Abstract
It has been suggested that polyandry allows females to increase offspring genetic diversity and reduce the prevalence and susceptibility of their offspring to infectious diseases. We tested this hypothesis in wild‐derived house mice (Mus musculus) by experimentally infecting the offspring from 15 single‐ and 15 multiple‐sired litters with two different strains of a mouse pathogen (Salmonella Typhimurium) and compared their ability to control infection. We found a high variation in individual infection resistance (measured with pathogen loads) and significant differences among families, suggesting genetic effects on Salmonella resistance, but we found no difference in prevalence or infection resistance between single‐ vs. multiple‐sired litters. We found a significant sex difference in infection resistance, but surprisingly, males were more resistant to infection than females. Also, infection resistance was correlated with weight loss during infection, although only for females, indicating that susceptibility to infection had more harmful health consequences for females than for males. To our knowledge, our findings provide the first evidence for sex‐dependent resistance to Salmonella infection in house mice. Our results do not support the hypothesis that multiple‐sired litters are more likely to survive infection than single‐sired litters; however, as we explain, additional studies are required before ruling out this hypothesis.
Keywords: bet‐hedging, genetic diversity, multiple paternity, Mus musculus, pathogen resistance, pathogen‐mediated sexual selection, polyandry, salmonella, sex differences in immunity
Introduction
Females may enhance the resistance of their offspring to infectious diseases by mating with disease‐resistant males (Hamilton–Zuk hypothesis) (Hamilton & Zuk, 1982), genetically compatible males (Penn et al., 2002) or multiple males (Jennions & Petrie, 2000). Numerous studies have investigated the Hamilton–Zuk hypothesis (Hamilton & Poulin, 1997; Møller et al., 1999), and a recent study provides direct experimental support. When female house mice (Mus musculus) are experimentally mated with their preferred males, their offspring are better able to survive an experimental Salmonella infection compared to offspring sired by nonpreferred males (Raveh et al., 2014). Yet, when female house mice are allowed to select their mates, they often mate with multiple males. For example, when females can choose to mate between one or two males, behavioural observations indicate that all females mate multiply (Rolland et al., 2003), and paternity analyses show that 29–46% of litters are multiply sired (Thonhauser et al., 2013b, 2014; Manser et al., 2015). Moreover, in wild populations of house mice, the frequency of multiple paternity ranges between 6% and 43% [mean 30%: M. musculus domesticus (Dean et al., 2006; Firman & Simmons, 2008c); M. musculus musculus (Thonhauser et al., 2013c)]. Several adaptive hypotheses have been proposed for multiple male mating (genetic polyandry) (Zeh & Zeh, 1996, 1997; Jennions & Petrie, 2000; Hosken & Stockley, 2003; Simmons, 2005), including the good genes and compatible genes hypotheses, which predict that paternity can be biased towards a high quality or a more compatible mate through sperm competition or cryptic female choice (Madsen et al., 1992; Tregenza & Wedell, 2002; Hosken et al., 2003; García‐González & Simmons, 2005; Fisher et al., 2006; Firman & Simmons, 2008a,b). In house mice, it has been shown that polyandry can facilitate inbreeding avoidance (Firman & Simmons, 2008b), enhance pup survival (Firman & Simmons, 2008a; Auclair et al., 2014) and reduce litter loss due to genetic incompatibilities (Manser et al., 2015; Sutter & Lindholm, 2015). Polyandry can also potentially provide genetic benefits by enhancing offspring diversity (Yasui, 1998), which may enhance resistance to infectious diseases (Jennions & Petrie, 2000). Surprisingly, however, no studies to our knowledge have tested whether multiple paternity affects offspring resistance to infectious diseases in any vertebrate species.
By increasing offspring genetic diversity, polyandry may provide two nonmutually exclusive types of indirect fitness benefits. First, it may increase the variation in fitness within while reducing the variation in fitness between litters, thereby enhancing the geometric mean fitness of females (bet‐hedging hypothesis) (Yasui, 1998; Fox & Rauter, 2003). Bet‐hedging can be selectively favoured (even if the arithmetic mean fitness of females is reduced) when environmental conditions are unpredictable, as increased genetic diversity ensures that at least some genotypes fit the prevailing circumstances. Second, increased genetic diversity within litters may enhance the arithmetic mean fitness of females (non‐bet‐hedging hypothesis). Such effects could be due to reduced competition and more effective resource utilization among half‐siblings or genetically based differences in disease resistance (Hamilton, 1987; Jennions & Petrie, 2000). For example, increased genetic diversity within insect colonies (either generated by increased multiple mating of queens or mixing broods from diverse backgrounds) reduced the intensity and prevalence of parasitic infection of colonies (Shykoff & Schmid‐Hempel, 1991; Liersch & Schmid‐Hempel, 1998; Baer & Schmid‐Hempel, 1999; Tarpy, 2003; Tarpy & Seeley, 2006; Seeley & Tarpy, 2007). It has been shown that genetic diversification provides disease control (Fuchs & Schade, 1994; Zhu et al., 2000; Hughes & Boomsma, 2004), and similarly, multiple male mating may allow females to increase the genetic diversity and disease resistance of their litters (McLeod & Marshall, 2009; Soper et al., 2012).
In wild house mice (M. musculus musculus), multiple‐sired litters have higher genetic diversity than single‐sired litters (Thonhauser et al., 2013c), but it is unknown whether increasing offspring genetic diversity provides disease resistance or any other indirect fitness benefits. Multiple mating in mice may depend upon the variation in quality of potential mates. Scent marking in mice is a secondary sexual trait that indicates male health and other aspects of quality (Kavaliers & Colwell, 1995; Penn et al., 1998; Zala et al., 2004). Interestingly, when female mice are able to select their mates, they are more likely to produce multiple‐sired litters when their potential mates show similar levels of scent marking, whereas they are more likely to produce single‐sired litters, fathered by the high‐marking male, when their available mates differ in their scent marking (Thonhauser et al., 2013a, b). The relationship between female multiple mating and parentage is not known for house mice, but it was suggested that females mate multiply when they detect no difference in male quality, or alternatively females may generally mate multiply and single paternity may be due to sperm competition or cryptic female choice. To understand the adaptive functions of multiple male mating, the fitness consequences of polyandry need to be evaluated – especially under infection, competition or other ecological challenges.
