Abstract
The use of probiotics for human and animal health is continuously increasing. The probiotics used in humans commonly come from dairy foods, whereas the sources of probiotics used in animals are often the animals’ own digestive tracts. Increasingly, probiotics from sources other than milk products are being selected for use in people who are lactose intolerant. These sources are non-dairy fermented foods and beverages, non-dairy and non-fermented foods such as fresh fruits and vegetables, feces of breast-fed infants and human breast milk. The probiotics that are used in both humans and animals are selected in stages; after the initial isolation of the appropriate culture medium, the probiotics must meet important qualifications, including being non-pathogenic acid and bile-tolerant strains that possess the ability to act against pathogens in the gastrointestinal tract and the safety-enhancing property of not being able to transfer any antibiotic resistance genes to other bacteria. The final stages of selection involve the accurate identification of the probiotic species.
Keywords: Probiotics, Unconventional sources, Lactic acid bacteria, Fermented food
Background
The term probiotic refers to live microorganisms that survive passage through the by improving its intestinal microbial balance [1, 2]. Recently, FAO/WHO has defined probiotics as living microorganisms that improve the health of humans and animals and must be safe and in sufficient quantity for bodily function [3]. For probiotic products to be identified as functioning, its concentration must be at least 106 viable cells (colony forming unit, CFU/g) of the product. The discovery process for new probiotics emphasizes strain selection and the survival of the culture during biomass production and storage.
For at least the past 10 years, probiotic microorganisms have been used continuously for health benefits in both humans and animals. The main reason for their use is that probiotics offer an alternative to antibiotics; such an alternative is proposed to decrease the drug resistance that occurs due to an overuse or prolonged use of antibiotics to treat infections in both humans and animals. In addition, foods of animal origin have been found to contain drug residues in their meat, which is a result of animals being reared in conditions of antibiotic misuse, including an excessive use of antibiotics for disease treatment, incorrect drug withdrawal times and the addition of antibiotics to feed. When humans consume the contaminated foods, the drug accumulates in the body and leads to drug resistance when an antibiotic is used to treat an infection.
Gut microflora can also be balanced by directly adding live microorganisms into the diet. Microbes were used as probiotics including bacteria, yeast and mold. The genera and species that have been used are Lactobacillus, Streptococcus, Leuconostoc, Pediococcus, Propionibacterium, Enterococcus, Bifidobacterium, Bacillus, Saccharomyces cerevisiae, Candida pintolopesii, Aspergillus niger and A. oryzae [4]. Lactic acid bacteria (LAB) are considered a major group of probiotic bacteria and are commonly used in both humans and animals [5]. The most commonly used LAB in humans are Lactobacillus and Bifidobacterium [6]. Spore-forming lactic acid bacteria, mostly of the genus Bacillus, are acid and bile tolerant strains that have been used as probiotics in both humans and animals. However, only Bacillus strains that have not been reported to be pathogenic, such as Bacillus lichenformis, B. cereus var. toyoi, B. clausii, B. coagulans, B. laterosporus, B. pumilus and B. racemilacticus can be used as probiotics [7–9]. The studies and developments related to probiotics include research on selecting probiotic strains with specific properties and, technologies that can be used to improve the probiotic production process. Therefore, probiotic markets are likely to increase continuously.
Unconventional sources of probiotics
The use of selected probiotics from alternative sources known as “unconventional sources” is likely to increase. One of the reasons why alternative sources for probiotic selection have increased in use is to avoid the consumption of dairy in lactose-intolerant individuals. Unconventional sources of microorganisms were screened for potential probiotics, which have been isolated from numerous different sources, including non-intestinal sources and non-dairy fermented food products, such as traditional fermented foods, traditional fermented drinks, vegetables, and fruit juices [10–13]. The differences in the raw materials and ingredients used to make non-fermented or fermented foods are the main factors that lead to the different available species or strains of probiotics in the food sources.
Probiotic microorganisms can be screened from non-intestinal sources, such as fruit juices [14], grains [15], honey-comb [16] and soil [17, 18]. Probiotic sources and selection criteria to apply in both humans and animals are summarized as the Fig. 1. LAB primarily Lactobacillus plantarum have been found in many types of fruit juices from both solid and citrus fruits whereas Leuconostoc mesenteroides is rarely found in these fruits but is the species that is most commonly found in tomatoes [14]. The alternative growth medium that is used to cultivate lactic acid bacteria can also be used to select probiotics from sources such as pineapple wastes [19] and tomato juice [20]. LAB can be found in food products stored at a low temperature (4 °C) such as vacuum-packaged beef and some beneficial isolates can be screened in a similar way as bacteria that produce bacteriocin-like substance [21].
Fig. 1.
