Summary
Sexual development in the filamentous model ascomycete T richoderma reesei (syn. H ypocrea jecorina) was described only a few years ago. In this study, we show a novel role for VELVET in fungi, which links light response, development and secondary metabolism. V el1 is required for mating in darkness, normal growth and conidiation. In light, vel1 was dispensable for male fertility but essential for female fertility in both mating types. VEL1 impacted regulation of the pheromone system (hpr1, hpr2, hpp1, ppg1) in a mating type‐dependent manner and depending on the mating partner of a given strain. These partner effects only occurred for hpp1 and hpr2, the pheromone precursor and receptor genes associated with the MAT1‐2 mating type and for the mating type gene mat1‐2‐1. Analysis of secondary metabolite patterns secreted by wild type and mutants under asexual and sexual conditions revealed that even in the wild type, the patterns change upon encounter of a mating partner, with again distinct differences for wild type and vel1 mutants. Hence, T . reesei applies a language of pheromones and secondary metabolites to communicate with mating partners and that this communication is at least in part mediated by VEL1.
Introduction
Sexual reproduction has developed and spread in evolution as a mechanism to survive challenging environmental circumstances and to improve competitiveness by increased phenotypic variability, despite the higher energetic costs and the risk to lose a successful phenotype due to recombination. Heterothallic ascomycetes, which need a mating partner of opposite mating type for sexual reproduction, can assume the male or female role in a cross and are hence considered hermaphroditic (Debuchy et al., 2010; Nieuwenhuis and Aanen, 2012). However, the distinction between male or female and sex determination by mating type are not equally clear in all fungi and consequently, it is a matter of debate whether mating type and sex can indeed be generally separated in fungi (Heitman et al., 2013). Assuming the female role thereby means a considerable investment in terms of resources to form female reproduction structures, which may or may not be fertilized by compatible males. Nevertheless, recombination during meiosis can also serve as the only escape strategy to survive decaying environmental conditions as a species (Aanen and Hoekstra, 2007). In fungi, it is debated if ‘real’ asexual species exist or if their sexual stage has just not been discovered yet, hence emphasizing the importance of sexual development (Taylor et al., 1999).
The filamentous ascomycete Trichoderma reesei (syn. Hypocrea jecorina) represents a workhorse for numerous biotechnological applications, which are nowadays mainly targeted at improvement of production of second‐generation biofuels (Schmoll et al., 2014). Decades of strain improvement with T. reesei resulted in detailed knowledge of nutrient requirements, regulation mechanisms and regulators of enzyme production (Kubicek et al., 2009; Schmoll et al., 2014). In recent years, also signal transduction pathways, especially those transmitting light signals, were investigated for their role in enzyme production and development in T. reesei (Schmoll et al., 2010a; 2014).
In T. reesei, the discovery of sexual development was reported a few years ago (for an overview, see Schmoll, 2013). The possibility to cross strains with different characteristics added a valuable tool for investigation of T. reesei. However, a QM6a derivative, which contained the other mating type, was not able to form fruiting bodies with QM6a, although the same experiment enabled fruiting body formation in the wild‐type strain CBS999.97. Hence, QM6a was proposed to be female sterile (Seidl et al., 2009). This finding that QM6a, the parent strain of all strains used in research and industry, is female sterile hampers applications of sexual development in strain improvement and is subject to continued research. So far, no conditions have been reported that would induce formation of protoperithecia with T. reesei. Therefore, combinations of female/male fertile and female/male sterile strains have to be used to test for fertility.
For accomplishment of sexual development, the mycelium is required to continuously function as an information network. Availability of nutrients, light or darkness and sensing of compatible partners are crucial for initiation of the sexual stage and hence represent important information that has to be transmitted and interpreted in the cell (Debuchy et al., 2010). One of the most crucial signaling systems for communication between potential mating partners, the pheromone system, shows an interesting deviation from other fungi in T. reesei. Besides normal orthologs of pheromone receptors (HPR1 and HPR2) and an alpha type peptide pheromone precursor (PPG1), an h‐type peptide pheromone precursor (HPP1) with characteristics of a‐ and alpha type assumes a‐type function (Schmoll et al., 2010b). For successful mating, a pair of receptor and cognate pheromone precursor is required. Consequently, either ppg1 and hpr2 or hpp1 and hpr1 are required for successful mating. Thereby, if one of the pheromones is missing from the genome, the other can take over in both mating types, as their transcription is not strictly mating type dependent (Seibel et al., 2012b). However, as in other fungi, lack of the mating type‐associated pheromone receptor (HPR1 in MAT1‐1 or HPR2 in MAT1‐2) leads to female sterility, while pheromone precursors are essential for male fertility in T. reesei (Kim and Borkovich, 2004; Seibel et al., 2012b). Hence, in contrast to the pheromone precursors, the effect of availability of pheromone receptors is mating type dependent. Transcript levels of hpp1, ppg1 and hpr1 increase under conditions of sexual development in the wild‐type strain CBS999.97, while for hpr2, this is only the case in MAT1‐2 upon fruiting body formation (Seibel et al., 2012b).
Sexual development in T. reesei is most efficient in daylight (light–dark cycles), but also occurs in darkness with some delay (Seidl et al., 2009; Chen et al., 2012; Seibel et al., 2012a). While the photoreceptors of T. reesei, BLR1, BLR2 and ENV1, influence many light responses (Castellanos et al., 2010), only ENV1 has an important influence on fruiting body formation. Two mutants of opposite mating types, both lacking env1, are not able to undergo sexual development in light, but do not show this defect in darkness. At the transcriptional level, these mutants show extreme overexpression of the pheromone precursor genes in light, especially of hpp1. Hence, their defect in light is assumed to be caused by misregulation of the pheromone system and subsequent loss of sexual identity (Seibel et al., 2012a).
Since the discovery of VELVET (VeA) in Aspergillus nidulans, its homologs were studied extensively and found to be crucial regulators of light‐dependent development in diverse fungi (Bayram and Braus, 2012). VeA is further known to be essential for normal secondary metabolism, particularly with respect to biosynthesis of sterigmatocystin in A. nidulans but also for diverse other metabolites in various fungi. This is due to regulation of downstream transcription factors (summarized in Calvo, 2008). The coordination of the light signal with development and secondary metabolism is accomplished by an intricate mechanism (Bayram et al., 2008a; Sarikaya‐Bayram et al., 2014). Thereby, VeA interacts with the phytochrome FphA in dependence of the presence of the chromophore in FphA, and this interaction is restricted to the nucleus. FphA in turn interacts with the photoreceptor complex (LreA‐LreB) via LreB (Purschwitz et al., 2008).
Interestingly, the function of VeA homologs in regulation of asexual development can be negative (Mooney and Yager, 1990; Bayram et al., 2008b; Jiang et al., 2011; Schumacher et al., 2012; Lopez‐Berges et al., 2013) or positive (Calvo et al., 2004; Hoff et al., 2010; Kopke et al., 2013; Kim et al., 2014). With respect to secondary metabolism, mostly positive regulation by VeA homologs is reported in the studies mentioned above, although in some cases, a negative function on individual metabolites was found (Wiemann et al., 2010; Schumacher et al., 2013).
In Trichoderma spp., the VeA ortholog VEL1 of Trichoderma virens was studied, which was found to act positively on secondary metabolism, biocontrol efficiency, morphogenesis and asexual development (Mukherjee and Kenerley, 2010). Recently, a positive effect on development was also observed in a female sterile strain of T. reesei, and VEL1 was found to be involved in regulation of cellulase gene expression (Karimi Aghcheh et al., 2014).
In this study, we aimed to investigate the role of VEL1 in light‐dependent development in T. reesei in detail. We show that VEL1 is essential for sexual development in darkness and required for female fertility in light. Investigation of the pheromone system as influenced by VEL1 suggested a role in communication between potential mating partners. Indeed, we found that VEL1 is involved in regulation of the response to another fungus encountered in terms of signal reception (regulation of pheromone receptor transcripts), signal transmission (pheromone precursors) and chemical communication.
Results
VEL1 is required for asexual development
The velvet family is represented in T. reesei with VEL1 (TR_122284), VEL2 (TR_40551) and VEL3 (TR_102737), which largely share the characteristics of their homologs in Aspergilli, but no VosA homolog is present in the genome (for details, see Supporting Information Appendix S1). For functional analysis, we deleted vel1 in the wild‐type strain QM6a. The resulting strain did not sporulate on various carbon sources and under different light and stress conditions. VEL1 was found to further impact growth and cellulase expression (Supporting Information Appendix S2). Hence, the phenotype of deletion of vel1 in the high cellulase mutant strain QM9414 as reported earlier (Karimi Aghcheh et al., 2014) was largely confirmed for its parental wild‐type strain QM6a.
VEL1 is essential for sexual development in darkness
In order to investigate the function of T. reesei VEL1 in sexual development in a female fertile strain background, we constructed strains (named FF1 and FF2) derived from female sterile QM6a of both mating types able to undergo mating with a female sterile partner by crossing with the female fertile wild‐type CBS999.97 and repeated backcrossing with QM6a as described earlier (Schuster et al., 2012). In agreement with earlier reports (Schuster et al., 2012), the difference in sporulation between growth in light and darkness was larger in these strains compared with QM6a. Otherwise, the phenotype was similar to QM6a.
With these female fertile strains in hand, we prepared strains lacking vel1 in the female fertile background as described above in both mating types [Δvel1F1 in MAT1‐1 (abbreviated as V1) and Δvel1F2 in MAT1‐2 (V2)]. The phenotype caused by deletion of vel1 in QM6a as described above was not significantly altered in these strains (data not shown).