The aim of this study was to compare the disease resistance of single‐ versus multiple‐sired litters produced by female house mice (M. musculus musculus) that were allowed to choose to mate with either one or two males (Thonhauser et al., 2013b). We investigated how the resulting offspring were able to clear (infection resistance) and cope (infection tolerance) with an experimental infection of Salmonella enterica serovar Typhimurium. S. Typhimurium is an enteric mouse pathogen that can cause medium to severe infections in hosts (Penn et al., 2002) as there is much variation in the virulence of different Salmonella strains (from benign to fatal). Infection resistance and tolerance to this pathogen are controlled by several genetic loci (Roy & Malo, 2002), including the highly polymorphic genes of the major histocompatibility complex (MHC) (Penn et al., 2002), and genetic homozygosity and inbreeding influence prevalence and reduce individual resistance of Salmonella infection (Penn et al., 2002; Ilmonen et al., 2008b). We infected mice with two strains of Salmonella enterica (a strain having very low virulence and another with higher virulence) and monitored changes in health (body mass) over 17 days and then measured pathogen loads to assess the ability to control infection (resistance/susceptibility). If multiple paternity functions as bet‐hedging, we predicted reduced variance in health and pathogen loads among multiple‐ versus single‐sired litters, although no differences in the mean load between the litters. If polyandry provides fitness benefits through a non‐bet‐hedging mechanism, then the mean Salmonella load should be lower in multiple‐ compared to single‐sired litters. On the other hand, if females mate singly when they can choose between high‐ and low‐quality males and mate preferences function to enhance offspring resistance or tolerance to infection (Raveh et al., 2014), then offspring from single‐sired litters may be equal or even more disease resistant than offspring from multiple‐sired litters. In addition, we compared the resistance of the sexes to Salmonella infection, as there have been only few such studies in wild‐derived house mice (Zala et al., 2004, 2008; Ilmonen et al., 2007, 2008b; Raveh et al., 2014).
Materials and methods
Experimental animals and housing
Experimental mice were second‐generation descendants of wild‐caught house mice (Mus musculus musculus) bred for a previous experiment, in which females were released into separate enclosures and could freely choose to mate with either one or both of two neighbouring males that were restricted to their own territories (Thonhauser et al., 2013b). We controlled for male harassment by providing a shelter within each male territory that was only accessible for females. We did not record female mating behaviour and conducted genetic paternity analyses to identify single‐ and multiple‐sired litters. We selected 15 single‐ and 15 multiple‐sired litters (213 offspring) from 30 different females. Experimental animals originated from 26 different families. Single‐sired litters were produced by 15 dams (all except three were unrelated to each other) and 15 sires (all except four were unrelated). Multiple‐sired litters were produced by 15 dams (all except three were unrelated) and 30 sires (all but nine were unrelated). Sires were always unrelated to dams. Litter size varied from three to eleven offspring and did not differ significantly between single‐ and multiple‐sired litters (t‐test: t = 0.562, df = 28, P = 0.579). Also, the sex ratio of offspring (the number of males and females) did not differ between single‐ and multiple‐sired litters (chi‐square test: χ² = 0.004; P = 0.949). Weaning occurred at the age of 21 ± 1 day and individuals were housed individually (type II mouse cages, 26.5 × 20.5 × 14 cm) under standard conditions (12‐h:12‐h light/dark cycle). Two d prior to infection, all experimental animals were moved into a separate room and placed individually into type IIL mouse cages (36.5 × 20.5 × 14 cm) with filter hoods on top. All cages were equipped with wooden bedding (ABEDD), wood shavings, a nest box and food (Altromin rodent diet 1324) and water ad libitum. Filter hoods were used to prevent mice from being exposed to other potential pathogens and to avoid the spread of the infection. At the start of the experiment, animals were between six and 9 months old.
Salmonella infection
We measured prevalence and resistance to infection by assessing individuals' ability to control and resolve an experimental infection of a mouse pathogen, Salmonella enterica serovar Typhimurium. Although there have been numerous studies on Salmonella infection in laboratory mice, there have only been a few studies on wild or wild‐derived house mice (Zala et al., 2004, 2008; Ilmonen et al., 2007, 2008b; Raveh et al., 2014). All experimental animals were infected with two different strains of Salmonella: a primary infection with an avirulent strain (AroA) and 10 day later a secondary infection with a more virulent strain (LT2). The primary infection ensured that all of the mice had been exposed to Salmonella and provided protection against the more virulent strain (Hoiseth & Stocker, 1981; Hormaeche et al., 1995). The bacteria from both strains were stored as slants at 4 °C (originated from frozen stocks at −80 °C) and cultured in 7.5 mL of heart–brain infusion at 37 °C (for 13 h while shaking at 170 rpm). We diluted the cultures with sterile phosphate‐buffered saline (PBS) solution until the desired concentration of 104 colony‐forming units (cfu) mL−1 for AroA and 103 cfu mL−1 for LT2. To verify inocula dosage, we used quantitative plate counts (three plates per dilution). All animals received intraperitoneal (IP) injections of 200 μL AroA inoculum and after 10 days 200 μL of LT2 inoculum. Due to space and time limitations, we divided the experimental animals into 15 groups, each containing one single‐ and one multiple‐sired litter. This design ensured that mice were all infected for the same duration (17 day for both strains). Fresh inocula were prepared for both infections in each group. AroA inocula concentrations varied from 4.8 × 104 to 1.0 × 105 and LT2 inocula from 8.2 × 102 to 2.1 × 103 between groups. All experimental animals were euthanized 7 days after the LT2 infection using CO2. The health condition of all mice was checked daily by visual inspection.
