Summary
Mycolic acids are unique long chain fatty acids found in the lipid‐rich cell walls of mycobacteria including the tubercle bacillus M ycobacterium tuberculosis. Essential for viability and virulence, enzymes involved in the biosynthesis of mycolic acids represent novel targets for drug development. This is particularly relevant to the impact on global health given the rise of multidrug resistant and extensively drug resistant strains of M . tuberculosis. In this review, we discuss recent advances in our understanding of how mycolic acid are synthesised, especially the potential role of specialised fatty acid synthase complexes. Also, we examine the role of a recently reported mycolic acid transporter MmpL3 with reference to several reports of the targeting of this transporter by diverse compounds with anti‐M . tuberculosis activity. Additionally, we consider recent findings that place mycolic acid biosynthesis in the context of the cell biology of the bacterium, viz its localisation and co‐ordination with the bacterial cytoskeleton, and its role beyond maintaining cell envelope integrity.
Introduction
The lipid‐rich cell wall of Mycobacterium tuberculosis, the bacterium that causes tuberculosis (TB), contains unique fatty acids termed mycolic acids (Minnikin and Polgar, 1966; Barry et al., 1998; Daffe and Draper, 1998). These long chain fatty acids, which have an alkyl side chain and a hydroxyl group at the α and β positions respectively (Fig. 1), are mainly found attached covalently to the distinctive peptidoglycan–arabinogalactan complex of the mycobacterial cell wall. Additionally, mycolic acids are constituents of outer cell envelope lipids including trehalose monomycolate (TMM), trehalose dimycolate (TDM) and glucose monomycolate, and they can also be found as free mycolic acids. Mycolic acids are derived from ‘housekeeping’ fatty acids, i.e. those important for fundamental cellular functions including cell membrane biosynthesis, which are synthesised by a ‘eukaryotic‐like’ modular type‐I fatty acid synthase termed FAS‐I (Bloch, 1977). Mycobacterial FAS‐I is bimodal and in addition to generating C18 fatty acids, M. tuberculosis FAS‐I also synthesises C24–26 fatty acids (Bloch, 1977). The former can also be fed into a second fatty acid synthase, the multienzyme, ‘prokaryotic‐like’ type‐II fatty acid synthase, FAS‐II, which eventually generates a long chain fatty acid termed the merochain. Malonyl‐CoA is the building block used by both FASs and its production requires acetyl‐CoA and an acetyl‐CoA carboxylase, AccD6 (Daniel et al., 2007). The malonate moiety from malonyl‐CoA is first transferred to an acyl carrier protein (ACP) by the enzyme FabD prior to entering the FAS‐II pathway for merochain biosynthesis (Kremer et al., 2001) via the proposed linking activity (so far only demonstrated in vitro) of the ketoacyl synthase FabH (Choi et al., 2000). FAS‐II consists of ‘core’ reductive cycle enzymes: a β‐ketoacyl‐ACP synthase, a β‐ketoacyl‐ACP‐reductase, a β‐hydroxyacyl‐ACP‐dehydratase and an enoyl‐ACP‐reductase that catalyse a reductive cycle using malonyl‐ACP as substrate to extend a growing acyl‐ACP chain by two carbons after each cycle, eventually yielding a merochain of chain length up to C60. Finally, the merochain is condensed with the FAS‐I derived C24–26 fatty acid by a polyketide synthase Pks13 (Gande et al., 2004; Portevin et al., 2004), to yield an oxo‐mycolic acid intermediate which is subsequently reduced by a mycolyl reductase to form a mature mycolic acid (Lea‐Smith et al., 2007; Bhatt et al., 2008). At some stage of this process, and it is not currently known when, unknown desaturase(s) introduce double bonds in the merochain, and subsequently modifying enzymes that include methyl transferases and cyclopropane synthases introduce modifications to the merochain resulting in the formation of different mycolic acid subclasses; M. tuberculosis produces three mycolic acid subclasses termed α, methoxy and keto mycolic acids (Fig. 1). The monomycolylated glycolipid TMM serves as both the carrier for mycolate transport (Grzegorzewicz et al., 2012; Varela et al., 2012), as well as the substrate for extracellular mycolyl transferases termed the Ag85 complex (Belisle et al., 1997; Puech et al., 2002). Enzymes of the Ag85 complex are responsible for the mycolylation of the arabinogalactan in the cell wall, as well as formation of TDM. While early research on mycolic acids was focussed on biochemical characterisation, the post genome sequencing era and the development of genetic tools for mycobacteria has resulted in a focus on identifying genes involved in the biosynthesis of these unique fatty acids. A vast majority of genes involved in mycolic acid biosynthesis are essential and thus cannot be studied via the generation of knockout strains. Though mycolic acids are unique, they are not exclusive to mycobacteria, but are also found in other related genera such as Nocardia, Rhodococcus and Corynebacterium. Three microbial species have particularly aided research into the genetic determinants of mycolic acid metabolism (Marrakchi et al., 2014). First, Corynebacterium glutamicum, by virtue of its ability to survive in the absence of mycolic acid production, has provided the opportunity to study viable mutants that lacked mycolic acids (Gande et al., 2004). Second, the fast growing saprophyte, Mycobacterium smegmatis has proven useful in the establishment of tools for generating conditional mutants, thus allowing us to study the loss of function of essential genes (Bhatt et al., 2005). M. smegmatis is also far more tolerant to mutations altering cell wall composition, which has been a particular advantage. And finally, the leprosy bacillus Mycobacterium leprae, which contains a ‘decayed’ genome, but produces mycolic acids, has aided the ‘bioinformatic filtering’ of candidate genes (Monot et al., 2005). However, some genes involved in mycolic acid biosynthesis are non‐essential in laboratory growth conditions and these are predominantly involved in merochain modification. M. tuberculosis mutants lacking these modifying enzymes have proved useful in outlining the importance of mycolic acids in immunomodulation, virulence and influencing host pathology (Dubnau et al., 2000; Glickman et al., 2000; Rao et al., 2005; 2006). Additionally, M. tuberculosis has two genes that each encode a β‐ketoacyl‐ACP synthase, a core FAS‐II enzyme. While one of these genes, kasA, is essential (Bhatt et al., 2005), the other, kasB, is not (Bhatt et al., 2007a). Interestingly, the M. tuberculosis kasB null mutant produces shorter mycolic acids, and while the mutant was severely attenuated, it was able to persist in mice without displaying any of the tissue pathology associated with infection (Bhatt et al., 2007a).
