson, 2014; Lambrus et al., 2015; Wong et al., 2015), and mitotic duration was extended ∼2.2-fold compared with centrosome-containing controls (Fig. 5 C). Approximately 20% of the TP53BP1Δ and USP28Δ cells, and 14% of the p53 shRNA–expressing cells, exhibited a severe segregation failure phenotype in which cells initiated what appeared to be anaphase, but the chromosome masses collapsed back together and division failed (Fig. 5, A and B; and Video 1). In contrast to the other cell lines, mean mitotic duration was only slightly increased after centrinone treatment in TRIM37Δ cells, even after 2 wk (1.2-fold; Fig. 5 C), and centrinone-treated TRIM37Δ cells did not exhibit a segregation failure phenotype. Instead, ∼30% of TRIM37Δ cells exhibited segregation figures consistent with extra spindle poles; about half of these resolved into bipolar configurations before segregation initiated, whereas multipolar segregation was observed for the remainder (Fig. 5 B).
Thus, whereas TP53BP1Δ and USP28Δ cells exhibit significant delays in chromosome alignment after centrosome loss and a high percentage of chromosome segregation failure, chromosomes align much more quickly, albeit often in a multipolar configuration, in TRIM37Δ cells (see Fig. S3 E for a comparison of timing in bipolar and multipolar mitoses). These results suggest that the ectopic centrosome component–containing foci that form after centrosome removal in TRIM37Δ cells can function like centrosomes to accelerate mitotic spindle assembly and chromosome alignment. That TRIM37Δ cells do not elevate p53 when centrosomes are removed could be because mitotic duration is reduced below the threshold for p53 activation; alternatively, it is possible that in addition to facilitating mitosis, the ectopic foci are sufficiently centrosome-like to be able to suppress p53 activation.
Analysis in a 20-d passaging assay after addition of centrinone revealed that TRIM37Δ cells continued to divide robustly (TRIM37Δ doubling time = 0.92 d (DMSO) and 1.27 d (centrinone); wild-type (DMSO) doubling time = 0.76 d; Fig. S3 G) and maintained a normal-looking morphology (Fig. 5, D and E). In contrast, the proliferative capacity of the TP53BP1 and USP28 knockout lines appeared to be more compromised, and many cells exhibited an aberrant morphology, presumably because of accumulated mitotic errors (Fig. 5, D and E). Thus, in RPE1 cells, the inability to sense centrosome loss or extended mitotic duration is likely not sufficient to confer the ability to proliferate robustly in the absence of centrosomes that is observed for many cancer cell lines.
TRIM37 is an E3 ubiquitin ligase of the TRIpartite motif (TRIM) protein family (Kallijärvi et al., 2005) previously implicated in restricting centriole number (Balestra et al., 2013). We show that when centriole assembly is suppressed by Plk4 inhibition, TRIM37 deletion promotes the formation of ectopic foci containing centrosomal components. It is unlikely that these foci contain bona fide centrioles, as they do not contain the essential outer centriole wall component CPAP, and Plk4 inhibition should block SAS-6 oligomerization to form the centriolar cartwheel (Brito et al., 2012; Gönczy, 2012; Fu et al., 2015). Cep192 is thought to act redundantly with Cep152 as a centriolar receptor for Plk4 (Cizmecioglu et al., 2010; Hatch et al., 2010; Kim et al., 2013; Sonnen et al., 2013; Park et al., 2014). We find that in centrosomeless cells generated by Plk4 inhibition, TRIM37 removal leads to the formation of ectopic foci containing Cep192 and Cep152 that recruit Plk4 in interphase and PCM components in mitotic cells. These results highlight a critical role for TRIM37 in ensuring that Plk4 recruitment and PCM assembly occur only on the scaffold provided by the outer centriole wall, thereby ensuring the singularity of centriole duplication and centrosome assembly.