Assessing prevalence, resistance and tolerance to infection
To assess prevalence and the ability to control and clear Salmonella infection (infection resistance), we measured the number of viable bacteria in the spleen (also known as ‘pathogen load’) following previous methods (Penn et al., 2002; Zala et al., 2004; Ilmonen et al., 2007, 2008b). Spleens from euthanized animals were immediately removed, weighed and homogenized (Dispergierstation, T 8.10, IKA®‐Werke) in 1 ml phosphate‐buffered saline (PBS) under sterile conditions. We plated 50 μL of spleen homogenates and their 10−1 and 10−2 dilutions on selective agar plates (Salmonella‐Shigella Agar, Roth) and incubated the plates for 18 h overnight at 36 °C. Pathogen loads per spleen were determined by quantitative plate counts using the mean of two replicate plates for each dilution. We confirmed that pathogen load was not affected by the variation in cfu in inocula (LMM: F 1,28 = 0.010, β = 0.00006, SE = 0.0006, P = 0.921). To estimate individual tolerance (the ability to cope with infection among mice with similar resistance), we measured individual body mass before each infection and directly after euthanasia to calculate body mass changes over the course of the experiment and to relate these changes to individual infection rates.
Statistical analyses
A Spearman rank correlation test was performed to assess the relationship between spleen mass and individual body mass and spleen mass and pathogen load. Differences in pathogen load among all litters were determined with a Kruskal–Wallis test, and the variance in pathogen load among all litters was compared with a Brown–Forsythe test. To test for differences in pathogen loads between single‐ and multiple‐sired litters, a Mann–Whitney U‐test was applied on the mean pathogen loads of single‐ and multiple‐sired litters. To compare the between‐litter variation in single‐ versus multiple‐sired litters, a Brown–Forsythe test was run on the mean values. To compare the within‐litter variation in single‐ and multiple‐sired litters, the standard deviation (SD) in pathogen loads of single‐ and multiple‐sired litters was compared with a Mann–Whitney U‐test. To assess which variables influence individual pathogen load, we applied a general linear mixed‐effects model (LMM) with individual pathogen load as the dependent variable, paternity (single or multiple) and sex as fixed factors, inocula dosage, individual body mass, body mass change during the experiment and age at the beginning of the experiment as a covariate. Family ID was included into the model as a random factor to control for nonindependence of individuals within families. The data were highly skewed and model residuals were not normally distributed. Log transformation was not sufficient to normalize residuals, and thus, we used a Box–Cox transformation to further improve the normal distribution of residuals. All statistical analyses were performed using ‘R' (version 2.14.1) (R Development Core Team, 2011). We implemented linear mixed‐effects models using the ‘lme’ function of the ‘nlme’ package (Pinheiro et al., 2012). We applied a backward stepwise removal procedure (Grafen & Hails, 2002) to avoid problems due to the inclusion of nonsignificant terms (Engqvist, 2005), and removed variables were re‐entered one by one to the final model to obtain relevant statistics.
Ethics statement
This study has been discussed by the Institutional Ethics Committee in accordance with Good Scientific Practice guidelines and has been approved by the Austrian Federal Ministry for Science and Research (Permit number: Zl.22/01/97/2012).
Results
We found no difference in prevalence of infection among litters, as only one individual cleared the infection (all but one mouse was still infected 17 days after primary and 7 days after secondary infection). Resistance to infection was highly variable among individuals, as pathogen loads ranged from 0 to 9.1 × 106 cfu mL−1. Spleen mass of individual mice was highly correlated with pathogen load (Spearman's rank correlation: ρ = 0.453, N = 213, P = 3.4 × 10−12) (Fig. 1), but not with body mass (Spearman's rank correlation: ρ = 0.022, N = 213, P = 0.748) or change in body mass over the course of the experiment (Spearman's rank correlation: ρ = 0.090, N = 213, P = 0.177). We found a significant difference in the pathogen loads among all litters (Kruskal–Wallis test: H = 49.11, df = 29, P = 0.011), but no significant difference in the variance among all litters (Brown–Forsythe test: F 29,183 = 0.870, P = 0.662) (Fig. 2). We found no significant difference in mean pathogen loads between single‐ and multiple‐sired litters (Mann–Whitney U: W = 94, P = 0.461) (Fig. 3) and no difference in the within‐litter (Mann–Whitney U: W = 103, P = 0.713) or between‐litter variance (Brown–Forsythe test: F 1,28 = 0.412, P = 0.527) in pathogen loads of single‐ and multiple‐sired litters.
Figure 1.
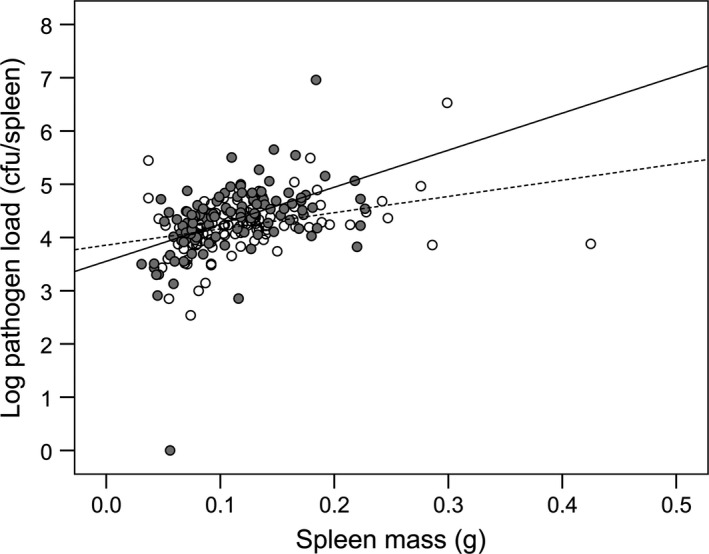
Relationship between spleen mass and pathogen load. Individual offspring spleen mass and pathogen load in single‐sired (white circles and dashed line, R² = 0.12) and multiple‐sired (grey circles and solid line, R² = 0.21) litters. Pathogen load is log‐transformed.