To analyze the role of vel1 in sexual development, Δvel1F1, Δvel1F2 and QM6a Δvel1 were crossed with wild‐type strains of CBS999.97 of the corresponding mating type under daylight (cycles of 12 h light–12 h dark) conditions, which are optimal for sexual development in T. reesei (Seidl et al., 2009; Chen et al., 2012). Crosses between the two wild‐type strains resulted in fruiting bodies 7 days after inoculation. However, mating between a wild‐type strain and a Δvel1 strain was delayed and only lower numbers of fruiting bodies appeared after 10–12 days (Fig. 1). Moreover, ascosporogenesis was delayed and strongly decreased compared with the wild‐type crosses as ascospores were hardly detectable on the lids of the petri dishes. Progeny of these crosses, which retained the vel1 deletion, were used as control to confirm that lack of vel1 causes the observed defects and showed the expected phenotype. Interestingly, the color of the fruiting bodies formed in crosses with vel1 deletions was somewhat lighter than of those formed in wild‐type crosses. Crosses among two strains, which both had a deletion in vel1, failed regardless of the strain background (Fig. 1). The same crossings as described above were repeated in constant darkness, where the wild type forms fruiting bodies with some delay and in lower numbers (Seibel et al., 2012a). We found that in every cross in which one partner had lost vel1, no fruiting bodies were formed, not even after prolonged incubation (Fig. 1). Hence, VEL1 is essential for sexual development in darkness.
Figure 1.
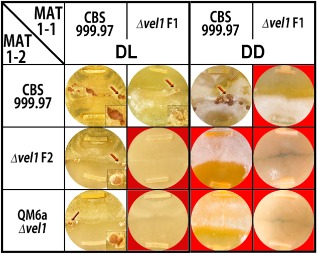
Analysis of sexual development of mutant strains lacking vel1. MAT1‐1 strains are inoculated on top; MAT1‐2 strains are inoculated on bottom. Fruiting bodies are indicated with arrows and shown enlarged in the right corner for inoculation in daylight (DL). Fruiting bodies from crosses with strains lacking VEL1 are pale compared with wild‐type crosses. Strains were grown on malt extract agar (MEX) as carbon source in darkness (DD) or daylight (DL), light–dark cycles) at 22°C for 10 days. Red background highlights abolishment of fruiting body formation.
Female fertility is perturbed in strains lacking VEL1
To determine whether VEL1 is important for male fertility, we used QM6a Δvel1. As this strain is female sterile because of the QM6a background, potential loss of male fertility due to the deletion of vel1 would result in an inability to mate with even a fully sexual competent mating partner. As crossing of QM6a Δvel1 (MAT1‐2) with CBS999.97 (MAT1‐1) was successful in light, albeit with some delay compared with wild‐type crosses (Fig. 1), we conclude that VEL1 is relevant, but not essential for male fertility.
As the presence of VEL1 in at least one mating partner is required for sexual development, we assumed that VEL1 might be involved in regulation of communication between mating partners and/or influence female fertility. Deletion of pheromone receptors leads to female sterility in a mating type‐dependent manner, and mutants of CBS999.97 Δhpr1 MAT1‐1 and CBS999.97 Δhpr2 MAT1‐2 were found to be female sterile (Seibel et al., 2012b). As these strains are still male fertile, they can be used to study the function of VEL1 in female fertility. If VEL1 perturbs female fertility, then sexual crossing with a female sterile partner would not be possible with the respective deletion strains.
Fruiting body formation did not occur between CBS999.97 Δhpr1 MAT1‐1 (female sterile) and QM6a (female sterile) or Δvel1F2, while a cross between CBS999.97 Δhpr1 MAT1‐2 (female fertile) and Δvel1F1 resulted in fruiting bodies, which is in agreement with the necessity of VEL1 for female fertility (Fig. 2A). In order to further validate this result, Δvel1F2 was crossed with a female sterile derivative of QM6a (QFS69, MAT1‐1), which had been obtained in parallel to FF1 and FF2 upon selection for female sterile strains. Mating failed between these two mating partners, which confirms that VEL1 is essential for female fertility (Fig. 2A). Tests with the other mating types yielded consistent results. Fruiting bodies of Δvel1 strains were smaller and fewer than in wild‐type crosses. As fruiting body formation does not occur at all upon lack of vel1 in one mating partner in darkness, we could only investigate these effects upon growth in daylight (12:12 cycles).
Figure 2.
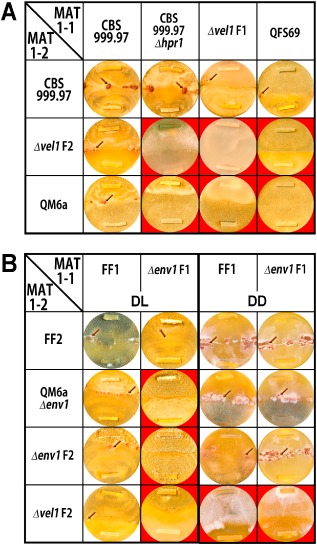
Analysis of female fertility of mutant strains lacking vel1. MAT1‐1 strains are inoculated on top; MAT1‐2 strains are inoculated on bottom. Fruiting bodies are indicated with arrows. Strains were grown on malt extract agar (MEX) as carbon source in darkness (DD) or daylight (DL), light–dark cycles) at 22°C for 10 days. Red background highlights abolishment of fruiting body formation.
As we assumed that mate recognition or sensing might be perturbed upon lack of vel1, we tested whether mating would be successful with Δenv1 strains, which strongly overexpress peptide pheromone precursor genes in light. Lack of ENV1 causes loss of sexual identity by deregulated transcription of pheromone receptors and precursors in light, but not darkness (Seibel et al., 2012a). Therefore, we used a Δenv1 strain with the QM6a background and prepared strains lacking ENV1 in a female fertile background with MAT1‐1 (Δenv1F1). In agreement with the female sterility of Δenv1 strains in light (Seibel et al., 2012a), no fruiting bodies emerged in a cross between Δvel1F2 and Δenv1F1 (Fig. 2B). As expected from our experiments described above, also in darkness, no fruiting bodies were formed in crosses between Δvel1F and Δenv1F (Fig. 2B). Consequently, the deregulated pheromone system in Δenv1F obviously could not overcome a potential sensing defect of Δvel1F.
Pheromone precursor genes hpp1 and ppg1 are differently regulated by VEL1
Several of our results described above indicate a function of VEL1 in environmental sensing and communication with mating partners. We therefore analyzed transcript abundance of pheromone receptor genes and peptide pheromone precursor genes in different mating partner combinations under optimal crossing conditions (12:12 light–dark cycles). Mycelia were consistently harvested upon contact (but not overgrowth) at subjective noon in order to avoid any alterations by circadian rhythms (Fig. 3). Several plates in equal growth stages were pooled and additionally two biological replicates were included. Contamination of total RNA of one sample with that of the respective mating partner on the same plate was analyzed by quantitative polymerase chain reaction (qPCR) from co‐precipitated DNA as described previously (Seibel et al., 2012b) and determined to be below 1.02%. This minor contamination did not interfere with reliable analysis of transcript patterns in the results shown. Asexual cultures without mating partners on the same plate were grown in parallel under equal conditions and harvested at the same time points as with sexual development.
Figure 3.
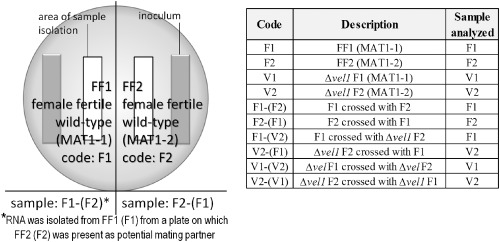
Schematic representation of sample preparation. Samples were harvested separately for both strains present on a plate close to the growth front, but avoiding overlap. Sample codes include the mating partner, which was present on the plate along with the strain from which mycelium was isolated, in brackets.
A significant effect of VEL1 on transcription of the pheromone precursor gene hpp1 was observed dependent on the mating type. Under asexual conditions, hpp1 was fivefold downregulated in Δvel1F1, whereas the deletion of vel1 in a MAT1‐2 background (Δvel1F2) only had a small influence on hpp1 transcript levels (Fig. 4A).
Figure 4.
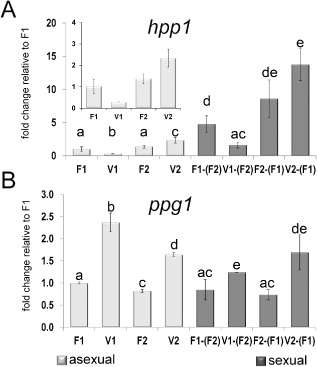
Analysis of pheromone precursor transcript levels. Transcript abundance is shown upon growth on malt extract agar plates at subjective noon as analyzed by qRT‐PCR for both mating types of analyzed strains. Sample codes are summarized in Fig. 6. Different letters indicate significantly different transcript levels.
A. Regulation of hpp1 under asexual conditions in wild type (F1, F2) and strains lacking vel1 (V1, V2) as well as regulation of hpp1 upon encounter of the compatible wild‐type mating partner (F1 or F2) under sexual conditions in wild type [F1 crossed with (F2), F2 crossed with (F1)] and strains lacking vel1 [V1 crossed with (F2), V2 crossed with (F1)]. The insert shows regulation of hpp1 under asexual conditions.
