Abstract
Pyramiding of alien-derived Wheat streak mosaic virus (WSMV) resistance and resistance enhancing genes in wheat is a cost-effective and environmentally safe strategy for disease control. PCR-based markers and cytogenetic analysis with genomic in situ hybridisation were applied to identify alien chromatin in four genetically diverse populations of wheat (Triticum aestivum) lines incorporating chromosome segments from Thinopyrum intermedium and Secale cereale (rye). Out of 20 experimental lines, 10 carried Th. intermedium chromatin as T4DL*4Ai#2S translocations, while, unexpectedly, 7 lines were positive for alien chromatin (Th. intermedium or rye) on chromosome 1B. The newly described rye 1RS chromatin, transmitted from early in the pedigree, was associated with enhanced WSMV resistance. Under field conditions, the 1RS chromatin alone showed some resistance, while together with the Th. intermedium 4Ai#2S offered superior resistance to that demonstrated by the known resistant cultivar Mace. Most alien wheat lines carry whole chromosome arms, and it is notable that these lines showed intra-arm recombination within the 1BS arm. The translocation breakpoints between 1BS and alien chromatin fell in three categories: (i) at or near to the centromere, (ii) intercalary between markers UL-Thin5 and Xgwm1130 and (iii) towards the telomere between Xgwm0911 and Xbarc194. Labelled genomic Th. intermedium DNA hybridised to the rye 1RS chromatin under high stringency conditions, indicating the presence of shared tandem repeats among the cereals. The novel small alien fragments may explain the difficulty in developing well-adapted lines carrying Wsm1 despite improved tolerance to the virus. The results will facilitate directed chromosome engineering producing agronomically desirable WSMV-resistant germplasm.
Introduction
Wheat streak mosaic virus (WSMV), transmitted by the wheat curl mite (WCM) Aceria tosichella Kiefer, is an important yield-limiting disease of wheat (Triticum aestivum, 2n=6x=42, AABBDD) (Graybosch et al., 2009). First recorded in the United States, it has since spread widely, infecting both winter and spring wheat cultivars. Infected plants show greenish yellow streaks and chlorosis with stunted growth, low root biomass, reduced water uptake efficiency and low yield (Thomas et al., 2004; Price et al., 2010). Both WSMV and the mite vector survive during the late summer on 'green bridges' provided by volunteer wheat and other susceptible wild and cultivated grasses (Divis et al., 2006). As viral diseases cannot be controlled directly by agrochemicals, management of insect vectors and agronomic conditions must be used. Some transgenic wheat lines have been developed with WSMV resistance in controlled environments (Fahim et al., 2010), but have not been tested in the field.
Genetic resistance to WSMV offers the most environmentally and economically desirable strategy for disease control and has been pursued since resistance to WSMV was discovered (Friebe et al., 1991; Graybosch et al., 2009; Mutti et al., 2011). Today, there is huge interest in both using genomic prediction to access genotype × environment interaction using marker and pedigree information in breeding programmes (Crossa et al., 2014), and in enhancement of the genetic variability available in wheat through wide hybridisation (see Heslop-Harrison and Schwarzacher, 2012). Among the natural sources, the gene pool of rye Secale cereale (2n=14, genomes RR) and the perennial wheat grasses, Thinopyrum intermedium (Host) Barkworth and Dewey syn. Agropyron intermedium (Host) P. Beauv. (2n=6x=42, JJJsJsSS) and Th. ponticum (Podp.) Barkworth and D.R. Dewey (2n=10x=70, JJJJJJJsJsJsJs) are of particular interest (Kim et al., 2004; Li and Wang, 2009). These sources provide large reservoirs of useful genes including WCM and WSMV resistance, and chromosomes or chromosome segments from Thinopyrum have been transferred into wheat backgrounds (Li et al., 2007; Graybosch et al., 2009). Although rye is susceptible to WSMV, there is evidence that rye genes in wheat background can delay the spread of the disease (Li et al., 2007) through resistance to either the vector or the virus itself. Many wheat lines carry a T1BL*1RS translocation, with the 1RS rye chromosome arm being reported to confer a range of biotic and abiotic stress resistances (Heslop-Harrison et al., 1990; Kim et al., 2004). Two genes for WSMV resistance, Wsm1 and Wsm2, have been used in wheat improvement (Graybosch et al., 2009; Haley et al., 2011). Wsm1 is present on the short arm of Th. intermedium chromosome 4Ai#2, and offers effective field resistance to WSMV in wheat (Chen et al., 2003; Friebe et al., 2009). The Wsm2 gene (of unknown origin, but perhaps from bread wheat itself) was mapped to the short arm of wheat chromosome 3B (Lu et al., 2011). A third Th. intermedium-derived gene, Wsm3, was mapped to chromosome T7BS*7S#3L (Liu et al., 2011), but it is yet to be exploited commercially.
The Wsm1, Wsm2 and Wsm3 resistances show temperature dependency (Liu et al., 2011; Seifers et al., 2013). Lines with Wsm1 gene from 4Ai#2S are resistant at 20 °C and delay symptoms of the disease up to 25 °C but are susceptible at 28 °C (Fahim et al., 2012). Wsm2 resistance was originally ineffective above 18 °C, but exposure to virus over several generations resulted in de novo resistance up to of 28 °C (Fahim et al., 2012; Seifers et al., 2013), whereas the Wsm3 derivatives displayed effective resistance at a temperature of 24 °C (Liu et al., 2011). Wheat-Th. intermedium substitution lines that carried entire chromosomes 4Ai#2 were resistant at 27 °C, suggesting the presence of further resistance genes on Th. intermedium 4Ai#2L (Fahim et al., 2012). Wsm1 might also be effective against other viruses, as the winter wheat cultivar ‘Mace' (Graybosch et al., 2009) with Wsm1 gene resists the co-infection of WSMV and the related Triticum mosaic virus up to 19 °C, indicating the effectiveness of Wsm1 selection in disease synergism (Liu et al., 2011).
Transfer of desirable genes and gene combinations into varieties with durable and non-race-specific resistance constitute core objectives of modern plant breeding (Tester and Langridge, 2010; Heslop-Harrison and Schwarzacher, 2012). The successful transfer of 4Ai#2S of Th. intermedium in the form of the T4DL*4Ai#2S translocation represents the most widely exploited source of WSMV resistance (Wells et al., 1982; Friebe et al., 2009). The present study used genomic in situ hybridisation (GISH; Schwarzacher et al., 1992) complemented by targeted DNA markers and field evaluation to characterize resistance in Nebraska-adapted winter wheat lines originating from four genetically diverse populations that include the Kansas WSMW-resistant Th. intermedium lines in their pedigree, and where resistant and susceptible sister lines are available within populations (Divis et al., 2006).
Materials and methods
Plant material
Table 1 lists the wheat lines used in this study and pedigrees where known. Reference WSMV-resistant (R) lines KS95H102 and KS96HW10-1 both carrying the Wsm1 gene (Divis et al., 2006; Graybosch et al., 2009) and susceptible (S) lines ‘Millennium' and ‘Tomahawk' were used as controls. Experimental lines previously classified as R or S to WSMV (Divis et al., 2006) were derived from four breeding populations, designated here as populations I, II, III and IV (Table 1). Mace (PI 651043; Graybosch et al., 2009) was derived from population III and was tested as N02Y5117. PCR marker screening involved selected experimental lines (Table 1) as well as Th. intermedium (cultivars Manaska, Beefmaker, Haymaker, Reliant), T. aestivum land race ‘Chinese Spring' wheat, ‘Beaver' wheat (T1BL*1RS wheat-rye translocation), S. cereale ‘Petkus' rye, aneuploids Chinese Spring wheat nullisomic-1B-tetrasomic-1A (CS N1B-T1A), nullisomic-4A-tetrasomic-4D (CS N4A-T4D) and nullisomic-4D-tetrasomic-4B (CS N4D-T4B) and the pedigree lines KS91H184, KS91H174, RioBlanco, MO8 Vista, Redland Tam107, Anton (see Supplementary Table S3). DNA was extracted using standard CTAB methods.