Figure 1.
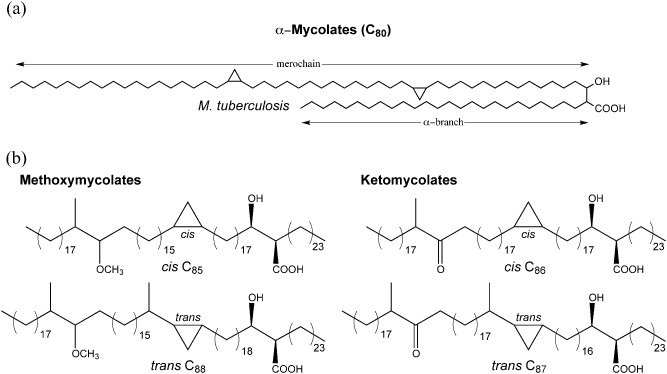
A. Fundamental structure of a mycolic acid shown using M . tuberculosis α‐mycolic acid as an example.
B. Structures of other mycolic acid subclasses from M . tuberculosis.
The essentiality of mycolic acids for viability and for virulence highlights the potential of components of the mycolic acid biosynthesis machinery as attractive drug targets. While isoniazid, the hallmark anti‐TB drug that targets the FAS‐II enoyl‐ACP‐reductase InhA has been in use for many years, mycolic acid biosynthesis pathways remain largely untapped as drug targets. This is especially timely and relevant given the rise of multidrug resistant and extensively drug resistant strains of M. tuberculosis (MDR‐TB and XDR‐TB), and more worryingly the relatively recent reports of ‘untreatable’ TB (Udwadia et al., 2012). While there have been numerous reviews on the cell wall of mycobacteria, very few have focussed exclusively on mycolic acids. Takayama and colleagues wrote the first comprehensive post‐genome sequence review that not only collated existing knowledge about mycobacterial mycolic acid biosynthesis but also formulated hypothesis about yet unidentified processes using bioinformatic searches (Takayama et al., 2005). The field has advanced rapidly since that review in 2005, and more recently Marrakchi et al. (2014) have offered a comprehensive review of these advances. The aim of this microreview is not to replicate the information contained in these earlier reviews. Instead we wish to reflect on advances made in the field since the latter half of 2013. We also consider the broader implications of these findings in terms of how they advance our understanding of mycolic acid biosynthesis in context of the biology of the bacterium, including higher order structures and integration with the cell biology of the bacterium.
Fatty acids and hotdogs: finding the elusive dehydratase
Core components of a FAS‐II complex typically contain four enzymes constituting a reductive cycle that catalyzes the extension of an acyl‐ACP chain by two carbons. While three of the four components (KasA/KasB, MabA and InhA) for the merochain producing mycobacterial FAS‐II had been identified by the late 1990s, until late the dehydratase catalysing conversion of β‐hydroxyacyl‐ACP to enoyl‐ACP remained unidentified. The two operons harbouring the genes encoding the other three core FAS‐II components (InhA and MabA, and KasA/KasB) do not include genes with homology to FabZ or FabA, the classic dehydratase or dehydratase–isomerase enzymes found in FAS‐II complexes from other bacteria (Rock and Cronan, 1996). Furthermore, no fabZ or fabA homologues were found in the genome of M. tuberculosis H37Rv, suggesting that the dehydratase activity of FAS‐II in M. tuberculosis and other mycobacteria may be resulting from a dehydratase with a different or alternative enzymatic motif. A breakthrough came from a bioinformatic study that queried the role of 11 genes encoding putative R‐specific hydratase/dehydratase family, in the biosynthesis of mycolic acids (Castell et al., 2005). All contained a ‘hot dog’ fold that resembled the structure of FabA/Z, but contained a distinct catalytic site. Of these, only one gene, Rv0636 was present in other mycolic acid producing genera and was demonstrated by Sacco et al. (2007) to be an essential gene and a dehydratase involved in mycolic acid biosynthesis. In a parallel study, Brown et al. (2007a, 2007b) used a drugs‐to‐target approach to demonstrate the role of Rv0636 by showing that mycobacterial strains overexpressing Rv0636 showed resistance to flavonoids known to target β‐hydroxyacyl‐ACP dehydratases in Escherichia coli and Plasmodium. Furthermore, the functional dehydratase component was shown to compose of a heterodimer of Rv0636 (HadB) with either Rv0635 (HadA) or Rv0637 (HadC). Both HadA and HadC also contain a hotdog fold and functional heterodimers thus have a ‘double hotdog’ fold. These asymmetric double hotdog folds are thought to play a dual role, with one (HadB) required for catalysis, and the other (from HadA or HadC) functioning as a stabiliser of long chain acyl groups that are typical of mycolic acids. This model has been lent further credence following the recently solved 3D structure of the HadA–HadB complex (Biswas et al., 2015). Furthermore, analogous to KasA and KasB (Kremer et al., 2002; Bhatt et al., 2007b), the two heterodimeric complexes HadA–HadB and HadB–HadC would be expected to differ in their specificities for different fatty acyl chain lengths, with the latter functioning to extend longer growing chains. Recently, Carrère‐Kremer et al (2015) demonstrated the presence of a second dehydratase, not found in M. tuberculosis, but present in non‐tuberculous mycobacteria (NTMs) including M. smegmatis (MSMEG_6754). The role of this non‐essential dehydratase, which is a ‘fused’ HadA–HadB‐like peptide, remains unclear but it was able to compensate for the loss of hadB in M. smegmatis. The authors were able to generate a mutant of hadB in M. smegmatis only when a chromosomally integrated, and constitutively expressed second copy of MSMEG_6754 was present in the strain, indicating a functional redundancy with hadB. Interestingly, the recombinant strain did display an accumulation of shorter mycolic acid precursors, in addition to mycolic acid subspecies usually found in this fast growing species. However, the inability of this second dehydratase to rescue viability during attempts to generate a hadB knockout suggested a low level of functional protein encoded by MSMEG_6754 was present in wild‐type M. smegmatis cells growing in laboratory conditions, and implied a regulatory mechanism controlling its expression. Indeed, MSMEG_6754 was essential for survival in amoebae suggesting a specialised environmental role for this dehydratase in NTMs (Carrère‐Kremer et al., 2015).