B. Regulation of ppg1 under asexual conditions in wild type and strains lacking vel1 as well as regulation of ppg1 upon encounter of the compatible wild‐type mating partner under sexual conditions in wild type and strains lacking vel1. Pooled samples from several plates and two independent biological replicates were considered. Statistical significance of differences was evaluated with the software qbase+ and applying analysis of variance with a P‐value threshold of < 0.05. Error bars show standard deviations. Contamination of total RNA of one sample with that of the respective mating partner on the same plate was determined to be below 1.02%.
A similar positive effect of VEL1 on hpp1 transcript abundance in strains with a MAT1‐1 background was observed under sexual conditions (Fig. 4A). However, for Δvel1F2, the hpp1 transcript levels showed no significant difference to wild‐type levels.
In contrast to hpp1, the pheromone precursor ppg1 seems to be negatively regulated by VEL1 independent of mating type in asexual conditions (Fig. 4B). ppg1 transcript levels increased in Δvel1 strains in both mating types (Fig. 4B; 2.0 fold ± 0.05 in MAT1‐2, 2.37 fold ± 0.26 in MAT1‐1). The same regulative pattern was observed in sexual conditions (Fig. 4B). We conclude that VEL1 differentially regulates both pheromone precursor genes in a mating type‐dependent manner.
VEL1 is a positive regulator for transcription of pheromone receptors
First, we checked mating type‐specific regulation in order to compare our data with earlier results. Both pheromone receptor transcripts (hpr1 and hpr2) show a mating type‐dependent regulation in the wild‐type strains FF1 or FF2 (Fig. 5). Transcription of hpr1 is enhanced in MAT1‐1, whereas hpr2 is strongly upregulated in MAT1‐2 background even in asexual conditions. A similar pattern was previously observed for pheromone receptor mutants in CBS999.97 (Seibel et al., 2012b). Hence, also considering transcription patterns of pheromone precursor genes, the pheromone system of T. reesei is consistently regulated in strains with fertile QM6a background (FF1 and FF2) and CBS999.97 strains.
Figure 5.
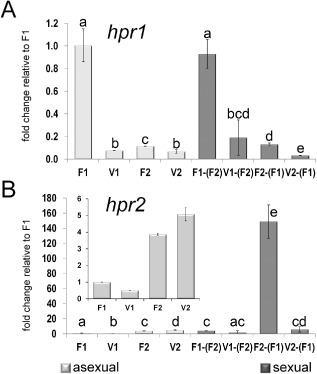
Analysis of pheromone receptor transcript levels. Transcript abundance is shown upon growth on malt extract agar plates at subjective noon as analyzed by qRT‐PCR for both mating types of analyzed strains. Sample codes are summarized in Fig. 6. Different letters indicate significantly different transcript levels.
A. Regulation of hpr1 under asexual conditions in wild type (F1, F2) and strains lacking vel1 (V1, V2) as well as regulation of hpr1 upon encounter of the compatible wild‐type mating partner (F1 or F2) under sexual conditions in wild type [F1 crossed with (F2), F2 crossed with (F1)] and strains lacking vel1 [V1 crossed with (F2), V2 crossed with (F1)].
B. Regulation of hpr2 under asexual conditions in wild type and strains lacking vel1 as well as regulation of hpr2 upon encounter of the compatible wild‐type mating partner under sexual conditions in wild type and strains lacking vel1. The insert shows regulation of hpr1 under asexual conditions. Pooled samples from several plates and two independent biological replicates were considered. Statistical significance of differences was evaluated with the software qbase+ and applying analysis of variance with a P‐value threshold of < 0.05. Error bars show standard deviations. Contamination of total RNA of one sample with that of the respective mating partner on the same plate was determined to be below 1.02%.
Under asexual conditions, VEL1 positively regulates hpr1 in MAT1‐1, with transcript levels of hpr1 dropping to MAT1‐2 levels in the deletion mutant. In MAT1‐2, only a minor positive effect of VEL1 was detected (Fig. 5A). Also upon encounter of a mating partner (sexual conditions), hpr1 levels were decreased in Δvel1F1 compared with wild type and drop to MAT1‐2 levels in this strain. In Δvel1F2 (MAT1‐2), transcript abundance drops even further below wild‐type levels (Fig. 5A).
Hpr2 was found to be downregulated in Δvel1F1 compared with wild type in MAT1‐1, whereas only a small alteration was observed in MAT1‐2 (Fig. 5B) under asexual conditions. A much stronger mating type‐dependent regulation of hpr2 was observed under sexual conditions. While in MAT1‐1 no significant difference was observed in hpr2 transcript levels between wild type and Δvel1F1 (both in the presence of wild type), we found a considerable decrease in transcript abundance of this gene upon deletion of vel1 in MAT1‐2 to levels around those in MAT1‐1 (Fig. 5B).
In summary, VEL1 is a positive regulator important for mating type‐specific regulation of pheromone receptor genes with substantial effects on hpr1 and hpr2 transcript abundance, especially upon encounter of a mating partner and in their cognate mating type.
The mating partner influences regulation of the pheromone system in Δvel1F
Analysis of the function of VEL1 in transcriptional regulation of the pheromone system clearly indicated a function in mating partner communication for VEL1. Consequently, we investigated the effect of different partners in sexual development on transcript levels. Hence, in addition to the wild type, we used the corresponding Δvel1F strain as mating partner and analyzed transcript levels of pheromone precursor and pheromone receptor genes. Fig. 6 compares fold changes of transcript levels if a vel1 mutant was the partner instead of wild type for hpp1, ppg1, hpr1 and hpr2 in both mating types. Values significantly different from 1 (1 = regulation independent of partner) represent an altered response to Δvel1F compared with the response to wild type.
Figure 6.
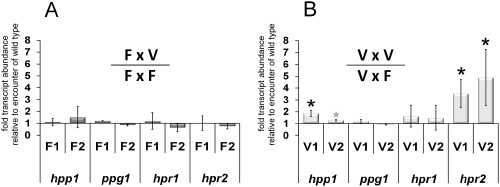
Comparison of transcript levels in wild‐type or vel1 mutants upon encounter of wild‐type or mutant mating partners. Transcript abundance is shown upon growth on malt extract agar plates at subjective noon as analyzed by qRT‐PCR for both mating types of analyzed strains. The results for every datapoint represent a comparison between encounter of the mutant strain (V) and the wild‐type strain (F). Therefore, whenever the result shown in one column including the error bar shows an increase above 1, the result is statistically significant.
A. Regulation of hpp1, ppg1, hpr1 and hpr2 in wild type (F1 or F2) if Δvel1F is the mating partner compared with transcript levels in wild type if the compatible wild type is the mating partner [e.g. ‘F1’ here represents (FF1xΔvel1F2)/(FF1xFF2)].
B. Regulation of hpp1, ppg1, hpr1 and hpr2 in Δvel1F (V1 or V2) if Δvel1F is the mating partner compared with transcript levels in Δvel1F if the compatible wild type is the mating partner [e.g. ‘V1’ here represents (V1xΔvel1F2)/(V1xFF2)]. Asterisks indicate significant difference between encounter of the wild‐type and encounter of the vel1 mutant strain on the plate. For hpp1 in Δvel1F2 (V2), the gray asterisk indicates only a minor effect. Pooled samples from several plates and two independent biological replicates were considered. Statistical significance of differences was evaluated with the software qbase+ and applying analysis of variance with a P‐value threshold of < 0.05. Error bars show standard deviations.
For crosses of wild‐type strains FF1 and FF2, we did not see any significant difference in expression of pheromone receptor or precursor genes regardless if the compatible wild type or a vel1 mutant was the mating partner (Fig. 6A). Regarding Δvel1F strains, no significant differential partner effects were found for ppg1 and hpr1, the cognate pheromone precursor and receptor of the MAT1‐1 mating type.
Interestingly, we found lower transcript levels of hpp1 in Δvel1F1 (MAT1‐1) if the wild‐type FF2 was the partner than if Δvel1F2 was the partner. With Δvel1F2 (MAT1‐2), no such partner‐dependent effect occurred for hpp1 (Fig. 6B). Hence, it appears that hpp1 levels are not only influenced by the mating type of a given strain, but also by the mating partner and that the underlying mechanism is altered in the absence of VEL1. As hpp1 is not the pheromone of MAT1‐1, its downregulation upon encounter of a mating partner is explainable and does not happen equally in a VEL1 mutant.
For hpr2, the cognate pheromone receptor of MAT1‐2, we found a similar trend. Transcript abundance for hpr2 in Δvel1F2 was clearly higher if Δvel1F1 was the partner compared with wild type as a partner. The same situation occurs if Δvel1F1 senses Δvel1F2 compared with sensing of wild type (Fig. 6B). Keeping in mind that transcript levels of hpr2 in Δvel1F2 were considerably lower than in wild‐type FF2 in the presence of a mating partner [Fig. 5B, F2‐(F1) compared with V2‐(F1)], the increased levels seen in the presence of Δvel1F1, but not wild‐type as the partner, reflect a situation closer to the reaction of the wild‐type strains to one lacking vel1. This result hints to a potentially perturbed feedback, which would likely involve pheromone signal perception in the other mating type.
VEL1 is involved in partner‐dependent mat1‐2‐1 transcription
hpp1 and hpr2, which appear to be subject to feedback regulation, encode the cognate pheromone precursor and pheromone receptor of mating type MAT1‐2.
This prompted us to screen for VEL1 regulation and partner effects on the respective mating type gene mat1‐2‐1. Under asexual conditions, mat1‐2‐1 is not regulated by VEL1 (Fig. 7). However, in sexual crosses, transcript levels of mat1‐2‐1 in Δvel1F2 were at wild‐type levels if Δvel1F1 was the crossing partner, but clearly decreased if wild‐type FF2 was the partner (to 44% of wild type; Fig. 7). Comparable with regulation of hpp1 and hpr2, the response of Δvel1F2 to wild type was altered compared with encounter of another strain lacking vel1. Consequently, VEL1 is involved in partner‐dependent regulation of mating type‐specific gene expression.