We identify 53BP1 and USP28 as critical components of the mechanism that leads to p53 elevation and G1 arrest in response to centrosome removal and prolonged mitotic duration, suggesting a central role for a 53BP1-USP28 module in communicating mitotic challenges to the p53 circuit. 53BP1 was first identified as a p53 binding partner (Iwabuchi et al., 1994), although the significance of this interaction is unknown (Panier and Boulton, 2014). USP28 is a deubiquitinase that interacts with the tandem BRCT domains of 53BP1 (Zhang et al., 2006; Knobel et al., 2014). USP28 has been proposed to promote the stability of proteins involved in the DNA damage response based on work in a human cell type (Zhang et al., 2006); however, Usp28-null mice do not show phenotypes characteristic of loss of a DNA damage response (Knobel et al., 2014). Understanding how 53BP1 and USP28 elevate p53 in response to centrosome loss and extended mitotic duration, and determining whether centrosome loss is an independent input into the p53 circuit or triggers p53 elevation because it leads to sequential prolonged mitoses, are important future goals arising from the results described here.
Materials and methods
Chemical inhibitors
The chemical inhibitors used in this study were centrinone (150 nM; LCR-263; synthesized by Sudia MediTech; Wong et al., 2015); MDM2 inhibitor ((2-(4-(tert-butyl)-2-ethoxyphenyl)-4,5-bis(4-chlorophenyl)-4,5-dimethyl-4,5-dihydro-1H-imidazol-1-yl)(4-(2-(methylsulfonyl)ethyl)piperazin-1-yl)methanone); 1 µM; synthesized by Sudia MediTech; Ding et al., 2009); doxorubicin (1 µM; Sigma-Aldrich); cytochalasin D (4 µM; Sigma-Aldrich); monastrol (100 µM; Tocris Bioscience); nocodazole (2.5 µg/ml; Sigma-Aldrich); and NMS-P715 (2 µM; EMD Millipore).
Antibodies
Antibodies against Cep192 (1–211 aa; used at 0.5 µg/ml), SAS-6 (501–657 aa; used at 0.5 µg/ml), and Plk4 (501–657 aa; used at 1 µg/ml) were previously described (Wong et al., 2015). The following antibodies were purchased from commercial sources, with their working concentrations indicated in parentheses: anti-Trim37 (1:2,000 for Western blotting; A301-174A; Bethyl Laboratories, Inc.), anti-Usp28 (1:1,000 for Western blot; ab126604; Abcam), anti-Usp28 (1:100 for immunofluorescence; HPA006778; Sigma-Aldrich), anti-53BP1 (1:5,000), anti-Cep152 (1:2,000; Abcam), anti-Cdk5rap2 (1:4,000; Abcam), GTU-88 (anti–γ-tubulin; 1:1,000; Sigma-Aldrich), anti-pericentrin (1 µg/ml; Abcam), anti-CPAP (1:400; Proteintech), anti-p53 (1:100 for Western blot; OP43; EMD Millipore), anti-p53 (1:500 for immunofluorescence; OP140; EMD Millipore), anti-p21 (1:1,000; #2947; Cell Signaling Technology), DM1A (anti–α-tubulin; 1:5,000; Sigma-Aldrich), anti-GAPDH (1:1,000; 14C10; Cell Signaling Technology), PCM-1 (1:400; #5259; Cell Signaling Technology), and Fab fragment (goat anti–rabbit IgG; 30 µg/ml; Jackson ImmunoResearch Laboratories, Inc.). Secondary antibodies were purchased from Jackson ImmunoResearch Laboratories, Inc.
Cell lines
RPE-1 cells were obtained from ATCC. RPE-1 cells and all derivative cell lines generated in this study (Table S1) were grown in DMEM/F12 media containing 10% FBS, 100 µg/ml streptomycin, and 100 U/ml penicillin.