Table 1. Pedigrees of reference lines and four populations of derived lines (from C. James Peterson, USDA-ARS) studied here with results of WSMV resistance field tests at different locations and years and summary of FISH/GISH results obtained with genomic Th. intermedium, rye and repetitive DNA probes.
| Reference/ population | Pedigree/source | Line/variety |
Wheat chromosome with alien segment |
Field response to WSMV
a |
|||||
|---|---|---|---|---|---|---|---|---|---|
| 4D | 1B | ?D |
2002b |
2012, Mead, NE |
|||||
| Sidney, NE | Hays, KS | SPAD c Mean (std) | Visual Score | ||||||
| KS102d | KS91H184/KS89H20//TAM107 | KS95H102 | + | − | R | R | |||
| KS10-1d | KS91HW29//RioBlanco/KS91H184 | KS96HW10-1 | + | − | R | R | |||
| Milleniumd | Arapahoe/Abilene//NE86488 | Millennium | − | − | S | S | |||
| Tomahawke | Agripro, WI88-083 | Tomahawk | 14.5 d (2.1) | S | |||||
| Population I | CO850034//T-57/5 *TAM107/3/(KS91H184/ RioBlanco/KS91HW29//VISTA) | N02Y5018 | + | BPI | R | R | |||
| N02Y5019 | − | BPI | S | S | 23.5 c (2.3) | MS | |||
| N02Y5021 | − | − | S | S | |||||
| N02Y5025 | + | − | R | R | |||||
| N02Y5003 | − | BPIII | R | R | 24.0 c (2.5) | MR | |||
| Population II | Yuma//T-57/3/LAMAR/4/4 *Yuma/5/(KS91H184/ArlinS/ KS91HW29//NE89526) | N02Y5057 | + | − | R | R | |||
| N02Y5075 | + | − | R | R | |||||
| N02Y5078 | + | − | R | R | |||||
| N02Y5082 | − | − | S | S | |||||
| N02Y5096 | − | − | S | S | |||||
| Population III | Yuma//T-57 /3/CO850034/4/4 *Yuma/5/(KS91H184/ArlinS/ KS91HW29)//NE89526) | N02Y5105 | − | − | S | MR | |||
| N02Y5106 | + | − | R | MR | |||||
| N02Y5109 | − | − | + | R | R | 15.4 d (1.7) | S | ||
| N02Y5117 (Mace) | + | − | R | R | 28.5 b (6.9) | R | |||
| N02Y5121 | − | − | S | S | |||||
| Population IV | MO8/Redland//KS91H184/3 *RioBlanco | N02Y5149 | + | BPIII | R | R | 36.3 a (3.5) | R | |
| N02Y5154 | + | − | R | R | |||||
| N02Y5156 | − | BPIII | S | S | |||||
| N02Y5163 | − | BPII | S | S | |||||
| N02Y2016 | + | BPI | R | R | |||||
Abbreviation: WSMV, Wheat streak mosaic virus. Recombinant chromosomes were disomic in all cases and included the T4DL*4Ai#2S Th. intermedium translocation (4D; see Figure 3a); an alien fragment on the short arm of 1B of classified into three breakpoint (BP) types by FISH and molecular markers (see Table 2 and Figures 3b and 4): BPI very small, BPII small and BPIII whole arm-alien fragment; and a small alien fragment on an unknown D-genome chromosome (?D; see Supplementary Figure S1).
Other Sources: Wells et al., 1982; Divis et al., 2006; Graybosch et al., 2009 and personal communication.
R=resistant or no symptoms, MR=moderately resistant or slight symptom severity, MS=moderately susceptible or moderate symptom severity, S=susceptible or severe. In terms of plant phenotype, a score of 1=no loss of chlorophyll, and no stunting; 2=random and occasional yellow foci or streaks, 3=50% loss of chlorophyll and 4=leaves complete yellow and plants severely stunted.
From Divis et al., 2006. Scores are visual ratings, based on degree of streaking/chlorosis and stunting.
Soil Plant Analytical Development metre readings for leaf chlorophyll content; Higher SPAD readings indicate higher intensity of greenness of leaf tissue (no or mild symptoms), while lower SPAD readings indicate increasing severity of virus symptoms. Means followed by the same letter were not significantly different at P=0.05.
Reference lines previously known for presence/absence of Th. intermedium chromatin and validated here.
Tomahawk was used as reference susceptible to WSMV in the 2012 trial only; it was selected from a bulk population of crosses of adapted parents.
Virus screening in greenhouse and field
Lines were originally scored in 2002 for response to natural infection by WSMV at Hays, KS USA and Sidney, NE, USA (Divis et al., 2006). Visual subjective assessment of the degree of chlorosis and plant stunting were used (Table 1 and footnotes). Selected lines were further evaluated in the field at the University of Nebraska Agricultural Research and Development Center, Mead, NE, USA. In 2011, individual spikes from each line were harvested and used to seedat least 20, 1.2-m replicate rows of single plant-derived lines in September 2011 as a block surrounded by early planted wheat that served as a ‘green bridge' for the development of WCM populations. Multiple plantings of the resistant cultivar Mace and the susceptible line Tomahawk were included as a control. Through the fall, WCM populations migrated from the early planted wheat to the experimental plants, providing a natural source of WSMV inoculation. The level of infestation was higher in 2012 than in 2002. Response to WSMV was rated using the visual scores (Table 1). In addition, at the flag leaf stage, chlorophyll content was assessed using a Soil Plant Analytical Development (SPAD) metre (model 502 Plus, Konica Minolta Sensing, Inc., Osaka, Japan). For each treatment, 10 SPAD readings were taken and averaged. Analysis of variance (Proc GLM, Version 9.4, SAS, Cary, NC, USA) followed by mean separation using Duncan's multiple range test was used to compare mean SPAD readings of each experimental line to each other, and to Mace and Tomahawk; significant and not significant differences at P=0.05 are indicated in Table 1.
Fluorescent in situ hybridisation (FISH)
Spread preparations of chromosomes were made from both seedling root tips or anthers from all 20 breeding lines and control Th. intermedium, normal wheats, and wheats containing the T1BL*1RS translocation using proteolytic digestions (see Schwarzacher and Heslop-Harrison, 2000). Probes (described in the studies by Forsström et al., 2002; Contento et al., 2005; Patokar et al., 2015) included the rDNA sequences pTa71 (9 kb complete repeat unit of 25S-5.8S-18S rDNA of T. aestivum) and pTa794 (410 bp fragment of 5S rDNA of T. aestivum), and the repetitive DNA sequences pSc119.2 (or CS13, 120 bp tandem repeat isolated from S. cereale) and dpTa1 (or Afa, 340 bp tandem repeat from T. aestivum). Small insert clones were amplified by PCR using M13 primers. Total genomic DNA from Th. intermedium, S. cereale and Aegilops (syn. Triticum) tauschii was sheared to 3–5 kb pieces by autoclaving. For labelling, biotin-16-dUTP and digoxigenin-11-dUTP (Roche Diagnostics, Basel, Switzerland) were incorporated in separate reactions using BioPrime Array CGH Genomic Labeling System (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer's instructions.
In situ hybridisation followed the technique of Schwarzacher and Heslop-Harrison (2000) with small modifications. A total of 40 μl probe mixture was applied per slide, containing 50% (v/v) formamide, 20% (w/v) dextran sulphate, 2 × saline sodium citrate (0.3 M NaCl, 0.03 M sodium citrate), 25–100 ng probe, 0.025 μg of salmon sperm DNA and 0.125% (w/v) sodium dodecyl sulphate and 0.125 mM ethylenediamine-tetraacetic acid. For GISH, autoclaved genomic DNA from Chinese Spring (20–30 × of the probe concentration) was added as blocking DNA. Probe and chromosomal DNA was denatured together on a Hybaid Omniblock (Thermo Fisher Scientific) at 75 °C for 7 min under plastic cover slips and slowly cooled to the hybridisation temperature of 37 °C overnight. Washes were carried out with 20% (v/v) formamide and 0.1 × saline sodium citrate at 42 °C, equivalent to 85% stringency. Hybridisation sites were detected with 2 μg ml−1 streptavidin conjugated to Alexa594 (Molecular Probes, Thermo Fisher Scientific) and 4 μg ml−1 antidigoxigenin conjugated to FITC (fluorescein isothiocyanate) (Roche Diagnostics). Chromosomes were counterstained with 4 μg ml−1 4′,6-diamidino-2-phenylindole diluted in McIlvaines buffer (pH 7.0) and mounted in antifade solution (Citifluor, London, UK). Photographs were taken on a Zeiss epifluorescence microscope with single band pass filters equipped with a CCD camera (ProgRes C12, Optronics, Milton Keynes, UK; model S97790) or Nikon Eclipse N80i fluorescent microscope equipped with a DS-QiMc monochromatic camera (Nikon, Tokyo, Japan). Each metaphase was captured in three different filter sets and then overlayed and analysed with Adobe Photoshop CS3 (Adobe Systems, San Jose, CA, USA) or NIS-Elements BR3.1 software (Nikon) using only cropping, and functions affecting the whole image equally.