Figure 7.
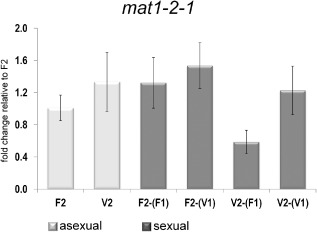
Analysis of mat1‐2‐1 transcript levels. Transcript abundance is shown upon growth on malt extract agar plates at subjective noon as analyzed by qRT‐PCR for both mating types of analyzed strains. Sample codes are summarized in Fig. 6. Regulation of mat1‐2‐1 under asexual conditions in wild type (F2) and Δvel1 F 2 (V2) along with regulation of mat1‐2‐1 upon encounter of the compatible wild‐type (F1) or Δvel1 F 1 (V1) mating partner under sexual conditions in wild type [F2 crossed with (F1)] and strains lacking vel1 [V2 crossed with (F1) or V2 crossed with (V1)]. Transcript levels between sexual and asexual conditions are not significantly different for the wild type [F2 vs. F2‐(F1)]. Only in Δvel1 F 2 upon encounter of the wild type [V2‐(F1)] mat1‐2‐1 is significantly downregulated compared with all other samples. Pooled samples from several plates and two independent biological replicates were considered. Statistical significance of differences was evaluated with the software qbase+ and applying analysis of variance with a P‐value threshold of < 0.05. Error bars show standard deviations. Contamination of total RNA of one sample with that of the respective mating partner on the same plate was determined to be below 1.02%.
VEL1 regulates communication between mating partners by secondary metabolites
The partner effects we observed did suggest an involvement of pheromone regulation in the response of wild‐type strains and those lacking vel1 to their partner. In wild type, pheromone and receptor expression appears to be well balanced, but in the absence of vel1, this balance is perturbed. Additionally, the strongly upregulated pheromone expression in Δenv1F could not compensate for the assumed sensing defect because of decreased transcript levels of pheromone receptors in Δvel1F strains. Consequently, we assumed that partner sensing as regulated by VEL1 is not limited to pheromones. Because of the well studied function of VELVET proteins in secondary metabolism, we figured that secreted metabolites might contribute to an altered response to an encountered mating partner. As we assume that these metabolites represent secondary metabolites, which are not required for growth or metabolism, we will refer to these compounds as such.
For that purpose, we applied semi‐quantitative high‐performance thin layer chromatography (HPTLC) using different visualization techniques to detect different substance classes. As secondary metabolism in T. reesei has hardly been studied yet, identity of the regulated compounds remains to be elucidated and work on this topic is in progress. In the following, we show selected results from the visualization techniques that most clearly showed the differences we found. Thereby, the different techniques are specific for different compounds. Agar without mycelium, but otherwise treated equally, was used as control. Pooled samples and two biological replicates were considered for interpretation of results. We want to note here that in some cases, compounds detected in the control sample are apparently degraded to different extent by different strains. Such degradation is not attributed to secondary metabolism, but rather reflects differential enzymatic activity between strains and is hence considered in our interpretation.
HPTLC revealed that secondary metabolites secreted by FF1 or FF2 in the absence of a mating partner were similar (Fig. 8A–C). Δvel1F strains showed altered secondary metabolite patterns compared with FF1 and FF2, which confirms a function in secondary metabolism in T. reesei as well. However, in contrast to what could be expected from other species, secretion of secondary metabolites was not simply diminished, but for some metabolites even increased in these mutants (Fig. 8A) and decreased for others (Fig. 8B and C).
Figure 8.
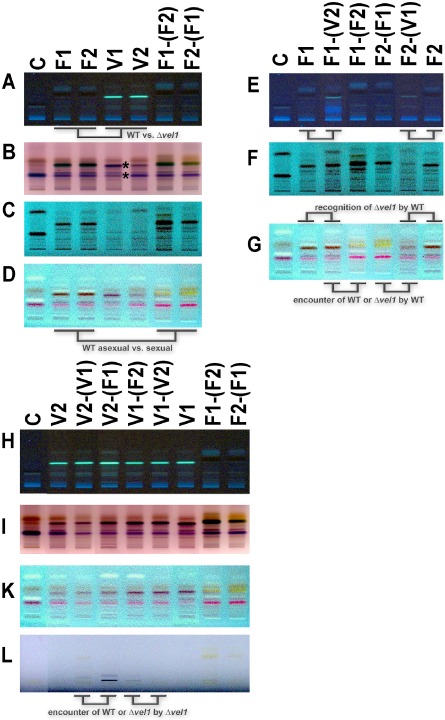
Secondary metabolite patterns of T richoderma reesei under asexual and sexual conditions. Sample codes are summarized in Fig. 6.
A–D. Secondary metabolite patterns of wild‐type (WT) (F1, F2) and vel1 deletion strains (V1, V2) in both mating types and of WT upon encounter of a compatible WT mating partner [F1‐(F2) or F2‐(F1)]. Asterisks in B show the bands differing between Δvel1F stains in different mating types.
E–G. Secondary metabolite patterns of WT under asexual conditions (F1 or F2) and upon response to a compatible WT strain [F1‐(F2) or F2‐(F1)] or to a strain lacking vel1 [(F1‐(V2) or F2‐(V1)].
H–L. Secondary metabolite patterns of Δvel1F under asexual conditions (V1 or V2) and upon response to a compatible WT strain [V1‐(F2) or V2‐(F1)] or to a strain lacking vel1 [(V1‐(V2) or V2‐(V1)]. Growth medium treated equally was used as control (‘C’). The different panels show different visualization techniques, which allow for detection of different substance classes in the sample. (A, E, H) Remission at 366 nm; (B, I) derivatized with anisaldehyde – sulfuric acid, transmission, visual light; (C, F) remission at 254 nm; (D, G, K) derivatized with anisaldehyde – sulfuric acid, remission at 254 nm; (L) remission, transmission at visual light. In order to enable optimal comparison between samples, pictures were reassembled. All samples shown in one panel are combined from the same HPTLC plate. All picture analyses were performed using the analysis software visionCATS 1.4.14017.1 (CAMAG) on the whole picture of the respective plate. Two biological replicates were made and considered for interpretation. The plates with original loading order are provided as Supporting Information material (Supporting Information Fig. S2) for comparison.
In order to evaluate if partner effects as seen in the transcript analysis also occurred for secondary metabolite patterns, we analyzed different partner combinations. The secondary metabolite pattern of wild‐type strains FF1 and FF2 clearly and consistently changed if a mating partner was present (Fig. 8C and D). Consequently, communication between mating partners or generally upon encounter of another fungus in the environment involves secondary metabolites in T. reesei as well. The derivatization method we used (p‐anisaldehyde/sulfuric acid) allows for detection of phenols, sugars, steroids and terpenes. Hence, an involvement of an isoprenoid compound related to trisporic acid, which has pheromone function in zygomycetes (Schimek and Wostemeyer, 2009), could contribute to this response in T. reesei.
We also tested the response of FF1 and FF2 to the respective compatible strains lacking vel1 (Δvel1F1 and Δvel1F2). Clear differences in metabolite patterns were observed for both FF1 and FF2 between encountering wild type or a mutant lacking vel1 (Fig. 8E–G). Although specific differences were also found (Fig. 8E), for most visualization techniques, the patterns secreted by FF1 and FF2 rather resemble the situation of those strains growing alone on the plate (Fig. 8F and G). These results suggest that recognition is hampered if vel1 is lacking in the partner's genome. Additionally, vel1 may be responsible for production of a metabolite (or metabolite combination) that elicits a specific response upon encounter.
In Δvel1F1 and Δvel1F2, the pattern observed upon encounter of a potential mating partner differed from that in the wild type (Fig. 8H). Our transcript analysis revealed that strains lacking vel1 respond differently depending on whether they encounter wild type or Δvel1F strains. For most metabolites, as reflected by different visualization techniques, only minor alterations between presence of wild type or a strain lacking vel1 were detected (Fig. 8H, I and K). Degradation of a compound in the medium upon encounter of strains lacking vel1 in contrast to wild type (uppermost band on Fig. 8K) suggests modulated enzymatic activities. Nevertheless, Δvel1F1 and in lower amounts also Δvel1F2 produce a secondary metabolite upon encounter of wild type but not vel1 mutants, which is not detected in wild type (Fig. 8L).
Discussion
Sexual development is of utmost importance in the physiology and evolution of fungi. With T. reesei, application of sexual crossing for industrial strain improvement adds another aspect for investigation of development. Thereby, communication between fungi prior to initiation of mating is crucial for the success of crossing.
Our study revealed a defect in female fertility in light, in mating in darkness as well as in conidiation to be caused by the lack of VEL1. While defects in conidiation upon deletion of vel1 have been observed in other fungi (Kim et al., 2002; Mukherjee and Kenerley, 2010; Kopke et al., 2013), a function in female fertility specifically in light has not been reported for VEL1 homologs before. In A. nidulans, VeA plays a crucial role in balancing asexual and sexual development (Kim et al., 2002). Apparently, the function of VEL1 in T. reesei targets both processes: upon deletion of vel1, conidiation is abolished and sexual development is severely perturbed. Similar defects in asexual development were also observed in T. virens strains lacking vel1 (Mukherjee and Kenerley, 2010).