Cells were maintained at 37°C and 5% CO2. For generation of CRISPR/Cas9-mediated knockout cell lines, specific gRNAs (TP53BP1Δ, 5′-CTGCTCAATGACCTGACTGA-3′; USP28Δ, 5′-TGAGCGTTTAGTTTCTGCAG-3′; TRIM37Δ, 5′-CTCCCCAAAGTGCACACTGA-3′) were cloned in lentiCRISPR v2 (#52961; Addgene; Sanjana et al., 2014) or PX459 (#48139; Addgene; Ran et al., 2013). RPE-1 cells were transiently transfected with gRNA and Cas9 containing plasmids using Lipofectamine 3000 (Thermo Fisher Scientific) according to the manufacturers’ guidelines. Transfected cells were grown in 150 nM centrinone for 2 wk, with media exchange every 5 d. Cells were grown for an additional 3–5 d in centrinone-free medium before colonies were isolated. Clones were tested by Western blotting. For generation of H2B-mRFP–expressing cell lines, H2B-mRFP was cloned into the lentiviral vector pCDH-EF1 with XbaI–NotI restriction sites. This plasmid and lentivirus packaging vectors (pCAG-HIVgp and pCMV-VSV-G-RSV-Rev from H. Miyoshi, RIKEN BioResource Center, Ibaraki, Japan) were cotransfected into HEK-293T cells using Fugene HD (Promega). 48 h after transfection, virus-containing culture supernatant was harvested and added to the growth medium of RPE-1 cells along with 8 µg/ml polybrene (EMD Millipore). Populations of each cell line expressing H2B-mRFP were selected by FACS. To knock down TP53 (p53), RPE-1 cells were infected with a lentivirus containing sh-p53 made using the plasmid shp53 pLKO.1 puro (19119; Addgene; Godar et al., 2008). Positive selection of sh-p53–expressing cells was performed 2 d after infection with 10 µg/ml puromycin.
CRISPR/Cas9 screen
The CRISPR/Cas9 screen was performed using the human GeCKO v2 library (#1000000048; Addgene; Sanjana et al., 2014). 3.2 million RPE-1 cells were infected with 12.8 million infectious virus particles in a 15-cm plate using 20 ml DMEM/F12 medium and 8 µg/ml polybrene. Cells were treated with 150 nM centrinone. After 2 d, cells were transferred to 32 15-cm plates (first screen) or 75 15-cm plates (second screen). Centrinone treatment was continued for 3 wk. After 3 wk, growing colonies were isolated. Isolated clones were further analyzed in a secondary Mdm2i screen (first and second screen) as well as pooled and analyzed by Illumina sequencing (first screen).
Mdm2 inhibitor screen
Wild-type RPE-1 cells and clones isolated from the centrinone CRISPR/Cas9 screen were plated as duplicates into 12-well plates and treated with either 1 µM Mdm2 inhibitor or DMSO vehicle control for 1 wk. Clones that arrested in 1 µM Mdm2 inhibitor but not in DMSO were harvested. The different gRNA sequences of each cell clone were amplified with oligos: (1) 5′-TCCGCTCGAGTGTGGGCGATGTGCGCTCTG-3′ and (2) 5′-GCGGGATCCGCAATGGACTATCATATGCTTACCGTAACTTGAAAGTATTTCG-3′. The PCR product, which contains a pool of different gRNAs, was cloned into a vector with BamHI–XhoI restriction sites. From each cell clone, the gRNA sequence of 10 plasmid clones was determined by Sanger sequencing.
Immunofluorescence and quantification of nuclear and centrosomal signals
RPE-1 control and the 53BP1Δ, USP28Δ, and TRIM37Δ knockout cell lines were treated for 5 d with 150 nM centrinone or DMSO. 1 day before fixation, 8,000 cells per well were plated into 96-well plates. Cells were fixed in ice-cold methanol for 7 min at −20°C, washed twice with washing buffer (PBS containing 0.1% Triton X-100), and blocked with blocking buffer (PBS containing 2% BSA, 0.1% Triton X-100, and 0.1% NaN3) for 1 h. Cells were washed three times with washing buffer and incubated for 1 h with the first antibody (concentrations as indicated earlier). Cells were washed three times with washing buffer, incubated for 1 h with the secondary antibody, and stained for DNA with Hoechst 33342 dye. For double labeling with primary antibodies from the same host species (Fig. S3 A), fixed cells were first incubated with the first primary antibody (rabbit anti-Cep192), which was then blocked with an AffiniPure Fab fragment (goat anti–rabbit IgG) before incubation with the second primary antibody (rabbit anti-PCM1). Efficient blocking of the first primary antibody by the Fab fragments was confirmed, because an anti-rabbit secondary antibody did not recognize the blocked first primary antibody. Cells were washed three times with washing buffer before inspection. Images were acquired on a CV7000 spinning disk confocal system (Yokogawa Electric Corporation) equipped with a 40× (0.95 NA) or 60× (water, 1.2 NA) U-PlanApo objective and 2,560 × 2,160-pixel sCMOS camera (Andor Technology) at 2 × 2 binning. Image acquisition was performed using CV7000 software. Nuclear signal intensity measurements were performed automatically using CV7000 analysis software. The nuclear area was determined using Hoechst 33342 staining as reference. To measure nuclear p53 signals, the mean gray value of the cytoplasmic signal, measured in a 2-µm area surrounding the nucleus, was subtracted from the mean gray value of the nuclear signal.