Molecular marker analysis
PCR markers were chosen from literature and appropriate databases, and the sequences along with their melting temperature, source, references and expected product size are given (Supplementary Table S1). PCR markers specific for 4Ai#2S chromatin were used to confirm the presence of the Wsm1 gene. 1BS- or 1RS-specific markers were used to detect the presence of rye chromatin and identify molecular breakpoints (BPs) along the 1BS wheat arm. Primer sequences of all markers except UL-Thin5 were obtained from published sources. Nucleotide sequences for some of the publically unavailable Gatersleben Wheat Microsatellites (GWM) markers were kindly provided by Marion S. Röder (IPK, Gatersleben, Germany). DNA amplification was carried out in a 15 μl reaction mixture containing 100 ng of template DNA, 1 × Kapa Biosystems buffer A, 1.5 mM MgCl, 200 μM of dNTPs (Bioline, London, UK), 0.6 μM of each primer and 0.5 U of Kapa Taq DNA polymerase (Kapa Biosystems, Wilmington, MA, USA). PCR conditions were: 94 °C for 4 min, followed by 30 cycles at 94 °C for 1 min, 50–60 °C, respectively (Supplementary Table S1), for 45 s, 72 °C for 2 min, and final extension of 72 °C for 7 min. PCR amplification and polymorphism of the products were analysed on 1.5–3% agarose gels.
Results
Twenty experimental lines from four breeding populations (Table 1) previously classified as resistant or susceptible to WSMV (Divis et al., 2006) were rescored in the field and analysed in detail by molecular cytogenetics and PCR markers to identify their genomic constitution, the alien chromatin present and nature of WSMV resistance.
Phenotypic responses to WSMV infection
Lines classified as resistant to WSMV in the 2002 season (Divis et al., 2006) were found to carry the T4DL*4Ai#2S translocation (Table 1 and below) except for N02Y5003 and N02Y5109. These two lines, along with the resistant Mace, the susceptible control Tomahawk and two other lines, N02Y5019 and N02Y5149, were selected for further phenotypic characterisation in 2012 (Table 1). The level of infection in 2012 was much higher than that seen in either location in 2002, due both to increased mite pressure, and unseasonably warm fall and spring growing conditions. N02Y5109 clearly was susceptible in 2012 (Table 1); its symptom development as measured by SPAD readings was not significantly different from that of the known susceptible Tomahawk. Some wheat lines possess tolerance to low levels of mite and virus infection, but, under higher viral loads, or under more severe environmental conditions, as observed in 2012, this tolerance breaks down, and may explain the inconsistent response of N02Y5109 that sometimes shows limited resistance. N02Y5003 and N02Y5109 were scored MR and MS, respectively, in 2012. Early in the season, both appeared resistant, but as temperatures increased, resistance diminished. Nonetheless, mean SPAD readings were significantly higher in these two lines than in Tomahawk (Table 1). Finally, N02Y5149 was scored as R in this trial, and had significantly higher SPAD readings than the R control line Mace (Figure 1, Table 1).
Figure 1.
Field response of highly resistant line N02Y5149 (a), resistant cultivar Mace (b) and susceptible check Tomahawk (c) to natural infection with WSMV, Mead, NE, 2012, showing yellow leaves due to loss of chlorophyll and stunting in susceptible plants and full green leavs in resistant plants.
Molecular cytogenetic characterisation
FISH with total genomic Th. intermedium DNA revealed a number of alien wheat translocation and recombinant chromosomes in the 20 lines analysed (Table 1; Figures 2 and 3, Supplementary Figures S2 and S3). The characteristic banding patterns of dpTa1/Afa (mainly on the D-genome), pSc119.2 (abundant in the B-genome with some sites on A and D-genome chromosomes), 5S rDNA (on the short arms of group 1 and 5 chromosomes) and 45S rDNA probes (on 1B, 6B, 5D, 1A and sometimes detected on 7D) were used and characteristic banding patterns (Forsström et al., 2002; Contento et al., 2005; Patokar et al., 2015) compared in normal chromosomes and those carrying alien segments. The recipient wheat chromosomes were identified as 4D (Figure 3a) and 1B present in some lines of Populations I and IV (Figure 3b); translocation and recombinant chromosomes are described in more detail below. Total genomic rye DNA was also used as probe to identify rye chromosome segments.
Figure 2.
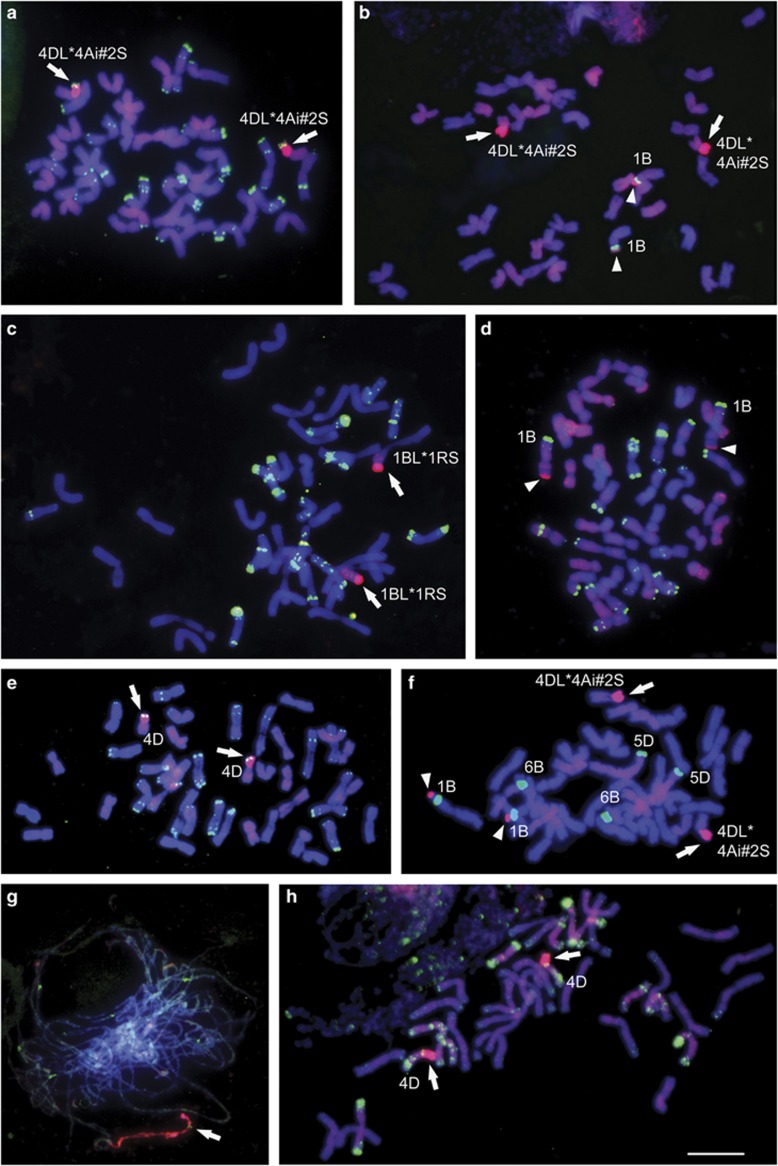
In situ hybridisation of example root-tip metaphase and meiotic pachytene chromosomes from WSMV-resistant (a–c, e, f and h) and WSMV-susceptible wheat lines (d and g). Hybridisation signal of total genomic DNA is shown in red and of repetitive DNA probes in green. Wheat chromosomes fluoresce blue with 4′,6-diamidino-2-phenylindole. Relevant chromosomes are identified. Whole alien chromosome arms are indicated with arrows and small segments or cross-hybridisation are indicated with arrow heads. Bar, 10 μm (a–f and h) and 20 μm (g). In situ hybridisation to further lines with alien fragment are shown in Supplementary Figure S3. (a) KS96HW10-1, (d) N02Y5163 and (e) N02Y5106: genomic Th. intermedium DNA (biotin, Alexa594, red) and 120 bp repeat pSc119.2 (digoxigenin, FITC, green). (b) N02Y5018: genomic Th. intermedium DNA (biotin, Alexa594, red) and 5S rDNA (digoxigenin, FITC, green). (c) N02Y5003: genomic rye DNA (biotin, Alexa594, red) and 340 bp repeat dpTa1/Afa (digoxigenin, FITC, green). (f) N02Y5149: Th. intermedium (digoxigenin, FITC shown in red) and 45S rDNA (biotin, Alex594 shown in green). (g) N02Y5156: Pachytene chromosomes: genomic rye DNA (biotin, Alexa594, red) and 5S rDNA (digoxigenin, FITC, green). (h) N02Y5057: genomic Th. intermedium DNA (biotin, Alexa594, red) and 340 bp repeat dpTa1/Afa (digoxigenin, FITC, green).
