Abstract
Somatic reprogramming by reexpression of the embryonic transcription factor T-box 18 (TBX18) converts cardiomyocytes into pacemaker cells. We hypothesized that this could be a viable therapeutic avenue for pacemaker-dependent patients afflicted with device-related complications, and therefore tested whether adenoviral TBX18 gene transfer could create biological pacemaker activity in vivo in a large-animal model of complete heart block. Biological pacemaker activity, originating from the intramyocardial injection site, was evident in TBX18-transduced animals starting at day 2 and persisted for the duration of the study (14 days) with minimal backup electronic pacemaker use. Relative to controls transduced with a reporter gene, TBX18-transduced animals exhibited enhanced autonomic responses and physiologically superior chronotropic support of physical activity. Induced sinoatrial node cells could be identified by their distinctive morphology at the site of injection in TBX18-transduced animals, but not in controls. No local or systemic safety concerns arose. Thus, minimally invasive TBX18 gene transfer creates physiologically relevant pacemaker activity in complete heart block, providing evidence for therapeutic somatic reprogramming in a clinically relevant disease model.
INTRODUCTION
Electronic pacemakers are devices that treat slow heart rhythms in patients afflicted with cardiac conduction system disorders causing slow heart rate (HR) and symptoms such as syncope. They have been successfully used for more than 50 years with continuous refinement (1), but are limited in some instances by complications ranging from inadequate autonomic support to lead fracture, infections, and adverse cardiac remodeling (2, 3). Biological pacemakers—that is, pacemakers that are created by cell or gene therapy—have been developed with the ultimate goal of replacing electronic devices. The prevalent approaches to date have used functional reengineering (overexpression of one or more ion channels) to create abnormal automaticity in working chamber myocytes; proof of concept has been primarily at the cellular and small-animal levels, with few follow-up translational studies (those designed in large animals and using clinically feasible delivery methods) (2, 4–8). In particular, previous studies have not shown physiologically relevant support of HR with biological pacemakers created using clinically realistic delivery methods (5–8). Translational barriers have been primarily related to open-chest or left-sided (intra-arterial) delivery methods, safety concerns regarding the gene delivery vector or cell type (in the case of cell-based biological pacemakers), and the fear of an increased risk of arrhythmias (9–11).
We recently showed that reexpression of the human embryonic transcription factor T-box 18 (TBX18) converts ordinary ventricular cardiomyocytes into pacemaker cells (12, 13), also called induced sinoatrial node (iSAN) cells. Such pacemaker conversion has been shown to effectively treat heart block in small animals (12). This somatic reprogramming approach differs from functional reengineering by providing a template for recapitulating genuine SAN function, with all its nuances. In nondisease conditions (in both rodents and humans), the SAN is a compact tissue composed of specialized pacemaker cells that fire spontaneous electrical impulses determining the individual's HR (2, 9, 14).
The present study was designed to develop a new approach to create a “bridge to device” in pacemaker-dependent patients with hardware infections, providing temporary hardware-free chronotropic support. These patients desperately need a pacemaker but have a contraindication to indwelling hardware (at least until the infection has been cleared by hardware removal and antibiotic therapy).
Here, we have used minimally invasive percutaneous gene delivery to achieve in vivo somatic reprogramming in a large-animal (porcine) model of complete heart block. This is an important translational step because of the minimally invasive delivery system used (5) and the ability of a single gene to recapitulate the key phenotypic features of the native SAN (12). Multiple physiological variables, as well as local and systemic safety profiles, were evaluated to show that this approach merits translation to humans.
RESULTS
TBX18 gene transfer creates a biological pacemaker
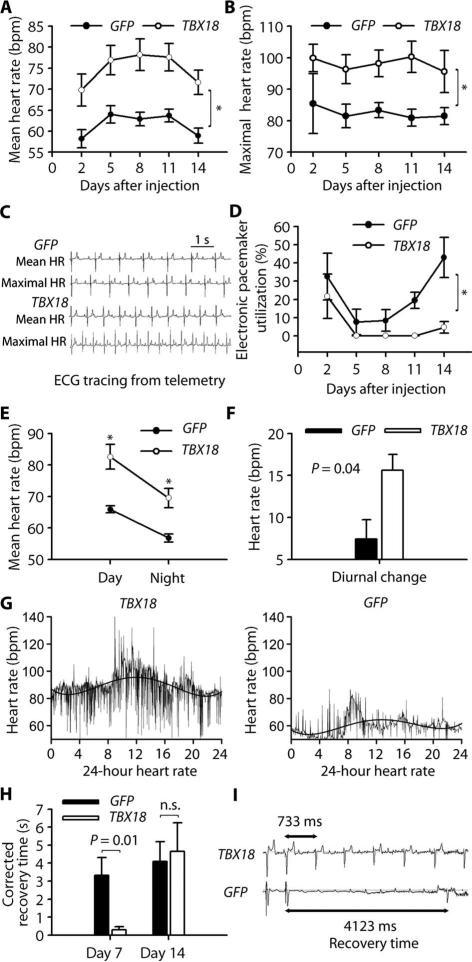
Twelve pigs were given percutaneous injections of either TBX18 (n = 7) or green fluorescent protein (GFP; control) (n = 5) and monitored for 14 days. The baseline clinical, electrocardiographic (ECG), and laboratory data showed no significant differences between animals (table S1). After gene delivery, mean HR was higher in TBX18-transduced animals compared to GFP-transduced controls starting at day 2 and persisting for 2 weeks (Fig. 1A). HR trended down after day 11 but remained significantly higher than in the control group. Additionally, TBX18-transduced animals achieved a higher maximal HR compared to controls (Fig. 1, B and C). Reliance upon a backup electronic pacemaker (inserted before gene delivery) was minimal in the TBX18 group (<1% from day 5 to day 11), unlike controls (8 to 40% throughout the 2 weeks of follow-up) (Fig. 1D). These findings indicate that TBX18 gene delivery successfully creates robust biological pacemaker activity, minimizing the need for backup electronic pacing.
Fig. 1. TBX18 gene transfer creates a biological pacemaker.
(A and B) Mean (A) and maximal (B) HR in TBX18- and GFP-transduced animals. Data are means ± SEM (n = 5 controls, 7 TBX18). *P < 0.05 for all time points by repeated-measures analysis of variance (ANOVA). bpm, beats per minute. (C) Representative telemetry tracings of mean and maximal HR from both experimental groups. (D) Electronic pacemaker use after gene transfer. Data are means ± SEM (n = 5 controls, 7 TBX18). *P < 0.05 for all time points by repeated-measures ANOVA. (E and F) Daytime and nighttime HR (E) and diurnal changes (F) in treatment groups. Data are means ± SEM (n = 5 controls, 7 TBX18). *P < 0.05 by two-tailed t test. (G) Representative 24-hour HR trend in TBX18- and GFP-transduced animals. HR is consistently higher in daytime than in nighttime in TBX18 group. (H and I) Electrophysiological (EP) evaluation of biological pacemaker function. Corrected recovery time was measured on days 8 and 14 (H). Data are means ± SEM (n = 3 controls, 5 TBX18 at day 8; n = 5 controls, 7 TBX18 on day 14). P values were determined by two-tailed t test. n.s., not significant. Representative recovery time tracings were taken on day 8 in TBX18 and GFP animals (I).
To test whether the TBX18 biological pacemaker experienced circadian variations similar to those in the native SAN (15), we analyzed the average HR in daytime (8 to 10 a.m., during feeding and activity) and nighttime (2 to 4 a.m., during sleep) over the 14 days after injection. Both daytime and nighttime HRs were higher in the TBX18-transduced group than in controls (Fig. 1E). The diurnal change (that is, the difference between daytime and nighttime HR) was also greater in the TBX18 group (Fig. 1F), resembling the behavior of the native SAN (15). Figure 1G illustrates the 24-hour HR trend for both experimental groups. HR from 8 a.m. to 4 p.m. in TBX18-transduced animals was consistently higher than that from 8 p.m. to 4 a.m. Such diurnal changes were blunted in GFP-transduced controls.