Analysis of mitosis and daughter cell fate
To study the correlation between mother cell prometaphase duration and daughter cell fate, RPE-1 control and TP53BP1Δ, USP28Δ, and TRIM37Δ knockout cell lines expressing H2B-mRFP were seeded into 96-well polystyrene plates at 2,000 cells/well, 12–14 h before imaging. Cells were treated with 100 µM monastrol and immediately imaged on the CV1000 or CQ1 spinning disk confocal systems (Yokogawa Electric Corporation) with a 20× 0.75 NA U-PlanApo objective at 37°C and 5% CO2. Image acquisition and data analysis were performed using CellVoyager software and ImageJ, respectively. 20–30 fields/well were imaged. 5 × 2-µm z-sections in RFP (25% power, 100 ms, 30% gain) were acquired in each field at 10-min intervals for 6 h. The plate was then removed from the microscope, and wells were washed twice with warm medium. The plate was returned to the microscope, and imaging resumed for 2 h at 10-min intervals and then for an additional 48 h at 20-min intervals. The mitotic fate of daughter cells was analyzed as well as clear chromosome missegregation events (formation of daughter cells with micronuclei).
To study mitosis of RPE-1 control and 53BP1Δ, USP28Δ, and TRIM37Δ knockout cells, cells expressing H2B-mRFP were treated for 5 d with DMSO or 150 nM centrinone before imaging. Cells were seeded into 96-well polystyrene plates at 10,000 cells/well, 24 h before imaging. Images were acquired on a CV7000 spinning disk confocal system (Yokogawa Electric Corporation) with a 40× 0.95 NA U-PlanApo objective and 2,560 × 2,160-pixel sCMOS camera (Andor Technology) at 2 × 2 binning. Image acquisition was performed using CV7000 software. 20–30 fields/well were imaged. 5 × 2-µm z-sections in RFP (30% power, 150 ms) channels were captured in each field at 4-min intervals for 8 h.
Tetraploid cell arrest assay
RPE-1 control and knockout cell lines expressing H2B-RFP were seeded into a 96-well polystyrene plate at 1,000 cells/well, 24 h before treatment. Cells were treated with 4 µM cytochalasin D (Sigma-Aldrich) for 24 h and washed five times with warm medium. After drug washout, the plate was imaged on the CQ1 system with a 20× 0.75 NA U-PlanApo objective. Image acquisition and data analysis were performed using CellVoyager software and ImageJ, respectively. 25–30 fields/well were imaged. For each field, five z-sections at 2-µm intervals were acquired in the RFP channel (25% power, 150 ms, and 30% gain) at 20-min intervals for 72 h. Tetraploid cells were identified as cells with two nuclei.
Proliferation assays
For each condition in the passaging assays, cells were seeded in triplicate into 10-cm plates at 100,000 cells/plate. Centrinone and Mdm2 inhibitor were added at the indicated concentrations. At 96-h intervals, plates were harvested, counted, and replated at 100,000 cells/plate. Cell counting was performed using a TC10 or TC20 automated cell counter (Bio-Rad Laboratories). For determination of doubling times, DMSO-treated control RPE1 and TRIM37Δ mutant cells, as well as TRIM37Δ mutant cells treated with centrinone for >2 wk, were plated into six-well dishes at 25,000 cells per well. For 3–4 d, at 24-h intervals, wells were harvested and counted using a TC20 automated cell counter (Bio-Rad Laboratories).
Microtubule regrowth assay
RPE1 control and TRIM37Δ mutant cells were treated for 5 d with DMSO or 150 nM centrinone. Cells were seeded into 96-well plates and treated for 2 h with 2.5 µg/ml nocodazole to depolymerize microtubules. Cells were washed five times with PBS and incubated for 4 min in fresh prewarmed growth medium at 37°C to allow microtubule polymerization. Cells were fixed with ice-cold methanol for 7 min at −20°C and stained for Cep192, α-tubulin, and DNA. Fixed cells were kept in PBS until imaging. Images were acquired on a CV7000 spinning disk confocal system (Yokogawa Electric Corporation) with a 60× (water, 1.2 NA) U-PlanApo objective and 2,560 × 2,160-pixel sCMOS camera (Andor Technology) at 1 × 1 binning. Image acquisition and processing was performed using CV7000 software and ImageJ, respectively.