Figure 3.
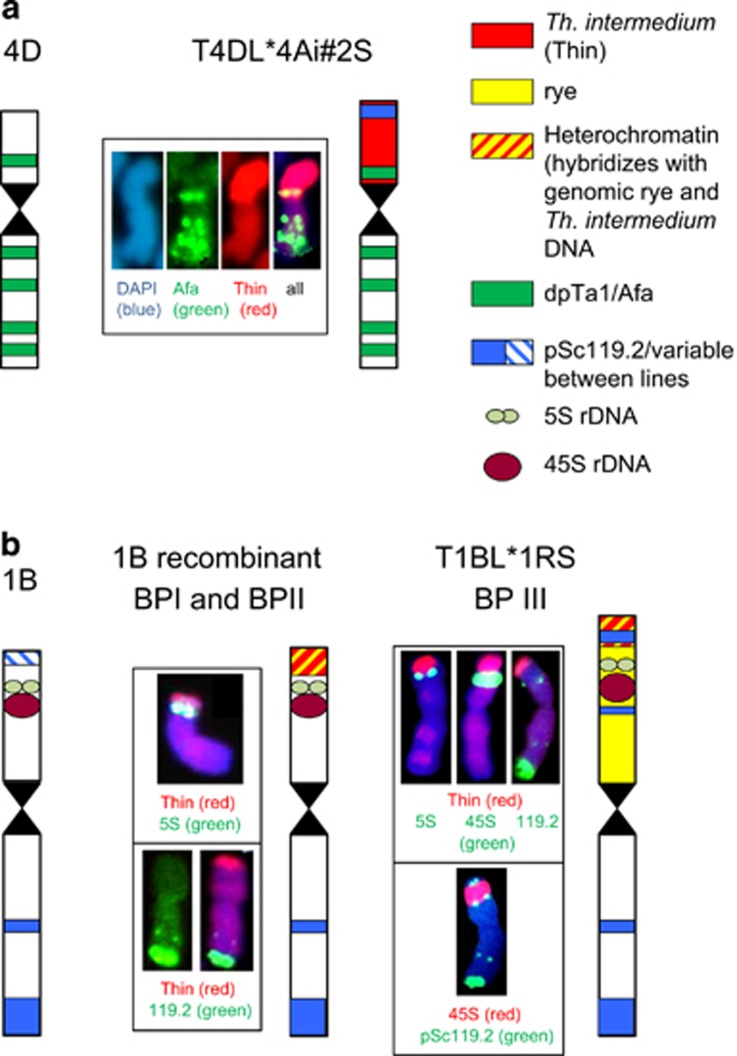
Diagram of alien translocation and recombinant chromosomes. Examples of FISH signals of two probes each (green and red) on blue 4′,6-diamidino-2-phenylindole-stained chromosomes are given. (a) Wheat 4D and T4DL*4Ai#2S chromosomes showing the dpTa1/Afa, pSc119.2 and genomic DNA hybridisation patterns. The Th. intermedium arm has a terminal pSc119.2 site (see Figure 2e) and the dpTa1 site is more proximal than on 4D. (b) Wheat chromosome 1B, 1B recombinant (BPI and PBII) and T1BL*1RS showing the characteristic arrangement of 5S (light green), 45S rDNA (brown) and pSc119.2 sites (blue). Chromosome arm 1BS has one telomeric pSc119.2 site that is not present in all lines, while 1RS is slightly longer and has two pSc119.2 sites distal and proximal of the NOR region. The BPI and BPII recombinant chromosomes characterised by molecular markers cannot be distinguished cytologically. Note that the subtelomeric heterochromatin hybridises strongly with both rye and Th. intermedium genomic DNA.
GISH with Th. intermedium genomic DNA
GISH to the resistant reference lines, known to include Th. intermedium DNA, KS96HW10-1 and KS95H102 (Figure 2a, Supplementary Figure S3a and Table 1), revealed a pair of small chromosome arms labelled with total genomic Th. intermedium DNA that also hybridised with weaker signal to the D-genome chromosomes of wheat (see also Figures 2b and h,Supplementary Figures S3b, e and g). This Th. intermedium chromatin has a strong terminal band with the repetitive DNA probe pSc119.2 (Figure 2a; see also in the derived line N02Y5106, Figure 2e, and N02Y2016, Supplementary Figure S3h) and was present as a centric translocation. The wheat chromosome arm was identified as 4DL by its hybridisation pattern with the repetitive DNA probe dpTa1/Afa (Figure 3a and shown in the derived line N02Y5057, Figure 2h, and Mace, Graybosch et al., 2009 and Supplementary Figure S3f). Millennium, the susceptible reference line, did not show any detectable Th. intermedium chromatin (Table 1).
Population I
The R-lines N02Y5018 (Figure 2b) and N02Y5025 (Supplementary Figure 3Sc) have the T4DL*4Ai#2S translocation (Table 1), but a further small fragment with strong Th. intermedium DNA hybridisation was detected at the distal end of the short arm of a large chromosome pair in line N02Y5018 (Figure 2b). This recombinant chromosome was identified as 1B owing to its hybridisation pattern with 5S rDNA, 45S rDNA and the repetitive probe pSc119.2 (Figure 3b; the origin of this recombinant chromosome is discussed below). Further alien fragments on 1B were detected in lines N0Y5019 (Supplementary Figure S3b) and N02Y5003 (Figure 2c) that both lack the T4DL*4Ai#2S translocation (Table 1). These two lines varied in their WSMV response in 2002, with line N0Y5019 showing more symptoms; however, SPAD readings in 2012 were similar. No Th. intermedium chromatin was detected in the S-line N02Y5021 (Table 1).
Population II and III
All R-lines, N02Y5057 (Figure 2h), N02Y5075 (Supplementary Figure S3d), N02Y5078 (Supplementary Figure S3e), N02Y5106 (Figure 2e) and Mace (N02Y5117; Supplementary Figure 2f), showed Th. intermedium chromatin in the form of the T4DL*4Ai#2S chromosomal translocation (while Th. intermedium origin chromatin was not detected in either S-lines N02Y5082 or N02Y5096 using FISH and no large Th. intermedium fragments were detected in N025105, N02Y5109 and N02Y5121 (Table 1)). Alien chromatin was detected on another D-genome chromosome identified by GISH with T. tauschii DNA (Supplementary Figure S2) in line N02Y5109 of population III that demonstrated inconsistent phenotypic response to WSMV (Table 1).
Population IV
The R-lines N02Y5149 (Figure 2f), N02Y5154 (Supplementary Figure S3g) and N02Y2016 (Supplementary Figure S3h) incorporated Th. intermedium chromatin in the form of T4DL*4Ai#2S chromosomal translocation (Table 1). Alien fragments on 1B hybridising with Th. intermedium DNA were also detected in R-lines N02Y5149 and N02Y2016 as well as in S-lines N02Y5156 (Figure 2g) and N02Y5163 (Figure 2d).