We further evaluated biological pacemaker function by performing a modification of standard SAN functional testing (SAN recovery time) (16). The heart was paced electronically at an HR of 120 for 1 min, and then electronic pacing was abruptly terminated to allow the endogenous pacemaker to emerge from overdrive suppression. Corrected recovery time after burst ventricular pacing at day 8 was 10-fold shorter in the TBX18 group (Fig. 1H). Representative ECG recordings from both experimental groups after electronic burst ventricular pacing show rapid recovery in a TBX18-transduced animal compared to GFP-transduced control (Fig. 1I). Although significant at day 8 (peak biological pacemaker activity), no difference was seen at 2 weeks, consistent with a progressive decline of biological pacemaker function over time with adenovirally mediated gene transfer (17, 18).
Autonomic regulation varies the rate of the TBX18 biological pacemaker
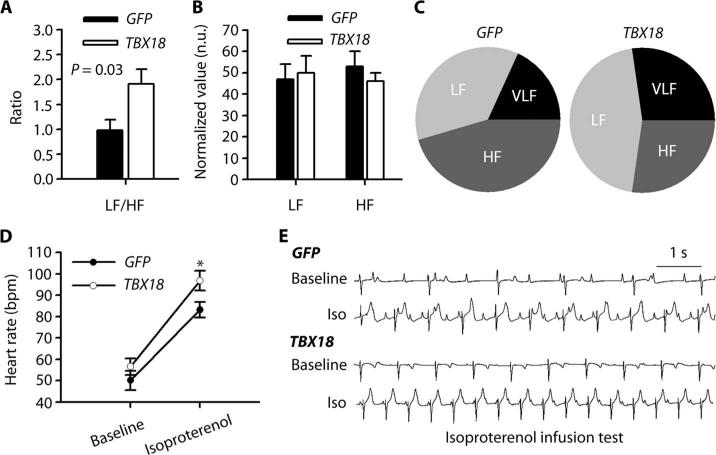
Autonomic regulation of the biological pacemaker was assessed by analysis of HR variability (HRV) (19, 20) and response to β-adrenergic stimulation (isoproterenol infusion, nonselective β-adrenergic agonist). HRV showed different low frequency (LF)/high frequency (HF) ratios (Fig. 2A). Normalized LF tended to be higher, and normalized HF lower, in the TBX18 group (Fig. 2, B and C, and table S2). These findings are compatible with sympathetic predominance in the TBX18-transduced group (19, 20). Isoproterenol infusion (8 μg/min) for 10 min increased HR by 71% in TBX18 animals, with more modest changes in GFP controls (Fig. 2, D and E). The HRV analysis and the response to β-adrenergic agonist (β-adrenergic sensitivity) indicate that the TBX18 biological pacemaker responds to endogenous and pharmacologic autonomic stimuli.
Fig. 2. Autonomic regulation of the TBX18 biological pacemaker.
(A) LF/HF ratio in TBX18 animals during maximal HR. P value was determined by two-tailed t test. (B) Normalized LF and HF (normalized to total power) in TBX18- and GFP-transduced controls. n.u., normalized units. (C) Frequency distribution in TBX18 and GFP groups. Very low frequency (VLF) is frequency ranging from 0 to 0.04 Hz. (D) Increase in HR after isoproterenol infusion while comparing two animal groups. *P < 0.05 compared to GFP groups, two-tailed t test. (E) Representative tracings of isoproterenol (Iso) response in both experimental groups. Data in (A), (B), and (D) are means ± SEM (GFP, n = 5; TBX18, n = 7).
TBX18 biological pacemaker supports daily physical activity
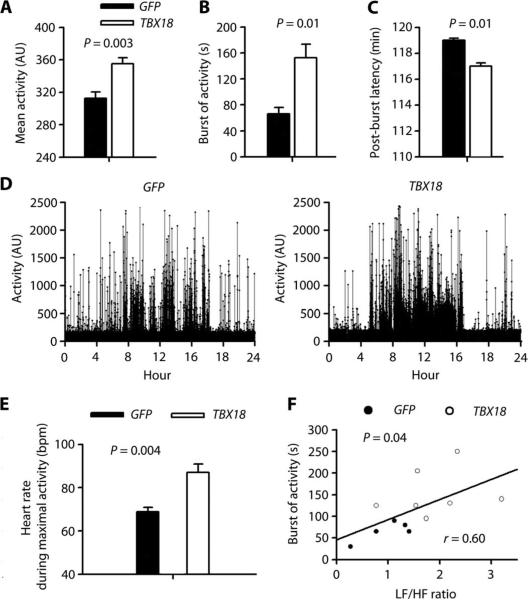
The animals’ physical activity was assessed using an implanted accelerometer. Mean daily activity during the 2-week follow-up after gene transfer was significantly greater in the TBX18 group than in controls (Fig. 3A). To further characterize the differences in activity pattern, we analyzed bursts of activity (brief episodes of activity >2000 arbitrary units) and the temporal variation of activity at day 8. Bursts of activity lasted longer during the daytime (8 to 10 a.m.) in the TBX18 group than in controls (Fig. 3B). The post-burst latency (time between bursts of activity) was likewise shorter in TBX18-transduced animals than in controls (Fig. 3C). The 24-hour summary showed different activity patterns between TBX18 and control groups (Fig. 3D): TBX18-transduced animals had persistent and stable activity with long bursts of activity during 8 a.m. to 4 p.m., in contrast to controls, which exhibited briefer periods of heightened activity.
Fig. 3. TBX18 biological pacemaker supports daily physical activity.
(A to C) Summary of daily physical activity, including mean activity during 2-week period after gene transfer (A), duration of burst activity [>2000 arbitrary units (AU)] (B), and post-burst activity (C). (D) Representative 24-hour activity pattern in GFP- and TBX18-transduced animals. (E) Mean HR during maximal activity. (F) HRV (LF/HF) was associated with the duration of bursts of activity in a linear regression model. Each point corresponds to the duration of burst of activity (y axis) and LF/HF ratio (x axis). Data in (A) to (C) and (E) are means ± SEM (GFP, n = 5; TBX18, n = 7), and P values were determined by two-tailed t test. In (F), P value was determined by the linear regression model.
A higher HR during maximal activity was seen in the TBX18 group compared to controls (Fig. 3E), indicating superior accommodation to hemodynamic and metabolic demands (chronotropic adaptability). In one animal, we further verified that TBX18 biological pacemaker can support both HR and physical activity even in the total absence of backup electronic pacing, with HR and activity patterns superimposable on the pooled data obtained with backup pacing (fig. S1). The duration of bursts of activity correlated significantly with the LF/HF ratio of HRV (Fig. 3F), linking the autonomic regulation of the biological pacemaker to physical activity. Thus, the TBX18 biological pacemaker, unlike the electronic backup pacing device, provides autonomically sensitive chronotropic adaptability appropriate to support physical activity.
