Abstract
The 1918 influenza pandemic caused ~50 million deaths. Many questions remain regarding the origin, pathogenicity, and mechanisms of human adaptation of this virus. Avian-adapted influenza A viruses preferentially bind α2,3-linked sialic acids (Sia) while human-adapted viruses preferentially bind α2,6-linked Sia. A change in Sia preference from α2,3 to α2,6 is thought to be a requirement for human adaptation of avian influenza viruses. Autopsy data from 1918 cases, however, suggest that factors other than Sia preference played a role in viral binding and entry to human airway cells. Here, we evaluated binding and entry of five 1918 influenza receptor binding domain variants in a primary human airway cell model along with control avian and human influenza viruses. We observed that all five variants bound and entered cells efficiently and that Sia preference did not predict entry of influenza A virus to primary human airway cells evaluated in this model.
Keywords: Influenza A virus, Pandemic, Receptor binding, hemagglutinin
INTRODUCTION
Influenza A viruses cause acute respiratory viral disease in humans in both annual epidemics and in infrequent pandemics (1). The 1918 “Spanish” influenza pandemic resulted in approximately 50 million deaths globally and is the most severe influenza pandemic on record (2, 3). Many questions remain regarding the origin, pathogenicity, and mechanisms of human host adaptation of this deadly virus (4). Influenza A viruses bind to terminal sialic acids (Sia) on target cell glycans and it is hypothesized that changes in the influenza A virus hemagglutinin (HA) protein receptor-binding domain (RBD) are important in the process of host adaptation, specifically allowing avian-origin influenza A viruses to adapt to humans.
The 1918 HA gene has been sequenced from multiple postmortem human lung samples, and several naturally occurring 1918 HA RBD sequence variants have been reported (5–7). These include A/South Carolina/1/1918 (SC), which has an aspartic acid at both positions 187 and 222 in HA1 (H1 subtype numbering) (5) conferring an α2,6 Sia receptor-binding specificity and A/NY/1/1918 (NY), which differs from SC by a single amino acid, encoding a glycine at position 222 that confers a mixed α2,3/α2,6 binding specificity (8–10). Sheng et al., 2011 reported two 1918 HA sequences with new RBD variants with yet to be confirmed binding specificities (Table 1). The HA RBD of A/Virginia/1/1918 (VA), which, in addition to aspartic acids at positions 187 and 222 has a change from glutamine to arginine at position 189 in the HA1 domain, may have an enhanced α2,6 binding specificity based on computational modeling (7, 9). No binding specificity data is available for the A/New York/3/1918 (NY3), although deep sequencing revealed a predominance of asparagine rather than glycine at position 222 (11). Finally, the ‘avianized’ laboratory-produced variant of the 1918 virus HA (AV), in which the aspartic acid at position 187 in NY was mutagenized back to the avian influenza virus consensus glycine, is reported to be exclusively α2,3 Sia binding (8–10).
Table 1.
1918 Pandemic Influenza HA Receptor Binding Domain Variants Amino Acid Sequences
It is not yet fully clear how influenza A virus Sia preferences, as predicted by in vitro glycan array analysis with limited numbers of synthetic oligosaccharides, relate to binding and entry of influenza viruses into the human respiratory tract, including the epithelium of the distal trachea and bronchi. Review of 1918 autopsy material, including correlative analyses of histopathology, distribution of influenza viral antigen by immunohistochemistry, and 1918 HA RBD variant gene sequencing, demonstrated no difference in cell tropism between the four naturally occurring 1918 RBD variants outlined above (7). Autopsy sections demonstrated that the 1918 virus, regardless of HA RBD sequence, infected the entirety of the respiratory tract, including ciliated cells and goblet cells of the tracheobronchial tree and of the bronchiolar epithelium, and alveolar lining type I and type II cells. Based on lectin histochemistry, however, the upper airway and distal trachea in humans is reported to display predominantly α2,6 Sia on apical epithelial cell surfaces (12–14). Mouse models of 1918 influenza viral infection also suggested that factors other than Sia preference play a role in influenza binding and entry to airway cells (15). Histopathological changes, cell tropism, and viral antigen distribution in lungs of mice infected with the 1918 RBD variants SC, NY, and AV were similar across viral variants and correlated with human autopsy findings.
Normal human bronchial epithelial (NHBE) cells, are primary human airway cells, and are variably reported to display exclusively α2,6 Sia or a mixture of α2,3/α2,6 Sia on their cell surfaces (14, 16–20). This laboratory has previously characterized the Sia distribution on a single lot of NHBE cells harvested from a healthy female donor and showed that this lot displays near exclusively α2,6 Sia on goblet and ciliated cell surfaces and rarely displays α2,3 Sia on goblet cell surfaces (14). Additionally it was shown that this lot of cells readily supports both human- and avian-adapted influenza virus growth to peak titers of 104-to-106 viral plaque forming units (pfu)/mL (14, 21). , Experiments described here were conducted to evaluate the binding, entry, and peak replication of 1918 HA RBD variants, utilizing this lot of well-characterized NHBE cells, and to compare their binding and entry to human-adapted and avian H1 subtype influenza virus controls. Prevailing hypotheses suggest that variants with α2,6 Sia preferences would bind and enter human airway epithelial cells more efficiently than those the α2,3 Sia or mixed α2,3/α2,6 Sia preferences. Here NHBE cells were infected at a constant multiplicity of infection (MOI). In multiple experiments binding and entry was examined by immunofluorescence at 5- and 20-minutes post-addition of virus and quantification of cell-associated virus was performed by quantitative reverse transcriptase polymerase chain reaction (qRT-PCR). Viral replication was assessed at 12, 24, and 36-hours post-infection in a subset of viruses representing the a range of Sia binding preference (AV α2,3; NY mixed α2,3 and 2,6; and SC α2,6).