Characterisation of lines with the 1B translocation or recombinant chromosomes
The parental R-lines (KS91H184 and KS91H174) carrying Th. intermedium chromatin in the form of T4DL*4Ai#2S translocation (Divis et al., 2006) have never been described as carriers of alien chromatin on 1B. However, labelled genomic Th. intermedium DNA probe identified seven 1B recombinants or translocations in populations I and IV (Table 1). Two approaches were followed to identify the nature and origin of the 1B alien fragments. First, FISH experiments carried out using labelled genomic DNA from Th. intermedium and the probe pSc119.2 on lines having the 4D translocation with and without the 1B recombinant chromosome were analysed, and a possible reciprocal translocation between the T4DL*4Ai#2S and wheat 1BS chromosome was ruled out as the pSc119.2 site remained constant (Figures 2a, e and 3b). Second, the 1BS recombinants were screened with GISH using labelled rye genomic DNA probe along with use of molecular markers characterising 1BS or 1RS (for example, Figure 2c, Table 2 and below).
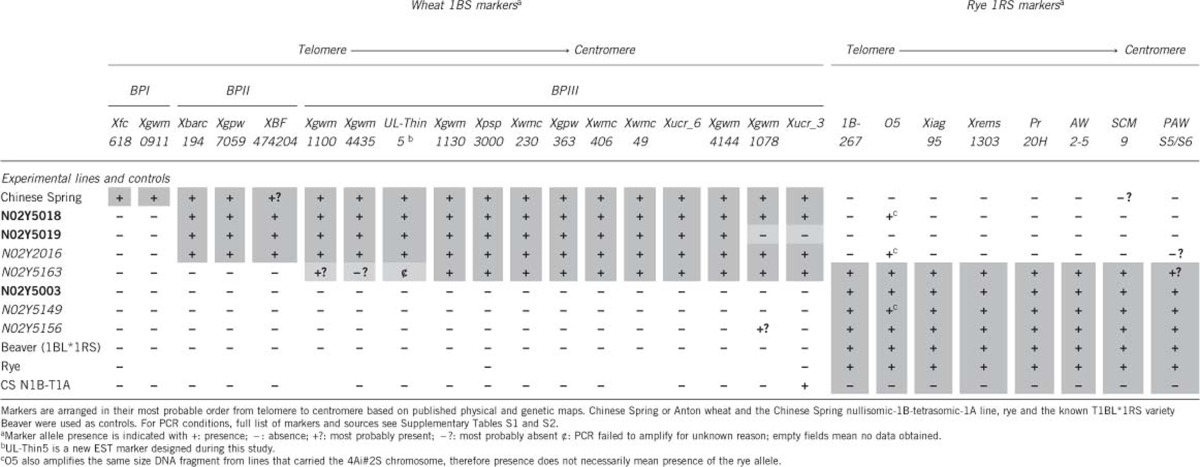
Table 2. Results of 1BS and 1RS polymorphic markers applied in breakpoint mapping of lines of Population I (bold) and Population IV (italic) carrying 1B alien chromatin.
GISH confirmed lines N02Y5003, N02Y5149 and N02Y5156 to have a centric T1BL*1RS (Type III) translocation, as revealed by the bright fluorescence of rye 1RS chromatin (for example, Figures 2c and g). Strong fluorescence of the terminal heterochromatic region of the alien rye chromatin was evident and also was detected by the Th. intermedium genomic probe (Figures 2c and f) indicating cross-hybridisation between rye and Th. intermedium DNA. N02Y5156 (susceptible) and N02Y5003 (moderately susceptible; Table 1) both lack the 4D-alien translocation. N02Y5149 (resistant, displaying the exceptionally high SPAD value) carries the Wsm1 gene of 4D and the 1RS arm, suggesting 1RS in this line enhances WSMV resistance (Table 1). Among the other resistant lines, N02Y5018 (Figure 2b) and N02Y2016 (Supplementary Figure S3h) also carried the additional small, Type I, 1BS alien chromatin along with Wsm1 of 4D (Table 1), but their WSMV field resistance response is typical of Wsm1 only. Small distal 1B alien fragments were detected in N02Y5018 (Figure 2b), N02Y5019 (Supplementary Figure S3b), N02Y5163 (Figure 2d) and N02Y2016 (Supplementary Figure S3h) with labelled Th. intermedium genomic DNA (Table 1), but GISH with rye DNA did not detect rye chromatin in these lines.
PCR marker screening
Detection of Th. intermedium chromatin
PCR markers STS-J15 and WSR11 produced characteristic amplicons of approximately 420 and 200 bp, respectively (Talbert et al., 1996; Fahim et al., 2012). Both markers detected polymorphism among the resistant and susceptible lines and the presence of a marker band related to the Wsm1 resistance gene (Supplementary Figure S1). Neither marker could identify the resistance mapped to chromosomes other than T4DL*4Ai#2S, as detected in line N02Y5003 (Table 1). Interestingly, WSR11 amplified a DNA band of the same size from two WSMV-susceptible lines N02Y5019 and N02Y5156 carrying the 1B recombinant chromosome (Table 1).
Detection of rye chromatin
1B recombinant lines (Table 1) along with positive and negative controls were analysed with nine 1RS PCR markers (Table 2; for references and conditions, see Supplementary Table S1). The rye telomeric repetitive DNA marker pAW161 amplified indiscriminately a monomorphic band of about 350 bp in all samples. Markers AW2–5, SCM9, pAWS5/S6, Xiag95, 1B-267, O5, Pr20H and Xrems1303 produced diagnostic bands from rye DNA and confirmed the presence of 1RS chromatin in lines N02Y5003, N02Y5149, N02Y5156, N02Y5163 and the control T1BL*1RS Beaver wheat, but not from lines N02Y5018, N02Y5019 and N02Y2016 (Table 2). The O5 marker also amplified DNA fragments of the same size from all lines with 4Ai#2S.
Breakpoint mapping of the 1B alien chromosome fragment
A total of 36 PCR markers from wheat 1B were applied for mapping the size of the alien chromatin seen on 1B in population I and IV and all markers successfully amplified DNA from wheat and/or rye chromatin (Supplementary Table S2). Only 18 markers (50%) produced polymorphic bands that could be assigned to the 1BS chromosome of wheat and were applied in BP mapping (Table 2). Few of these polymorphic markers amplified multiple loci, but only bands for the expected product size were scored (Supplementary Table S2). The BP between wheat and rye chromatin was identified by the appearance of wheat markers on the recombinant 1BS arm of wheat after taking into account its presence or absence in the control Chinese Spring wheat nullisomic-1B-tetrasomic-1A (CS N1B-T1A) line, Beaver and rye. The order of markers and BP localisation along the 1BS recombinants is based on published genetic and physical maps (Table 2). Overall, the molecular marker results revealed good agreement with the original sources for marker size and relative positions along the 1BS (Supplementary Table S1).
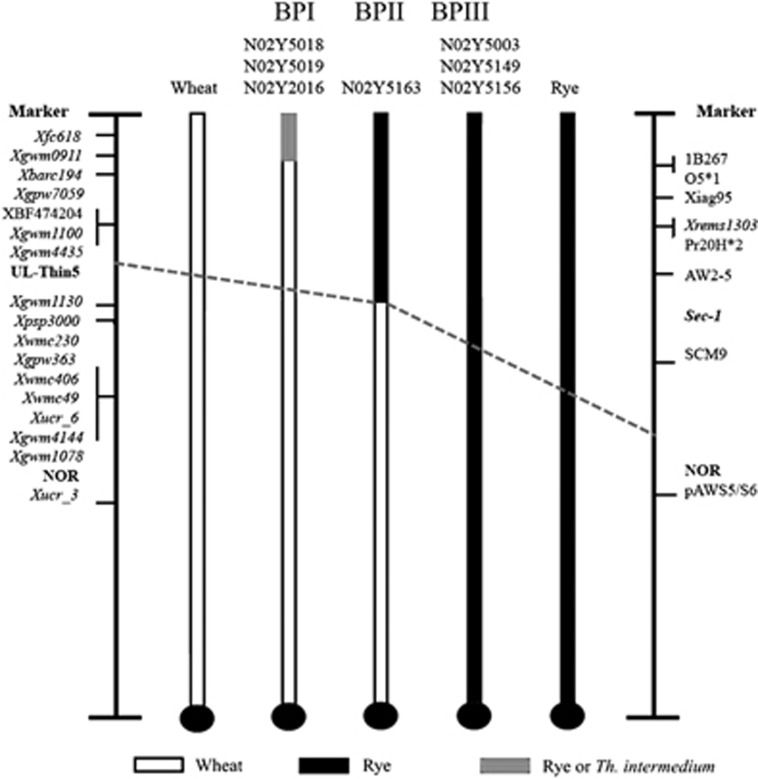
On the basis of in situ hybridisation data (Table 1) and wheat 1BS-specific markers, the seven 1BS recombinants identified were divided into three groups by their BP (Table 2) that identify three types of 1B recombinants. Line N02Y5018, N02Y5019 and N02Y2016 incorporated the smallest, Type I; N02Y5163 with intermediate, Type II; and N02Y5003, N02Y5149 and N02Y5156 have the complete 1RS chromatin, Type III (Table 1, Figure 4). Lines N02Y5018 (Figure 2b), N02Y2016 and N02Y5019 have lost only two distal wheat markers Xfc618, Xgwm0911 and are grouped in BPI. The size of the lost 1BS arm in N02Y5163 is larger than BPI having lost five distal wheat markers Xfc618, Xgwm0911, Xbarc194, Xgpw7059 and XBF474204 but retains Xpsp3000 a marker linked to Gli-1 gene and is placed in BPII (Table 2). GISH with rye genomic DNA revealed the centromeric BP in line N02Y5003 (Figure 2c), N02Y5149 and N02Y5156 (Figure 2g) and all tested 1BS-specific wheat markers were absent placing the lines in BPIII (Table 2). Physical and genetic map-based position of the 1BS and 1RS polymorphic markers is based on comparative map position from published sources and the most likely order of these markers here is given (Figure 4).