Figure 2.
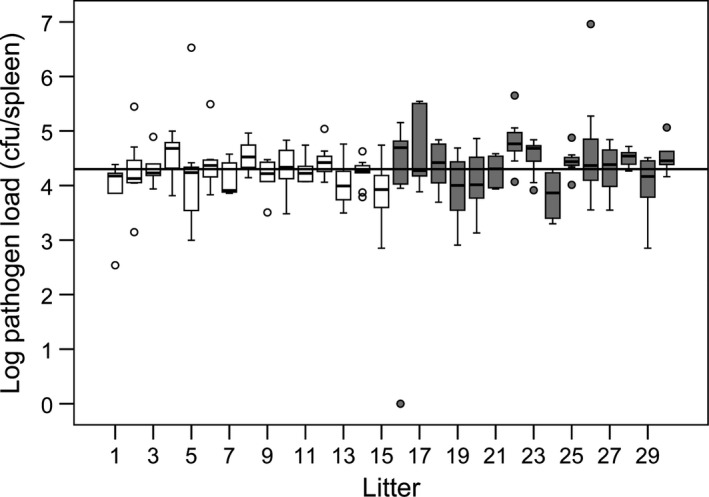
Differences in pathogen loads between litters. Single‐sired litters are depicted in white, and multiple‐sired litters are in grey. The solid line shows the median pathogen load of all litters. Pathogen load is log‐transformed.
Figure 3.
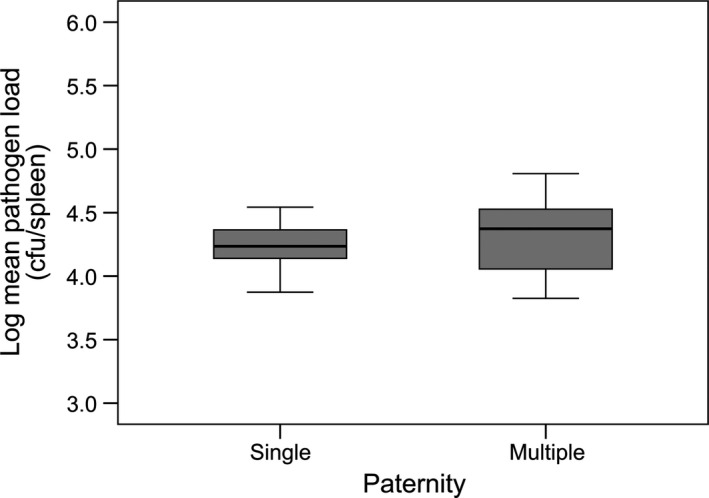
Pathogen load in single‐ versus multiple‐sired litters. Differences in the mean pathogen load of single (N = 15)‐ vs. multiple (N = 15)‐sired litters. Pathogen load is log‐transformed.
Individual pathogen load did not differ between offspring from single‐ vs. multiple‐sired litters (LMM: F 1,28 = 1.264, P = 0.271) and was not influenced by age (LMM: F 1,28 = 0.220, β = −0.004, SE = 0.008, P = 0.643) or body mass (LMM: F 1,178 = 1.197, β = 0.028, SE = 0.062, P = 0.658). Interestingly, we found a significantly higher pathogen load in females than in males (LMM: F 1,179 = 8.134, P = 0.005) (Fig. 4) and a significant interaction between individual body mass change over the course of the experiment and sex (LMM: F 1,179 = 12.974, β = −0.812, SE = 0.225, P = 0.0004): female body mass significantly reduced with increasing pathogen load during infection, whereas male body mass change was not affected by pathogen load (Fig. 5). The main effect of body mass change on pathogen load was not significant (LMM: F 1,179 = 1.197, β = −0.059, SE = 0.164, P = 0.722).
Figure 4.
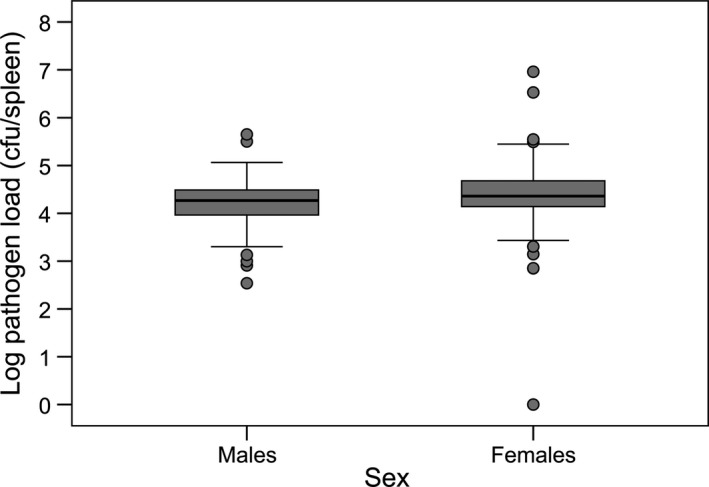
Sex difference in pathogen load. Pathogen load of males (N = 108) and females (N = 105). Pathogen load is log‐transformed.
Figure 5.