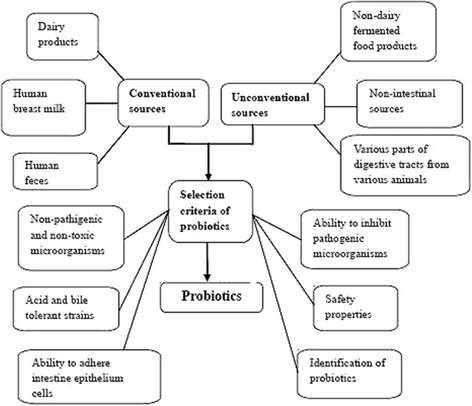
Diagram of probiotic sources and selection criteria to apply in both humans and animals
Probiotics isolated from non-intestinal sources are likely to not be strains that produce a bacteriocin-like substance. For example, Lactobacillus plantarum isolated from fermented foods containing fish or pork were resistant to low pH, tolerated bile and exhibited strong antimicrobial activity against pathogenic bacteria when using the normal supernatant of the strain, whereas the neutralized supernatant showed weak antagonistic activity [22, 23]. Several fresh fruits and vegetables, including dragon, durian, ginger, papaya, star fruits and guava, can be screened to find beneficial LAB that produce antimicrobial substances other than bacteriocin, such as hydrogen peroxide and lactic and propionic acids, to combat against pathogenic bacteria and pathogenic fungi of chilies [22]. These LAB can be used for starter cultures in human fermented foods and can have probiotic effects in humans. However, some studies have reported that LAB from some fruits and vegetables showed bacteriocin-like activity [24]. Tajabadi et al. [16] have screened LAB from the honey of giant honey bees. Most of these LAB isolates were Lactobacillus spp., mainly Lactobacillus kunkeei. This bacterial species has been reported to have antagonistic effects against yeast growth and the spoilage-related effects of yeast in honey [25].
Siddiqee et al. [10] reported that animal intestines comprise the most potential sources of LAB but that other sources, including fruit juice, flesh, long grass and vegetables, can also be screened to find LAB. Milk whey as dairy waste from cheese production industries has been used to cultivate LAB to produce more lactic acid compared with the conventional Luria-Bertani medium. Thus, it may be the one of alternative sources from which beneficial LAB can be isolated [26].
Other unconventional sources from which high potential probiotics have been isolated are the environments around food products, such as the air surrounding environments for preparing sourdoughs and the air of the storage and working rooms of a bakery. These air samples have been found to contain Lactobacillus plantarum, a similar species to the species that was isolated from the dough [27].
Probiotic isolates from non-dairy fermented foods
Fermented food products that are screened for potential probiotics are made from two main materials: plant and animal matter. Several types of fermented foods for human consumption differ in the materials and ingredients used to make them depending on the culture and traditions of the local people from each country. The difference in raw materials and ingredients used in fermented foods is a main factor that has led to the identification of different species and strains of probiotics. The various traditional fermented foods that have been found to contain LAB are made from a variety of raw materials, including fish [28, 29], beef [30], pork [31], salted crab [32], seafood [33], soybeans [34] and vegetables [12].
LAB species have been isolated from fermented foods from several countries. In Thailand, six genera of LAB species have been identified: Aerococcus, Enterococcus, Lactobacillus, Pediococcus, Tetragenococcus and Weissella. Most LAB genera isolated from fermented fishes and crustaceans is Enterococcus, while the LAB genera most commonly isolated from fermented meats and fermented plants is Lactobacillus. These strains are the most halophilic LAB and grew under a NaCl concentration of more than 22 %. However, the main species isolated from fermented plant materials are L. fermentum and L. plantarum. These species grew under NaCl concentrations of less than 6 % [35]. NaCl-like LAB has also been found in Thai traditional salted crab (Poo-Khem). Out of 306 isolates, four probiotic LAB species can be used as starter cultures in Poo-Khem foods for humans because they showed probiotic properties including acid and bile tolerance, antagonistic effects against food-borne pathogenic bacteria and hydrophobic activity. These four species were identified as one strain each of Enterococcus thailandensis and L. plantarum and two strains of L. fermentum [32]. In addition, Siripornadulsil et al. [31] reported that Pediococcus pentosaceous strains were the LAB most often isolated from various traditional Thai fermented foods containing fish and pork. They were tolerant to acidic conditions at pH 2, 0.3–0.5 % bile salt and 1–14 % NaCl. They also inhibited the growth of some pathogenic bacteria, including Pseudomonas aeruginosa, Salmonella typhimurium, Vibrio cholera, E. coli, Bacillus cereus and Staphylococcus epidermidis. Other beneficial effects of LAB, such as the ability to convert starch to lactic acid, have been observed in species isolated from Thai fermented rice noodles [36].