MATERIALS AND METHODS
Growth and differentiation of cells
Primary Normal Human Bronchial/Tracheal Epithelial (NHBE) cells (CC-2541, Lonza; Walkersville, MD) from a single donor were grown as per manufacturer’s instructions as described in detail previously (14). Briefly, NHBE cells were grown submerged in vendor-supplied medium on transwell-clear membrane supports coated with rat-tail collagen until fully confluent, at which point the apical medium was removed, creating an air-liquid interface and the media type was changed. Cells were then grown until they formed a mature pseudostratified epithelium, ~28 days total time.
Construction and rescue of chimeric viruses
Five variants of 1918 influenza A HA RBD virus were generated including four previously reported and a fully avianized version (5, 7, 8). Table 1 shows the amino acid sequences of each HA RBD variant’s critical amino acid mutations, its hemagglutinin Sia binding preference (if known), and the abbreviated name as used in this study. In summary, all viruses but AV encode an aspartic acid (D) at position 187 [H1 numbering used throughout]; AV was engineered to encode the avian H1 influenza virus consensus glutamic acid (E) at this position. At position 222, SC and VA have D, NY and AV have glycine (G), and the HA of NY3 encodes an asparagine (N) (7). VA encodes a D at positions 187 and 222 as does SC but additionally encodes an arginine (R) at position 189 instead of the consensus glutamine (Q).
The fully reconstructed 1918 H1N1 influenza viruses were isogenic except for the above HA RBD polymorphisms, and were prepared by reverse genetics as previously described (15). RBD mutations in the 1918 HA gene for each of the other viruses were generated with a site-directed mutagenesis kit following the manufacturer's instructions (Stratagene; La Jolla, CA). All rescued viruses were propagated in Madin-Darby canine kidney (MDCK) cells (ATCC; Manassas, VA). The genomic sequence of each rescued virus was then confirmed by sequence analysis of the inoculum prior to performing the experiments. All viruses and infectious samples were handled under BSL3+ conditions in accordance with the Select Agent guidelines of the National Institutes of Health (NIH) and the Centers for Disease Control and Prevention under the supervision of the NIH Select Agent and Biosurety Programs and the NIH Department of Health and Safety.
Hemagglutinination Assay
Viral stocks were titered by plaque assay as previously described (15) and NHBE infections were normalized by multiplicity of infection (MOI). Given a primary experimental focus on viral binding and entry, viral stocks were also quantitated by hemagglutinin units using a standard hemagglutination assay for influenza A virus with turkey red blood cells (22).
Viral infections
Uninfected histologic control inserts were immersed in 10% neutral buffered formalin (NBF) prior to transporting the rest of the cells into the select agent suite where infections were conducted. In order to examine variation in binding and entry patterns of the five 1918 viruses, infections were initially conducted as a single experiment with MOIs of 1.0 and 3.0 in triplicate for all five viruses (three inserts per MOI/virus combination). Following aspiration of mucus from each well, adherent mucus was solubilized by incubation in 200 μL of PBS for 1 hr, removed by aspiration, and then infected with 100 μLvirus at the appropriate MOI (diluted in sterile PBS). Cells were incubated with virus for 20 min at 37 °C in an atmosphere of 5% CO2, followed by aspiration of the virus supernatant from the apical aspect of the upper chamber. Cells were then washed once with sterile PBS, and inserts were placed in NBF. After 48 hr, fixed noninfectious inserts were removed from the Select Agent suite. Subsequent experiments were conducted as above but with variations to the MOI and incubation times. These experimental conditions included infections with MOIs of 0.1 and 0.01 for a 20 min incubation period, and infections with MOIs of 1.0, 0.1, and 0.01 for a 5 min period.
Quantitative reverse transcriptase PCR on cell lysates
We determined viral load (intracytoplasmic and plasma membrane bound) by qRT-PCR for the influenza A virus matrix protein 1 (M1) gene using RNA isolated from cell lysates from parallel experiments with all 5 1918 RBD variants at MOI of 0.1 and 0.01 for both 5- and 20- minute incubation time periods, conducted in triplicate. Further, we compared 1918 RBD variants AV, NY, and SC to a set of control influenza viruses, including human seasonal influenza A/New York/312/2001 (H1N1) [NY312] (14), 2009 H1N1 pandemic influenza A/Mexico/4108/09 (H1N1) [Mex09] (23), and low-pathogenic avian influenza A/mallard/Ohio/265/1987 (H1N9) (24) [LPAI]. These infections were performed at an MOI of 0.01 at 5 and 20 minutes incubations in sextuplicate. Briefly, total RNA was isolated from individual infected NHBE samples and their viral loads estimated as previously described (23). Results were graphed as a threshold cycle threshold (CT) ratio of the human housekeeping gene glyceraldehyde-3-phosphate-dehydrogenase (GAPDH) over influenza A viral gene Matrix. Two-way ANOVAs with Tukey’s Multiple Comparison Tests were used for statistical analysis of both cell lysate real-time PCR data and replication kinetics and with alpha set to 0.05. Graphs and analyses were performed in Prism 6.0c (GraphPad; La Jolla, CA) and graphs further annotated in Adobe Illustrator CC 2014 (Adobe; San Jose, CA).
Viral replication kinetics
Replication kinetics including the 12, 24, and 36-hour time-points were determined for the AV, SC, and NY 1918 viruses. These viruses were selected to represent the range of known Sia binding preferences (AV α2,3; NY mixed α2,3 and 2,6; and SC α2,6). Timepoints were selected to cover peak viral infection as seen in prior experiments with human and avian influenza A viruses in this lot of NHBE cells (14, 21). These experiments had also indicated that live-virus could not be detected in cell supernatants prior to 8 hours post-infection and that cell viability drops off between 24 and 48 hours post-viral infection (14, 21). Cells were infected with 100 μL of virus at an MOI of 0.1 and wells were cultured at 37°C in an atmosphere of 5% CO2 until each time-point was reached. This experiment was conducted in triplicate for each time-point/virus combination. Apical media was collected at the appropriate time for each infected insert by addition of 400 μL PBS (200 μL X 2) to each biological replicate and then collecting all fluid at the apical aspect. The remaining insert was placed into NBF. All supernatants were stored individually in cryovials at −80 °C for subsequent evaluation of viral titers by standard influenza A virus plaque assays (15).