Figure 4.
Diagrammatic representation of most likely marker positions and translocation BPs along the recombinant 1B lines N02Y5018, N02Y5019, N02Y2016, N02Y5163, N02Y5003, N02Y5149 and N02Y5156 based on published marker order for 1BS and 1RS (Anugrahwati et al., 2008; Reddy et al., 2008; and references and data from Supplementary Table S1). White bars represent wheat 1BS, black represents rye 1RS and red bars represent the distal 1B alien chromatin that may be rye or Th. intermedium chromosomal segments; centromeres are represented by dark circles. Sec-1 and NOR position are based on Lei et al. (2012). Asterisks (*) represent repetitive DNA markers (Tang et al., 2011) for which chromosomal positions are proposed here; UL-Thin5 is a new marker. Line N02Y5018, N02Y5019 and N02Y2016 has the translocation BP between Xgwm0911 and Xbarc194. Physical length of this alien chromatin may not be the same but this BP is identified by the same markers (Table 2). Line N02Y5163 has the translocation BP between UL-Thin5 and Xgwm1130, while line N02Y5003, N02Y5149 and N02Y5156 has a centric T1BL*1RS translocation (Table 1).
Origin of rye fragments in wheat–Th. intermedium hybrid lines
The origin of the 1B alien chromatin was investigated by analysing available lines in the pedigree of populations I and IV with specific markers for 1BS and 1RS. Parental WSMV-resistance lines KS91H174 and KS91H184 have an intact 1BS of wheat origin, while RioBlanco and M08 tested positive for 1B alien chromatin. Further, wheat 1BS-specific markers Xfc618 and Xpsp3000 revealed the size of the 1B alien chromatin to be smaller in RioBlanco compared with M08 (Supplementary Table S3). Marker profile and negative PCR with rye marker of RioBlanco was similar to N02Y5018, N02Y5019 and N02Y2016 that may be the donor parent of 1BS alien chromatin of these lines (Table 1). Both RioBlanco and M08 are among the parents of population IV (Table 1), but marker profile and FISH results make M08 the potential donor of the 1RS chromatin seen in lines N02Y5149 and N02Y5156 (Figure 2g). The origin of the whole arm 1RS chromatin in R-line N02Y5003 (Figure 2c) and medium size in S-line N02Y5163 (Figure 2d) could not be assigned to M08 or RioBlanco (compare Table 1 and Table 2). Seed of three parents of population I (CO850034, T-57 and KS91HW29) was not available and thus origin and introgression of the whole 1RS in N02Y5003 (Figure 2c) from these lines could not be tested. TAM107 with a T1AL*1RS translocation (Villareal et al., 1996) is also in the pedigree of population I, but no alien chromatin was found on the 1A chromosomes in the lines investigated here, but it cannot be excluded that the 1RS has transferred to 1B.
Discussion
Many current WSMV-resistant wheat lines including the hard winter wheat variety Mace (Figure 1) have benefited from the Wsm1 gene of 4Ai#2 origin. Some of the initial lines carrying Wsm1 were associated with undesirable traits such as reduced yield and poor bread-making quality, but subsequent crosses and selection eliminated many negative effects from the introgressed Th. intermedium chromatin (Divis et al., 2006; Graybosch et al., 2009). We could demonstrate that the reference Kansas resistance lines KS95H102 and KS96HW10-1 have alien material in the form of T4DL*4Ai#2S translocation (Figures 2 and 3, Supplementary Figures S2 and S3). Further, in four populations of the Nebraska USDA-ARS crossing programme (Divis et al., 2006) that gave rise to Mace, the 4Ai#2S Wsm1 origin resistance was identified in 10 (including Mace) of the adapted winter wheat lines. Sensitive sister control lines in each population did not carry the T4DL*4Ai#2S translocation (Table 1). The source of resistance in all R-lines is most likely either KS91H184 or KS91H174, both being selected from populations that had been randomly mated for several generations and derived from CI 17884, a WSMV-resistance line carrying the Wsm1 gene on a chromosome arm translocated from Th. intermedium (Wells et al., 1982).
In addition, we detected rye chromatin transferred to chromosome 1B in some of the lines and resistance enhancement associated with rye was identified in the Th. intermedium alien wheat introgression lines tested. It is generally thought that rye itself is probably susceptible to WSMV and that 1RS in wheat–rye translocation does not confer resistance, but that rye can resist colonisation of both greenbug and WCM, the vector of WSMV (Thomas et al., 2004; Divis et al., 2006). Our results revealed that the parental WSMV-resistance lines KS91H174 and KS91H184 have intact 1B chromosomes and the 1RS fragments identified in the lines investigated must originate from other lines within the pedigrees. The pedigree of population IV contains the line M08 a 1R(1B) substitution or T1BL*1RS translocation line (Zhang et al., 1998) derived from anther culture after hybridisation of hexaploid wheat and hexaploid Triticale and identified as having a recombinant 1B (Supplementary Table S3). More importantly, M08 was noted in preliminary observations made at the University of Nebraska (P.S. Baenziger, personal communication, 2008) as potentially harbouring genes conferring some tolerance to WSMV. The 1RS arm of population IV is most likely from M08 and together with the Wsm1 gene of T4DL*4Ai#2S translocation in line N02Y5149 resulted in field resistance to WSMV superior to any other line (Figure 1, Table 1). This enhanced resistance to WSMV in the 2012 field study might very well have arisen from increased tolerance to the vector, WCM. On its own, 1RS in line N02Y5163 was, however, not able to confer WSMV resistance. The pedigree of population I includes TAM107 with T1AL*1RS (Villareal et al., 1996) and RioBlanco with an incomplete 1RS arm (Supplementary Table S3), and thus the origin of the whole 1RS arm of N02Y5003, probably different to the one of N02Y5149, is not clear; however, it seems to be responsible for the enhanced resistance of this line that lacks the T4DL*4Ai#2S translocation (Tables 1 and 2).
Variable behaviour of 1RS resistance supports different origins, but also emphasises the potential role of epigenetic modification and chromatin remodelling in gene expression (see Slotkin and Martienssen, 2007). Our lines (Table 1) have undergone interspecific hybridisation stress that may cause heritable changes to epigenetic marks. Development of resistance in originally WSMV-susceptible lines has shown that WSMV-resistance genes in wheat cultivars might be present but unexpressed (Lu et al., 2011; Seifers et al., 2013). The expression of these genes could be modulated and controlled by genes, transcription factors or chromatin regulators from the alien chromosome segments.
Within the lines studied, a very small distal alien fragment was identified on 1B in some lines (Table 2 and Figures 2,3 and 4), although it was not able to confer WSMV resistance. Wheat–alien translocations with minimal alien chromatin per se are of importance, as they are predicted to have less likelihood of linkage drag (Forsström et al., 2002; Niu et al., 2011), although linkage drag is less pronounced or buffered in polyploid wheat compared with the diploid progenitors. Not all large alien fragments are disadvantageous; they may potentially introduce more variation and assure a wider variety of resistances (Friebe et al., 2009). The presence of cryptic alien chromatin, as discovered here on chromosome 1B, and the associated loss of some important wheat genes might explain some of the negative effects and undesirable traits in the lines studied and we cannot exclude that the small alien fragments interfered in developing novel well-adapted lines with Wsm1.