Interestingly, although VEL1 was found to be involved in regulation of the pheromone system under asexual conditions as well, the most striking regulation was detected for the pheromone receptors in their cognate mating type under sexual conditions (Fig. 9). Hpr1 transcript levels are 5‐fold decreased in MAT1‐1 and hpr2 is 25‐fold downregulated in MAT1‐2 under sexual conditions if vel1 is deleted. As these pheromone receptors in their cognate mating type are important for female fertility (Seibel et al., 2012b), it seems possible that this regulation is responsible for the defect in female fertility of Δvel1 strains or at least contributes to this defect.
Figure 9.
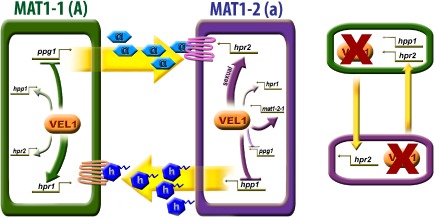
Model of VEL1 regulation of the pheromone system. VEL1 is involved in regulation of both pheromone receptors and pheromone precursors in both mating types under sexual and asexual conditions. The most striking regulation is thereby exerted on the pheromone receptors in their cognate mating types (hpr1 in MAT1‐1 and hpr2 in MAT1‐2). Considering the function of pheromone receptors in their cognate mating type in female fertility, this regulation is likely to contribute to female fertility of vel1 mutants. Strains lacking vel1 react to the presence of another vel1 mutant by upregulation of hpr2 in MAT1‐2 and of hpr2 and hpp1 in MAT1‐1 compared with encounter of a wild‐type strain, where this response does not occur. Only effects with statistically significant regulation in the respective cognate mating type are shown in the scheme with arrows for positive and plungers for negative regulation.
Studying partner effects in regulation of transcript levels of pheromone precursor and receptor genes, we found that strains lacking vel1 respond differently to the presence of a vel1 deletion strain than to wild‐type strains in sexual crosses. This different response is restricted to hpp1 and hpr2, the pheromone precursor and receptor genes associated with the MAT1‐2 mating type. Accordingly, a similar partner effect was also observed for mat1‐2‐1, the mating type gene of the MAT1‐2 mating type, upon lack of vel1. mat1‐2‐1 is also regulated by VEL1 under sexual conditions, hence supporting a function of VEL1 in mating type‐dependent regulation of sexual development.
One explanation of the partner effect we saw would be the operation of a feedback mechanism, which responds to altered levels of pheromones. These pheromones are assumed to be secreted by Δvel1F strains based on their altered transcript levels and are then sensed in the environment as well as by the fungus secreting them itself. Sensing and secreting the same signaling molecule is a ubiquitous mechanism for communication among fungi (Youk and Lim, 2014) and reflects an adaptation to a changing environment. Feedback regulation of pheromone expression has been shown previously and involves heterotrimeric G‐protein signaling (particularly also through beta and gamma subunits) and mitogen‐activated protein (MAP) kinase pathways (Feng and Davis, 2000). It has been shown in A. nidulans that the MAP kinase pathway impacting development and secondary metabolite production targets VeA (Bayram et al., 2012).
The G‐protein pathway is known to sense pheromones and to adjust downstream gene regulation. This is in accordance with our findings that lack of the G‐protein beta subunit decreases hpp1 transcript levels and abolishes ascosporogenesis in T. reesei (Tisch et al., 2011b). As the presence of pheromones in the environment is sensed by pheromone sensors, which are not transcribed in a strictly mating type‐dependent manner in T. reesei (Seibel et al., 2012b), a feedback mechanism is conceivable also in this fungus. However, our analysis of partner effects with strains lacking ENV1, in which the pheromone system is more strongly deregulated than in Δvel1F (Seibel et al., 2012a), did not show significant differences upon encounter of different strains. This led us to assume that pheromone regulation alone is unlikely to cause this phenomenon.
A connection between secondary metabolism and development is well known in fungi (Kück and Böhm, 2013). We were hence interested whether the function of VELVET proteins in fungi is involved in mating partner recognition in T. reesei. Hardly anything is currently known on secondary metabolites secreted by T. reesei (Blumenthal, 2004; Lehner et al., 2013) although its genome comprises numerous genes associated with secondary metabolism (Mukherjee et al., 2012). Harmful compounds are not known to be secreted by T. reesei and hence it achieved GRAS (generally regarded as safe) status by the Food and Drug Administration (21 Code of Federal Regulations §184,1250). However, production of trichodermin (Watts et al., 1988) and the peptide antibiotic paracelsin (Brückner et al., 1984) was reported.
As expected, VEL1 was found to be involved in regulation of secondary metabolism as well. Altered secondary metabolite secretion in wild‐type and Δvel1 strains is summarized in Fig. 10 along with partner effects upon recognition of wild type or mutant on the plate. Strains lacking VEL1 secrete a different set of metabolites as the wild type already when grown alone on a plate (Fig. 10A). Interestingly, we found that the wild type clearly responds to encounter of another fungus on the plate with altered secondary metabolite profiles. This pattern is again changed if a strain lacking vel1 is the partner (Fig. 10B), and also Δvel1F reacts differently to wild type or mutant (Fig. 10C). Currently, the nature of these metabolites is not known, but under investigation in our lab.
Figure 10.
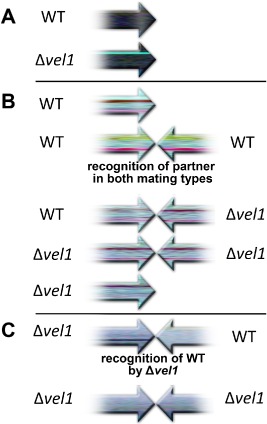
Schematic representation of partner effects in wild‐type (WT) and Δvel1. Metabolite patterns observed in HPTLC are shown as arrows, which are meant to show mutual influence on metabolite patterns. Opposing arrows represent sexual conditions with patterns of both partners on the plates.
A. Different metabolite patterns in WT and Δvel1 indicate that VEL1 is involved in regulation of secondary metabolism (Fig. 8A; remission at 366 nm).
B. WT recognizes the presence of a WT partner on the plate as reflected in altered metabolite patterns compared with growth alone. Presence of Δvel1 causes hardly any response by the WT, because metabolite patterns show only minor changes. The same is the case for two Δvel1 mutants on the plate (Fig. 8D, G and K; derivatized with anisaldehyde – sulfuric acid, remission at 254 nm).
C. Δvel1 recognizes WT and alters secondary metabolite production. This does not occur in the presence of another Δvel1 strain (Fig. 8L; remission, transmission at visual light). Mycelia were harvested individually as shown in Fig. 3. Bands from the visualization technique, which shows the effect most clearly, were used for the scheme.
While extracellular metabolites including oxylipins are known to coordinate a switch between asexual and sexual development (summarized in Christensen and Kolomiets, 2011; Leeder et al., 2011), to the best of our knowledge, effects similar to those we observed have not been reported so far from other fungi. However, such effects are to some extent reminiscent of quorum sensing. Additionally, subtle differences in band intensity suggest that the mating type of stains lacking vel1 influences secondary metabolite production (Fig. 8B). As the lower of the two differing bands appears in the control as well, altered enzymatic activity for degradation of this compound in these strains is likely. Alternatively, true secondary metabolites at this position might have been masked by the background of the medium. Nevertheless, further analyses will be necessary to substantiate this difference.
Quorum sensing has been studied in Aspergillus flavus and is reported to involve oxylipin signaling (Tsitsigiannis et al., 2004; 2005). Oxylipin signaling has been suggested to be involved in injury response in Trichoderma atroviride based on transcriptome data (Hernandez‐Onate et al., 2012). In A. flavus, this mechanism is perturbed if veA or laeA is deleted (Amaike and Keller, 2009; Amare and Keller, 2014). However, the specific sensing of a wild‐type strain versus a mutant strain clearly goes beyond quorum sensing, but indicates that VELVET contributes to transmission/interpretation of a more sophisticated communication between fungi. In‐depth studies of secondary metabolism in T. reesei are in progress and will reveal whether oxylipin signaling plays a role in this communication.
In summary, T. reesei applies a complex chemical language for communication with fungi in their environment, in which vel1 plays a role in both sending and receiving signals. Hence, the regulation of secondary metabolites by VEL1 goes beyond an involvement in simple defense mechanisms, but is responsible for a sophisticated and fine‐tuned chemical communication.
Experimental procedures
Microbial strains and culture conditions
Trichoderma reesei (H. jecorina) QM6a wild‐type strain (ATCC 13631) was used as the parental strain to construct deletions of Δvel1. CBS999.97 MAT1‐1 and MAT1‐2 strains and several other wild‐type and mutant strains from different sources were included in this study to set up informative crosses (Table 1). Female fertile strains FF1 and FF2 with QM6a background were prepared as described previously (Schuster et al., 2012), but with 10 instead of 5 crosses.
Table 1.