Western blotting
Asynchronously growing cells from 10-cm plates were harvested at 50–80% confluence and lysed by sonication in RIPA buffer plus protease and phosphatase inhibitor cocktail (Thermo Fisher Scientific). Cell extracts were stored at −80°C. Before use, extract concentrations were normalized based on a Bio-Rad Protein Assay (Bio-Rad Laboratories). For every sample, 30 µg protein/lane was run on Mini-PROTEAN gels (Bio-Rad Laboratories) and transferred to PVDF membranes using a TransBlot Turbo system (Bio-Rad Laboratories). Blocking and antibody incubations were performed in TBS-T plus 5% nonfat dry milk or in TBS-T plus 5% BSA. Detection was performed using HRP-conjugated secondary antibodies (GE Healthcare) with WesternBright Sirius (Advansta) or SuperSignal West Femto (Thermo Fisher Scientific) substrates. Membranes were imaged on a ChemiDoc MP system (Bio-Rad Laboratories). Membranes were stripped and reprobed with antibodies against α-tubulin or GAPDH as loading controls.
Analysis of p53 expression by Western blotting
The expression of p53 upon exposure to different type of stresses (centrosome depletion, DNA damage, and Mdm2 inhibition) was analyzed by Western blotting. Control RPE1 and 53BP1Δ, USP28Δ, and TRIM37Δ knockout cells were incubated in 10-cm plates for 24 h with 1 µM doxorubicin (DNA damage), for 24 h with 1 µM Mdm2 inhibitor, or for 48 h with 150 nM centrinone before harvesting. Western blotting was performed with the indicated antibodies as described earlier.
Online supplemental material
Fig. S1, which is related to Fig. 1, shows the results from the CRISPR/Cas9 screen. Fig. S1 A lists all gRNAs identified in the isolated clones. Fig. S1 B lists all gRNAs that were identified by deep sequencing of the pooled clones from screen 1. Fig. S2 is related to Fig. 2. Fig. S2 A shows the locations of the gRNAs used to generate the single deletion mutants in the TP53BP1, USP28, and TRIM37 genes, along with the locations in the corresponding proteins. Fig. S2 B shows the absence of 53BP1 and USP28 from nuclei in the TP53BP1Δ and USP28Δ knockouts, respectively. Fig. S2 C shows the efficiency of centrosome depletion in the TP53BP1Δ, USP28Δ, and TRIM37Δ mutants compared with RPE1 control. Fig. S2 D shows the quantification in two additional experiments of nuclear p53 in TP53BP1Δ, USP28Δ, and TRIM37Δ knockout cells compared with RPE1 control after 5 d of centrinone treatment. Fig. S2 E shows p53 levels after 12, 24, and 48 h of centrinone treatment. Fig. S2 F shows results of experiments performed to rescue mitotic timing in centrinone-treated cells with the Mps1 inhibitor NMS-P715, including a Western blot of p53 levels of centrinone- and NMS-P715–treated cells, mitotic duration in centrinone- and NMS-P715–treated cells, and a plot of the percentage of cells that fail chromosome segregation and cytokinesis upon centrinone and NMS-P715 treatment. Fig. S3 is related to Figs. 4 and 5. Fig. S3 A shows colabeling of PCM-1 and Cep192 in DMSO- and centrinone-treated TRIM37Δ cells. Fig. S3 B shows quantification of ectopic foci staining for centrosomal markers in interphase cells. Fig. S3 C shows staining of centrosomal markers in mitotic DMSO-treated TRIM37Δ cells, along with centrinone-treated TP53BP1Δ and USP28Δ cells. Fig. S3 D shows the number of mitotic cells that were examined for the analysis shown in Figs. 4 D and S3 C as well as the quantification of centrosomal-component foci in DMSO- and centrinone-treated mitotic TRIM37Δ cells. Fig. S3 E plots mitotic duration for bipolar and multipolar mitosis in DMSO- and centrinone-treated TRIM37Δ cells. Fig. S3 F shows a table of p-values for the experiment shown in Fig. 5 C. Fig. S3 G shows doubling times for long-term DMSO- or centrinone-treated TRIM37Δ cells. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201604081/DC1.