TBX18 converts pig ventricular myocytes to iSAN cells in situ
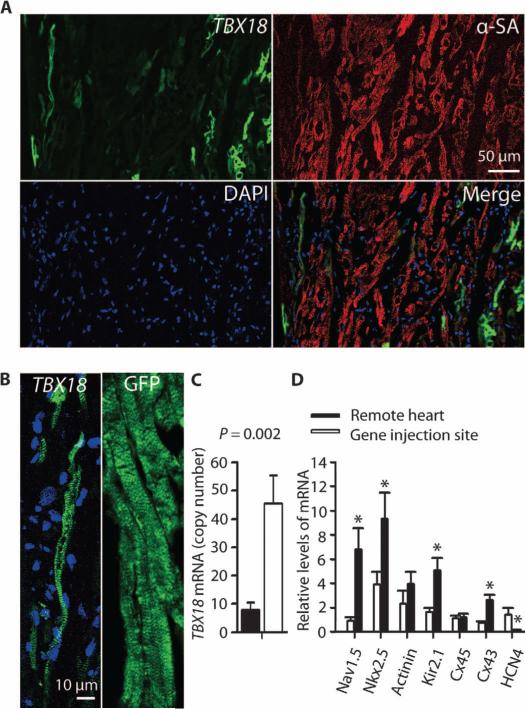
Because the TBX18 vector coexpressed GFP, and the control vector expressed only GFP, the presence of focal green fluorescence at the injection site further verified transduction for both TBX18 and control vector. Positive staining for α-sarcomeric actinin indicated that the transduced cells were cardiomyocytes (Fig. 4A). Ventricular cardiomyocytes transduced with GFP only showed typical brick-like morphology with well-organized striations, whereas many TBX18-transduced cardiomyocytes were thinner, with a distinctive spindle shape like that of endogenous SAN cells (Fig. 4B) (12). Such thin, tapering myocytes are not normally found in the mammalian ventricle.
Fig. 4. TBX18 converts ventricular myocytes to iSAN cells in situ.
(A) Confocal microscopy image showing focal tissue expression of TBX18 (green fluorescence). Cardiomyocyte sarcomeres were stained with α-sarcomeric actinin (α-SA); 4′,6-diamidino-2-phenylindole (DAPI) indicates nuclei. Scale bar, 50 μm. (B) TBX18-transduced ventricular myocytes are smaller in size and spindle-shaped compared with the morphology of GFP-transduced cardiomyocyte cells. Images are representative of the injection site. (C) TBX18 mRNA expression at the gene injection site and in the left ventricular free wall (remote site). Data are means ± SEM (TBX18, n = 3, one sample per site for each animal). P value was determined by two-tailed t test. (D) Relative expression (compared to the housekeeping gene ACTA1) of iSAN-associated mRNA at gene injection site compared to the left ventricular free wall (remote site). Data are means ± SEM (TBX18, n = 6, one sample per site for each animal). *P < 0.05, determined by two-tailed t test.
Quantification of tissue block images from TBX18-transduced hearts (fig. S2) yielded estimates of ~15,000 GFP-positive cells at or near the injection site, 14 days after injection. This number is a lower-limit estimate owing to the fact that iSAN cells, once reprogrammed, may no longer express GFP (12). The morphology of 556 sequentially counted GFP-positive cells was iSAN-like (long, lean, and tapering) in 24.5%, ventricular (brick-like cells with angular ends) in only 1.2%, or indeterminate (tangentially sliced) in the remaining 74.3% of cases. Perhaps not coincidentally, the number of GFP-positive cells per TBX18-transduced heart is not dissimilar to the estimate of ~10,000 genuine pacemaker cells in the native mammalian SAN (21).
TBX18 gene expression was assessed by reverse transcription, real-time quantitative polymerase chain reaction (PCR) at both the gene injection site (posterior septum) and in remote myocardium (left ventricular free wall). Levels of TBX18 mRNA were significantly higher at the gene injection site than in remote myocardium (Fig. 4C). We also looked for changes in the expression levels of various genes that characterize iSAN (and genuine SAN) cells. Transcript levels of Cx43, Kir2.1, Nkx2.5, and Nav1.5 were down-regulated, whereas HCN4 was increased, in the gene injection site compared to the remote heart area (Fig. 4D). The expression levels of Cx45 and actinin did not differ. These findings are compatible with the gene expression profile in iSAN cells (12).
By immunostaining, TBX18-transduced cardiomyocytes displayed sparse Cx43 compared to nontransduced ones, where Cx43 was plentiful (fig. S3). This again replicates the effects of TBX18 gene transfer on cardiomyocytes (12). Attempts to visualize HCN4 by immunostaining were unsuccessful using various antibodies and staining protocols (table S3). Nevertheless, the notion that we have achieved in situ conversion of ventricular myocytes into iSAN cells is supported by three findings at the TBX18 injection site: transduced cells with histologically distinctive morphology, characteristic mRNA changes, and focal down-regulation of Cx43.
TBX18 biological pacemaker activity originates at the focal injection site
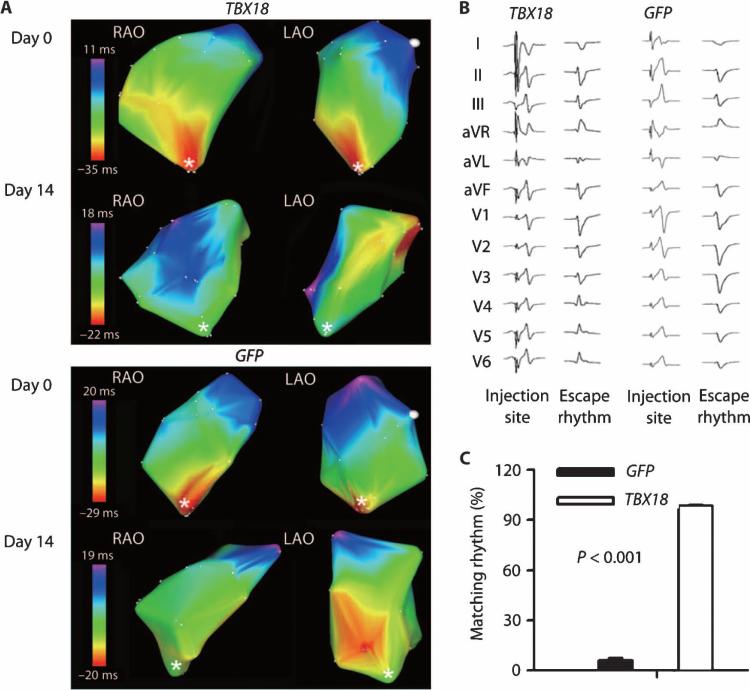
To determine the site of origin of biological pacemaker activity, we used in vivo electroanatomic mapping of the right ventricle (RV) and pace mapping from the injection site. Electroanatomic mapping showed that, at day 0, the earliest activation site (red color) was at the right ventricular apex, where the electronic device paces the heart (asterisk in Fig. 5A). At day 14 after gene delivery, activation arose in the high septal region, colocalized with the injection site in TBX18-transduced animals (Fig. 5A, white dot) but not in control animals.
Fig. 5. TBX18 biological pacemaker activity originates at the focal injection site.
(A) RV activation pattern from in vivo three-dimensional (3D) electroanatomic mapping in TBX18-transduced animals and controls at days 0 and 14. Earliest activation site is shown in red. RV apex, where electronic pacing device was implanted, is denoted by a white asterisk. The gene injection site is noted by a white dot. The 3D electroanatomic maps are shown by two different views of projection: right anterior oblique (RAO) and left anterior oblique (LAO). Images are representative of n = 3 for each group. (B) Twelve-lead ECG morphology from paced beat from injection site was compared to those from escape rhythms in both groups. Traces are representative of n = 3 for each group. (C) Single-lead ECG morphology match (%) from gene injection site. Data are means ± SEM (GFP, n = 5; TBX18, n = 7). P value was determined by two-tailed t test.