There are many FAS‐IIs
Takayama et al. (2005) first suggested the presence of three FAS‐II ‘modules’ including a core elongation FAS‐II consisting of FabD, a β‐ketoacyl‐ACP synthase (KasA/KasB), MabA, InhA and, at the time unidentified, dehydratase component. Additionally, specialised FAS‐IIA and FAS‐IIB modules capable of elongation were hypothesised to be involved in the addition of cis unsaturations at distal and proximal position on the meromycolate chain respectively. The authors suggested that mycolic acid biosynthesis proceeds along linear ‘assembly line’ arrays similar to type‐II polyketide synthases, and that there are five such linear arrays, each terminating with a final condensation reaction catalysed Pks13. Each of these arrays, containing differing combinations of FAS‐II, FAS‐IIA, FAS‐IIB and modifying enzymes, was dedicated for the biosynthesis of one subclass of M. tuberculosis mycolic acids. The five arrays were thus involved in the biosynthesis of α mycolic acids, cis‐methoxy mycolic acids, trans‐methoxy mycolic acids, cis‐keto mycolic acids and trans‐keto mycolic acids. Experimental proof indicating the presence of such complexes came from protein–protein interaction studies conducted using the yeast two‐hybrid and three‐hybrid systems, and in vitro co‐immunoprecipitation studies using individual FAS‐II components (Veyron‐Churlet et al., 2004; 2005; Cantaloube et al., 2011). Cantaloube et al. (2011) extended earlier studies to define a ‘mycolic acid biosynthesis interactome’ consisting of different specialised FAS‐II elongation complexes, all of which contained a ‘core’ consisting of MabA, InhA and FabD (Fig. 2). An initiation FAS‐II (I‐FAS‐II) contains, in addition to the core, FabH and links FAS‐I to FAS‐II (Fig. 2). Two elongation complexes contain the core and either KasA and HadA–HadB (E1‐FAS‐II), or KasB and HadB–HadC (E2‐FAS‐II). E1‐FAS‐II is proposed to carry out the initial elongation cycles, with E2‐FAS‐II acting as unit for further elongation of meroacyl chains produced by E1‐FAS‐II (Fig. 2). Finally, a ‘termination’ FAS‐II complex involves Pks13 (Fig. 2), resulting in the Claisen condensation of a meromycolate chain with a FAS‐I derived C26 fatty acid to yield an oxo‐mycolic acid which is subsequently reduced by the mycolyl reductase encoded by Rv2509, to yield a mature mycolic acid moiety (Fig. 2). Furthermore, these studies have also shown that enzymes introducing modifications, such as methyltransferases, isomerases and cyclopropane synthases also interact with the elongation complex, suggesting that merochain modification occurs during elongation. Additionally, these modification enzymes show a preference for a particular type of Had heterodimer and consequently the type of elongation FAS‐II complex. CmaA2 and PcaA that introduce modifications to the proximal position of the merochain interact preferentially with HadBC, whereas MmaA3, the enzyme involved in the introduction of a methoxy group at the distal position, interacts preferentially with the HadA–HadB heterodimer.
Figure 2.
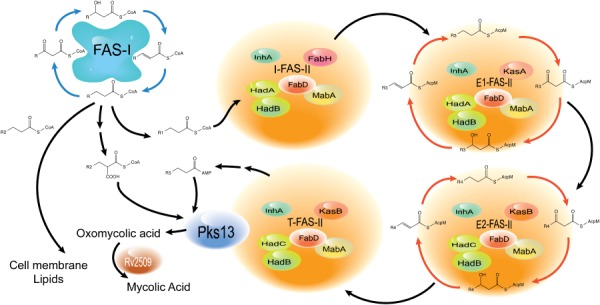
Schematic illustrating the biosynthesis pathway for mycolic acids in M . tuberculosis in the context of proposed specialised FAS‐II complexes. R1; C 16–C 18, R2; C 24–C 26, R3 represents a range of intermediate length meroacyl chains, R4 represents a range of longer meroacyl chains, R5 is the longest meroacyl chain. I‐FAS‐II; initiator FAS‐II, T‐FAS‐II; termination FAS‐II, and E1‐FAS‐II and E2‐FAS‐II represent elongation complexes.
Mycolic acid transporters
While there has been considerable progress in the past two decades towards identifying genes involved in the biosynthesis of mycolic acids, mechanisms for their transport to the outside of the mycobacterial cell remained unknown. Takayama et al. (2005) had hypothesised that mycolic acids were likely transported in the form of TMM. The M. tuberculosis H37Rv genome encodes a class of membrane proteins termed mycobacterial membrane proteins large (MmpL), which are part of the resistance‐nodulation‐division (RND) family of efflux pumps (Cole et al., 1998; Domenech et al., 2005). Many of the 14 mmpL genes of M. tuberculosis are located adjacent to biosynthesis clusters for cell wall‐associated glycolipids, and have been shown to be transporters for the cognate lipids (Cox et al., 1999; Converse et al., 2003; Domenech et al., 2004; Belardinelli et al., 2014). We reckoned that TMM, also a glycolipid, was likely transported by a MmpL, and given the essentiality of mycolic acids for viability, the gene encoding this MmpL would be essential. Previous, unsuccessful attempts to generate a mutant of the gene mmpL3 (Rv0205c) (Domenech et al., 2005), and its presence in the decayed M. leprae genome suggested that if an MmpL was involved in TMM transport, MmpL3 was the likely candidate for this function. Indeed we were able to demonstrate that mmpL3 is essential in mycobacteria, and that conditional depletion of MmpL3 in M. smegmatis led to the intracellular accumulation of TMM (Varela et al., 2012). In parallel to our studies, Grzegorzewicz et al (2012) and Tahlan et al. (2012) identified MmpL3 as a target for novel anti‐TB drug candidates and subsequently demonstrated its role in as a translocator of TMM across the mycobacterial membrane. Whether MmpL3 is sufficient for the transport of TMM remains to be determined. However, recent findings from C. glutamicum indicate that transport of mycolates may be a complex process. Yamaryo‐Botte et al. (2015) showed that transient acetylation of the mycolyl moiety of trehalose monocorynomycolate (TMCM) was necessary for its subsequent transport across the corynebacterial membrane. Homologues of the TMCM acetyltransferase are present in mycobacteria, suggesting a similar process may occur in mycobacterial TMM transport.