Probiotic Pediococcus pentosaceous strains are also the most commonly found strains in Wakalim, a traditional Ethiopian fermented beef sausage. These strains were tolerant to a pH of 3 and a 0.3 % bile salt concentration [37]. A fish sauce product is also a fermented food made from different raw materials such as fish and shellfish. It was found probiotic isolates such as Lactobacillus plantarum, Saccharomyces cerevisiae and Staphylococcus arlettae. They possessed inhibitory effect against S. aureus and Listeria monocytogenes. [38]. In India, L. plantarum isolated from fermented idli batter has been tested successfully as a co-aggregation with pathogens like Listeria monocytogenes and Escherichia coli [39]. In addition, LAB genera, including genera of Enterococcus, Lactobacillus, Lactococcus, Vagococcus and Weissella, have been isolated from a traditional fermented soybean food in India. These isolates, excluding Vagococcus sp. and Weissella sp., showed antibacterial activity against some pathogenic bacteria (B. cereus, E. coli and Salmonella paratyphi) [40]. In many European countries, fermented foods of plant origins, such as fermented olives, are highly interesting as health-promoting, functional foods that could replace fermented dairy food products for lactose-intolerant humans. The fermented olives have been screened for probiotic lactic acid bacteria. The selected probiotic strains possessed probiotic properties in vitro, including an ability to resist low pH levels and high bile concentrations and an ability to adhere to Caco-2 cells. However, the probiotics did not inhibit the growth of pathogens [41]. The main isolates from green olives were L. pentosus, L. plantarum and L. paracasei and from black olives were L. pentosus and Leuconostoc mesenteroides [42, 43]. One strain of LAB isolated from kitchen waste of fermented vegetables was identified as L. delbrueckii, which inhibited the growth of some pathogens, including Proteus vulgaris, Bacillus subtilis, Pseudomonas aeruginosa, E. coli, and Klebsiella pneumoniae [44]. Naturally fermented Croatian dry fermented sausage has been screened to find LAB; L. plantarum and L. brevis were the main species found [45]. In addition, in Middle Eastern countries, different fermented foods containing different raw materials, including parboiled dried wheat, garlic, parsley and olives are rich sources of LAB and can be screened for potential probiotics [46].
Several traditional non-dairy fermented beverages are also good sources of probiotics. These beverages are made from a variety of raw materials, including cereals, millets, legumes, fruits and vegetables [47]. These raw materials have been used to make traditional fermented beverages such as Boza, Pozol, Bushera, Mahewu, and Togwa in several countries in Europe, America and Africa [48]. Cereal grains are a highly nutritional source of protein, carbohydrates, vitamins, minerals and water-soluble fiber, which are materials well-suited to act as prebiotics. Most LAB isolates of Lactobacillus plantarum have been isolated from a Turkish traditional fermented drink (Boza). These isolates showed antagonistic activity by producing substances (mainly organic acids and hydrogen peroxide) to combat pathogenic bacteria such as Listeria monocytogenes, Bacillus cereus, B. subtilis, Yersinia enterocolitica, E. coli, Pseudomonas aeruginosa, Salmonella typhimurium and Klebsiella pneumonia [49]. In addition, Oluwajoba et al.[50] isolated probiotic LAB from Kunu-zaki, a Nigerian traditional fermented drink made from non-germinated sorghum and millet cereal grains. These LAB species showed promising probiotic properties, including resistance to a pH 3 and to 3 % bile and antimicrobial activity against the referent strains of Staphylococcus aureus, Escherichia coli, Pseudomonas aeruginosa and Enterococcus faecalis. These species were identified as Lactobacillus, Pediococcus and Lactococcus species but were primarily Lactobacillus species.
Probiotic isolates used in humans
For the last two decades, the probiotics that were selected for use in humans originated from the human body, mainly feces or breast milk, or from human foods, which were usually fermented dairy products. The probiotics used in animals, in contrast, often came from animals’ own digestive tract or from probiotic strains originating in human subjects. Probiotic strains can be isolated directly from natural fermented milk products or milk and can then be added as starter cultures for fermentation in products such as cheese, yogurt, and butter. Potential probiotics have been isolated from human sources from different parts of the human body, including the human feces of both healthy adults [51] and breast-fed infants [52], as well as human breast milk [53, 54].