Pace mapping from the gene injection site showed a 100% match (12 of 12 ECG leads) with the TBX18-induced biological pacemaker rhythm (Fig. 5B). In contrast, a 0% match (0 of 12) was seen with the nonspecific escape rhythms observed in GFP-transduced controls. The QRS duration of the TBX18-induced rhythm was significantly shorter than that in controls (55.4 ± 3.1 ms versus 67.4 ± 1.1 ms, P = 0.01 by two-tailed t test), consistent with conduction from the high septal region (22). Additionally, the telemetric QRS morphology template from the injection site (confirmed by pace mapping and activation maps) was obtained and compared to the QRS morphology of the spontaneous rhythms using an automated morphology recognition algorithm. The spontaneous rhythms in the TBX18 group had a 98.5% match compared to only 6.2% in GFP-transduced controls (Fig. 5C). Together, these findings support the notion that TBX18-induced rhythms originate from the gene delivery site, whereas spontaneous rhythms in control animals rarely did.
TBX18 biological pacemaker does not increase arrhythmic risk
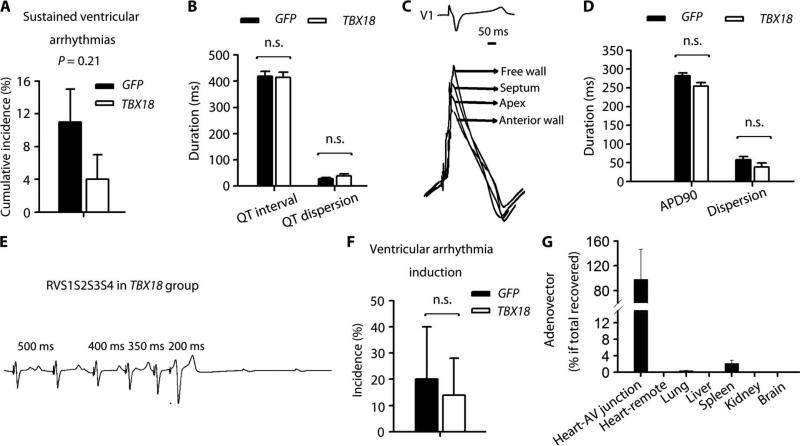
Continuous rhythm recordings from the implanted telemetry system were digitally stored for subsequent analysis. During the 2-week follow-up period, the cumulative incidence of sustained ventricular arrhythmias did not differ significantly between the two experimental groups, although the TBX18 animals tended to have fewer such events (Fig. 6A). Corrected QT interval and dispersion from 12-lead ECG (Fig. 6B) and APD90 dispersion and duration (Fig. 6, C and D) did not differ in the two treatment groups. Programmed ventricular stimulation with and without isoproterenol infusion elicited no episodes of inducible ventricular tachycardia in either control or TBX18-transduced animals. One episode of inducible ventricular fibrillation was evoked in each of the TBX18- and GFP-transduced groups (Fig. 6, E and F).
Fig. 6. Local and systemic safety profiles.
(A) Cumulative incidence of sustained ventricular arrhythmias during 14-day follow-up. (B) QTc interval and QT dispersion 2 weeks after gene delivery. QTc interval: QT interval corrected by Bazett's formula. (C) Representative APD recordings from different locations in a TBX18-transduced animal. (D) Average APD90 and APD90 dispersion. (E) Representative ventricular arrhythmia induction protocol from TBX18 animal. RVS1S2S3S4 indicates that the stimulation protocol for ventricular arrhythmia induction consisted of right ventricular pacing with three extrastimulations. (F) Ventricular arrhythmia induction after programmed stimulation with or without isoproterenol. (G) Viral biodistribution in heart and remote organs (n = 6 in each organ). Data in (A), (B), (D), (F), and (G) are means ± SEM (GFP, n = 5; TBX18, n = 7). P values were determined by two-tailed t test.
Biodistribution and systemic safety profile
Real-time PCR analysis indicated that 98.2% of the adenoviral vector was localized at the gene injection site, 2.1% in the spleen, and 0.3% in the lung 2 weeks after gene delivery (Fig. 6G). Viral expression was undetectable in remote myocardium (free wall of left ventricle) and in other organs (brain, liver, and kidney). Blood-borne biomarkers (cardiac, hepatic, renal, pancreatic, and inflammatory markers) remained unchanged during follow-up, supporting the absence of systemic toxicity (Table 1).
Table 1.
Biochemistry profile of TBX18- and GFP-transduced animals 2 weeks after gene delivery.
| GFP | TBX78 | P | |
|---|---|---|---|
| Liver function | |||
| AST (IU/liter) | 22.0 ± 2.6 | 23.6 ± 3.4 | 0.74 |
| ALT (IU/liter) | 21.8 ± 2.3 | 26.3 ± 2.8 | 0.28 |
| Total bilirubin (mg/dl) | 0.12 ± 0.02 | 0.11 ± 0.01 | 0.80 |
| Alkaline phosphatase (IU/liter) | 98.6 ± 22.1 | 107.0 ± 12.3 | 0.73 |
| Renal function | |||
| BUN (mg/dl) | 9.6 ± 0.8 | 9.0 ± 0.4 | 0.48 |
| Creatinine (mg/dl) | 1.34 ± 0.08 | 1.39 ± 0.05 | 0.63 |
| Pancreas function | |||
| Amylase (IU/liter) | 1304.4 ± 78.4 | 1112.1 ± 138.7 | 0.31 |
| Cardiac enzyme | |||
| CPK (ng/ml) | 410.3 ± 59.3 | 463.9 ± 69.7 | 0.62 |
| CKMB (%) | 2.74 ± 0.56 | 1.97 ± 0.40 | 0.28 |
| Troponin I (ng/ml) | 0.03 ± 0.01 | 0.04 ± 0.01 | 0.42 |
| Hematology | |||
| WBC (103/μl) | 15.2 ± 1.3 | 18.4 ± 2.4 | 0.32 |
| HGB (g/dl) | 9.0 ± 0.3 | 8.5 ± 0.3 | 0.31 |
| Platelets (103/μl) | 447.2 ± 69.7 | 375.6 ± 35.8 | 0.34 |
| Lymphocytes (103/μl) | 9.3 ± 0.7 | 9.4 ± 0.6 | 0.92 |
| Neutrophils (103/μl) | 5.2 ± 0.5 | 7.8 ± 1.7 | 0.25 |
| Eosinophil (103/μl) | 0.2 ± 0.1 | 0.4 ± 0.1 | 0.32 |
Data are means ± SEM (n = 5 GFP, 7 TBX18). P values were determined by two-tailed t test. ALT, alanine aminotransferase; AST, aspartate aminotransferase; BUN, blood urea nitrogen; CPK, creatine phosphokinase; CKMB, MB isoenzyme of CPK; HGB, hemoglobin; WBC, white blood cell count.
DISCUSSION
The present study demonstrates that in situ somatic reprogramming can stimulate proper heart function in a large-animal model of complete heart block. Our gene delivery techniques were consistent with routine clinical practice in which venous access is considered the safest method for electronic pacemaker implantation (23). TBX18 gene transfer successfully created stable biological pacemaker activity for 2 weeks, with minimal backup electronic pacemaker use. The TBX18 biological pacemaker was superior to an electronic pacemaker in its ability to support physical activity and was responsive to autonomic regulation, showing features of the native SAN. Neither arrhythmic risk nor systemic toxicity was noted, providing at least preliminary evidence for favorable local and systemic safety profiles. Until now, no biological pacemaker therapy (gene- or cell-based) has been shown to offer physiologically relevant chronotropic support and an appropriate safety profile for a potential clinical application (2, 24). These important characteristics lay the groundwork for translation to humans.