Located in the same cluster as mmpL3 is another mmpL gene, mmpL11, which was shown to be non‐essential (Domenech et al., 2005; Owens et al., 2013). However, a mutant of mmpL11 in M. smegmatis was deficient in the transport of the less abundant monomeromycolyl diacylglycerol and a mycolate containing wax ester, and showed accumulation of MycPL (a proposed intracellular carrier of mycolic acids), indicating a role for MmpL11 in transport processes related to mycolic acid containing lipids (Pacheco et al., 2013). Interestingly both MmpL3 and MmpL11 have also been shown to play a role in heme transport, suggesting that they may have multiple roles in M. tuberculosis (Owens et al., 2013).
Co‐ordination with polar growth
Mycobacteria, unlike other rod shaped bacteria such as Bacillus, exhibit polar growth where nascent peptidoglycan is synthesised and deposited at the poles during cell elongation (Kang et al., 2008). The tropomyosin‐like protein Wag31 (orthologue of Bacillus DivIVA) plays an important role in co‐ordinating polar growth; the protein localises to the tip of growing poles of mycobacterial cells and interacts with enzymes involved in cell wall biosynthesis (Kang et al., 2008; Jani et al., 2010). Two key papers have now shown that this interaction also involves components of the mycolate synthesising machinery. Carel et al. (2014) used GFP‐fusions of the FAS‐II components MabA, InhA and KasA to show that enzymes of the reductive cycle co‐localise with Wag31 at the ‘old’ growing poles. The authors also showed a fivefold enrichment of the mycolate transporter MmpL3 at the polar membranes. Meniche et al. (2014) took a different approach to identify additional components of the mycolic acid biosynthesis machinery that localise at the growing tips. Co‐purification experiments with endogenously tagged Wag31 and Pks13 in M. smegmatis revealed a polar complex consisting of AccA3, AccD4 and AccD5, members of an acyl‐CoA carboxylase (ACC) complex, Pks13 and FadD32, all enzymes required for the final stages of production of a nascent mycolic acid. The positions of the ACC components coincided with Wag31. However, Pks13 and FadD32 were found in a ‘subpolar’ location, suggesting that the Wag31‐associated complex and the terminal mycolate biosynthesis enzymes occupied exclusive regions of the polar tip, with new mycolic acids being deposited at a subpolar location in a growing tip.
Beyond the wall
Given the abundance of mycolic acids in the lipid‐rich mycobacterial cell wall, it is tempting to assume that their role is limited to the structural integrity of the cell envelope. However, we now have a better understanding of the extended role played by mycolic acids, particularly TDM, in manipulating the host immune system and driving pathology during infection (reviewed extensively by Marrakchi et al., 2014). Mycolic acids have also been shown to play a role in the formation of biofilms often referred to as pellicular growth. The first link between pellicle formation and mycolic acids came from studies on a mutant strain of M. smegmatis that was deficient in the production of a GroEL1, a non‐essential chaperone in mycobacteria. The GroEL1 mutant was unable to form a pellicle on a liquid–air interface in a laboratory broth medium and displayed an altered mycolic acid profile (Ojha et al., 2005). Furthermore, GroEL1 was shown to directly interact with the FAS‐II enzyme, KasA. Mycolic acid composition also affects pellicle formation; a strain deficient in keto‐mycolic acid formation was unable to form pellicles (Sambandan et al., 2013). Furthermore, the mmpL11 mutant of M. smegmatis that failed to transport monomeromycolyl diacylglycerol and a mycolate containing wax ester was defective in biofilm formation (Pacheco et al., 2013). Mycolates are not the only lipids to influence pellicle/biofilm formation; however, the enrichment of free mycolic acids in pellicles of mycobacteria showed that they play a significant role in the formation of this structure (Ojha et al., 2008).
Targeting the Achilles' heel: developing drugs that inhibit mycolate metabolism
Unique components of bacteria that are also essential represent ideal drug targets as they allow development of drugs that specifically inhibit the aetiological agent of disease, while leaving the normal flora intact. In this regard, M. tuberculosis mycolic acids fulfil both these criteria. Indeed, the hallmark anti‐TB drug targets InhA, the core FAS‐II component (Banerjee et al., 1994). However, in the past 15 years a number of other inhibitors targeting other mycolate biosynthesis components have been described (reviewed by Marrakchi et al., 2014). These include compounds that target enzymes involved in mycolic acid condensation (Pks13, FadD32), and those involved in the biosynthesis/elongation of the fatty acyl chain (AccD5, AccD6, KasA/KasB, InhA, HadA, HadB, HadC, Ag85). Surprisingly, some targets have also included ‘non‐essential’ components; the compound dioctylamine inhibited multiple methyltransferases involved in mycolic acid modification, resulting in a loss of all mycolic acid cyclopropanation and cell death (Barkan et al., 2009). This was surprising as individual genes involved in mycolic acid cyclopropanation were not essential for growth, leading to the authors to suggest that loss of viability was likely due to the deleterious effect of loss of cyclopropanation on membrane fluidity. Similarly, thiacetazone was also shown to inhibit cyclopropanation of mycolic acids (Alahari et al., 2009). However, more recently, the focus of anti‐TB drug discovery has been on the TMM transporter, MmpL3. This is because spontaneous mutants resistant to a diverse group of compounds with anti‐M. tuberculosis activity were found to contain mutations in the mmpL3 gene (Li et al., 2014). The first report was that of SQ109, a diamine, developed from the combinatorial library of ethambutol, which showed potent activity against drug‐sensitive and drug‐resistant mycobacteria (Protopopova et al., 2005). Subsequently, Tahlan et al. (2012) showed that SQ109 treatment led to TMM accumulation in M. tuberculosis and spontaneous SQ109‐resistant mutants contained single nucleotide polymorphisms (SNPs) in the mmpL3 gene. Shortly thereafter, there were two reports of drugs targeting M. tuberculosis, the pyrrole derivative BM212 and the adamantly urea compound AU1235 (Grzegorzewicz et al., 2012; La Rosa et al., 2012). Mutants resistant to either compound were shown to harbour a mutated mmpL3 allele. However, the divergent scaffolds from which the different inhibitors were derived, and their broad‐spectrum activity against non‐mycolate producing bacteria and fungal species, suggested that MmpL3 was probably not the direct target of these inhibitors. As mentioned earlier, MmpL3 and other mycobacterial MmpLs are members of the RND superfamily of proteins that require a proton motive force (PMF) for function. Li et al. (2014) have shown that SQ109 causes a dissipation of PMF in M. tuberculosis, thus disrupting MmpL3 function. Furthermore, exposure to SQ109 also led to reduction in lipid export by MmpL8 and MmpL10, indicating that effects of SQ109 were not MmpL3‐specific. This finding of a broader effect of SQ109 on multiple targets was at odds with the consistent isolation of resistant mutants with SNPs in mmpL3. The authors suggested that spontaneous mutants that are resistant to SQ109 (and other compounds) result from mutations in mmpL3 as an early response to counteract toxic effects of these compounds (Li et al., 2014). Indeed these mutations mapped close to those predicted to participate in the proton gradient required for translocation of substrate. In light of several other reports of SNPs in mmpL3 leading to resistance to diverse compounds, the idea that these are exclusively MmpL3 (and thus mycolic acid transport) targeting compounds needs to be revisited. The mycolic acid biosynthesis complex represents a unique target, one that is currently rendered unexploited in MDR‐TB and XDR‐TB strains (due to resistance to isoniazid). Further work on current known inhibitors of the enzymes mentioned earlier in this section will pave the way towards new inhibitors of mycolic acid biosynthesis.