Strains used in this study
| Strain | Code | Characteristics | Source/reference |
|---|---|---|---|
| QM6a | Wild‐type MAT1‐2 | Martinez et al. (2008) | |
| CBS999.97 MAT1‐1 | Wild‐type MAT1‐1 | Seidl et al. (2009) | |
| CBS999.97 MAT1‐2 | Wild‐type MAT1‐2 | Seidl et al. (2009) | |
| CBS999.97 MAT1‐1 Δhpr1 | Δhpr1::hph+ MAT1‐1 | Seibel et al. (2012b) | |
| CBS999.97 MAT1‐2 Δhpr1 | Δhpr1::hph+ MAT1‐2 | Seibel et al. (2012b) | |
| FF1 | F1 | Female fertile derivative of QM6a MAT1‐1 | This study |
| FF2 | F2 | Female fertile derivative of QM6a MAT1‐2 | This study |
| QFS69 MAT1‐1 | Female sterile derivative of QM6a MAT1‐1 | This study | |
| QM6a Δvel1 | Δvel1::amds + MAT1‐2, female sterile background (QM6a) | This study | |
| QM6a Δenv1 | Δenv1::hph + MAT1‐2, female sterile background (QM6a) | This study | |
| Δvel1F1 | V1 | Δvel1::amds + MAT1‐1, female fertile background | This study |
| Δvel1F2 | V2 | Δvel1::amds + MAT1‐2, female fertile background | This study |
| Δenv1F1 | E1 | Δenv1::hph + MAT1‐1, female fertile background | This study |
| Δenv1F2 | E2 | Δenv1::hph + MAT1‐2, female fertile background | This study |
Propagation of strains was performed on 3% (w/v) malt extract agar (Merck, Darmstadt, Germany). Crossing experiments were made on 2% (w/v) malt extract agar at 22°C in daylight (cycles of 12 h light–12 h dark) or constant darkness. Therefore, strains were grown on opposite sides of petri dishes and were evaluated for fruiting body formation and ascospore discharge 7 and 20 days after inoculation respectively. For evaluation of conidiation in QM6a Δvel1, Mandels–Andreotti (MA) medium (Mandels and Andreotti, 1978) supplemented with 1% (w/v) carbon source and 0.1% (w/v) peptone (Merck) to induce germination was used. D‐galactose (Sigma Aldrich, St. Louis, USA), D‐sorbitol (ALFA AESAR, Karlsruhe, Germany), D‐mannitol (Fluka, St. Gallen, Switzerland), D‐arabinose (Sigma Aldrich), meso‐erythritol (ALFA AESAR) and γ‐amino butyric acid (GABA) (Sigma Aldrich) or D(+)‐xylose (Sigma Aldrich) were used as carbon sources in sporulation assays.
For transcriptional analysis, the strains were pre‐cultured on 3% (w/v) malt extract agar in constant darkness for at least 3 days. Inoculation of strains was performed on 2% (w/v) malt extract agar plates covered with cellophane to facilitate harvesting. To study gene expression in the course of sexual development, the strains were inoculated on both sides of the petri dishes and the mycelia of the partner strains were harvested at the stage of contact (3 days after inoculation). As the mutant strains have growth defects compared with wild‐type strains, they were inoculated in different distances from the opposite mating strain on petri dishes in order to obtain the contact stage of mycelia for the mutant strains at consistent times. Five plates at the stage of contact were pooled and at least two biological replicates were used for quantitative reverse transcription polymerase chain reaction (qRT‐PCR) analysis. In order to compare the obtained data with asexual growth, strains were grown alone on plates and the mycelia were harvested at the time points corresponding to the contact of strains in sexual development. The strains were grown under cycles of 12 h light–12 h darkness at 22°C (1800 lux).
For cellulase screening, strains were grown on plates with MA medium containing 1% (w/v) carboxymethylcellulose (CMC) (Sigma Aldrich) and after 2 days were stained with Congo red (0.1% w/v solution in water) (Roth, Karlsruhe, Germany), which stains cellulose (Carder, 1986). After washing of the plate with 1 M NaCl, halos indicate cellulase production. Addition of 1% (w/v) lactose or glucose (both from Sigma Aldrich) was used as additional controls. Escherichia coli JM109 was used for cloning (Yanisch‐Perron et al., 1985).
Nucleic acid isolation and transcript analysis
Isolation of total RNA was performed using the RNAeasy plant mini kit (QIAGEN, Hilden, Germany) as described previously (Tisch et al., 2011a). Estimation of crossing partner contamination was evaluated by the relative abundance of mating type genes in co‐precipitated chromosomal DNA in the RNA samples (Seibel et al., 2012b). Contamination was generally lower than 1.02% and did not influence the results presented. The integrity of extracted RNA was first evaluated by agarose gel electrophoresis and staining with SYBR® Safe DNA (Invitrogen, Carlsbad, USA). The concentration and purity of extracted RNA were analyzed using the Nanodrop ND‐1000 spectrophotometer (PEQLAB, Erlangen, Germany). The quality of samples was further confirmed on the Agilent 2100 Bioanalyzer platform using the RNA 6000 Nano Kit (Agilent, Santa Clara, USA) according to the manufacturer's instructions. Only samples with a RIN (RNA integrity number) > 9 were taken for cDNA preparation (Fleige and Pfaffl, 2006). One microgram of total RNA was treated with DNAaseI (Thermo Fisher/Fermentas, St. Leon‐Rot, Germany) for 30 min. The DNAaseI treatment was terminated by adding EDTA to a final concentration of 2.5 mM and incubation at 65°C for 10 min. The RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher/Fermentas) was used for first‐strand cDNA synthesis according to the manufacturer's protocol, and cDNA was diluted prior to qRT‐PCR. No‐template controls and RT‐minus controls (reverse transcription omitted) were included. qPCR reactions were performed in a CFX96 Real‐Time PCR Detection System (Bio‐Rad Laboratories GmbH, Hercules, USA) using the iQ SYBR Green Supermix (Bio‐Rad Laboratories GmbH). Gene‐specific primers used in this study are summarized in Supporting Information Table S1. rpl6e encoding the ribosomal protein RPL6e was used as reference gene. Transcription of rpl6e was demonstrated to be unaffected by light or light regulators (Tisch et al., 2011a). Melting curves after the qPCR were evaluated to confirm that the signal was the result of single‐product amplification and not due to primer dimers or arbitrary amplification. Cycle threshold values were determined for a minimum of two biological replicates and three technical replicates. All data were evaluated and analyzed with the qbase+ software package (Biogazelle, Gent, Belgium) (Hellemans et al., 2007). Statistical analysis of the qPCR data was performed with the qbase + (analysis of variance, Tukey–Kramer post‐test, P < 0.05).
Construction of deletion strains
In order to delete env1, plasmid pDELENV2 (Castellanos et al., 2010) was used. The deletion cassette was released from pDELENV2, using restriction enzymes Acc65I and BamHI (Thermo Fisher/Fermentas), and was used to transform protoplasts (Gruber et al., 1990). Selection of colonies was performed on 3% (w/v) malt extract agar plates containing 100 μg ml−1 hygromycin B (Roth). The deletion strains were checked using PCR primers binding outside the deleted region (ENVScreen_F1 and ENVScreenR1) resulting in considerably longer fragments in mutant strains compared with the wild type.
To construct pDELVEL1 for deletion of vel1, a 1644 bp fragment spanning 5′ region of the gene was amplified using primers vel5F and vel5R. Similarly, a 922 bp fragment flanking the 3′ region of the gene was amplified using primers vel3F and vel3R. The marker construct containing the amdS gene (Penttila et al., 1987) was amplified using primers amdSF and amdSR. The prepared fragments were transformed into yeast and the construction of the vector performed by yeast recombination (Colot et al., 2006). T. reesei transformation was done as described above with the deletion cassette amplified from the vector using vel5F and vel3R. Successful homologous integration in transformants was analyzed using primers ScreenVel_AF1 and ScreenVel_AR1. The copy number of the deletion cassettes in the deletion strains was analyzed using qPCR as described previously (Tisch et al., 2011b). Strains comprising one deletion cassette on the homologous locus were used for further analysis.
Microscopy analysis
Agar block microscopy was performed as described previously (Woo et al., 2010). Briefly, strains were grown on 3% (w/v) malt extract agar at 22°C. Agar blocks were cut using a dissecting knife from the edge of the colony and were subsequently placed on a glass slide. A drop of lactophenol blue stain (Sigma) and a coverslip were put onto the agar block. Samples were immediately examined under a Nikon Eclipse E200 light microscope (Nikon, Tokyo, Japan).
Analysis of secondary metabolites
Thin‐layer chromatography (TLC) analysis was performed as described earlier (Bok and Keller, 2004; Reyes‐Dominguez et al., 2010) with several modifications: Briefly, samples were collected from agar plates in the same size, growth stage and location as used for qRT‐PCR (Fig. 3). Agar pieces from at least three plates were pooled and two independent biological replicates were made. Samples were homogenized in liquid nitrogen and resuspended in acetone : water (1:1; v/v; chemicals from Roth). For extraction of metabolites from mycelia and agar, samples were stored at 4°C for several hours and thereafter chloroform was added. The organic phase (chloroform) was transferred to a conical tube. After complete evaporation of the solvent, the samples were resuspended in 160 μl chloroform and 8 μl was spotted on a TLC plate (HPTLC silica gel 60 F254s, Merck 1.15696.0001) using the CAMAG Automatic TLC sampler 4 (CAMAG, Muttenz, Switzerland). This amount was chosen in order to detect smaller changes in metabolite patterns without overloading the plate. Separation was performed in a saturated twin trough chamber with chloroform : formic acid 7:1 (v/v). The plates were analyzed under ultraviolet light (254 nm and 366 nm) using a CAMAG visualizer (CAMAG). Additionally, the plates were derivatized with p‐anisaldehyde : sulfuric acid and evaluated again with white and ultraviolet light. Results were visualized using the software visionCATS 1.4.14017.1 (CAMAG).
Bioinformatic analyses
Blast analyses were done using the NCBI Blast homepage (http://blast.st‐va.ncbi.nlm.nih.gov/Blast.cgi), AspGD (http://www.aspgd.org/) and JGI MycoCosm (http://genome.jgi‐psf.org/programs/fungi/index.jsf). PEST motifs (Rechsteiner and Rogers, 1996) were determined using ePESTfind (http://emboss.bioinformatics.nl/cgi‐bin/emboss/epestfind) and nuclear export sequences were searched using the NetNES1.1 server (la Cour et al., 2004); http://www.cbs.dtu.dk/services/NetNES/). PsortII (http://psort.hgc.jp/form2.html) was used to identify nuclear localization sequences.