Immunofluorescence (IF)
Fixed, differentiated infected and uninfected cells on their transwell membranes were individually embedded on edge in paraffin. Approximately 5 μm thick cross sections of differentiated cells were cut and placed on positively charged slides, two sections per slide (NC State University College of Veterinary Medicine Histopathology Lab; Raleigh, NC). Each slide included two serial sections from a given well. In order to determine which epithelial cell types facilitated binding and entry of the influenza A viruses, sections for each MOI/virus combination were labeled using immunofluorescence for influenza A antigen as well as NHBE ciliated, goblet, and basal epithelial cell types as previously described (14) with the following changes. The donkey serum block was reduced to 5% strength as the lot of the anti-influenza antibody lacked the background issues documented in Davis et al., 2015 (14). Goblet cells were detected with 2 μg/mL biotinylated Jacalin (B-1155, Vector Labs; Burlingame, CA) and visualized with 10 μg/ml of Streptavidin Alexa Fluor-488 conjugate (S-11223, Molecular Probes; Eugene, OR), instead of fluorochrome conjugated Jacalin. Ciliated and basal cells were detected as before but visualized with Dylight-549 AffiniPure Donkey Anti-Mouse (discontinued, Jackson Immunoresearch (JI); West Grove, PA) and Dylight-649 AffiniPure Donkey Anti-Rabbit (discontinued, JI) respectively. Sia distribution by lectin histochemistry was not repeated in this experiment given prior detailed characterization of this in the same NHBE donor and cell lot (14).
Immunofluorescence for endocytic pathway markers was used to determine the cellular compartment in which the influenza antigen signal was located. Slides were prepared for antibody application as described previously (14). Dual labels were prepared with influenza A virus antibody, and an early endosome marker, rabbit polyclonal Anti-Rab5 antibody (ab13253, Abcam; Cambridge, MA), or the lysosome marker mouse monoclonal antibody Anti-LAMP1 [H4A3] (ab25630, Abcam). Additionally, a triple label was prepared with both endosome markers and influenza A virus antibody. Rab5 was applied at a 1:50 dilution and Lamp1 at 2 ug/mL, with both dilutions made in TBS. Appropriate secondary antibody pairings were selected for each of the markers in accordance with the previously described immunofluorescence work (14). All slides were mounted in Prolong Gold (P36930, Life Technologies; Grand Island, NY) as per vendor instructions. Slides were visualized and images captured on a Leica TCS SP5 X White Light Laser confocal system (Leica Microsystems; Buffalo Grove, IL) configured with Argon laser, 405 nm dioide and photomultiplier tube detectors.
Image capture and analysis
Individual confocal sessions focused on an entire batch, defined as a single MOI/incubation time. In this manner, intra-batch image capture conditions were as constant as possible. Also, settings were reused between sessions, re-checking the thresholding of the influenza antigen signal against batch-specific uninfected control slides at each session start. Slides from the first experiment, MOI 3.0 and 1.0 with 20 min incubation, were reviewed manually with representative images taken for each virus/MOI combination.
For the second experiment, each well for each virus/MOI combination was reviewed manually for all virus/MOI combinations, visually scanning both strips of cells on the slide. Minimally, three representative 63x oil objective at zoom 1, 1024 x 1024 pixels full-thickness z-stacks were taken. Additionally, a 63x oil objective at zoom 2.5, 1024 x 1024 pixels representative full-thickness z-stack was captured for each virus/MOI combination was taken. As there was little difference between MOI 0.1 and 0.01, subsequent analyses focused on infections at an MOI of 0.01 at the 5 minute time point.
Image post-processing and analysis were done with the Leica Application Suite (Leica Microsystems) and Imaris 7.6.3 (Bitplane; Zurich, Switzerland). Maximum projected fluorescent images for influenza antigen with and without cell type markers were created. Within an experimental batch, intensity levels were adjusted consistently across all samples. Additionally, the differential interference contrast (DIC) slice, wherein ciliated and goblet cells were near equivalently in focus for the largest number of cells, was selected from each z-stack and merged with a max projected image of multi-label fluorescence. These images were reviewed for presence of influenza A virus antigen by cell type. Final figure compilation was done in Adobe Photoshop CS6 Extended (Adobe).
RESULTS
1918 RBD variants bound and entered NHBE cells efficiently
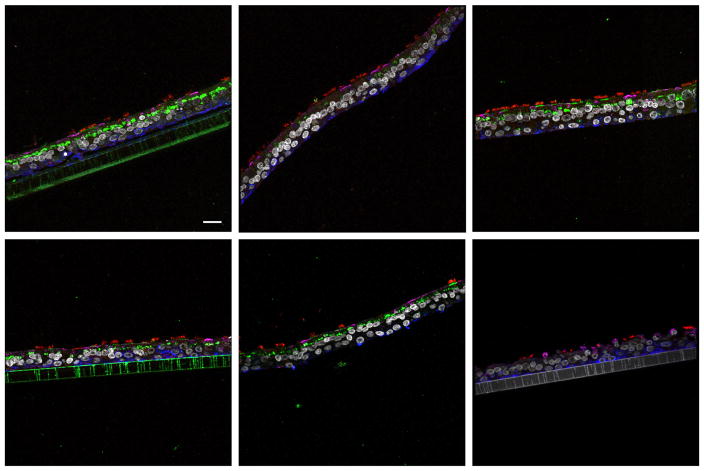
Infections at all MOI and incubation time combinations resulted in binding and entry of influenza virus in all samples. Immunofluorescence (IF) analysis of virus infections at MOI 3.0 or 1.0 incubated for 20 minutes demonstrated intracytoplasmic influenza antigen from nearly every apical epithelial cell (data not shown). To look for more subtle differences between the 1918 virus RBD variants (Table 1), experiments with lower MOI and shorter incubation periods were performed. Variance in the virus entry patterns at MOI 0.1, especially for the 5 min incubation time, was noted. The results from the MOI 0.01, 5 min incubation time combination reinforced these findings. Therefore the main focus of the subsequent detailed analyses were infections at MOI 0.01 for 5 min along with cross checks with the 20 min incubations from the same MOI and the MOI 0.1 triplicates. Representative images for each 1918 virus at MOI 0.01 at the 5 min timepoint are shown in Figure 1.