Future perspectives
A majority of the enzymatic components required for mycolic acid biosynthesis have now been identified; however, there are still some stages in mycolate metabolism that remain poorly understood. These include the process of introducing double bonds in merochains, and the enzymes involved in post‐Pks13 processing of mycolic acids that ultimately leads to the formation of TMM. However, in parallel, our understanding of mycolic acid biosynthesis and transport in context of a higher order of structures has been initiated by in vitro studies to detect interactions between FAS‐II components and other enzymes involved in the biosynthesis process. With the advance of microscopy techniques that enable visualisation of complexes in single bacterial cells, it is likely that such complexes may one day be demonstrated in vivo. Such studies could also indicate whether biosynthesis occurs at localised complexes that also integrate transport. A better understanding is also required of the co‐ordination of this process with polar growth, and with peptidoglycan and arabinogalactan deposition in the cell wall during cell division. These studies could potentially also open alternative avenues for targeting mycolic acid assembly via the design of complex‐disrupting inhibitors. Furthermore, the regulation of mycolic acid biosynthesis has only just begun to be unravelled. Regulation of activities of FAS‐II enzymes following post translation modification by Ser/Thr protein kinase‐mediated phosphorylation has been studied extensively in vitro (Veyron‐Churlet et al., 2009; Khan et al., 2010; Molle and Kremer, 2010; Molle et al., 2010; Slama et al., 2011; Corrales et al., 2012), and advances in ‘knock in’ methodologies have initiated studies to conduct the same in vivo (Vilcheze et al., 2014). While the transcriptional regulator MabR is known to regulate mycolic acid biosynthesis in vitro (Salzman et al., 2010), it is not known if transcription of merochain modification enzymes is regulated in vivo, resulting in changes to ratios of mycolic acid subclasses. In summary, this waxy ‘brick’ in the mycobacterial cell wall continues to be a focus of research on the tubercle bacillus, as we attempt to develop new and better therapies for this ancient disease.
Acknowledgements
A.B. acknowledges funding from the Medical Research Council (UK), the Medical Research Foundation and the Royal Society, particularly for their support for projects and equipment related to research on mycolic acids. C.V. and V.N. were supported by PhD studentships from the Darwin Trust of Edinburgh.
References
- Alahari, A. , Alibaud, L. , Trivelli, X. , Gupta, R. , Lamichhane, G. , Reynolds, R.C. , et al (2009) Mycolic acid methyltransferase, MmaA4, is necessary for thiacetazone susceptibility in Mycobacterium tuberculosis . Mol Microbiol 71: 1263–1277. [DOI] [PubMed] [Google Scholar]
- Banerjee, A. , Dubnau, E. , Quemard, A. , Balasubramanian, V. , Um, K.S. , Wilson, T. , et al (1994) inhA, a gene encoding a target for isoniazid and ethionamide in Mycobacterium tuberculosis . Science 263: 227–230. [DOI] [PubMed] [Google Scholar]
- Barkan, D. , Liu, Z. , Sacchettini, J.C. , and Glickman, M.S. (2009) Mycolic acid cyclopropanation is essential for viability, drug resistance, and cell wall integrity of Mycobacterium tuberculosis . Chem Biol 16: 499–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry, C.E., 3rd , Lee, R.E. , Mdluli, K. , Sampson, A.E. , Schroeder, B.G. , Slayden, R.A. , et al (1998) Mycolic acids: structure, biosynthesis and physiological functions. Prog Lipid Res 37: 143–179. [DOI] [PubMed] [Google Scholar]
- Belardinelli, J.M. , Larrouy‐Maumus, G. , Jones, V. , Sorio de Carvalho, L.P. , McNeil, M.R. , and Jackson, M. (2014) Biosynthesis and translocation of unsulfated acyltrehaloses in Mycobacterium tuberculosis . J Biol Chem 289: 27952–27965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belisle, J.T. , Vissa, V.D. , Sievert, T. , Takayama, K. , Brennan, P.J. , and Besra, G.S. (1997) Role of the major antigen of Mycobacterium tuberculosis in cell wall biogenesis. Science 276: 1420–1422. [DOI] [PubMed] [Google Scholar]
- Bhatt, A. , Kremer, L. , Dai, A.Z. , Sacchettini, J.C. , and Jacobs, W.R., Jr (2005) Conditional depletion of KasA, a key enzyme of mycolic acid biosynthesis, leads to mycobacterial cell lysis. J Bact 187: 7596–7606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt, A. , Fujiwara, N. , Bhatt, K. , Gurcha, S.S. , Kremer, L. , Chen, B. , et al (2007a) Deletion of kasB in Mycobacterium tuberculosis causes loss of acid‐fastness and subclinical latent tuberculosis in immunocompetent mice. Proc Natl Acad Sci 104: 5157–5162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt, A. , Molle, V. , Besra, G.S. , Jacobs, W.R., Jr , and Kremer, L. (2007b) The Mycobacterium tuberculosis FAS‐II condensing enzymes: their role in mycolic acid biosynthesis, acid‐fastness, pathogenesis and in future drug development. Mol Microbiol 64: 1442–1454. [DOI] [PubMed] [Google Scholar]