For phylogenetic analysis, the alignment was done with clustalX, and minimum evolution analysis was performed using MEGA4 (2007) with 500 bootstrap cycles.
Supporting information
Supporting information
Acknowledgements
We want to thank Joseph Strauss for critically reading the manuscript.
Work of HB, CD, DT, ES and MS was supported by grant of the Austrian Science Fund (FWF; www.fwf.ac.at), projects no. V152‐B20 and P22511 to MS.
The authors declare that they have no conflict of interest.
References
- Aanen, D.K. , and Hoekstra, R.F. (2007) Why sex is good In Sex in Fungi – Molecular Determination and Evolutionary Implications. Heitman J., Kronstad J.W., Taylor J.W., and Casselton L. (eds). Washington DC: ASM Press, pp. 527–534. [Google Scholar]
- Amaike, S. , and Keller, N.P. (2009) Distinct roles for VeA and LaeA in development and pathogenesis of Aspergillus flavus . Eukaryot Cell 8: 1051–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amare, M.G. , and Keller, N.P. (2014) Molecular mechanisms of Aspergillus flavus secondary metabolism and development. Fungal Genet Biol 66: 11–18. [DOI] [PubMed] [Google Scholar]
- Bayram, O. , and Braus, G.H. (2012) Coordination of secondary metabolism and development in fungi: the velvet family of regulatory proteins. FEMS Microbiol Rev 36: 1–24. [DOI] [PubMed] [Google Scholar]
- Bayram, O. , Krappmann, S. , Ni, M. , Bok, J.W. , Helmstaedt, K. , Valerius, O. , et al (2008a) VelB/VeA/LaeA complex coordinates light signal with fungal development and secondary metabolism. Science 320: 1504–1506. [DOI] [PubMed] [Google Scholar]
- Bayram, O. , Krappmann, S. , Seiler, S. , Vogt, N. , and Braus, G.H. (2008b) Neurospora crassa ve‐1 affects asexual conidiation. Fungal Genet Biol 45: 127–138. [DOI] [PubMed] [Google Scholar]
- Bayram, O. , Bayram, O.S. , Ahmed, Y.L. , Maruyama, J. , Valerius, O. , Rizzoli, S.O. , et al (2012) The Aspergillus nidulans MAPK module AnSte11‐Ste50‐Ste7‐Fus3 controls development and secondary metabolism. PLoS Genet 8: e1002816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenthal, C.Z. (2004) Production of toxic metabolites in Aspergillus niger, Aspergillus oryzae, and Trichoderma reesei: justification of mycotoxin testing in food grade enzyme preparations derived from the three fungi. Regul Toxicol Pharmacol 39: 214–228. [DOI] [PubMed] [Google Scholar]
- Bok, J.W. , and Keller, N.P. (2004) LaeA, a regulator of secondary metabolism in Aspergillus spp. Eukaryot Cell 3: 527–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brückner, H. , Graf, H. , and Bokel, M. (1984) Paracelsin; characterization by NMR spectroscopy and circular dichroism, and hemolytic properties of a peptaibol antibiotic from the cellulolytically active mold Trichoderma reesei. Part B. Experientia 40: 1189–1197. [DOI] [PubMed] [Google Scholar]
- Calvo, A.M. (2008) The VeA regulatory system and its role in morphological and chemical development in fungi. Fungal Genet Biol 45: 1053–1061. [DOI] [PubMed] [Google Scholar]
- Calvo, A.M. , Bok, J. , Brooks, W. , and Keller, N.P. (2004) veA is required for toxin and sclerotial production in Aspergillus parasiticus . Appl Environ Microbiol 70: 4733–4739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carder, J.H. (1986) Detection and quantitation of cellulase by Congo red staining of substrates in a cup‐plate diffusion assay. Anal Biochem 153: 75–79. [DOI] [PubMed] [Google Scholar]
- Castellanos, F. , Schmoll, M. , Martinez, P. , Tisch, D. , Kubicek, C.P. , Herrera‐Estrella, A. , and Esquivel‐Naranjo, E.U. (2010) Crucial factors of the light perception machinery and their impact on growth and cellulase gene transcription in Trichoderma reesei . Fungal Genet Biol 47: 468–476. [DOI] [PubMed] [Google Scholar]
- Chen, C.L. , Kuo, H.C. , Tung, S.Y. , Hsu, P.W. , Wang, C.L. , Seibel, C. , et al (2012) Blue light acts as a double‐edged sword in regulating sexual development of Hypocrea jecorina (Trichoderma reesei) . PLoS ONE 7: e44969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen, S.A. , and Kolomiets, M.V. (2011) The lipid language of plant‐fungal interactions. Fungal Genet Biol 48: 4–14. [DOI] [PubMed] [Google Scholar]
- Colot, H.V. , Park, G. , Turner, G.E. , Ringelberg, C. , Crew, C.M. , Litvinkova, L. , et al (2006) A high‐throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc Natl Acad Sci USA 103: 10352–10357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- la Cour, T. , Kiemer, L. , Molgaard, A. , Gupta, R. , Skriver, K. , and Brunak, S. (2004) Analysis and prediction of leucine‐rich nuclear export signals. Protein Eng Des Sel 17: 527–536. [DOI] [PubMed] [Google Scholar]
- Debuchy, R. , Berteaux‐Lecellier, V. , and Silar, P. (2010) Mating systems and sexual morphogenesis in ascomycetes In Cellular and Molecular Biology of Filamentous Fungi. Borkovich K.A., and Ebbole D.J. (eds). Washington, DC: ASM Press, pp. 501–535. [Google Scholar]
- Feng, Y. , and Davis, N.G. (2000) Feedback phosphorylation of the yeast a‐factor receptor requires activation of the downstream signaling pathway from G protein through mitogen‐activated protein kinase. Mol Cell Biol 20: 563–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleige, S. , and Pfaffl, M.W. (2006) RNA integrity and the effect on the real‐time qRT‐PCR performance. Mol Aspects Med 27: 126–139. [DOI] [PubMed] [Google Scholar]
- Gruber, F. , Visser, J. , Kubicek, C.P. , and de Graaff, L.H. (1990) The development of a heterologous transformation system for the cellulolytic fungus Trichoderma reesei based on a pyrG‐negative mutant strain. Curr Genet 18: 71–76. [DOI] [PubMed] [Google Scholar]
- Heitman, J. , Sun, S. , and James, T.Y. (2013) Evolution of fungal sexual reproduction. Mycologia 105: 1–27. [DOI] [PubMed] [Google Scholar]
- Hellemans, J. , Mortier, G. , De Paepe, A. , Speleman, F. , and Vandesompele, J. (2007) qBase relative quantification framework and software for management and automated analysis of real‐time quantitative PCR data. Genome Biol 8: R19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez‐Onate, M.A. , Esquivel‐Naranjo, E.U. , Mendoza‐Mendoza, A. , Stewart, A. , and Herrera‐Estrella, A.H. (2012) An injury‐response mechanism conserved across kingdoms determines entry of the fungus Trichoderma atroviride into development. Proc Natl Acad Sci USA 109: 14918–14923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoff, B. , Kamerewerd, J. , Sigl, C. , Mitterbauer, R. , Zadra, I. , Kurnsteiner, H. , and Kuck, U. (2010) Two components of a velvet‐like complex control hyphal morphogenesis, conidiophore development, and penicillin biosynthesis in Penicillium chrysogenum . Eukaryot Cell 9: 1236–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, J. , Liu, X. , Yin, Y. , and Ma, Z. (2011) Involvement of a velvet protein FgVeA in the regulation of asexual development, lipid and secondary metabolisms and virulence in Fusarium graminearum . PLoS ONE 6: e28291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi Aghcheh, R. , Nemeth, Z. , Atanasova, L. , Fekete, E. , Paholcsek, M. , Sandor, E. , et al (2014) The VELVET A orthologue VEL1 of Trichoderma reesei regulates fungal development and is essential for cellulase gene expression. PLoS ONE 9: e112799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H. , and Borkovich, K.A. (2004) A pheromone receptor gene, pre‐1, is essential for mating type‐specific directional growth and fusion of trichogynes and female fertility in Neurospora crassa . Mol Microbiol 52: 1781–1798. [DOI] [PubMed] [Google Scholar]
- Kim, H. , Han, K. , Kim, K. , Han, D. , Jahng, K. , and Chae, K. (2002) The veA gene activates sexual development in Aspergillus nidulans . Fungal Genet Biol 37: 72–80. [DOI] [PubMed] [Google Scholar]
- Kim, H.J. , Han, J.H. , Kim, K.S. , and Lee, Y.H. (2014) Comparative functional analysis of the velvet gene family reveals unique roles in fungal development and pathogenicity in Magnaporthe oryzae . Fungal Genet Biol 66: 33–43. [DOI] [PubMed] [Google Scholar]
- Kopke, K. , Hoff, B. , Bloemendal, S. , Katschorowski, A. , Kamerewerd, J. , and Kuck, U. (2013) Members of the Penicillium chrysogenum velvet complex play functionally opposing roles in the regulation of penicillin biosynthesis and conidiation. Eukaryot Cell 12: 299–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubicek, C.P. , Mikus, M. , Schuster, A. , Schmoll, M. , and Seiboth, B. (2009) Metabolic engineering strategies for improvement of cellulase production by Hypocrea jecorina . Biotechnol Biofuels 2: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kück, U. , and Böhm, J. (2013) Mating type genes and cryptic sexuality as tools for genetically manipulating industrial molds. Appl Microbiol Biotechnol 97: 9609–9620. [DOI] [PubMed] [Google Scholar]