Previous gene-based biological pacemaker approaches have focused on functional reengineering by overexpression of wild-type or mutant ion channels (2, 5–7). Functional reengineering by ion channel manipulation induces artificial automaticity in normally quiescent ventricular myocytes. Here, we used an alternative strategy designed to achieve focal somatic reprogramming of working cardiomyocytes into iSANs (12). The present study not only confirmed the ability of TBX18 to convert cardiomyocytes to iSANs in pigs but also comprehensively investigated the physiological relevance of such conversion. Ideal pacemakers, either biological or electronic, should respond to the complex interaction between autonomic regulation and physical activity (2, 25, 26). Until now, no study has comprehensively addressed this important limitation of biological pacemaker strategies. Characterizing physical activity by a built-in accelerometer (27, 28), we found that the TBX18-induced biological pacemaker supported greater daily physical activity, a stable and sustained daytime pattern, and more frequent, sustained bursts of activity compared with control animals. These findings were linked to better HR adaptation to physical activity and corresponding autonomic responsiveness, as demonstrated by HRV and β-adrenergic sensitivity. In summary, TBX18-induced biological pacemakers resemble the native SAN in terms of physiological homeostasis, HR–physical activity interaction, and autonomic regulation.
One primary concern of gene therapy–driven automaticity in the heart is the potential occurrence of ventricular arrhythmias (2, 24). Here, no evidence of increased ventricular arrhythmias was seen during 2 weeks of continuous monitoring. Repolarization parameters (QT and APD) and repolarization gradients (QT and APD90 dispersion) that can promote arrhythmogenesis (29, 30) were not abnormally elevated in TBX18-transduced animals. Moreover, no inducible ventricular tachycardia was seen by programmed ventricular stimulation 2 weeks after TBX18 gene delivery. Together, these findings support the absence of pro-arrhythmic risk of the TBX18-induced biological pacemaker, at least in the short term. Viral biodistribution analysis indicated that the adenoviral vector was primarily localized at the injection site, with minimal systemic distribution. Further, there was no evidence of systemic toxicity after adenoviral TBX18 gene transfer.
The ease of gene delivery here further improves the prospects for clinical translation. The injection catheter applied here (NOGA MyoStar) has been extensively used in multiple human biological therapy trials (31–33). By introducing this catheter transvenously, we were able to deliver adenoviral vectors to the atrioventricular (AV) junction without the need for open-chest surgery or arterial access. This gene delivery technique has many potential advantages: low invasiveness, minimal pain/distress, minimal blood loss, low risk of stroke (no access to the left-sided circulation), and the ability to deliver the transgene focally into the AV junction area, resulting in potential antegrade conduction through the specialized His-Purkinje system.
The use of nongutted adenoviral vectors in immunocompetent animals, as in the present study, is expected to lead to fairly rapid clearance of transduced cells (17, 18). Indeed, we found that the biological pacemaker activity peaks at 8 days and persists, but with a measurable decline, by 2 weeks, a pattern consistent with what is known about clearance of adenovirally transduced cells in mammals (17). Adenoviral vectors are well suited for temporary applications that do not require persistent gene expression or long-term transduced cell survival. A temporary “bridge” biological pacemaker would enable pacemaker-dependent patients with device-related infections to be treated with systemic antibiotics after complete removal of all hardware until they become infection-free (34). Routine management now requires implantation of a temporary transvenous pacing device while antibiotic treatment clears the infection, which typically requires ~2 weeks. Unfortunately, the presence of an indwelling catheter can potentially undermine the ability of systemic antibiotics to clear the infection. A “hardware-free” temporary pacing alternative would be desirable in such patients in the interval after removal of the infected hardware and before implantation of a new permanent electronic pacemaker. Thus, the innate immunogenicity of adenoviral expression systems can be used to advantage in creating a temporary biological pacemaker. The eventual clearance of all transduced cells is also an important safety feature of adenoviral vectors for first-in-human studies, in that persistent transgene expression, with its potential for long-term complications, can be avoided.
The following lines of evidence support the notion that TBX18 gene transfer creates iSAN cells in situ: (i) the finding of transduced cells with morphology distinctive for iSAN cells, (ii) characteristic mRNA changes including up-regulation of HCN4 and suppression of Cx43, (iii) focal down-regulation of Cx43 by immunostaining, and (iv) verification that the impulse originates at the injection site in TBX18-transduced animals. Nevertheless, our study has a number of limitations. The evidence for in situ reprogramming is not as extensive as in our previous rodent work, where we had isolated single cells for EP characterization (12). Such experimentation is difficult to perform in the expensive and labor-intensive porcine model, which is designed to assess safety and functional efficacy. Although recordings from isolated cells are lacking in the pig model, the totality of the evidence, coupled with the lack of a plausible alternative mechanism, gives good reason to believe that the biological pacemaker arises from somatic reprogramming. We also do not show long-term safety and efficacy data, which will be required as the next step in the translational pathway.
Success with a focused temporary biological pacemaker strategy—such as that demonstrated here—in selected patients would justify more ambitious efforts to replace electronic pacemakers altogether. Different viral (or nonviral) expression systems may be necessary for applications requiring long-term survival of transduced cells. For common clinical applications where devices are tractable and effective, head-to-head comparisons between electronic and biological pacemakers will eventually be required to assess the relative merits of the two approaches.
MATERIALS AND METHODS
Study design
Minimally invasive percutaneous gene delivery was used to achieve in vivo somatic reprogramming in a large-animal model of complete heart block. Farm pigs were randomized to GFP as control (n = 5) or TBX18 gene delivery (n = 7). The sample size was estimated to achieve a power of 0.8 and α = 0.05 using a two-tailed t test. Multiple physiological variables, as well as local and systemic safety profiles, were evaluated in an open-label design. Animals that developed acute procedural complications such as death, cardiac tamponade, or signs of distress during follow-up that compromised animal welfare were eliminated from the study. We observed one acute procedural death in each experimental group, and those animals were excluded from the analysis. All the eligible experimental animals were included without defining or excluding outliers. The endpoint of the experiment was the completion of 2-week experimental follow-up.
Porcine model of complete heart lock
We implemented a modified version of our previously reported protocol (5), schematically depicted in fig. S4. All experiments were approved by the Cedars-Sinai Institutional Animal Care and Use Committee and performed in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals. In brief, female farm pigs (30 to 40 kg, 3 to 4 months old) were randomized to GFP as control or TBX18 gene delivery.
Animals were premedicated with intramuscular acepromazine (0.25 mg/kg), atropine (0.05 mg/kg), and ketamine (20 mg/kg). Anesthesia was induced with intravenous propofol (4 to 6 mg/kg), followed by intubation and maintenance on isoflurane during positive pressure ventilation. A baseline 12-lead electrocardiogram (ECG) was recorded. The right internal jugular vein was cannulated by open cut-down technique, and blood was sampled through a venous catheter before any drug infusion or experimental manipulation. A ventricular pacemaker lead was advanced and positioned under fluoroscopic guidance into the RV apex (fig. S4). A single-chamber electronic pacemaker (St. Jude Medical) was inserted and programmed in ventricular demand pacing (VVI) mode at a rate of 50 beats/min. Monophasic action potentials (MAPs) were recorded from the posteroseptal, mid-anterior wall, mid-free wall, and apex of RV during temporary pacing at 120 beats/min by carefully positioning a 7-French MAP catheter (Synaptic Medical). The average of action potential duration at 90% (APD90) from four locations was calculated. APD90 dispersion was defined as maximum APD90 interval minus minimum APD90 interval.
After MAP recordings, radio frequency (RF) ablation was performed at the atrioventricular nodal region to achieve complete heart block using a 5-mm tip nonirrigated RF catheter (Boston Scientific). Persistence of complete heart block was confirmed by electronic pacemaker dependency immediately after ablation (VVI at 50 beats/min) and persistent AV dissociation for 30 min. During the RF procedure, prophylactic lidocaine (40 μg/kg per minute) was infused temporarily to prevent ventricular tachycardia and fibrillation.