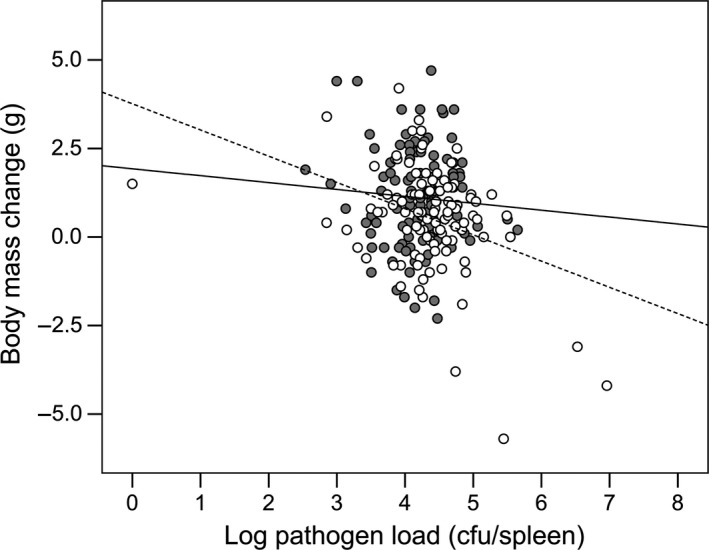
Body mass change in relation to pathogen load. Individual pathogen load and body mass in males (grey circles and solid line; R² = 0.005) and females (white circles and dashed line; R² = 0.134). Pathogen load is log‐transformed.
Discussion
We found a high variation in infection resistance (pathogen loads) among individuals and a significant difference in the mean load between different litters, which is consistent with other evidence that resistance to Salmonella is genetically influenced. Salmonella is known to cause splenomegaly, and we found that spleen size increased with increasing pathogen loads, which confirms that resistance affects the pathogenicity. However, we found no difference in the mean or variance in pathogen loads between single‐ and multiple‐sired litters. Therefore, our data do not support the hypothesis that increased litter genetic diversity enhances offspring disease resistance through either a bet‐hedging or a non‐bet‐hedging mechanism in our study population. Our study may have lacked sufficient power to detect small differences in resistance, although our sample size was larger than previous studies that found genetic diversity enhances disease resistance and colony survival (Liersch & Schmid‐Hempel, 1998; Baer & Schmid‐Hempel, 1999; Tarpy & Seeley, 2006) and applying a one‐tailed test, as used in these previous studies, would not change our results. Nonetheless, our results do not necessarily rule out the possibility that multiple male mating affects offspring disease resistance for the following reasons. First, genetic diversity of litters may have been insufficient in our study to reduce the prevalence of infection. Multiple‐sired litters are more genetically diverse compared to single‐sired litters in wild populations of house mice (Thonhauser et al., 2013c), but we did not investigate genetic diversity in this study. In addition, multiple‐sired litters in our study had only two sires, as in wild populations (Dean et al., 2006; Firman & Simmons, 2008c; Thonhauser et al., 2013c), whereas previous studies with bees inseminated queens with sperm from 10 different drones (Tarpy & Seeley, 2006; Seeley & Tarpy, 2007). Furthermore, social insects produce larger broods than mice (honeybee queens produce up to 250,000 eggs per year), and therefore, they have greater potential to increase the genetic diversity of their broods. Second, we measured pathogen resistance against two strains of Salmonella, whereas the benefits of genetic diversity may only apply to more pathogens, either simultaneously or over time. In particular, bet‐hedging benefits likely only occur when hosts are infected with different pathogens having different resistance/susceptibility profiles (McClelland et al., 2003). Third, we assessed how mice were able to control Salmonella infections (pathogen load 7 days after the second infection), and although many studies have found that this measure predicts disease resistance (Roy & Malo, 2002) and survival in laboratory mice (Penn et al., 2002), the fitness effects of resistance are less clear in wild mice. We did not find any difference in the prevalence of infection, as only one mouse cleared the infection, and thus, further studies are needed to investigate prevalence over a longer period of time. Also, studies are needed to test the ability to tolerate and survive virulent infections (Raveh et al., 2014). As we experimentally infected the mice, our study design does not allow to test whether genetic diversity helps to prevent infections or reduce their spread within litters, and thus, additional studies are needed to test for such effects. Fourth, we aimed to compare the consequences of producing single‐ versus multiple‐paternity litters and we assume that females that mate multiply are more likely to have multiple paternity than other females, but studies are needed that experimentally assign paternity to control for potential differences in female quality and ensure that any benefits of multiple paternity are not masked by benefits of mate choice for a single, high‐quality male.
We found a significant sex difference in infection resistance and that males and females tolerate high bacterial loads differently, which is the first such evidence with this pathogen to our knowledge. Females had significantly higher pathogen loads (lower resistance) than males, and lower resistance (increasing pathogen loads) correlated with loss in body mass during infection in females, but not males. Thus, females were less resistant and less tolerant of infection than males. Resistance to infection is sex dependent in many species, and although there are many exceptions, males are generally more susceptible than females (Zuk & McKean, 1996). Previous studies on sex differences in disease resistance have mainly measured immune responses to novel antigens (immunocompetence) or observed parasite burdens, but our findings emphasize the importance of measuring hosts' ability to cope with infection (Schneider & Ayres, 2008). There have been numerous experimental Salmonella infection studies on laboratory mice (Mittrücker & Kaufmann, 2000), as they are the most common host‐pathogen model for typhoid fever, yet surprisingly few studies have examined sex differences. Early studies on laboratory mice detected no sex differences in pathogen loads (Plant & Glynn, 1976), but since then, most studies focus on one sex or the sexes are not reported. There have been only a few studies on Salmonella resistance on wild‐derived house mice, and only some of these compared the sexes. One study found sex differences in pathogen prevalence (females showed lower prevalence than males following experimental infection) (Ilmonen et al., 2008a) and another found interactions between sex and genetic control of resistance (Ilmonen et al., 2008b). Another study found that females had better survival following a primary Salmonella infection compared to males, but no sex differences in pathogen clearance, indicating a sex differences in tolerance to primary infection (Raveh et al., 2014). Taken together, these findings suggest that sex differences in resistance and tolerance to Salmonella depend upon whether the infection is a primary or secondary challenge. They also suggest that sex differences in Salmonella resistance and tolerance may be more pronounced in wild compared to inbred laboratory mice. It has been reported that, among other loci, an X‐linked locus (btk) controls resistance to Salmonella in mice through B‐cell functions (O'Brien et al., 1979, 1981), but it is not known whether this locus contributes to sex differences in resistance. Sex differences in resistance to infection are generally thought to be due to immunosuppressive effects of testosterone or other steroid hormones (Klein, 2000, 2004). In house mice, males are more resistant than females to some parasites, including Toxoplasma gondii, Babesia microti, Schistosoma mansoni and Taenia crassips (Klein, 2004; Morales‐Montor et al., 2004). The harmful effects of T. gondii infection depend on estradiol as ovariectomy reduced and administration of estradiol enhanced the development of tissue cysts caused by T. gondii (Pung & Luster, 1986). Estradiol reduces CD8+ T cells (Boll & Reimann, 1996), and CD8+ T cells control protection against virulent S. Typhimurium (at least in secondary infections) (Mastroeni et al., 1992; Mittrücker & Kaufmann, 2000). Also, mice deficient in CD8+ T cells that survived an attenuated S. Typhimurium infection were more susceptible to a virulent S. Typhimurium strain compared to control animals (Lo et al., 1999). Given that testosterone enhances CD4+ and CD8+ T cells and CD4+ T cells are also highly important in protection against Salmonella infection (Mittrücker & Kaufmann, 2000), the sex difference in Salmonella load in our study may be explained by differences in steroid hormone concentrations.