GISH is a powerful technique that allows the identification of alien wheat recombinant chromosomes, particularly when combined with repetitive DNA probes (for example, Forsström et al., 2002; Molnar et al., 2015; Patokar et al., 2015), and thus subsequent selection of molecular markers could concentrate on the chromosome arm (or segment) identified, rather than a genome-wide marker screen (Niu et al., 2011). The markers used here classified the seven 1BS recombinants into three BP categories (Figure 4,Table 2). The order and location of these markers along the 1BS was largely as reported, although a few, for example, Xgwm1130 and Xgwm0911 (Ganal and Röder, 2007) were inverted. This discrepancy is not a rare phenomenon when wheat cultivars from different pedigrees are compared (Akhunov et al., 2013). The recombination BPs were all distal to the NOR on 1BS (Figure 4) suggesting the presence of recombination hot-spots in this region (Reddy et al., 2008).
Although the T1BL*1RS whole arm translocation (BPIII) was identified clearly by GISH in N02Y5003, N02Y5149 and N02Y5156, and rye-specific markers distal to the NOR were present (Table 2), both in situ hybridisation with rye genomic DNA and 1RS-specific PCR markers failed to identify the small alien BPI fragments on 1BS in lines N02Y518, N02Y5019 and N02Y2016 that lost only the most distal wheat 1BS-specific markers Xfc618 and Xpsp3000 (Table 2 and Figure 4). However, the alien fragment is not below the resolution of GISH, as genomic Th. intermedium DNA hybridised to these fragments consistently (Table 1 and Figure 2). Therefore, the possibility of a non-rye alien fragment, possibly from Th. intermedium itself cannot be ruled out although Th. intermedium genomic DNA can cross-hybridise to the distal tandem repeats present at the telomere of a normal 1RS (Figures 2d and f). Detection of very small alien chromatin segments can be difficult both cytologically and with molecular markers: GISH screening using labelled rye genomic DNA probe and wheat DNA as a block failed to visualise the presence of rye chromatin in known recombinants that carried the SrR gene (Anugrahwati et al., 2008) and negative PCR results have been reported for several wheat–rye translocation lines that contained 1RS chromatin (Mago et al., 2002; Anugrahwati et al., 2008).
The variable PCR results with some 1RS markers (Table 2) could be due to DNA changes as a consequence of intensive crossing and back-crossing involved in modern wheat lines, because rearrangements, shuffling or even loss of marker positions is not a rare phenomenon and previously undetected interstitial deletions can occur; mutation in the priming sites may also result in no PCR amplification. Alternatively, the small alien fragment is not of rye origin as is indicated by the GISH, or the markers amplify from Th. intermedium chromatin not on 1B (Table 1).
The presence of rye material and the value of 1RS in combination with Th. intermedium WSMV resistance is new (Tables 1 and 2). The short arm of rye chromosome 1 (Figure 2c) is the most widespread alien chromatin in wheat-breeding programmes and has been used to incorporate new genes for stress tolerance and yield potential into wheat including several important genes, such as Lr26, Sr31, Yr9, Pm8 and Pm17 (Heslop-Harrison et al., 1990; Forsström et al., 2002; Mago et al., 2002; Reddy et al., 2008). Th. intermedium is a member of the tertiary gene pool of wheat, and since the 1960s, at least 15 genes for fungal or viral resistance have originated from Th. intermedium and Th. ponticum as chromosomal segments (Sepsi et al., 2008; Li and Wang, 2009). The 4Ai#2S alien arms, which carry WSMV resistance, also harbour resistance for the fungal pathogen Tapesia yallundae (Chen et al., 2003). In wheat backgrounds, the 4Ai#2S does not cause meiotic instability, and there is considerably more potential for exploitation of Th. intermedium introgression in wheat breeding.
Significant yield losses are associated with WSMV infections (Graybosch et al., 2009) and at least three additional viruses have been reported as being transmitted by the WCM vector (Navia et al., 2013). There is huge interest in deploying multiple effective genes in combination as a means of disease control and improving the durability of resistance (Liu et al., 2011; Heslop-Harrison and Schwarzacher, 2012). A good means to improve resistance would be to stack the novel 1RS enhancer in germplasm carrying other known WSMV resistances to achieve the desired goals of deploying combinations of effective genes. Line N02Y5149 is potentially the first R-line with multiple WSMV resistance affecting genes and the direct utilisation of this selection should improve the durability of WSMV resistance. Therefore, the rye chromatin and increased WSMV resistance are important, especially for gene-pyramiding approaches integrating genomic and molecular cytogenetic approaches for chromosome segment identification with traditional breeding (Tester and Langridge, 2010; Heslop-Harrison and Schwarzacher, 2012).
Data archiving
Relevant data are given in the Supplementary Information and in the Leicester Research Archive http://hdl.handle.net/2381/10951. It is also available from the Dryad Digital Repository: http://dx.doi.org/10.5061/dryad.82f5q.
Acknowledgments
We thank Hazara University, Mansehra and Higher Education Commission (HEC), Pakistan for PhD fellowship to NA. RAG and GLH acknowledge the United States Wheat Board for support. We thank Steve Reader, John Innes Centre, Norwich, UK, for providing nullisomic-tetrasomic wheat lines, Marion S. Röder (IPK, Gatersleben Germany) for providing sequences of unpublished oligonucleotides.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Akhunov ED, Sehgal S, Liang H, Wang S, Akhunova AR, Kaur G et al. (2013). Comparative analysis of syntenic genes in grass genomes reveals accelerated rates of gene structure and coding sequence evolution in polyploid wheat. Plant Physiol 161: 252–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anugrahwati DR, Shepherd KW, Verlin DC, Zhang P, Mirzaghaderi G, Walker E et al. (2008). Isolation of wheat-rye 1RS recombinants that break the linkage between the stem rust resistance gene SrR and secalin. Genome 51: 341–349. [DOI] [PubMed] [Google Scholar]
- Chen Q, Conner R, Li H, Sun S, Ahmad F, Laroche A et al. (2003). Molecular cytogenetic discrimination and reaction to Wheat streak mosaic virus and the wheat curl mite in Zhong series of wheat Thinopyrum intermedium partial amphiploids. Genome 46: 135–145. [DOI] [PubMed] [Google Scholar]
- Contento A, Heslop-Harrison JS, Schwarzacher T. (2005). Diversity of a major repetitive DNA sequence in diploid and polyploid Triticeae. Cytogenet Genome Res 109: 34–42. [DOI] [PubMed] [Google Scholar]
- Crossa J, Pérez P, Hickey J, Burgueño J, Ornella L, Cerón-Rojas J et al. (2014). Genomic prediction in CIMMYT maize and wheat breeding programs. Heredity 112: 48–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divis L, Graybosch R, Peterson C, Baenziger P, Hein G, Beecher B et al. (2006). Agronomic and quality effects in winter wheat of a gene conditioning resistance to Wheat streak mosaic virus. Euphytica 152: 41–49. [Google Scholar]
- Fahim M, Ayala-Navarrete L, Millar AA, Larkin PJ. (2010). Hairpin RNA derived from viral NIa gene confers immunity to Wheat streak mosaic virus infection in transgenic wheat plants. Plant Biotec J 8: 821–834. [DOI] [PubMed] [Google Scholar]
- Fahim M, Mechanicos A, Ayala-Navarrete L, Haber S, Larkin P. (2012). Resistance to Wheat streak mosaic virus-a survey of resources and development of molecular markers. Plant Pathol 61: 425–440. [Google Scholar]
- Friebe B, Mukai Y, Dhaliwal H, Martin T, Gill B. (1991). Identification of alien chromatin specifying resistance to Wheat streak mosaic and greenbug in wheat germ plasm by C-banding and in situ hybridization. Theor Appl Genet 81: 381–389. [DOI] [PubMed] [Google Scholar]
- Friebe B, Qi L, Wilson D, Chang Z, Seifers D, Martin T et al. (2009). Wheat-Thinopyrum intermedium recombinants resistant to Wheat streak mosaic virus and Triticum mosaic virus. Crop Sci 49: 1221–1226. [Google Scholar]
- Forsström P, Merker A, Schwarzacher T. (2002). Characterisation of mildew resistant wheat-rye substitution lines and identification of an inverted chromosome by fluorescent in situ hybridisation. Heredity 88: 349–355. [DOI] [PubMed] [Google Scholar]
- Ganal MW, Röder MS. (2007). Microsatellite and SNP markers in wheat breeding. In Genomics-Assisted Crop Improvement Vol 2 Genomics Applications in Crops; Varshney RK, Tuberosa R (eds) Springer Netherlands: Dordrechtm Netherlands. pp 1–2. [Google Scholar]
- Graybosch RA, Peterson C, Baenziger PS, Baltensperger DD, Nelson LA, Jin Y et al. (2009). Registration of ‘Mace' hard red winter wheat. Journal of Plant Reg 3: 51–56. [Google Scholar]
- Haley SD, Johnson JJ, Peairs FB, Stromberger JA, Heaton EE, Seifert SA et al. (2011). Registration of ‘Snowmass' wheat. J Plant Reg 5: 87–90. [Google Scholar]
- Heslop-Harrison JS, Leitch A, Schwarzacher T, Anamthawat-Jonsson K. (1990). Detection and characterization of 1B/1R translocations in hexaploid wheat. Heredity 65: 385–392. [Google Scholar]
- Heslop-Harrison JS JS, Schwarzacher T (2012). Genetics and genomics of crop domestication. In: Altman A, Hasegawa PM (eds). Plant Biotechnology and Agriculture: Prospects for the 21st Century. Academic Press: San Diego, CA, USA, pp 3–18..