Figure 1.
Comparison of influenza viral antigen distribution by cell type for 1918 receptor binding domain variants. Leica SP5 white light laser confocal maximum projection images of full thickness zstacks of immunofluorescence labeled formalin-fixed paraffin embedded NHBE cells. All imaging was done with a 63x oil objective at zoom 1, 1024 x 1024 pixels. (A) 1918 AV, (B) 1918 SC, (C) 1918 NY, (D) 1918 NY3, (E) 1918 VA and (F) media control (no virus); all were 5 min incubations with virus at MOI 0.01. The pseudo-colors are influenza viral antigen (green), goblet cells (magenta), ciliated cells (red), basal cells (blue), and nuclei (gray). Presented images are representative of the relative influenza viral antigen intensity across three individually infected wells for each virus and where there was inter- or intra-well variance in influenza antigen intensity, a field representative of the median relative intensity was selected. The influenza antigen signal in SC (B) is less than the other viruses and AV (A) has the strongest signal. Scale, 20 μm.
1918 Influenza viral antigen was intracytoplasmic as well as bound to the apical epithelial surface of both goblet and ciliated cells. The relative intensity of influenza antigen labeling across the entire epithelium of the NHBE cells was highest for AV (Figure 1A), lowest for SC (Figure 1B), and intermediary for the other three viruses (Figure 1C–E; no virus control shown in Figure 1F). For each of the 1918 variants examined, intracytoplasmic influenza viral antigen signal was more prominent in goblet cells than in ciliated cells. Overall, patterns of 1918 viral antigen in NHBE cells at MOI 0.01 after incubation for 5 minutes were very similar for the naturally occurring 1918 RBD variants (Figure 1B–E), with more prominent labeling with the AV variant (Figure 1A). When the intracytoplasmic location of the influenza antigen was explored using endosomal markers (Fig. 2), there was occasional co-localization with early, Rab5 (Fig. 2D, yellow color), and more co-localization with late, Lamp1 (Fig. 2D, white), endosomal markers as well as a noticeable portion of intracytoplasmic influenza antigen signal that failed to co-localize with either marker (Fig. 2D, green color), suggesting that the majority of intracytoplasmic influenza virus was not present in endosomes at 20 minutes.
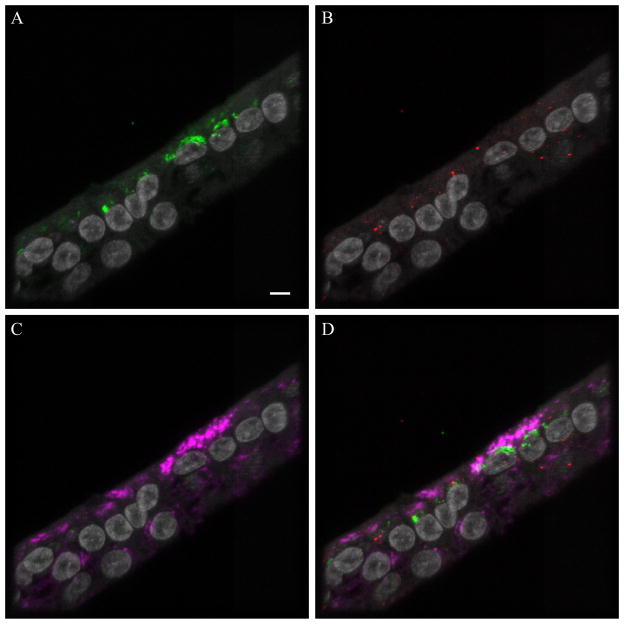
Figure 2.
Endosomal markers with influenza antigen. Leica SP5 white light laser confocal maximum projection images of full thickness zstacks of immunofluorescence labeled formalin-fixed paraffin embedded NHBE cells infected with AV at MOI 1.0 for 20 min. All imaging was done with a 63x oil objective at zoom 3.5, 1024 x 1024 pixels. All images have nuclei (gray): (A) influenza viral antigen (green), (B) early endosomal marker Rab5 (red), (C) late endosomal marker Lamp1 (magenta), and (D) the fluorescence merge. Yellow or orange indicates colocalization of influenza viral antigen with the early endosomal marker Rab5. White indicates co-localization of influenza antigen with late endosomal marker LAMP1. Bar = 5 μm.
Evaluation of hemagglutination by three 1918 RBD variant viruses
We selected the 1918 viral variants AV, NY, and SC, representing the diversity of Sia preferences and conducted hemagglutination assays in order to determine if HA binding capacity varied across viral stocks. AV viral stock titer by plaque assay and HA units were approximately 4 times lower than those of NY and SC. After adjusting for differences in viral stock titer, however, HA units were equivalent across the three viruses (Table 2).
Table 2.
Hemagglutination assay results for AV, NY, and SC
| Virus | Titer of viral stock by plaque assay (pfu/ml) | HA unit (measured) | HA unit (adjusted for viral stock titer) |
|---|---|---|---|
| SC | 7.6 x 107 | 1:256 | 1:256 |
| NY | 8.0 x 107 | 1:256 | 1:256 |
| AV | 2.0 x 107 | 1:64 | 1:256 |
Quantitative reverse transcriptase PCR for viral RNA in NHBE cells
1918 RBD variant viral RNA was detected in lysates from cells infected at MOIs of 0.1 and 0.01 following 5 and 20 min incubations. The relative amounts of viral RNA were not statistically different across all viruses when infections were performed at an MOI of 0.1 for either incubation time (data not shown). When performed at an MOI of 0.01 at 5 minutes, AV viral RNA was present in statistically significantly higher amounts than the other four viral variants at 5 minutes (p<0.05) (Figure 3). At 20 min after MOI 0.01 infection, AV viral RNA was still present in statistically significantly higher amounts than NY (p<0.05) and SC (p<0.05), but not as compared to NY3 or VA (Figure 3).