In summary, we found significant differences among litters in their resistance to Salmonella infection, which is consistent with genetic influence on immune resistance, but we found no evidence that multiple paternity affects pathogen prevalence or immune resistance to this pathogen. Increasing the number of sires might provide different results, but because the number of sires only rarely exceeds two in litters of wild house mice, previous findings from insects may not apply to mice or other vertebrates. Future studies are needed to investigate resistance and tolerance to infection in wild mice and to test the effects of a variety of different parasites on multiple‐ vs. single‐sired litters. We also found that females were less resistant to secondary Salmonella infection than males. This result was unexpected because males are usually more susceptible to infectious diseases than females, and because numerous studies on Salmonella infection with laboratory mice have not reported such sex differences.
Conflict of interests
The authors declare that they have no conflicts of interest.
Acknowledgments
This work was funded by the Austrian Science Fund (FWF: P 24711‐B21), and S.R. was supported by the Swiss National Science Foundation fellowships (PBNEP3‐132801 and PBNEP3‐140190). The authors thank Sarah Zala and three anonymous reviewers that helped to improve earlier versions of this manuscript.
Data deposited at Dryad: doi: 10.5061/dryad.s4t8k
References
- Auclair, Y. , König, B. & Lindholm, A.K. 2014. Socially mediated polyandry: a new benefit of communal nesting in mammals. Behav. Ecol. 25: 1467–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baer, B. & Schmid‐Hempel, P. 1999. Experimental variation in polyandry affects parasite loads and fitness in a bumble‐bee. Nature 397: 151–154. [Google Scholar]
- Boll, G. & Reimann, J. 1996. Oestrogen treatment depletes extrathymic T cells from intestinal lymphoid tissues. Scand. J. Immunol. 43: 345–350. [DOI] [PubMed] [Google Scholar]
- Dean, M. , Ardlie, G. & Nachman, M. 2006. The frequency of multiple paternity suggests that sperm competition is common in house mice (Mus domesticus). Mol. Ecol. 15: 4141–4151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engqvist, L. 2005. The mistreatment of covariate interaction terms in linear model analyses of behavioural and evolutionary ecology studies. Anim. Behav. 70: 967–971. [Google Scholar]
- Firman, R.C. & Simmons, L.W. 2008a. Polyandry, sperm competition, and reproductive success in mice. Behav. Ecol. 19: 695–702. [Google Scholar]
- Firman, R.C. & Simmons, L.W. 2008b. Polyandry facilitates postcopulatory inbreeding avoidance in house mice. Evolution 62: 603–611. [DOI] [PubMed] [Google Scholar]
- Firman, R.C. & Simmons, L.W. 2008c. The frequency of multiple paternity predicts variation in testes size among island populations of house mice. J. Evol. Biol. 21: 1524–1533. [DOI] [PubMed] [Google Scholar]
- Fisher, D.O. , Double, M.C. , Blomberg, S.P. , Jennions, M.D. & Cockburn, A. 2006. Post‐mating sexual selection increases lifetime fitness of polyandrous females in the wild. Nature 444: 89–92. [DOI] [PubMed] [Google Scholar]
- Fox, C.W. & Rauter, C.M. 2003. Bet‐hedging and the evolution of multiple mating. Evol. Ecol. Res. 5: 273–286. [Google Scholar]
- Fuchs, S. & Schade, V. 1994. Lower performance in honeybee colonies of uniform paternity. Apidologie 25: 155–168. [Google Scholar]
- García‐González, F. & Simmons, L.W. 2005. The evolution of polyandry: intrinsic sire effects contribute to embryo viability. J. Evol. Biol. 18: 1097–1103. [DOI] [PubMed] [Google Scholar]
- Grafen, A. & Hails, R. 2002. Modern Statistics for the Life Sciences. Learn to analyse your own data. Oxford University Press, New York. [Google Scholar]
- Hamilton, W.D. 1987. Kinship, recognition, disease and intelligence: constraints of social evolution In: Animal Societies: Theory and Facts (Itô Y., Brown J.L. & Kikkawa J., eds), pp. 81–102. Japan Scientific Societies Press, Tokyo. [Google Scholar]
- Hamilton, W. & Poulin, R. 1997. The Hamilton and Zuk hypothesis revisited: a meta‐analytical approach. Behaviour 134: 299–320. [Google Scholar]
- Hamilton, W.D. & Zuk, M. 1982. Heritable true fitness and bright birds: a role for parasites? Science 218: 384–387. [DOI] [PubMed] [Google Scholar]
- Hoiseth, S.K. & Stocker, B.A. 1981. Aromatic‐dependent Salmonella typhimurium are non‐virulent and effective as live vaccines. Nature 291: 238–239. [DOI] [PubMed] [Google Scholar]
- Hormaeche, C.E. , Khan, C.M.A. , Mastroeni, P. , Viia‐real, B. , Dougan, G. , Roberts, M. et al 1995. Salmonella vaccines: mechanisms of immunity and their use as carriers of recombinant antigens In: Molecular and Clinical Aspects of Vaccine Development (Ala'Aldeen D. & Hormaeche C.E., eds), pp. 119–153. John Wiley and sons, London. [Google Scholar]
- Hosken, D.J. & Stockley, P. 2003. Benefits of polyandry: a life history perspective In: Evolutionary Biology. (Macintyre R.J., Clegg M.T., eds), pp. 173–194. Springer, Bosten. [Google Scholar]