A 3D electroanatomic map of the RV was created using a NOGA mapping system (Biologics Delivery Systems, Johnson & Johnson). Viral vectors were delivered into the upper posteroseptum of the RV through a steerable NOGA MyoStar injection catheter. An iodine contrast pre-test was used to confirm the injection site as shown in fig. S5 (200 μl per injection, five injections per animal). Pace mapping through the NOGA catheter before gene delivery was performed to acquire a 12-lead ECG template from the injection site, to be compared to the 12-lead ECG morphology of the biological pacemaker rhythm.
Subcutaneous telemetry transmitters (Data Sciences International) were implanted in the left posterior upper thoracic region, with both subcutaneous electrodes placed in the anterior chest. Animals were followed continuously for 14 days by continuous telemetry recordings and serial electronic pacemaker interrogations. EP study was performed at 7 days and 2 weeks after gene delivery (fig. S3).
A nonsurvival surgery was conducted 2 weeks after gene delivery. Anesthesia, vascular access, 12-lead ECG, blood sampling, MAP recordings, and 3D electroanatomic maps were performed as described for the baseline procedures. EP study with programmed ventricular stimulation was also conducted. At the end of the procedure, animals were euthanized and organs were harvested for PCR and immunohistochemistry.
Molecular cloning, adenovirus production, and purification
Human TBX18 cloning and purification have been described (12). In brief, human TBX18 with the GFP (ZsGreen1) reporter gene was inserted into an adenovirus vector backbone (Invitrogen) to create the final adenoviral expression construct, pAd-CMV-TBX18-IRES-GFP. An adenoviral vector expressing GFP alone under the cytomegalovirus (CMV) promoter (AdGFP) was used as control. A total of 5.0 × 1011 viral particles of each vector (AdGFP and Ad-TBX18-IRES-GFP) in the buffer (Puresyn Inc.) was injected into each animal.
Electroanatomic mapping
Endocardial activation patterns were assessed by detailed 3D electroanatomic mapping of the RV before and 14 days after the injection procedure. A magnetic electroanatomic system (NOGA, Biologics Delivery Systems, Johnson & Johnson) was used to measure the spatial distribution of local endocardial activation times relative to a reference electrogram. By moving the mapping catheter to different locations in the RV during spontaneous or ventricular paced beats, a color-coded 3D endocardial activation map was constructed (red, earliest activation; purple, latest activation). Additionally, 12-lead ECG comparison during pace mapping was performed to compare the morphology of the biological pacemaker rhythm and pacing from the injection site.
EP studies
Evaluation of biological pacemaker function
Sinus node recovery time is used clinically to evaluate sinus node function. The difference between the time to first beat after rapid stimulation and baseline cycle length (corrected recovery time) is associated with pacemaker function of the native sinus node (16). A similar method was implemented to evaluate the function of biological pacemaker 7 and 14 days after gene delivery. Burst stimulation was performed on the RV through either a venous EP catheter or the implanted electronic pacemaker at a pacing cycle length of 500 ms for 60 s. The biological pacemaker recovery time was defined as the time interval between the last paced beat to the first spontaneous ventricular beat. Prolonged pauses (that is, secondary pauses) occurring after the initial recovery interval were also included. To adjust for HR, the spontaneous ventricular cycle length before pacing was measured and subtracted from the recovery time by which the corrected recovery time is obtained.
Ventricular arrhythmia induction protocol
Stimulation was performed through a venous EP catheter positioned at the RV apex 14 days after gene delivery with and without isoproterenol infusion (20 μg/min). A 500-ms drive train was used followed by one to four ventricular extrastimuli (pulse duration: 2 ms, current amplitude: 10 mA). Extrastimuli were decremented down to a coupling interval until the effective refractory period was reached. Rapid ventricular pacing from 500 ms in 10-ms decrements to 200 ms was performed subsequently. The ventricular arrhythmias included the induction of a sustained monomorphic ventricular tachycardia, polymorphic ventricular tachycardia, or ventricular fibrillation. A sustained ventricular arrhythmia was defined as one lasting ≥30 s.
Continuous ECG and activity recordings
Continuous digital ECG and physical activity recordings were performed and analyzed at a sampling frequency of 500 Hz using a Ponemah Physiology Platform (Data Sciences International). The telemetry implants included a built-in triaxial accelerometer and ECG recorders. Single-lead ECG and physical activity could be recorded simultaneously and subsequently analyzed.
Morphology matching from a template QRS obtained by pace mapping from the injection site and the biological pacemaker rhythms was performed using Ponemah ECG Pro template-based analysis software. This semiautomated analysis allowed for a comparison with all the QRS complexes of the data set, providing an overall percent of cycles that matched the preacquired template.
Activity data were obtained from a triaxial accelerometer (x, y, and z axes), and the activity value, in arbitrary units (AU), was calculated using the following equation:
where C is a constant based on the δ time for the accelerometer sampling rate (C = interpolation frequency × 3.5347).
Animals were observed as per institutional guidelines in a post-procedure recovery room for 1 day after surgery without telemetry monitoring; thus, ECG and daily activity in the first 24 hours were not available for analysis.
HR variability
To evaluate autonomic modulation during dominant biological pacemaker activity, we analyzed HRV from 5-min recordings during maximal HR at day 8 after gene delivery when peak gene expression would be expected. HRV was analyzed by frequency domain methods using Ponemah analysis software. RR intervals from the 5-min recordings were digitized and analyzed by fast Fourier transformation. Power spectra were quantified by measuring the area in three frequency bands: VLF from 0 to 0.04 Hz, LF from 0.04 to 0.15 Hz, and HF from 0.15 to 0.40 Hz. Measurement of VLF, LF, and HF power components was made not only in absolute values of power (ms2) but also in normalized units (20).
Real-time quantitative PCR
For biodistribution analysis of the adenoviral vector, postmortem heart samples were obtained, including a ~0.5-cm × 0.5-cm endocardial sample at the injection site (identified by the needle tracks) and another from the left ventricular epicardium (remote site). Tissue samples were also obtained from lung, liver, spleen, kidney, and brain. Total DNA was isolated using DNeasy (Qiagen) per the manufacturer's instructions. Real-time PCR primers designed to amplify human adenovirus type 5 genome at E2 region were as follows: forward primer (5′-GGCTAGGACGGGTTACAACA-3′), reverse primer (5′-ACGAGGAGGCACTAAAGCAA-3′), and probe (56-FAM/GACACCCAG/ZEN/CAGAAACCTGT/3IABkFQ). Real-time PCR was performed by using the TaqMan assay and a 7900HT Fast Real-Time PCR System (Applied Biosystems/Life Technologies Corp.) according to the manufacturer's recommendations. To perform absolute quantification of viral vector, viral DNA extracted from TBX18 adenoviral vector was serially diluted to create an appropriate standard curve. The viral DNA standard curve, tissue DNA samples, and no template controls were simultaneously expanded and subjected to analysis.
To quantify TBX18 RNA in the injection site, heart samples from injection and remote sites were obtained and stored in RNAlater stabilization reagent (Qiagen). The mRNA was extracted (Qiagen RNeasy Fibrous Tissue Mini Kit). mRNA (3 μg per sample) was converted to first-strand complementary DNA (cDNA) using SuperScript III first-strand synthesis system (Invitrogen). Serial dilutions of the plasmid DNA template (pLV-TBX18-IRES-ZsGreen) with known copy numbers were used to create the standard curve. Real-time PCR was performed using human TBX18 primer sets (assay ID: Hs01385457_m1, Applied Biosystems).
To check the expression of genes that characterize iSANs, the same methods to extract RNA and cDNA were used as those for TBX18. However, actin was used as the housekeeping gene and relative expression was calculated. The primer sets are provided in table S4.