Figure 3.
Real-time PCR results for all five viruses at a multiplicity of infection of 0.01. Real-time PCR data for MOI 0.01 both 5 min (left side) and 20 min (right side) post-viral infection. The data are presented as GAPDH cycle time (CT)/Influenza viral Matrix CT as previously described (23). At 5 min (*) denotes AV cell associated viral RNA level is significantly higher as compared to all other viruses and at 20 min is significantly higher than SC and NY by ANOVA with an alpha of 0.05.
To determine the significance of these observations an additional experiment was performed comparing 1918 AV, NY, and SC RBD variant binding and entry to a set of control influenza viruses, including human seasonal influenza A/New York/312/2001 (H1N1) [NY312] (14), 2009 H1N1 pandemic influenza A/Mexico/4108/09 (H1N1) [Mex09] (23), and low-pathogenic avian influenza A/mallard/Ohio/265/1987 (H1N9) (24) [LPAI] as described above. At 5 and 20 minutes after addition of virus, the relative amounts of cell-associated viral RNA were generally comparable between human seasonal, pandemic, and avian H1 subtype influenza viruses with the seasonal H1N1 strain NY312 showing the highest amount of cell-associated viral RNA at both time points (p <0.05). Otherwise, viral RNA levels were comparable between the different 1918 RBD strains, Mex09, and LPAI H1 viruses (Figure 4).
Figure 4.
Real-time PCR results for Mex09, NY312, LPAI virus and three1918 RBD variants AV, NY, SC. Real-time PCR data for MOI 0.01 both 5 minutes (left panel) and 20 minutes (right panel) post-viral infection. The data are presented as GAPDH cycle time (CT)/Influenza viral Matrix CT as previously described (23). At 5 min (*) denotes NY312 cell-associated viral RNA level is significantly higher as compared to all other viruses, except the LPAI virus. At 20 min (*) denotes NY312 cell-associated viral RNA level is significantly higher as compared to all other viruses by ANOVA with an alpha of 0.05.
Viral replication of selected 1918 viral variants
Viral replication was measured in NHBE cells at 12, 24, and 36 hours following infection with the 1918 SC, NY, and AV viruses (Fig. 5). There was no statistical difference between viral titers among these groups at 12 hours, but at 24 and 36 hours, the AV variant replicated to slightly higher titers as compared to NY and SC (p<0.05), with AV replicating ~1 log10 pfu/mL higher than SC and NY.
Figure 5.
Replications kinetics for viruses AV, NY and SC. Plaque forming units at 12-, 24- and 36-hours post-infection with MOI 0.1 of NY, SC and AV. (*) denotes that AV is significantly different than both NY and SC at both 24 and 36 hours post-viral infection by ANOVA with an alpha of 0.05.
DISCUSSION
In this study we evaluated whether 1918 influenza viral variants with polymorphisms in their HA RBD differed in binding and entry to primary human airway cells from a single donor source. During preliminary studies performed at high MOIs (3.0 and 1.0), it was observed that the majority of luminal NHBE cells were positive for intracytoplasmic influenza antigen by IF labeling. Consequently we decreased MOIs to 0.1 and 0.01 in an effort to detect subtle differences in viral binding and entry under more discerning conditions. Following a 20-minute incubation at these lower MOIs, no difference in viral binding and entry could be detected by IF. At an MOI of 0.01 and 5-minute incubation, the “avianized” 1918 virus AV, with an α2,3 Sia receptor-binding specificity, was found to bind and enter NHBE cells more efficiently than the other four variants as assessed by IF (Figure 1).
The extent of antigen signal by IF was greater than predicted for MOIs of 0.1 (i.e., 1 viral particle per 10 cells) and 0.01 (i.e., 1 viral particle per 100 cells) for all viruses. Replication competent virus is believed to compose only a small fraction of viral stocks, which are otherwise largely composed of replication incompetent viral-like particles (25). Given this we sought to confirm that the proportion of HA binding particles did not vary significantly across viral stocks. We performed HA assays on AV, SC and NY, representing the range of binding configurations, and after adjusting for baseline viral stock titers, we confirmed that these infections were performed with equivalent HA units (Table 2). Additionally, we sequence-confirmed all rescued viral variants to rule out the possibility that observed differences in viral binding and entry might be attributable to human error during viral rescue or viral mutations during cell passage.
The observation that AV infections at MOI 0.01 following a 5-minute incubation resulted in slightly enhanced binding and entry to NHBE cells as compared with the four other 1918 RBD variants was further assessed by qRT-PCR for cell-associated viral RNA. In independent parallel experiments performed at MOI 0.01, we observed that cell-associated AV viral RNA was detected in slightly greater proportions than the 4 other RBD variants following both a 5- and a 20-minute incubation (Figure 3). To further validate these results an additional experiment was performed comparing a seasonal H1N1 virus [NY312], a 2009 pandemic H1N1 virus [Mex09], and a low-pathogenic avian influenza H1N9 virus [LPAI] to three 1918 RBD variants - AV, NY, and SC (Figure 4). As shown, all viruses tested bound and entered NHBE cells, including an avian H1 virus with an α2,3 Sia preference. These data support the primary observation that avian viruses with an α2,3 Sia preference bound and entered NHBE cells in our system efficiently.
All five 1918 RBD variants were detected on the apical surface and within the cytoplasm of both ciliated and goblet cells with no discernable qualitative or quantitative difference in cell-type tropism between viruses. The potential for subtle quantitative differences in cell-type tropism between variants exists and could be evaluated in future work. While we did not repeat lectin histochemical analysis of cell surface Sia distribution in this study, prior detailed studies of the same NHBE cell donor and lot grown in the same conditions indicate that α2,6 Sia are predominantly displayed on ciliated and goblet cell surfaces and rarely α2,3 Sia are displayed on goblet cell surfaces (14). Similarly, we did not repeat glycan-binding arrays on the 1918 RBD variants AV, NY, and SC given that these have been previously well characterized and that we sequence-confirmed the 1918 RBD variants used in these experiments. Consequently in the present study cell surface Sia distribution did not predict binding and entry or cell-type preference of the five 1918 RBD variants in the primary human airway epithelial cells examined here.