- Bhatt, A. , Brown, A.K. , Singh, A. , Minnikin, D.E. , and Besra, G.S. (2008) Loss of a mycobacterial gene encoding a reductase leads to an altered cell wall containing beta‐oxo‐mycolic acid analogs and accumulation of ketones. Chem Biol 15: 930–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas, R. , Dutta, A. , Dutta, D. , Hazra, D. , Banerjee, D.R. , Basak, A. , et al (2015) Crystal structure of dehydratase component HadAB complex of mycobacterial FAS‐II pathway. Biochem Biophy Res Commun 458: 369–374. [DOI] [PubMed] [Google Scholar]
- Bloch, K. (1977) Control mechanisms for fatty acid synthesis in Mycobacterium smegmatis . Adv Enzymol Relat Areas Mol Biol 45: 1–84. [DOI] [PubMed] [Google Scholar]
- Brown, A.K. , Bhatt, A. , Singh, A. , Saparia, E. , Evans, A.F. , and Besra, G.S. (2007a) Identification of the dehydratase component of the mycobacterial mycolic acid‐synthesizing fatty acid synthase‐II complex. Microbiol 153: 4166–4173. [DOI] [PubMed] [Google Scholar]
- Brown, A.K. , Papaemmanouil, A. , Bhowruth, V. , Bhatt, A. , Dover, L.G. , and Besra, G.S. (2007b) Flavonoid inhibitors as novel antimycobacterial agents targeting Rv0636, a putative dehydratase enzyme involved in Mycobacterium tuberculosis fatty acid synthase II. Microbiol 153: 3314–3322. [DOI] [PubMed] [Google Scholar]
- Cantaloube, S. , Veyron‐Churlet, R. , Haddache, N. , Daffe, M. , and Zerbib, D. (2011) The Mycobacterium tuberculosis FAS‐II dehydratases and methyltransferases define the specificity of the mycolic acid elongation complexes. PLoS ONE 6: e29564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carel, C. , Nukdee, K. , Cantaloube, S. , Bonne, M. , Diagne, C.T. , Laval, F. , et al (2014) Mycobacterium tuberculosis proteins involved in mycolic acid synthesis and transport localize dynamically to the old growing pole and septum. PLoS ONE 9: e97148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrère‐Kremer, S. , Blaise, M. , Singh, V.K. , Alibaud, L. , Tuaillon, E. , Halloum, I. , et al (2015) A new dehydratase conferring innate resistance to thiacetazone and intra‐amoebal survival of Mycobacterium smegmatis . Mol Microbiol 96: 1085–1102. [DOI] [PubMed] [Google Scholar]
- Castell, A. , Johansson, P. , Unge, T. , Jones, T.A. , and Backbro, K. (2005) Rv0216, a conserved hypothetical protein from Mycobacterium tuberculosis that is essential for bacterial survival during infection, has a double hotdog fold. Prot Sci 14: 1850–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, K.H. , Kremer, L. , Besra, G.S. , and Rock, C.O. (2000) Identification and substrate specificity of beta ‐ketoacyl (acyl carrier protein) synthase III (mtFabH) from Mycobacterium tuberculosis . J Biol Chem 275: 28201–28207. [DOI] [PubMed] [Google Scholar]
- Cole, S.T. , Brosch, R. , Parkhill, J. , Garnier, T. , Churcher, C. , Harris, D. , et al (1998) Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393: 537–544. [DOI] [PubMed] [Google Scholar]
- Converse, S.E. , Mougous, J.D. , Leavell, M.D. , Leary, J.A. , Bertozzi, C.R. , and Cox, J.S. (2003) MmpL8 is required for sulfolipid‐1 biosynthesis and Mycobacterium tuberculosis virulence. Proc Natl Acad Sci 100: 6121–6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrales, R.M. , Molle, V. , Leiba, J. , Mourey, L. , de Chastellier, C. , and Kremer, L. (2012) Phosphorylation of mycobacterial PcaA inhibits mycolic acid cyclopropanation: consequences for intracellular survival and for phagosome maturation block. J Biol Chem 287: 26187–26199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox, J.S. , Chen, B. , McNeil, M. , and Jacobs, W.R., Jr (1999) Complex lipid determines tissue‐specific replication of Mycobacterium tuberculosis in mice. Nature 402: 79–83. [DOI] [PubMed] [Google Scholar]
- Daffe, M. , and Draper, P. (1998) The envelope layers of mycobacteria with reference to their pathogenicity. Adv Microbial Physiol 39: 131–203. [DOI] [PubMed] [Google Scholar]
- Daniel, J. , Oh, T.J. , Lee, C.M. , and Kolattukudy, P.E. (2007) AccD6, a member of the Fas II locus, is a functional carboxyltransferase subunit of the acyl‐coenzyme A carboxylase in Mycobacterium tuberculosis . J Bacteriol 189: 911–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domenech, P. , Reed, M.B. , Dowd, C.S. , Manca, C. , Kaplan, G. , and Barry, C.E., 3rd (2004) The role of MmpL8 in sulfatide biogenesis and virulence of Mycobacterium tuberculosis . J Biol Chem 279: 21257–21265. [DOI] [PubMed] [Google Scholar]
- Domenech, P. , Reed, M.B. , and Barry, C.E., 3rd (2005) Contribution of the Mycobacterium tuberculosis MmpL protein family to virulence and drug resistance. Infect Immun 73: 3492–3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnau, E. , Chan, J. , Raynaud, C. , Mohan, V.P. , Laneelle, M.A. , Yu, K. , et al (2000) Oxygenated mycolic acids are necessary for virulence of Mycobacterium tuberculosis in mice. Mol Microbiol 36: 630–637. [DOI] [PubMed] [Google Scholar]
- Gande, R. , Gibson, K.J. , Brown, A.K. , Krumbach, K. , Dover, L.G. , Sahm, H. , et al (2004) Acyl‐CoA carboxylases (accD2 and accD3), together with a unique polyketide synthase (Cg‐pks), are key to mycolic acid biosynthesis in Corynebacterianeae such as Corynebacterium glutamicum and Mycobacterium tuberculosis . J Biol Chem 279: 44847–44857. [DOI] [PubMed] [Google Scholar]