- Hosken, D.J. , Garner, T.W.J. , Tregenza, T. , Wedell, N. & Ward, P.I. 2003. Superior sperm competitors sire higher‐quality young. Proc. R. Soc. Lond. B 270: 1933–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, W.O.H. & Boomsma, J.J. 2004. Genetic diversity and disease resistance in leaf‐cutting ant societies. Evolution 58: 1251–1260. [DOI] [PubMed] [Google Scholar]
- Ilmonen, P. , Penn, D.J. , Damjanovich, K. , Morrison, L. , Ghotbi, L. & Potts, W.K. 2007. Major histocompatibility complex heterozygosity reduces fitness in experimentally infected mice. Genetics 176: 2501–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilmonen, P. , Kotrschal, A. & Penn, D.J. 2008a. Telomere attrition due to infection. PLoS One 3: e2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilmonen, P. , Penn, D.J. , Damjanovich, K. , Clarke, J. , Lamborn, D. , Morrison, L. et al 2008b. Experimental infection magnifies inbreeding depression in house mice. J. Evol. Biol. 21: 834–841. [DOI] [PubMed] [Google Scholar]
- Jennions, M.D. & Petrie, M. 2000. Why do females mate multiply? A review of the genetic benefits. Biol. Rev. 75: 21–64. [DOI] [PubMed] [Google Scholar]
- Kavaliers, M. & Colwell, D.D. 1995. Odours of parasitized males induce aversive response in female mice. Anim. Behav. 50: 1161–1169. [Google Scholar]
- Klein, S.L. 2000. The effects of hormones on sex differences in infection: from genes to behavior. Neurosci. Biobehav. Rev. 24: 627–638. [DOI] [PubMed] [Google Scholar]
- Klein, S.L. 2004. Hormonal and immunological mechanisms mediating sex differences in parasite infection. Parasite Immunol. 26: 247–264. [DOI] [PubMed] [Google Scholar]
- Liersch, S. & Schmid‐Hempel, P. 1998. Genetic variation within social insect colonies reduces parasite load. Proc. R. Soc. Lond. B 265: 221–225. [Google Scholar]
- Lo, W.‐F. , Ong, H. , Metcalf, E.S. & Soloski, M.J. 1999. T cell responses to gram‐negative intracellular bacterial pathogens: a role for CD8+ T cells in immunity to Salmonella infection and the involvement of MHC class Ib molecules. J Immunol. 162: 5398–5406. [PubMed] [Google Scholar]
- Madsen, T. , Shine, R. , Loman, J. & Håkansson, T. 1992. Why do female adders copulate so frequently? Nature 355: 440–441. [Google Scholar]
- Manser, A. , König, B. & Lindholm, A.K. 2015. Female house mice avoid fertilization by t haplotype incompatible males in a mate choice experiment. J. Evol. Biol. 28: 54–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastroeni, P. , Villarreal‐Ramos, B. & Hormaeche, C.E. 1992. Role of T cells, TNFα and IFNγ in recall of immunity to oral challenge with virulent salmonellae in mice vaccinated with live attenuated aro− salmonella vaccines. Microb. Pathog. 13: 477–491. [DOI] [PubMed] [Google Scholar]
- McClelland, E.E. , Penn, D.J. & Potts, W.K. 2003. Major histocompatibility complex heterozygote superiority during coinfection. Infect. Immun. 71: 2079–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLeod, L. & Marshall, D.J. 2009. Do genetic diversity effects drive the benefits associated with multiple mating? a test in a marine invertebrate. PLoS One 4: E6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittrücker, H.W. & Kaufmann, S.H. 2000. Immune response to infection with Salmonella typhimurium in mice. J. Leukoc. Biol. 67: 457–463. [DOI] [PubMed] [Google Scholar]
- Møller, A.P. , Christe, P. & Lux, E. 1999. Parasitism, host immune function, and sexual selection. Q. Rev. Biol. 74: 3–20. [DOI] [PubMed] [Google Scholar]
- Morales‐Montor, J. , Chavarria, A. , De León, M.A. , Del Castillo, L.I. , Escobedo, E.G. , Sanchez, E.N. et al 2004. Host gender in parasitic infections of mammals: an evaluation of the female host supremacy paradigm. J. Parasitol. 90: 531–546. [DOI] [PubMed] [Google Scholar]
- O'Brien, A.D. , Scher, I. , Campbell, G.H. , MacDermott, R.P. & Formal, S.B. 1979. Susceptibility of CBA/N mice to infection with Salmonella Typhimurium: influence of the X‐linked gene controlling B lymphocyte function. J. Immunol. 123: 720–724. [PubMed] [Google Scholar]
- O'Brien, A.D. , Scher, I. & Metcalf, E.S. 1981. Genetically conferred defect in anti‐Salmonella antibody formation renders CBA/N mice innately susceptible to Salmonella typhimurium infection. J. Immunol. 126: 1368–1372. [PubMed] [Google Scholar]
- Penn, D. , Schneider, G. , White, K. , Slev, P. & Potts, W. 1998. Influenza infection neutralizes the attractiveness of male odour to female mice (Mus musculus). Ethology 104: 685–694. [Google Scholar]
- Penn, D.J. , Damjanovich, K. & Potts, W.K. 2002. MHC heterozygosity confers a selective advantage against multiple‐strain infections. Proc. Natl. Acad. Sci. USA 99: 11260–11264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro, J. , Bates, D. , DebRoy, S. & Sarkar, D . & and the R Development Core team 2012. nlme: Linear and Nonlinear Mixed Effects Models (R Package Version 3.1‐103).