Previously published data support that influenza viruses with α2,3 Sia preferences successfully infect NHBE cells. Oshansky et al., 2011 showed that low pathogenic avian influenza viruses with α2,3 binding preference infected differentiated NHBE cells displaying only α2,6 Sia receptors (26). Similar findings have been observed in ex vivo human respiratory tissue using an H5N1 avian virus (26–28). Our data fit with these prior observations. Sia-independent pathways for influenza viral entry into airway cells likely exist (29, 30). For example, when NHBE cells are treated with neuraminidase to remove the Sia, it has been observed that both avian and human influenza viruses readily infect them (26, 28). Mice lacking the enzyme ST6Gal I sialyltransferase, and thus unable to attach α2,6 Sia to cell surface N-linked glycoproteins, had similar viral titers in lung tissue as wild-type mice following influenza infection (31).
In an effort to discern intracellular location of influenza virus in our study, immunohistochemistry multi-labeling analysis for both viral antigen with early (Rab5) and late (Lamp1) endosomal markers was employed. Influenza viral antigen co-localized with early and late endosomal markers but was predominantly observed independent of endosomes within the cytoplasm (Fig 2). Influenza viruses are thought capable of entering cells through a variety of mechanisms including macropinocytosis, calveolar entry, clathrin-mediated endocytosis, as well as a non-calveolar, non-clathrin dependent pathways (32, 33). In this study, while co-localization of virus with early and late endosomal markers supports viral entry via endocytosis, the preponderance of virus independent of endosomes also suggests a non-endocytic entry (Figure 2). Future studies in primary human airway cell lines may further characterize mechanisms of influenza A viral binding and entry.
Finally at 24 and 36 hours post-infection, 1918 AV with α2,3 Sia-preference, replicated to higher titers than NY with mixed α2,3/α2,6 preference and SC with α2,6 preference. It was previously shown that seasonal and pandemic H1N1 influenza viruses replicated to peak viral titers of ~104 pfu/mL following MOI 0.1 infection in this NHBE cell lot (14) and multiple avian influenza viruses following MOI 1.0 infection replicate to peak titers between 104 and105 pfu/mL at similar time-points post-infection (21). Similar to these prior observations, in this study two naturally occurring human 1918 influenza A RBD variants NY and SC replicated to peak titers of 104 pfu/mL while the avianized variant AV replicated to peak titers of ~105 pfu/mL.
Understanding viral and host factors contributing to influenza virus infection of human airway cells is necessary to elucidate the process of host-switch events and thus pandemic viral emergence. While cell surface Sia specificity may be important, it is conceivable that influenza viruses with α2,6 Sia receptor-binding specificity are better adapted to humans not due to an advantage in viral binding and entry at the cell surface but due to an advantage in evading decoy receptors present in mucus lining the human airway. Mucus is known to be rich in O-linked α2,3 sialic acids that efficiently bind influenza viruses with an α2,3 Sia binding preference (34). In our NHBE model, mucus was systematically solubilized and removed from the NHBE cell surfaces prior to infection so as not to interfere with viral infection. This process may have eliminated any selective advantage of influenza variants with α2,6 Sia receptor-binding specificity exposing subtle binding and entry advantages of the AV virus.
It is worth noting that in vitro glycan array hybridization has been relied upon to predict in vivo influenza virus Sia preference. However, in vitro glycan arrays likely do not reproduce the spectrum of Sia receptors present on human airway epithelia. A spectrum of Sia receptors on the human airway was recently characterized by mass spectrometry, confirming a wide variety of both N- and O-linked glycans, both α2,3 and α2,6, that correlated poorly with those found on the glycan arrays currently employed to determine viral binding preferences (35). Consequently Sia preference as predicted by glycan array hybridization may not accurately predict Sia preference in vivo. Additionally, the variability of airway Sia receptors with co-factors such as age, co-morbid medical conditions, etc. remains unknown. Host factors that result in differential expression of Sia receptors on cell surfaces may in part account for observed variability in host susceptibility to respiratory viral infections. Finally, plant lectins, which have been used to characterize the distribution of α2,3 Sia versus α2,6 Sia receptors on cell surfaces, have variable sensitivity and specificity. It is possible that in our model AV preferentially bound α2,3 Sia receptors distributed on NHBE cell surfaces not detected by standard lectins previously employed (14).
The primary limitation of this work is that all experiments were performed using a single NHBE cell donor and lot and a limited subset of viruses. Generalizability of our observations to other primary human airway cell lines and viruses would need to be evaluated in future work. Additionally in vitro models, even with primary human airway cell lines, cannot replicate the full spectrum of pathogen-host interactions that influence viral binding and entry in vivo. The strength of utilizing a single lot of NHBE cells in this study, however, relates to our prior detailed characterization of α2,3 versus α2,6 Sia distribution over multiple experiments from this donor and cell lot (14). Despite a clear predominance of α2,6 Sia on cell surfaces we have shown that 1918 influenza viral variants with α2,3 and mixed α2,3/α2,6 Sia preferences readily bind, enter, and replicate within primary human airway cells. These findings suggest that predicted Sia preferences alone do not dictate influenza A viral binding and entry to human airway cells.
In summary, five 1918 RBD influenza viral variants efficiently bound and infected primary human airway cells comparably to control human and avian influenza A viruses. The experimental findings in this study correlated with the post-mortem observations of lung specimens from fatal 1918 influenza infections, which show no differences in viral distribution along the airway for the four naturally occurring 1918 RBD viral variants studied here (7). Taken together, these observations support that viral and host factors unrelated to Sia binding preference likely contribute to influenza binding and entry into human tracheobronchial airway epithelial cells, or perhaps Sia preferences restrict α2,3 Sia binding and entry in the nasopharyngeal epithelium. Further probing of these factors is needed to unravel the complexities of host switch events, the essential first step in the initiation of a pandemic.