- Glickman, M.S. , Cox, J.S. , and Jacobs, W.R., Jr (2000) A novel mycolic acid cyclopropane synthetase is required for cording, persistence, and virulence of Mycobacterium tuberculosis . Mol Cell 5: 717–727. [DOI] [PubMed] [Google Scholar]
- Grzegorzewicz, A.E. , Pham, H. , Gundi, V.A. , Scherman, M.S. , North, E.J. , Hess, T. , et al (2012) Inhibition of mycolic acid transport across the Mycobacterium tuberculosis plasma membrane. Nature Chem Biol 8: 334–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jani, C. , Eoh, H. , Lee, J.J. , Hamasha, K. , Sahana, M.B. , Han, J.S. , et al (2010) Regulation of polar peptidoglycan biosynthesis by Wag31 phosphorylation in mycobacteria. BMC Microbiol 10: 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, C.M. , Nyayapathy, S. , Lee, J.Y. , Suh, J.W. , and Husson, R.N. (2008) Wag31, a homologue of the cell division protein DivIVA, regulates growth, morphology and polar cell wall synthesis in mycobacteria. Microbiol 154: 725–735. [DOI] [PubMed] [Google Scholar]
- Khan, S. , Nagarajan, S.N. , Parikh, A. , Samantaray, S. , Singh, A. , Kumar, D. , et al (2010) Phosphorylation of enoyl‐acyl carrier protein reductase InhA impacts mycobacterial growth and survival. J Biol Chem 285: 37860–37871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer, L. , Nampoothiri, K.M. , Lesjean, S. , Dover, L.G. , Graham, S. , Betts, J. , et al (2001) Biochemical characterization of acyl carrier protein (AcpM) and malonyl‐CoA:AcpM transacylase (mtFabD), two major components of Mycobacterium tuberculosis fatty acid synthase II. J Biol 276: 27967–27974. [DOI] [PubMed] [Google Scholar]
- Kremer, L. , Dover, L.G. , Carrere, S. , Nampoothiri, K.M. , Lesjean, S. , Brown, A.K. , et al (2002) Mycolic acid biosynthesis and enzymic characterization of the beta‐ketoacyl‐ACP synthase A‐condensing enzyme from Mycobacterium tuberculosis . Biochem J 364: 423–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa, V. , Poce, G. , Canseco, J.O. , Buroni, S. , Pasca, M.R. , Biava, M. , et al (2012) MmpL3 is the cellular target of the antitubercular pyrrole derivative BM212. Antimicrob Agents Chemother 56: 324–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lea‐Smith, D.J. , Pyke, J.S. , Tull, D. , McConville, M.J. , Coppel, R.L. , and Crellin, P.K. (2007) The reductase that catalyzes mycolic motif synthesis is required for efficient attachment of mycolic acids to arabinogalactan. J Biol Chem 282: 11000–11008. [DOI] [PubMed] [Google Scholar]
- Li, W. , Upadhyay, A. , Fontes, F.L. , North, E.J. , Wang, Y. , Crans, D.C. , et al (2014) Novel insights into the mechanism of inhibition of MmpL3, a target of multiple pharmacophores in Mycobacterium tuberculosis . Antimicrob Agents Chemother 58: 6413–6423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrakchi, H. , Laneelle, M.A. , and Daffe, M. (2014) Mycolic acids: structures, biosynthesis, and beyond. Chem Biol 21: 67–85. [DOI] [PubMed] [Google Scholar]
- Meniche, X. , Otten, R. , Siegrist, M.S. , Baer, C.E. , Murphy, K.C. , Bertozzi, C.R. , et al (2014) Subpolar addition of new cell wall is directed by DivIVA in mycobacteria. Proc Natl Acad Sci 111: E3243–E3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minnikin, D.E. , and Polgar, N. (1966) Studies on the mycolic acids from human tubercle bacilli. Tetrahedron Lett 23: 2643–2647. [DOI] [PubMed] [Google Scholar]
- Molle, V. , and Kremer, L. (2010) Division and cell envelope regulation by Ser/Thr phosphorylation: mycobacterium shows the way. Mol Microbiol 75: 1064–1077. [DOI] [PubMed] [Google Scholar]
- Molle, V. , Gulten, G. , Vilcheze, C. , Veyron‐Churlet, R. , Zanella‐Cleon, I. , Sacchettini, J.C. , et al (2010) Phosphorylation of InhA inhibits mycolic acid biosynthesis and growth of Mycobacterium tuberculosis . Mol Microbiol 78: 1591–1605. [DOI] [PubMed] [Google Scholar]
- Monot, M. , Honore, N. , Garnier, T. , Araoz, R. , Coppee, J.Y. , Lacroix, C. , et al (2005) On the origin of leprosy. Science 308: 1040–1042. [DOI] [PubMed] [Google Scholar]
- Ojha, A. , Anand, M. , Bhatt, A. , Kremer, L. , Jacobs, W.R., Jr , and Hatfull, G.F. (2005) GroEL1: a dedicated chaperone involved in mycolic acid biosynthesis during biofilm formation in mycobacteria. Cell 123: 861–873. [DOI] [PubMed] [Google Scholar]
- Ojha, A.K. , Baughn, A.D. , Sambandan, D. , Hsu, T. , Trivelli, X. , Guerardel, Y. , et al (2008) Growth of Mycobacterium tuberculosis biofilms containing free mycolic acids and harbouring drug‐tolerant bacteria. Mol Microbiol 69: 164–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens, C.P. , Chim, N. , Graves, A.B. , Harmston, C.A. , Iniguez, A. , Contreras, H. , et al (2013) The Mycobacterium tuberculosis secreted protein Rv0203 transfers heme to membrane proteins MmpL3 and MmpL11. J Biol Chem 288: 21714–21728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacheco, S.A. , Hsu, F.F. , Powers, K.M. , and Purdy, G.E. (2013) MmpL11 protein transports mycolic acid‐containing lipids to the mycobacterial cell wall and contributes to biofilm formation in Mycobacterium smegmatis . J Biol Chem 288: 24213–24222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portevin, D. , De Sousa‐D'Auria, C. , Houssin, C. , Grimaldi, C. , Chami, M. , Daffe, M. , et al (2004) A polyketide synthase catalyzes the last condensation step of mycolic acid biosynthesis in mycobacteria and related organisms. Proc Natl Acad Sci 101: 314–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protopopova, M. , Hanrahan, C. , Nikonenko, B. , Samala, R. , Chen, P. , Gearhart, J. , et al (2005) Identification of a new antitubercular drug candidate, SQ109, from a combinatorial library of 1,2‐ethylenediamines. J Antimicrob Chemother 56: 968–974. [DOI] [PubMed] [Google Scholar]