Immunohistochemistry and confocal microscopy
Frozen sections of heart tissue (14 days after adenoviral transduction) were fixed with 4% paraformaldehyde and permeabilized with 0.1% Triton X-100 and then incubated with the appropriate primary antibody—sarcomeric α-actinin (Abcam, ab9465, 1:200), Cx43 polyclonal antibody (Sigma, C6219, 1:100), ZsGreen polyclonal antibody (Clontech, 632474, 1:200) for TBX18 group, and GFP antibody (Abcam, ab290, 1:200) for GFP group—and Alexa Fluor–conjugated secondary antibodies (Invitrogen). Counterstaining with DAPI (Molecular Probes) was performed. Sections were imaged using a confocal laser scan microscope (Leica Microsystems), and images were processed by Leica LAS software suite.
Statistical analysis
Data are presented as means ± SEM. The two-tailed t test was used (comparison of two groups only) as appropriate. Differences in continuous variables between mean or maximal HR and pacing ratio were analyzed using repeated-measures ANOVA with subsequent Bonferroni multiple comparison tests. The correlation between the duration of bursts of activity and LF/HF was analyzed by linear regression model. P < 0.05 was considered statistically significant. Statistical analyses were performed using SPSS software version 17 (SPSS Inc.).
Supplementary Material
Acknowledgments
Funding: Cedars-Sinai Board of Governors and Cedars-Sinai Medical Center.
Footnotes
Author contributions: All authors contributed to the concepts in this study. Y.-F.H., J.F.D., and E.C. performed large-animal experiments. Y.-F.H. performed adenoviral vector production, data analysis, tissue staining, and PCR. Y.-F.H. and E.C. performed statistical analyses. Writing of the paper was done in close collaboration by Y.-F.H., H.C.C., E.M., and E.C.
Competing interests: A patent, assigned to the Cedars-Sinai Medical Center, is pending on the use of TBX18 as a biological pacemaker. The authors declare no other competing financial interests.
Data and materials availability: The sequence of TBX18 primer set can be found as National Center for Biotechnology Information reference sequence: NM_001080508.2.
SUPPLEMENTARY MATERIALS
www.sciencetranslationalmedicine.org/cgi/content/full/6/245/245ra94/DC1
Fig. S1. HR trends, diurnal changes, and physical activity data from a TBX18-transduced animal without backup electronic pacing, superimposed on the corresponding pooled data for the routine protocol.
Fig. S2. TBX18-transduced cardiomyocytes for quantification.
Fig. S3. Cx43 expression in iSAN cells.
Fig. S4. Schematic of the experimental protocol in the porcine model of complete heart block.
Fig. S5. Injection site images.
Table S1. Baseline characteristics of GFP and TBX18 groups.
Table S2. Comparison of HRV between TBX18 and GFP animals.
Table S3. Staining conditions for HCN4.
Table S4. Primer sequences for the different genes studied in reprogrammed cells.
REFERENCES AND NOTES
- 1.Chardack WM, Gage AA, Greatbatch W. A transistorized, self-contained, implantable pacemaker for the long-term correction of complete heart block. Surgery. 1960;48:643–654. [PubMed] [Google Scholar]
- 2.Cho HC, Marbán E. Biological therapies for cardiac arrhythmias: Can genes and cells replace drugs and devices? Circ. Res. 2010;106:674–685. doi: 10.1161/CIRCRESAHA.109.212936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.2012 Writing Group Members. Tracy CM, Epstein AE, Darbar D, DiMarco JP, Dunbar SB, Estes NA, III, Ferguson TB, Jr., Hammill SC, Karasik PE, Link MS, Marine JE, Schoenfeld MH, Shanker AJ, Silka MJ, Stevenson LW, Stevenson WG, Varosy PD, 2008 Writing Committee Members. Epstein AE, DiMarco JP, Ellenbogen KA, Estes NA, III, Freedman RA, Gettes LS, Gillinov AM, Gregoratos G, Hammill SC, Hayes DL, Hlatky MA, Newby LK, Page RL, Schoenfeld MH, Silka MJ, Stevenson LW, Sweeney MO, ACCF/AHA Task Force Members. Anderson JL, Jacobs AK, Halperin JL, Albert NM, Creager MA, DeMets D, Ettinger SM, Guyton RA, Hochman JS, Kushner FG, Ohman EM, Stevenson W, Yancy CW, American College of Cardiology Foundation; American Heart Association Task Force on Practice Guidelines; American Association for Thoracic Surgery; Heart Failure Society of America; Society of Thoracic Surgeons 2012 ACCF/AHA/HRS focused update of the 2008 guidelines for device-based therapy of cardiac rhythm abnormalities: A report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. J. Thorac. Cardiovasc. Surg. 2012;144:e127–e145. doi: 10.1016/j.jtcvs.2012.08.032. [DOI] [PubMed] [Google Scholar]
- 4.Miake J, Marbán E, Nuss HB. Biological pacemaker created by gene transfer. Nature. 2002;419:132–133. doi: 10.1038/419132b. [DOI] [PubMed] [Google Scholar]
- 5.Cingolani E, Yee K, Shehata M, Chugh SS, Marbán E, Cho HC. Biological pacemaker created by percutaneous gene delivery via venous catheters in a porcine model of complete heart block. Heart Rhythm. 2012;9:1310–1318. doi: 10.1016/j.hrthm.2012.04.020. [DOI] [PubMed] [Google Scholar]
- 6.Tse HF, Xue T, Lau CP, Siu CW, Wang K, Zhang QY, Tomaselli GF, Akar FG, Li RA. Bioartificial sinus node constructed via in vivo gene transfer of an engineered pacemaker HCN Channel reduces the dependence on electronic pacemaker in a sick-sinus syndrome model. Circulation. 2006;114:1000–1011. doi: 10.1161/CIRCULATIONAHA.106.615385. [DOI] [PubMed] [Google Scholar]
- 7.Bucchi A, Plotnikov AN, Shlapakova I, Danilo P, Jr., Kryukova Y, Qu J, Lu Z, Liu H, Pan Z, Potapova I, KenKnight B, Girouard S, Cohen IS, Brink PR, Robinson RB, Rosen MR. Wild-type and mutant HCN channels in a tandem biological-electronic cardiac pacemaker. Circulation. 2006;114:992–999. doi: 10.1161/CIRCULATIONAHA.106.617613. [DOI] [PubMed] [Google Scholar]
- 8.Boink GJ, Nearing BD, Shlapakova IN, Duan L, Kryukova Y, Bobkov Y, Tan HL, Cohen IS, Danilo P, Jr., Robinson RB, Verrier RL, Rosen MR. Ca2+-stimulated adenylyl cyclase AC1 generates efficient biological pacing as single gene therapy and in combination with HCN2. Circulation. 2012;126:528–536. doi: 10.1161/CIRCULATIONAHA.111.083584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marbán E, Cho HC. Creation of a biological pacemaker by gene- or cell-based approaches. Med. Biol. Eng. Comput. 2007;45:133–144. doi: 10.1007/s11517-007-0165-2. [DOI] [PubMed] [Google Scholar]
- 10.Plotnikov AN, Sosunov EA, Qu J, Shlapakova IN, Anyukhovsky EP, Liu L, Janse MJ, Brink PR, Cohen IS, Robinson RB, Danilo P, Jr., Rosen MR. Biological pacemaker implanted in canine left bundle branch provides ventricular escape rhythms that have physiologically acceptable rates. Circulation. 2004;109:506–512. doi: 10.1161/01.CIR.0000114527.10764.CC. [DOI] [PubMed] [Google Scholar]