Highlights.
1918 influenza viral receptor-binding variants all bound and entered primary human airway cells.
All 1918 variants were detected on the apical surface and cytoplasm of ciliated and goblet cells.
Viral RNA levels in cells were comparable between human, pandemic, and avian influenza viruses.
Sialic acid preference predicted by glycan array may not accurately predict preference in vivo.
Acknowledgments
The authors would like to acknowledge the staff at the NIAID RTB Bioimaging Section for their assistance with confocal microscopy and image post-processing. We thank Sandra Horton and her staff at the North Carolina State University College of Veterinary Medicine Histopathology Laboratory for their preparation of tissue and embedded NHBE cell sections for immunofluorescence and histological analysis. This work was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institute of Health. ASD and JKT are further thankful for the support of the NIH Comparative Molecular Pathology Research Training Program. This work was done in partial fulfillment of A. Sally Davis’ dissertation work towards a Ph.D. in Comparative Biomedical Sciences at the North Carolina State University College of Veterinary Medicine.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wright PFNG, Kawaoka Y. Orthomyxoviruses. Fields virology. (5) 2007;2:1691–1740. [Google Scholar]
- 2.Johnson NP, Mueller J. Updating the accounts: global mortality of the 1918–1920 "Spanish" influenza pandemic. Bull Hist Med. 2002;76:105–115. doi: 10.1353/bhm.2002.0022. [DOI] [PubMed] [Google Scholar]
- 3.Taubenberger JK, Morens DM. 1918 Influenza: the mother of all pandemics. Emerg Infect Dis. 2006;12:15–22. doi: 10.3201/eid1201.050979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taubenberger JK, Kash JC. Insights on influenza pathogenesis from the grave. Virus Res. 2011;162:2–7. doi: 10.1016/j.virusres.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reid AH, Fanning TG, Hultin JV, Taubenberger JK. Origin and evolution of the 1918 "Spanish" influenza virus hemagglutinin gene. Proc Natl Acad Sci U S A. 1999;96:1651–1656. doi: 10.1073/pnas.96.4.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reid AH, Janczewski TA, Lourens RM, Elliot AJ, Daniels RS, Berry CL, Oxford JS, Taubenberger JK. 1918 influenza pandemic caused by highly conserved viruses with two receptor-binding variants. Emerg Infect Dis. 2003;9:1249–1253. doi: 10.3201/eid0910.020789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sheng ZM, Chertow DS, Ambroggio X, McCall S, Przygodzki RM, Cunningham RE, Maximova OA, Kash JC, Morens DM, Taubenberger JK. Autopsy series of 68 cases dying before and during the 1918 influenza pandemic peak. Proc Natl Acad Sci U S A. 2011;108:16416–16421. doi: 10.1073/pnas.1111179108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Glaser L, Stevens J, Zamarin D, Wilson IA, Garcia-Sastre A, Tumpey TM, Basler CF, Taubenberger JK, Palese P. A single amino acid substitution in 1918 influenza virus hemagglutinin changes receptor binding specificity. J Virol. 2005;79:11533–11536. doi: 10.1128/JVI.79.17.11533-11536.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stevens J, Blixt O, Glaser L, Taubenberger JK, Palese P, Paulson JC, Wilson IA. Glycan microarray analysis of the hemagglutinins from modern and pandemic influenza viruses reveals different receptor specificities. J Mol Biol. 2006;355:1143–1155. doi: 10.1016/j.jmb.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 10.Stevens J, Corper AL, Basler CF, Taubenberger JK, Palese P, Wilson IA. Structure of the uncleaved human H1 hemagglutinin from the extinct 1918 influenza virus. Science. 2004;303:1866–1870. doi: 10.1126/science.1093373. [DOI] [PubMed] [Google Scholar]
- 11.Xiao YL, Kash JC, Beres SB, Sheng ZM, Musser JM, Taubenberger JK. High-throughput RNA sequencing of a formalin-fixed, paraffin-embedded autopsy lung tissue sample from the 1918 influenza pandemic. J Pathol. 2013;229:535–545. doi: 10.1002/path.4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shinya K, Ebina M, Yamada S, Ono M, Kasai N, Kawaoka Y. Influenza virus receptors in the human airway. Nature. 2006;440:435–436. doi: 10.1038/440435a. [DOI] [PubMed] [Google Scholar]
- 13.Nicholls JM, Chan RWY, Russell RJ, Air GM, Peiris JSM. Evolving complexities of influenza virus and its receptors. Trends in Microbiology. 2008;16:149–157. doi: 10.1016/j.tim.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 14.Davis AS, Chertow DS, Moyer JE, Suzich J, Sandouk A, Dorward DW, Logun C, Shelhamer JH, Taubenberger JK. Validation of Normal Human Bronchial Epithelial Cells as a Model for Influenza A Infections in Human Distal Trachea. J Histochem Cytochem. 2015 doi: 10.1369/0022155415570968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qi L, Kash JC, Dugan VG, Wang R, Jin G, Cunningham RE, Taubenberger JK. Role of sialic acid binding specificity of the 1918 influenza virus hemagglutinin protein in virulence and pathogenesis for mice. J Virol. 2009;83:3754–3761. doi: 10.1128/JVI.02596-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matrosovich MN, Matrosovich TY, Gray T, Roberts NA, Klenk HD. Human and avian influenza viruses target different cell types in cultures of human airway epithelium. Proc Natl Acad Sci U S A. 2004;101:4620–4624. doi: 10.1073/pnas.0308001101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ibricevic A, Pekosz A, Walter MJ, Newby C, Battaile JT, Brown EG, Holtzman MJ, Brody SL. Influenza virus receptor specificity and cell tropism in mouse and human airway epithelial cells. J Virol. 