- Kim W, Johnson JW, Baenziger PS, Lukaszewski AJ, Gaines CS. (2004). Agronomic effect of wheat-rye translocation carrying rye chromatin (1R) from different sources. Crop Sci 44: 1254–1258. [Google Scholar]
- Lei M, Li G, Liu C, Yang Z, Scoles G. (2012). Characterization of wheat-Secale africanum introgression lines reveals evolutionary aspects of chromosome 1R in rye. Genome 55: 765–774. [DOI] [PubMed] [Google Scholar]
- Li H, Conner RL, Liu Z, Li Y, Chen Y, Zhou Y et al. (2007). Characterization of wheat-triticale lines resistant to powdery mildew, stem rust, stripe rust, wheat curl mite, and limitation on spread of WSMV. Plant Dis 914: 368–374. [DOI] [PubMed] [Google Scholar]
- Li H, Wang X. (2009). Thinopyrum ponticum and Th. intermedium: the promising source of resistance to fungal and viral diseases of wheat. J Genet Genomics 36: 557–565. [DOI] [PubMed] [Google Scholar]
- Liu W, Seifers D, Qi L, Friebe B, Gill B. (2011). A compensating wheat-Thinopyrum intermedium Robertsonian translocation conferring resistance to Wheat Streak Mosaic Virus and Triticum Mosaic Virus. Crop Sci 51: 2382–2390. [Google Scholar]
- Lu H, Price J, Devkota R, Rush C, Rudd J. (2011). A dominant gene for resistance to Wheat Streak Mosaic Virus in winter wheat line CO960293-2. Crop Sci 51: 5–12. [Google Scholar]
- Mago R, Spielmeyer W, Lawrence G, Lagudah E, Ellis J, Pryor A. (2002). Identification and mapping of molecular markers linked to rust resistance genes located on chromosome 1RS of rye using wheat-rye translocation lines. Theor Appl Genet 104: 1317–1324. [DOI] [PubMed] [Google Scholar]
- Molnar I, Vrana J, Farkas A, Kubakakova M, Cseh A, Molnar Lang M et al. (2015). Flow sorting of C-genome chromosomes from wild relatives of wheat Aegilops markgrafii, Ae. triuncialis and Ae. cylindrica, and their molecular organization. Ann Bot 116: 189–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutti JS, Baenziger PS, Graybosch RA, French R, Gill KS. (2011). Registration of seven winter wheat germplasm lines carrying the gene for resistance. J Plant Reg 5: 414–417. [Google Scholar]
- Navia D, de Mendonça RS, Skoracka A, Szydło W, Knihinicki D, Hein GL et al. (2013). Wheat curl mite, Aceria tosichella, and transmitted viruses: an expanding pest complex affecting cereal crops. Exp Appl Acara 59: 95–143. [DOI] [PubMed] [Google Scholar]
- Niu Z, Klindworth DL, Friesen TL, Chao S, Jin Y, Cai X et al. (2011). Targeted introgression of a wheat stem rust resistance gene by DNA marker-assisted chromosome engineering. Genetics 187: 1011–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patokar C, Sepsi A, Schwarzacher T, Kishii M, Heslop-Harrison JS. (2015). Molecular cytogenetic characterization of novel wheat-Thinopyrum bessarabicum recombinant lines carrying intercalary translocations. Chromosoma 125: 163–172. [DOI] [PubMed] [Google Scholar]
- Price J, Workneh F, Evett S, Jones D, Arthur J, Rush C. (2010). Effects of Wheat streak mosaic virus on root development and water-use efficiency of hard red winter wheat. Plant Dis 94: 766–770. [DOI] [PubMed] [Google Scholar]
- Reddy L, Friesen TL, Meinhardt SW, Chao S, Faris JD. (2008). Genomic analysis of the Snn1 locus on wheat chromosome arm 1BS and the identification of candidate genes. The Plant Genome 1: 55–66. [Google Scholar]
- Schwarzacher T, Anamthawat-Jonsson K, Harrison G, Islam A, Jia J, King I et al. (1992). Genomic in situ hybridization to identify alien chromosomes and chromosome segments in wheat. Theor Appl Genet 84: 778–786. [DOI] [PubMed] [Google Scholar]
- Schwarzacher T, Heslop-Harrison JS. (2000) Practical In Situ Hybridization. BIOS Scientific Publishers Ltd: Abindgdon, UK. now published by Garland Science, New York, USA. [Google Scholar]
- Seifers D, Martin T, Haber S. (2013). Temperature-sensitive resistance to Wheat streak mosaic virus in CO960333 and KS06HW79 wheat. Plant Dis 97: 983–987. [DOI] [PubMed] [Google Scholar]
- Sepsi A, Molnár I, Szalay D, Molnár-Láng M. (2008). Characterization of a leaf rust-resistant wheat Thinopyrum ponticum partial amphiploid BE-1, using sequential multicolor GISH and FISH. Theor Appl Genet 116: 825–834. [DOI] [PubMed] [Google Scholar]
- Slotkin RK, Martienssen R. (2007). Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet 8: 272–285. [DOI] [PubMed] [Google Scholar]
- Talbert L, Bruckner P, Smith L, Sears R, Martin T. (1996). Development of PCR markers linked to resistance to Wheat streak mosaic virus in wheat. Theor Appl Genet 93: 463–467. [DOI] [PubMed] [Google Scholar]
- Tang Z, Fu S, Ren Z, Zhang T, Zou Y, Yang Z et al. (2011). Diversity and evolution of four dispersed repetitive DNA sequences in the genus. Secale. Genome 54: 285–300. [DOI] [PubMed] [Google Scholar]
- Tester M, Langridge P. (2010). Breeding technologies to increase crop production in a changing world. Science 327: 818–822. [DOI] [PubMed] [Google Scholar]
- Thomas J, Conner R, Graf R. (2004). Comparison of different sources of vector resistance for controlling Wheat streak mosaic in winter wheat. Crop Sci 44: 125–130. [Google Scholar]
- Villareal RL, Del Toro E, Rajaram S, Mujeeb-Kazi A. (1996). The effect of chromosome 1AL/1RS translocation on agronomic performance of 85 F2-derived F6 lines from three Triticum aestivum L. crosses. Euphytica 89: 363–369. [Google Scholar]
- Wells DG, Kota R, Sandhu H, Gardner W, Finney K. (1982). Registration of one disomic substitution line and five translocation lines of winter wheat germplasm Resistant to Wheat streak mosaic virus1 (Reg. No. GP 199 to GP 204). Crop Sci 22: 1277–1278. [Google Scholar]
- Zhang X, Wang X, Jing J, Ross K, Hu H, Gustafson J. (1998). Characterization of wheat-triticale doubled haploid lines by cytological and biochemical markers. Plant Breeding 117: 7–12. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.