- 11.Rosen MR, Brink PR, Cohen IS, Robinson RB. Genes, stem cells and biological pacemakers. Cardiovasc. Res. 2004;64:12–23. doi: 10.1016/j.cardiores.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 12.Kapoor N, Liang W, Marbán E, Cho HC. Direct conversion of quiescent cardiomyocytes to pacemaker cells by expression of Tbx18. Nat. Biotechnol. 2013;31:54–62. doi: 10.1038/nbt.2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kapoor N, Galang G, Marbán E, Cho HC. Transcriptional suppression of connexin43 by TBX18 undermines cell-cell electrical coupling in postnatal cardiomyocytes. J. Biol. Chem. 2011;286:14073–14079. doi: 10.1074/jbc.M110.185298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fedorov VV, Glukhov AV, Chang R, Kostecki G, Aferol H, Hucker WJ, Wuskell JP, Loew LM, Schuessler RB, Moazami N, Efimov IR. Optical mapping of the isolated coronary-perfused human sinus node. J. Am. Coll. Cardiol. 2010;56:1386–1394. doi: 10.1016/j.jacc.2010.03.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cinca J, Moya A, Figueras J, Roma F, Rius J. Circadian variations in the electrical properties of the human heart assessed by sequential bedside electrophysiologic testing. Am. Heart J. 1986;112:315–321. doi: 10.1016/0002-8703(86)90268-1. [DOI] [PubMed] [Google Scholar]
- 16.Narula OS, Samet P, Javier RP. Significance of the sinus-node recovery time. Circulation. 1972;45:140–158. doi: 10.1161/01.cir.45.1.140. [DOI] [PubMed] [Google Scholar]
- 17.Lusky M, Christ M, Rittner K, Dieterle A, Dreyer D, Mourot B, Schultz H, Stoeckel F, Pavirani A, Mehtali M. In vitro and in vivo biology of recombinant adenovirus vectors with E1, E1/E2A, or E1/E4 deleted. J. Virol. 1998;72:2022–2032. doi: 10.1128/jvi.72.3.2022-2032.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gorziglia MI, Kadan MJ, Yei S, Lim J, Lee GM, Luthra R, Trapnell BC. Elimination of both E1 and E2 from adenovirus vectors further improves prospects for in vivo human gene therapy. J. Virol. 1996;70:4173–4178. doi: 10.1128/jvi.70.6.4173-4178.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Montano N, Ruscone TG, Porta A, Lombardi F, Pagani M, Malliani A. Power spectrum analysis of heart rate variability to assess the changes in sympathovagal balance during graded orthostatic tilt. Circulation. 1994;90:1826–1831. doi: 10.1161/01.cir.90.4.1826. [DOI] [PubMed] [Google Scholar]
- 20.Heart rate variability: Standards of measurement, physiological interpretation and clinical use. Task Force of the European Society of Cardiology and the North American Society of Pacing and Electrophysiology. Circulation. 1996;93:1043–1065. [PubMed] [Google Scholar]
- 21.Bleeker WK, Mackaay AJ, Masson-Pévet M, Bouman LN, Becker AE. Functional and morphological organization of the rabbit sinus node. Circ. Res. 1980;46:11–22. doi: 10.1161/01.res.46.1.11. [DOI] [PubMed] [Google Scholar]
- 22.McGavigan AD, Roberts-Thomson KC, Hillock RJ, Stevenson IH, Mond HG. Right ventricular outflow tract pacing: Radiographic and electrocardiographic correlates of lead position. Pacing Clin. Electrophysiol. 2006;29:1063–1068. doi: 10.1111/j.1540-8159.2006.00499.x. [DOI] [PubMed] [Google Scholar]
- 23.Rajappan K. Permanent pacemaker implantation technique: Part I: Arrhythmias. Heart. 2009;95:259–264. doi: 10.1136/hrt.2007.132753. [DOI] [PubMed] [Google Scholar]
- 24.Rosen MR, Robinson RB, Brink PR, Cohen IS. The road to biological pacing. Nat. Rev. Cardiol. 2011;8:656–666. doi: 10.1038/nrcardio.2011.120. [DOI] [PubMed] [Google Scholar]
- 25.Occhetta E, Bortnik M, Marino P. Usefulness of hemodynamic sensors for physiologic cardiac pacing in heart failure patients. Cardiol. Res. Pract. 2011;2011:925653. doi: 10.4061/2011/925653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brubaker PH, Kitzman DW. Chronotropic incompetence: Causes, consequences, and management. Circulation. 2011;123:1010–1020. doi: 10.1161/CIRCULATIONAHA.110.940577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moreau M, Siebert S, Buerkert A, Schlecht E. Use of a tri-axial accelerometer for automated recording and classification of goats’ grazing behaviour. Appl. Anim. Behav. Sci. 2009;119:158–170. [Google Scholar]
- 28.Westerterp KR. Physical activity assessment with accelerometers. Int. J. Obes. Relat. Metab. Disord. 1999;23(Suppl. 3):S45–S49. doi: 10.1038/sj.ijo.0800883. [DOI] [PubMed] [Google Scholar]
- 29.Kozhevnikov D, Caref EB, El-Sherif N. Mechanisms of enhanced arrhythmogenicity of regional ischemia in the hypertrophied heart. Heart Rhythm. 2009;6:522–527. doi: 10.1016/j.hrthm.2008.12.021. [DOI] [PubMed] [Google Scholar]
- 30.Antzelevitch C, Oliva A. Amplification of spatial dispersion of repolarization underlies sudden cardiac death associated with catecholaminergic polymorphic VT, long QT, short QT and Brugada syndromes. J. Intern. Med. 2006;259:48–58. doi: 10.1111/j.1365-2796.2005.01587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perin EC, Dohmann HF, Borojevic R, Silva SA, Sousa AL, Mesquita CT, Rossi MI, Carvalho AC, Dutra HS, Dohmann HJ, Silva GV, Belém L, Vivacqua R, Rangel FO, Esporcatte R, Geng YJ, Vaughn WK, Assad JA, Mesquita ET, Willerson JT. Transendocardial, autologous bone marrow cell transplantation for severe, chronic ischemic heart failure. Circulation. 2003;107:2294–2302. doi: 10.1161/01.CIR.0000070596.30552.8B. [DOI] [PubMed] [Google Scholar]
- 32.Perin EC, Dohmann HF, Borojevic R, Silva SA, Sousa AL, Silva GV, Mesquita CT, Belém L, Vaughn WK, Rangel FO, Assad JA, Carvalho AC, Branco RV, Rossi MI, Dohmann HJ, Willerson JT. Improved exercise capacity and ischemia 6 and 12 months after transendocardial injection of autologous bone marrow mononuclear cells for ischemic cardiomyopathy. Circulation. 2004;110:II213–II218. doi: 10.1161/01.CIR.0000138398.77550.62. [DOI] [PubMed] [Google Scholar]
- 33.Losordo DW, Henry TD, Davidson C, Sup Lee J, Costa MA, Bass T, Mendelsohn F, Fortuin FD, Pepine CJ, Traverse JH, Amrani D, Ewenstein BM, Riedel N, Story K, Barker K, Povsic TJ, Harrington RA, Schatz RA, Investigators AC. Intramyocardial, autologous CD34+ cell therapy for refractory angina. Circ. Res. 2011;109:428–436. doi: 10.1161/CIRCRESAHA.111.245993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sohail MR, Uslan DZ, Khan AH, Friedman PA, Hayes DL, Wilson WR, Steckelberg JM, Stoner S, Baddour LM. Management and outcome of permanent pacemaker and implantable cardioverter-defibrillator infections. J. Am. Coll. Cardiol. 2007;49:1851–1859. doi: 10.1016/j.jacc.2007.01.072. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.