Supplementary Material
ACKNOWLEDGMENTS
We thank Robert Davis and David Jenkins for advice and assistance with cell culture and imaging, respectively, and Hyun J. Lee for help with centrosome counting.
This work was supported by a National Institutes of Health grant to K. Oegema (GM074207) and by funds from the Hilton Ludwig Cancer Prevention Initiative to A.K. Shiau. F. Meitinger was supported by the Deutsche Forschungsgemeinschaft (ME 4713/1-1). F. Meitinger, J.V. Anzola, A. Desai, A.K. Shiau, and K. Oegema receive salary and other support from the Ludwig Institute for Cancer Research.
The authors declare no additional competing financial interests.
Footnotes
Abbreviations used in this paper:
- gRNA
- guide RNA
- PCM
- pericentriolar material
References
- Balczon R., Bao L., and Zimmer W.E.. 1994. PCM-1, A 228-kD centrosome autoantigen with a distinct cell cycle distribution. J. Cell Biol. 124:783–793. 10.1083/jcb.124.5.783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balestra F.R., Strnad P., Flückiger I., and Gönczy P.. 2013. Discovering regulators of centriole biogenesis through siRNA-based functional genomics in human cells. Dev. Cell. 25:555–571. 10.1016/j.devcel.2013.05.016 [DOI] [PubMed] [Google Scholar]
- Bazzi H., and Anderson K.V.. 2014. Acentriolar mitosis activates a p53-dependent apoptosis pathway in the mouse embryo. Proc. Natl. Acad. Sci. USA. 111:E1491–E1500. 10.1073/pnas.1400568111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brito D.A., Gouveia S.M., and Bettencourt-Dias M.. 2012. Deconstructing the centriole: Structure and number control. Curr. Opin. Cell Biol. 24:4–13. 10.1016/j.ceb.2012.01.003 [DOI] [PubMed] [Google Scholar]
- Cizmecioglu O., Arnold M., Bahtz R., Settele F., Ehret L., Haselmann-Weiss U., Antony C., and Hoffmann I.. 2010. Cep152 acts as a scaffold for recruitment of Plk4 and CPAP to the centrosome. J. Cell Biol. 191:731–739. 10.1083/jcb.201007107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conduit P.T., Wainman A., and Raff J.W.. 2015. Centrosome function and assembly in animal cells. Nat. Rev. Mol. Cell Biol. 16:611–624. 10.1038/nrm4062 [DOI] [PubMed] [Google Scholar]
- Dammermann A., and Merdes A.. 2002. Assembly of centrosomal proteins and microtubule organization depends on PCM-1. J. Cell Biol. 159:255–266. 10.1083/jcb.200204023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q., Graves B.J., Kong N.J.L., Lovey A.J., Pizzolato G., Roberts J.L., So S., Vu B.T., and Wovkulich P.M., 2009. 2,4,5-Triphenyl imidazoline derivatives as inhibitors of the interaction between P53 and Mdm2 proteins for use as anticancer agents. Switzerland patent WO 2007063013. Filed November 22, 2006, and issued December 9, 2009. [Google Scholar]
- Firat-Karalar E.N., Rauniyar N., Yates J.R. III, and Stearns T.. 2014. Proximity interactions among centrosome components identify regulators of centriole duplication. Curr. Biol. 24:664–670. 10.1016/j.cub.2014.01.067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J., Hagan I.M., and Glover D.M.. 2015. The centrosome and its duplication cycle. Cold Spring Harb. Perspect. Biol. 7:a015800 10.1101/cshperspect.a015800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem N.J., Cornils H., Chiu S.Y., O’Rourke K.P., Arnaud J., Yimlamai D., Théry M., Camargo F.D., and Pellman D.. 2014. Cytokinesis failure triggers hippo tumor suppressor pathway activation. Cell. 158:833–848. 10.1016/j.cell.2014.06.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godar S., Ince T.A., Bell G.W., Feldser D., Donaher J.L., Bergh J., Liu A., Miu K., Watnick R.S., Reinhardt F., et al. . 2008. Growth-inhibitory and tumor-suppressive functions of p53 depend on its repression of CD44 expression. Cell. 134:62–73. 10.1016/j.cell.2008.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gönczy P. 2012. Towards a molecular architecture of centriole assembly. Nat. Rev. Mol. Cell Biol. 13:425–435. 10.1038/nrm3373 [DOI] [PubMed] [Google Scholar]