- Leeder, A.C. , Palma‐Guerrero, J. , and Glass, N.L. (2011) The social network: deciphering fungal language. Nat Rev Microbiol 9: 440–451. [DOI] [PubMed] [Google Scholar]
- Lehner, S.M. , Atanasova, L. , Neumann, N.K. , Krska, R. , Lemmens, M. , Druzhinina, I.S. , and Schuhmacher, R. (2013) Isotope‐assisted screening for iron‐containing metabolites reveals a high degree of diversity among known and unknown siderophores produced by Trichoderma spp. Appl Environ Microbiol 79: 18–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez‐Berges, M.S. , Hera, C. , Sulyok, M. , Schafer, K. , Capilla, J. , Guarro, J. , and Di Pietro, A. (2013) The velvet complex governs mycotoxin production and virulence of Fusarium oxysporum on plant and mammalian hosts. Mol Microbiol 87: 49–65. [DOI] [PubMed] [Google Scholar]
- Mandels, M. , and Andreotti, R. (1978) Problems and challenges in the cellulose to cellulase fermentation. Proc Biochem 13: 6–13. [Google Scholar]
- Martinez, D. , Berka, R.M. , Henrissat, B. , Saloheimo, M. , Arvas, M. , Baker, S.E. , et al (2008) Genome sequencing and analysis of the biomass‐degrading fungus Trichoderma reesei (syn. Hypocrea jecorina). Nat Biotechnol 26: 553–560. [DOI] [PubMed] [Google Scholar]
- Mooney, J.L. , and Yager, L.N. (1990) Light is required for conidiation in Aspergillus nidulans . Genes Dev 4: 1473–1482. [DOI] [PubMed] [Google Scholar]
- Mukherjee, P.K. , and Kenerley, C.M. (2010) Regulation of morphogenesis and biocontrol properties in Trichoderma virens by a VELVET protein, Vel1. Appl Environ Microbiol 76: 2345–2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee, P.K. , Horwitz, B.A. , and Kenerley, C.M. (2012) Secondary metabolism in Trichoderma ‐ a genomic perspective. Microbiology 158: 35–45. [DOI] [PubMed] [Google Scholar]
- Nieuwenhuis, B.P. , and Aanen, D.K. (2012) Sexual selection in fungi. J Evol Biol 25: 2397–2411. [DOI] [PubMed] [Google Scholar]
- Penttila, M. , Nevalainen, H. , Ratto, M. , Salminen, E. , and Knowles, J. (1987) A versatile transformation system for the cellulolytic filamentous fungus Trichoderma reesei . Gene 61: 155–164. [DOI] [PubMed] [Google Scholar]
- Purschwitz, J. , Muller, S. , Kastner, C. , Schoser, M. , Haas, H. , Espeso, E.A. , et al (2008) Functional and physical interaction of blue‐ and red‐light sensors in Aspergillus nidulans . Curr Biol 18: 255–259. [DOI] [PubMed] [Google Scholar]
- Rechsteiner, M. , and Rogers, S.W. (1996) PEST sequences and regulation by proteolysis. Trends Biochem Sci 21: 267–271. [PubMed] [Google Scholar]
- Reyes‐Dominguez, Y. , Bok, J.W. , Berger, H. , Shwab, E.K. , Basheer, A. , Gallmetzer, A. , et al (2010) Heterochromatic marks are associated with the repression of secondary metabolism clusters in Aspergillus nidulans . Mol Microbiol 76: 1376–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarikaya‐Bayram, O. , Bayram, O. , Feussner, K. , Kim, J.H. , Kim, H.S. , Kaever, A. , et al (2014) Membrane‐bound methyltransferase complex VapA‐VipC‐VapB guides epigenetic control of fungal development. Dev Cell 29: 406–420. [DOI] [PubMed] [Google Scholar]
- Schimek, C. , and Wostemeyer, J. (2009) Carotene derivatives in sexual communication of zygomycete fungi. Phytochemistry 70: 1867–1875. [DOI] [PubMed] [Google Scholar]
- Schmoll, M. (2013) Sexual development in Trichoderma – scrutinizing the aspired phenomenon In Trichoderma – Biology and Applications. Mukherjee P.K., Horwitz B.A., Singh U.S., Mukherjee M., and Schmoll M. (eds). Oxfordshire, UK: CAB International, pp. 67–86. [Google Scholar]
- Schmoll, M. , Esquivel‐Naranjo, E.U. , and Herrera‐Estrella, A. (2010a) Trichoderma in the light of day – physiology and development. Fungal Genet Biol 47: 909–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmoll, M. , Seibel, C. , Tisch, D. , Dorrer, M. , and Kubicek, C.P. (2010b) A novel class of peptide pheromone precursors in ascomycetous fungi. Mol Microbiol 77: 1483–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmoll, M. , Seiboth, B. , Druzhinina, I. , and Kubicek, C.P. (2014) Genomics analysis of biocontrol species and industrial enzyme producers from the genus Trichoderma In The Mycota, Volume XIII, Fungal Genomics. Nowrousian M. (ed.). Springer: Heidelberg Berlin, pp. 233–264. [Google Scholar]
- Schumacher, J. , Pradier, J.M. , Simon, A. , Traeger, S. , Moraga, J. , Collado, I.G. , et al (2012) Natural variation in the VELVET gene bcvel1 affects virulence and light‐dependent differentiation in Botrytis cinerea . PLoS ONE 7: e47840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher, J. , Gautier, A. , Morgant, G. , Studt, L. , Ducrot, P.H. , Le Pecheur, P. , et al (2013) A functional bikaverin biosynthesis gene cluster in rare strains of Botrytis cinerea is positively controlled by VELVET. PLoS ONE 8: e53729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster, A. , Bruno, K.S. , Collett, J.R. , Baker, S.E. , Seiboth, B. , Kubicek, C.P. , and Schmoll, M. (2012) A versatile toolkit for high throughput functional genomics with Trichoderma reesei . Biotechnol Biofuels 5: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seibel, C. , Tisch, D. , Kubicek, C.P. , and Schmoll, M. (2012a) ENVOY is a major determinant in regulation of sexual development in Hypocrea jecorina (Trichoderma reesei). Eukaryot Cell 11: 885–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seibel, C. , Tisch, D. , Kubicek, C.P. , and Schmoll, M. (2012b) The role of pheromone receptors for communication and mating in Hypocrea jecorina (Trichoderma reesei) . Fungal Genet Biol 49: 814–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidl, V. , Seibel, C. , Kubicek, C.P. , and Schmoll, M. (2009) Sexual development in the industrial workhorse Trichoderma reesei . Proc Natl Acad Sci USA 106: 13909–13914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura, K. , Dudley, J. , Nei, M. , and Kumar, S. (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596–1599. [DOI] [PubMed] [Google Scholar]
- Taylor, J. , Jacobson, D. , and Fisher, M. (1999) THE EVOLUTION OF ASEXUAL FUNGI: reproduction, speciation and classification. Annu Rev Phytopathol 37: 197–246. [DOI] [PubMed] [Google Scholar]
- Tisch, D. , Kubicek, C.P. , and Schmoll, M. (2011a) New insights into the mechanism of light modulated signaling by heterotrimeric G‐proteins: ENVOY acts on gna1 and gna3 and adjusts cAMP levels in Trichoderma reesei (Hypocrea jecorina). Fungal Genet Biol 48: 631–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tisch, D. , Kubicek, C.P. , and Schmoll, M. (2011b) The phosducin‐like protein PhLP1 impacts regulation of glycoside hydrolases and light response in Trichoderma reesei . BMC Genomics 12: 613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsitsigiannis, D.I. , Zarnowski, R. , and Keller, N.P. (2004) The lipid body protein, PpoA, coordinates sexual and asexual sporulation in Aspergillus nidulans . J Biol Chem 279: 11344–11353. [DOI] [PubMed] [Google Scholar]
- Tsitsigiannis, D.I. , Kowieski, T.M. , Zarnowski, R. , and Keller, N.P. (2005) Three putative oxylipin biosynthetic genes integrate sexual and asexual development in Aspergillus nidulans . Microbiology 151: 1809–1821. [DOI] [PubMed] [Google Scholar]
- Watts, R. , Dahiya, J. , Chandhary, K. , and Tauro, P. (1988) Isolation and characterization of a new antifungal metabolite of Trichoderma reesei . Plant Soil 107: 81–84. [Google Scholar]
- Wiemann, P. , Brown, D.W. , Kleigrewe, K. , Bok, J.W. , Keller, N.P. , Humpf, H.U. , and Tudzynski, B. (2010) FfVel1 and FfLae1, components of a velvet‐like complex in Fusarium fujikuroi, affect differentiation, secondary metabolism and virulence. Mol Microbiol 77: 972–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo, P.C. , Ngan, A.H. , Chui, H.K. , Lau, S.K. , and Yuen, K.Y. (2010) Agar block smear preparation: a novel method of slide preparation for preservation of native fungal structures for microscopic examination and long‐term storage. J Clin Microbiol 48: 3053–3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch‐Perron, C. , Vieira, J. , and Messing, J. (1985) Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33: 103–119. [DOI] [PubMed] [Google Scholar]
- Youk, H. , and Lim, W.A. (2014) Secreting and sensing the same molecule allows cells to achieve versatile social behaviors. Science 343: 1242782. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information