2006;80:7469–7480. doi: 10.1128/JVI.02677-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chan RW, Yuen KM, Yu WC, Ho CC, Nicholls JM, Peiris JS, Chan MC. Influenza H5N1 and H1N1 virus replication and innate immune responses in bronchial epithelial cells are influenced by the state of differentiation. PLoS One. 2010;5:e8713. doi: 10.1371/journal.pone.0008713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chan RW, Chan MC, Nicholls JM, Malik Peiris JS. Use of ex vivo and in vitro cultures of the human respiratory tract to study the tropism and host responses of highly pathogenic avian influenza A (H5N1) and other influenza viruses. Virus Res. 2013;178:133–145. doi: 10.1016/j.virusres.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kogure T, Suzuki T, Takahashi T, Miyamoto D, Hidari KI, Guo CT, Ito T, Kawaoka Y, Suzuki Y. Human trachea primary epithelial cells express both sialyl(alpha2-3)Gal receptor for human parainfluenza virus type 1 and avian influenza viruses, and sialyl(alpha2-6)Gal receptor for human influenza viruses. Glycoconj J. 2006;23:101–106. doi: 10.1007/s10719-006-5442-z. [DOI] [PubMed] [Google Scholar]
- 21.Qi L, Pujanauski LM, Davis AS, Schwartzman LM, Chertow DS, Baxter D, Scherler K, Hartshorn KL, Slemons RD, Walters KA, Kash JC, Taubenberger JK. Contemporary avian influenza A virus subtype H1, H6, H7, H10, and H15 hemagglutinin genes encode a mammalian virulence factor similar to the 1918 pandemic virus H1 hemagglutinin. MBio. 2014;5:e02116. doi: 10.1128/mBio.02116-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirst GK. The Quantitative Determination of Influenza Virus and Antibodies by means of Red Cell Agglutination. J Exp Med. 1942;75:49–64. doi: 10.1084/jem.75.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kash JC, Walters KA, Davis AS, Sandouk A, Schwartzman LM, Jagger BW, Chertow DS, Li Q, Kuestner RE, Ozinsky A, Taubenberger JK. Lethal synergism of 2009 pandemic H1N1 influenza virus and Streptococcus pneumoniae coinfection is associated with loss of murine lung repair responses. mBio. 2011;2:e00172–11. doi: 10.1128/mBio.00172-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qi L, Davis SA, Jagger BW, Schwartzman LM, Dunham EJ, Kash JC, Taubenberger JK. Analysis by Single-Gene Reassortment Demonstrates that the 1918 Influenza Virus Is Functionally Compatible with a Low-Pathogenicity Avian Influenza Virus in Mice. J Virol. 2012;86:9211–9220. doi: 10.1128/JVI.00887-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marriott AC, Dimmock NJ. Defective interfering viruses and their potential as antiviral agents. Rev Med Virol. 2010;20:51–62. doi: 10.1002/rmv.641. [DOI] [PubMed] [Google Scholar]
- 26.Oshansky CM, Pickens JA, Bradley KC, Jones LP, Saavedra-Ebner GM, Barber JP, Crabtree JM, Steinhauer DA, Tompkins SM, Tripp RA. Avian influenza viruses infect primary human bronchial epithelial cells unconstrained by sialic acid alpha2,3 residues. PLoS One. 2011;6:e21183. doi: 10.1371/journal.pone.0021183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nicholls JM, Chan MC, Chan WY, Wong HK, Cheung CY, Kwong DL, Wong MP, Chui WH, Poon LL, Tsao SW, Guan Y, Peiris JS. Tropism of avian influenza A (H5N1) in the upper and lower respiratory tract. Nat Med. 2007;13:147–149. doi: 10.1038/nm1529. [DOI] [PubMed] [Google Scholar]
- 28.Thompson CI, Barclay WS, Zambon MC, Pickles RJ. Infection of human airway epithelium by human and avian strains of influenza a virus. J Virol. 2006;80:8060–8068. doi: 10.1128/JVI.00384-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Vries E, de Vries RP, Wienholts MJ, Floris CE, Jacobs MS, van den Heuvel A, Rottier PJ, de Haan CA. Influenza A virus entry into cells lacking sialylated N-glycans. Proc Natl Acad Sci U S A. 2012;109:7457–7462. doi: 10.1073/pnas.1200987109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chu VC, Whittaker GR. Influenza virus entry and infection require host cell N-linked glycoprotein. Proc Natl Acad Sci U S A. 2004;101:18153–18158. doi: 10.1073/pnas.0405172102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Glaser L, Conenello G, Paulson J, Palese P. Effective replication of human influenza viruses in mice lacking a major alpha2,6 sialyltransferase. Virus Res. 2007;126:9–18. doi: 10.1016/j.virusres.2007.01.011. [DOI] [PubMed] [Google Scholar]
- 32.Sieczkarski SB, Whittaker GR. Influenza virus can enter and infect cells in the absence of clathrin-mediated endocytosis. J Virol. 2002;76:10455–10464. doi: 10.1128/JVI.76.20.10455-10464.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Vries E, Tscherne DM, Wienholts MJ, Cobos-Jimenez V, Scholte F, Garcia-Sastre A, Rottier PJ, de Haan CA. Dissection of the influenza A virus endocytic routes reveals macropinocytosis as an alternative entry pathway. PLoS Pathog. 2011;7:e1001329. doi: 10.1371/journal.ppat.1001329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baum LG, Paulson JC. Sialyloligosaccharides of the respiratory epithelium in the selection of human influenza virus receptor specificity. Acta Histochem Suppl. 1990;40:35–38. [PubMed] [Google Scholar]
- 35.Walther T, Karamanska R, Chan RW, Chan MC, Jia N, Air G, Hopton C, Wong MP, Dell A, Malik Peiris JS, Haslam SM, Nicholls JM. Glycomic analysis of human respiratory tract tissues and correlation with influenza virus infection. PLoS Pathog. 2013;9:e1003223. doi: 10.1371/journal.ppat.1003223. [DOI] [PMC free article] [PubMed] [Google Scholar]