- Plant, J. & Glynn, A.A. 1976. Genetics of Resistance to Infection with Salmonella typhimurium in mice. J. Infect. Dis. 133: 72–78. [DOI] [PubMed] [Google Scholar]
- Pung, O.J. & Luster, M.I. 1986. Toxoplasma gondii: decreased resistance to infection in mice due to estrogen. Exp. Parasitol. 61: 48–56. [DOI] [PubMed] [Google Scholar]
- R Development Core Team 2011. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- Raveh, S. , Sutalo, S. , Thonhauser, K.E. , Thoß, M. , Hettyey, A. , Winkelser, F. et al 2014. Female partner preferences enhance offspring ability to survive an infection. BMC Evol. Biol. 14: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland, C. , MacDonald, D.W. , de Fraipont, M. & Berdoy, M. 2003. Free female choice in house mice: leaving best for last. Behaviour 140: 1371–1388. [Google Scholar]
- Roy, M.F. & Malo, D. 2002. Genetic regulation of host responses to Salmonella infection in mice. Genes Immun. 3: 381–393. [DOI] [PubMed] [Google Scholar]
- Schneider, D.S. & Ayres, J.S. 2008. Two ways to survive infection: what resistance and tolerance can teach us about treating infectious diseases. Nat. Rev. Immunol. 8: 889–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeley, T.D. & Tarpy, D.R. 2007. Queen promiscuity lowers disease within honeybee colonies. Proc. R. Soc. B 274: 67–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shykoff, J. & Schmid‐Hempel, P. 1991. Parasites and the advantage of genetic variability within social insect colonies. Proc. R. Soc. Lond. B 243: 55–58. [Google Scholar]
- Simmons, L.W. 2005. The evolution of polyandry: sperm competition, sperm selection, and offspring viability. Annu. Rev. Ecol. Evol. Syst. 36: 125–146. [Google Scholar]
- Soper, D.M. , Delph, L.F. & Lively, C.M. 2012. Multiple paternity in the freshwater snail, Potamopyrgus antipodarum . Ecol. Evol. 2: 3179–3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutter, A. & Lindholm, A.K. 2015. Detrimental effects of an autosomal selfish genetic element on sperm competitiveness in house mice. Proc. R. Soc. B 282: 20150974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarpy, D.R. 2003. Genetic diversity within honeybee colonies prevents severe infections and promotes colony growth. Proc. R. Soc. Lond. B 270: 99–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarpy, D.R. & Seeley, T.D. 2006. Lower disease infections in honeybee (Apis mellifera) colonies headed by polyandrous vs monandrous queens. Naturwissenschaften 93: 195–199. [DOI] [PubMed] [Google Scholar]
- Thonhauser, K.E. , Raveh, S. , Hettyey, A. , Beissmann, H. & Penn, D.J. 2013a. Scent marking enhances male reproductive success in wild house mice. Anim. Behav. 86: 1013–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thonhauser, K.E. , Raveh, S. , Hettyey, A. , Beissmann, H. & Penn, D.J. 2013b. Why do female mice mate with multiple males? Behav. Ecol. Sociobiol. 67: 1961–1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thonhauser, K.E. , Thoß, M. , Musolf, K. , Klaus, T. & Penn, D.J. 2013c. Multiple paternity in wild house mice (Mus musculus musculus): effects on offspring genetic diversity and body mass. Ecol. Evol. 4: 200–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thonhauser, K.E. , Raveh, S. & Penn, D.J. 2014. Multiple paternity does not depend on male genetic diversity. Anim. Behav. 93: 135–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tregenza, T. & Wedell, N. 2002. Polyandrous females avoid costs of inbreeding. Nature 415: 71–73. [DOI] [PubMed] [Google Scholar]
- Yasui, Y. 1998. The ‘genetic benefits’ of female multiple mating reconsidered. Trends Ecol. Evol. 13: 246–250. [DOI] [PubMed] [Google Scholar]
- Zala, S.M. , Potts, W.K. & Penn, D.J. 2004. Scent‐marking displays provide honest signals of health and infection. Behav. Ecol. 15: 338–344. [Google Scholar]
- Zala, S.M. , Potts, W.K. & Penn, D.J. 2008. Exposing males to female scent increases the cost of controlling Salmonella infection in wild house mice. Behav. Ecol. Sociobiol. 62: 895–900. [Google Scholar]
- Zeh, J.A. & Zeh, D.W. 1996. The Evolution of Polyandry I: intragenomic Conflict and Genetic Incompatibility. Proc. R. Soc. Lond. B 263: 1711–1717. [Google Scholar]
- Zeh, J.A. & Zeh, D.W. 1997. The evolution of polyandry II: post‐copulatory defences against genetic incompatibility. Proc. R. Soc. Lond. B 264: 69–75. [Google Scholar]
- Zhu, Y.Y. , Chen, H.R. , Fan, J.H. , Wang, Y.Y. , Li, Y. , Chen, J.B. et al 2000. Genetic diversity and disease control in rice. Nature 406: 718–722. [DOI] [PubMed] [Google Scholar]
- Zuk, M. & McKean, K.A. 1996. Sex differences in parasite infections: patterns and processes. Int. J. Parasitol. 26: 1009–1023. [PubMed] [Google Scholar]