- Gu B., and Zhu W.G.. 2012. Surf the post-translational modification network of p53 regulation. Int. J. Biol. Sci. 8:672–684. 10.7150/ijbs.4283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurpinar E., and Vousden K.H.. 2015. Hitting cancers’ weak spots: Vulnerabilities imposed by p53 mutation. Trends Cell Biol. 25:486–495. 10.1016/j.tcb.2015.04.001 [DOI] [PubMed] [Google Scholar]
- Hatch E.M., Kulukian A., Holland A.J., Cleveland D.W., and Stearns T.. 2010. Cep152 interacts with Plk4 and is required for centriole duplication. J. Cell Biol. 191:721–729. 10.1083/jcb.201006049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwabuchi K., Bartel P.L., Li B., Marraccino R., and Fields S.. 1994. Two cellular proteins that bind to wild-type but not mutant p53. Proc. Natl. Acad. Sci. USA. 91:6098–6102. 10.1073/pnas.91.13.6098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson S.P., and Bartek J.. 2009. The DNA-damage response in human biology and disease. Nature. 461:1071–1078. 10.1038/nature08467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins L.M., Durell S.R., Mazur S.J., and Appella E.. 2012. p53 N-terminal phosphorylation: A defining layer of complex regulation. Carcinogenesis. 33:1441–1449. 10.1093/carcin/bgs145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallijärvi J., Lahtinen U., Hämäläinen R., Lipsanen-Nyman M., Palvimo J.J., and Lehesjoki A.E.. 2005. TRIM37 defective in mulibrey nanism is a novel RING finger ubiquitin E3 ligase. Exp. Cell Res. 308:146–155. 10.1016/j.yexcr.2005.04.001 [DOI] [PubMed] [Google Scholar]
- Karni-Schmidt O., Lokshin M., and Prives C.. 2016. The roles of MDM2 and MDMX in cancer. Annu. Rev. Pathol. 11:617–644. 10.1146/annurev-pathol-012414-040349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov A., and Rieder C.L.. 2001. Centrosomes enhance the fidelity of cytokinesis in vertebrates and are required for cell cycle progression. J. Cell Biol. 153:237–242. 10.1083/jcb.153.1.237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoo K.H., Verma C.S., and Lane D.P.. 2014. Drugging the p53 pathway: Understanding the route to clinical efficacy. Nat. Rev. Drug Discov. 13:217–236. 10.1038/nrd4288 [DOI] [PubMed] [Google Scholar]
- Kim T.S., Park J.E., Shukla A., Choi S., Murugan R.N., Lee J.H., Ahn M., Rhee K., Bang J.K., Kim B.Y., et al. . 2013. Hierarchical recruitment of Plk4 and regulation of centriole biogenesis by two centrosomal scaffolds, Cep192 and Cep152. Proc. Natl. Acad. Sci. USA. 110:E4849–E4857. 10.1073/pnas.1319656110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knobel P.A., Belotserkovskaya R., Galanty Y., Schmidt C.K., Jackson S.P., and Stracker T.H.. 2014. USP28 is recruited to sites of DNA damage by the tandem BRCT domains of 53BP1 but plays a minor role in double-strand break metabolism. Mol. Cell. Biol. 34:2062–2074. 10.1128/MCB.00197-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruiswijk F., Labuschagne C.F., and Vousden K.H.. 2015. p53 in survival, death and metabolic health: A lifeguard with a licence to kill. Nat. Rev. Mol. Cell Biol. 16:393–405. 10.1038/nrm4007 [DOI] [PubMed] [Google Scholar]
- Kubo A., Sasaki H., Yuba-Kubo A., Tsukita S., and Shiina N.. 1999. Centriolar satellites: Molecular characterization, ATP-dependent movement toward centrioles and possible involvement in ciliogenesis. J. Cell Biol. 147:969–980. 10.1083/jcb.147.5.969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrus B.G., Uetake Y., Clutario K.M., Daggubati V., Snyder M., Sluder G., and Holland A.J.. 2015. p53 protects against genome instability following centriole duplication failure. J. Cell Biol. 210:63–77. 10.1083/jcb.201502089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levav-Cohen Y., Goldberg Z., Tan K.H., Alsheich-Bartok O., Zuckerman V., Haupt S., and Haupt Y.. 2014. The p53-Mdm2 loop: A critical juncture of stress response. Subcell. Biochem. 85:161–186. 10.1007/978-94-017-9211-0_9 [DOI] [PubMed] [Google Scholar]