- Puech, V. , Guilhot, C. , Perez, E. , Tropis, M. , Armitige, L.Y. , Gicquel, B. , et al (2002) Evidence for a partial redundancy of the fibronectin‐binding proteins for the transfer of mycoloyl residues onto the cell wall arabinogalactan termini of Mycobacterium tuberculosis . Mol Microbiol 44: 1109–1122. [DOI] [PubMed] [Google Scholar]
- Rao, V. , Fujiwara, N. , Porcelli, S.A. , and Glickman, M.S. (2005) Mycobacterium tuberculosis controls host innate immune activation through cyclopropane modification of a glycolipid effector molecule. J Exp Med 201: 535–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao, V. , Gao, F. , Chen, B. , Jacobs, W.R., Jr , and Glickman, M.S. (2006) Trans‐cyclopropanation of mycolic acids on trehalose dimycolate suppresses Mycobacterium tuberculosis ‐induced inflammation and virulence. J Clin Inv 116: 1660–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock, C.O. , and Cronan, J.E. (1996) Escherichia coli as a model for the regulation of dissociable (type II) fatty acid biosynthesis. Biochim Biophys Acta 1302: 1–16. [DOI] [PubMed] [Google Scholar]
- Sacco, E. , Covarrubias, A.S. , O'Hare, H.M. , Carroll, P. , Eynard, N. , Jones, T.A. , et al (2007) The missing piece of the type II fatty acid synthase system from Mycobacterium tuberculosis . Proc Natl Acad Sci 104: 14628–14633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzman, V. , Mondino, S. , Sala, C. , Cole, S.T. , Gago, G. , and Gramajo, H. (2010) Transcriptional regulation of lipid homeostasis in mycobacteria. Mol Microbiol 78: 64–77. [DOI] [PubMed] [Google Scholar]
- Sambandan, D. , Dao, D.N. , Weinrick, B.C. , Vilcheze, C. , Gurcha, S.S. , Ojha, A. , et al (2013) Keto‐mycolic acid‐dependent pellicle formation confers tolerance to drug‐sensitive Mycobacterium tuberculosis . MBio 4: e00222‐13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slama, N. , Leiba, J. , Eynard, N. , Daffe, M. , Kremer, L. , Quemard, A. , et al (2011) Negative regulation by Ser/Thr phosphorylation of HadAB and HadBC dehydratases from Mycobacterium tuberculosis type II fatty acid synthase system. Biochem Biophys Res Commun 412: 401–406. [DOI] [PubMed] [Google Scholar]
- Tahlan, K. , Wilson, R. , Kastrinsky, D.B. , Arora, K. , Nair, V. , Fischer, E. , et al (2012) SQ109 targets MmpL3, a membrane transporter of trehalose monomycolate involved in mycolic acid donation to the cell wall core of Mycobacterium tuberculosis . Antimicrob Agents Chemother 56: 1797–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama, K. , Wang, C. , and Besra, G.S. (2005) Pathway to synthesis and processing of mycolic acids in Mycobacterium tuberculosis . Clin Microbiol Rev 18: 81–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udwadia, Z.F. , Amale, R.A. , Ajbani, K.K. , and Rodrigues, C. (2012) Totally drug‐resistant tuberculosis in India. Clin Infect Dis 54: 579–581. [DOI] [PubMed] [Google Scholar]
- Varela, C. , Rittmann, D. , Singh, A. , Krumbach, K. , Bhatt, K. , Eggeling, L. , et al (2012) MmpL genes are associated with mycolic acid metabolism in mycobacteria and corynebacteria. Chem Biol 19: 498–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veyron‐Churlet, R. , Guerrini, O. , Mourey, L. , Daffe, M. , and Zerbib, D. (2004) Protein‐protein interactions within the Fatty Acid Synthase‐II system of Mycobacterium tuberculosis are essential for mycobacterial viability. Mol Microbiol 54: 1161–1172. [DOI] [PubMed] [Google Scholar]
- Veyron‐Churlet, R. , Bigot, S. , Guerrini, O. , Verdoux, S. , Malaga, W. , Daffe, M. , et al (2005) The biosynthesis of mycolic acids in Mycobacterium tuberculosis relies on multiple specialized elongation complexes interconnected by specific protein‐protein interactions. J Mol Biol 353: 847–858. [DOI] [PubMed] [Google Scholar]
- Veyron‐Churlet, R. , Molle, V. , Taylor, R.C. , Brown, A.K. , Besra, G.S. , Zanella‐Cleon, I. , et al (2009) The Mycobacterium tuberculosis beta‐ketoacyl‐acyl carrier protein synthase III activity is inhibited by phosphorylation on a single threonine residue. J Biol Chem 284: 6414–6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilcheze, C. , Molle, V. , Carrère‐Kremer, S. , Leiba, J. , Mourey, L. , Shenai, S. , et al (2014) Phosphorylation of KasB regulates virulence and acid‐fastness in Mycobacterium tuberculosis . PLoS Pathog 10: e1004115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaryo‐Botte, Y. , Rainczuk, A.K. , Lea‐Smith, D.J. , Brammananth, R. , van der Peet, P.L. , Meikle, P. , et al (2015) Acetylation of trehalose mycolates is required for efficient MmpL‐mediated membrane transport in corynebacterineae. ACS Chem Biol 10: 734–746. [DOI] [PubMed] [Google Scholar]