- Levine A.J., and Oren M.. 2009. The first 30 years of p53: Growing ever more complex. Nat. Rev. Cancer. 9:749–758. 10.1038/nrc2723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T.A., Menendez D., Resnick M.A., and Anderson C.W.. 2014. Mutant TP53 posttranslational modifications: Challenges and opportunities. Hum. Mutat. 35:738–755. 10.1002/humu.22506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panier S., and Boulton S.J.. 2014. Double-strand break repair: 53BP1 comes into focus. Nat. Rev. Mol. Cell Biol. 15:7–18. 10.1038/nrm3719 [DOI] [PubMed] [Google Scholar]
- Park S.Y., Park J.E., Kim T.S., Kim J.H., Kwak M.J., Ku B., Tian L., Murugan R.N., Ahn M., Komiya S., et al. . 2014. Molecular basis for unidirectional scaffold switching of human Plk4 in centriole biogenesis. Nat. Struct. Mol. Biol. 21:696–703. 10.1038/nsmb.2846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., and Zhang F.. 2013. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8:2281–2308. 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana N.E., Shalem O., and Zhang F.. 2014. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 11:783–784. 10.1038/nmeth.3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sir J.H., Pütz M., Daly O., Morrison C.G., Dunning M., Kilmartin J.V., and Gergely F.. 2013. Loss of centrioles causes chromosomal instability in vertebrate somatic cells. J. Cell Biol. 203:747–756. 10.1083/jcb.201309038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnen K.F., Gabryjonczyk A.M., Anselm E., Stierhof Y.D., and Nigg E.A.. 2013. Human Cep192 and Cep152 cooperate in Plk4 recruitment and centriole duplication. J. Cell Sci. 126:3223–3233. 10.1242/jcs.129502 [DOI] [PubMed] [Google Scholar]
- Uetake Y., and Sluder G.. 2010. Prolonged prometaphase blocks daughter cell proliferation despite normal completion of mitosis. Curr. Biol. 20:1666–1671. 10.1016/j.cub.2010.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams A.B., and Schumacher B.. 2016. p53 in the DNA-damage-repair process. Cold Spring Harb. Perspect. Med. 6:a026070 10.1101/cshperspect.a026070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong Y.L., Anzola J.V., Davis R.L., Yoon M., Motamedi A., Kroll A., Seo C.P., Hsia J.E., Kim S.K., Mitchell J.W., et al. . 2015. Cell biology. Reversible centriole depletion with an inhibitor of Polo-like kinase 4. Science. 348:1155–1160. 10.1126/science.aaa5111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff J.B., Wueseke O., and Hyman A.A.. 2014. Pericentriolar material structure and dynamics. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369:20130459. 10.1098/rstb.2013.0459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Zaugg K., Mak T.W., and Elledge S.J.. 2006. A role for the deubiquitinating enzyme USP28 in control of the DNA-damage response. Cell. 126:529–542. 10.1016/j.cell.2006.06.039 [DOI] [PubMed] [Google Scholar]
- Zitouni S., Nabais C., Jana S.C., Guerrero A., and Bettencourt-Dias M.. 2014. Polo-like kinases: Structural variations lead to multiple functions. Nat. Rev. Mol. Cell Biol. 15:433–452. 10.1038/nrm3819 [DOI] [PubMed] [Google Scholar]
- Wong Y.L., Anzola J.V., Davis R.L., Yoon M., Motamedi A., Kroll A., Seo C.P., Hsia J.E., Kim S.K., Mitchell J.W., et al. . 2015. Cell biology. Reversible centriole depletion with an inhibitor of Polo-like kinase 4. Science. 348:1155–1160. 10.1126/science.aaa5111 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.