Probiotics isolated from human breast milk have usually been of the Lactobacillus genus [54, 55], while probiotics from the feces of healthy human adults and breast-fed infants have been from at least two genera: Lactobacillus [55–57] and Bifidobacterium [52]. A few studies have reported probiotic Enterococcus faecalis found in human feces [58]. L. salivarius has been found in human milk and infant feces in individuals of a mother-child pair [59]. L. rhamnosus and L. casei were isolated from human breast milk and showed resistance to low pH (pH 3), tolerance against a 0.3 % bile concentration and antimicrobial activity against Escherichia coli, Bacillus cereus and Staphylococcus aureus [54]. Another genus of probiotic, Pediococcus, which produces a bacteriocin-like substance, can be isolated from healthy human breast milk [60]. Lactobacilli have been isolated from the feces of children aged 4–15 years. One of the 20 lactobacilli isolated from these specimens was identified as L. pentosus and possessed basic probiotic properties of acid and -bile tolerance and antimicrobial activity. This isolate also had other probiotic properties, including the abilities to produce and aggregate exopolysaccharides (EPS) and to provide a cholesterol removal effect [61].
Probiotic isolates used in farm animals
The direct fed microbial (DFM) supplementation concept involves microorganisms mixed with feed to benefit the animals. It is mainly used in the US. In European countries, probiotics have been developed for use mainly in animal production. This supplementation is also based on the administration of one or several live microorganisms, usually yeast or bacteria. In the last two decades, most Lactobacillus strains used in humans have also been used as probiotics in animals, but Bifidobacterium strains isolated from a human origin were used as probiotics only in humans. Over the past 20 years, the probiotic strains widely used in animals, especially those used in Europe and Japan, are spore forming bacteria of the genus Bacillus [22]. Currently, most of the probiotics used in animal farming are lactic acid bacteria (LAB). Sources of probiotics for use in various animal species, including poultry [62], pigs [63] and ruminants [64], are mainly the gastrointestinal (GI) tracts of the same animal species. These probiotics can also be isolated from the feces of different animal species, including chickens [65], pigs [66] and ruminants [67]. Probiotics isolated from an animal species have also been used in another animal species. They may also come from other sources, including fermentation products of plants and animal origin. B. pumilus WIT 588 isolated from sea water has been tested in animals and exhibits an ability to inhibit the growth of Escherichia coli [68]. Propionibacterium freudenreichii isolated from dairy products has been used to reduce enteritis and to improve health in pigs [69]. Several researchers have isolated probiotics from different sources and used in animals as summarized in Table 1.
Table 1.
Probiotic strains used in farm animals
| Probiotic strains | Sources | Identification techniques | Activities | References |
|---|---|---|---|---|
| Streptomyces sp. JD9 (KF878075) | Indigenous and broiler chickens | PCR with universal primers and 16S rRNA gene sequencing | Enhanced broiler production | Latha et al. [70] |
| Wickerhamomyces anomalus LV-6 | Broiler chickens | PCR-fingerprinting technique and 26S rRNA gene sequencing | Enhanced broiler production | García-Hernández et al. [71] |
| Lactobacillus salivarius 15 K | Chickens | PCR with specific primers and16S- 23S rRNA gene sequencing | Against Klebsiella and Escherichia coli. | Bujnakova et al. [72] |
| Lactobacillus plantarum TN8 | Indigenous Poultry | PCR with specific primers and 16S rRNA gene sequencing | Imunomodulation in vitro | Ben Salah et al. [73] |
| Propionibacterium acidipropionici LET 105 | Laying hens | PCR with universal primers and 16S rRNA gene sequencing | Produced short chain fatty acids (SCFA) and against Salmonella | Argañaraz-Martínez et al. [74] |
| Lactobacillus plantarum P-8 | Traditional fermented dairy products | PCR with universal primers and 16S rRNA gene sequencing | Imunomodulation in broilers | Wang et al. [75] |
| Lactobacillus salivarius DSPV 001P | Broiler chickens | PCR with universal primers and 16S rRNA gene sequencing | Colonization in intestinal broilers | Blajman et al. [76] |
| L. plantarum (strain P6), L. paraplantarum (strain P25), and L. reuteri (strain P30) | Cows, pigs, chickens, and ducks. | PCR with universal primers and 16S rRNA gene sequencing | Anti-pathogens in vitro | Pringsulaka et al. [77] |
|
Lactobacillus johnsonii,
L. salivarius, L. murinus, L. mucosae, L. amylovorus, L. mucosae |
Young calves | PCR with universal primers and 16S rRNA gene sequencing | -Anti-pathogens in vitro -Adhesion property |
Maldonado et al. [78] |
| Lactobacillus reuteri DDL 19, Lactobacillus alimentarius DDL 48, Enterococcus faecium DDE 39, and Bifidobacterium bifidum DDBA | Goat | PCR with universal primers and 16S rRNA gene sequencing | -Increased milk production and polyunsaturated fatty acid -Antimutagenic activity |
Apás et al. [79] |
| Bacillus subtilis KN-42 | Weaned pig | PCR with the primers for denaturing gradient gel electrophoresis and 16S rRNA gene sequencing | -Reduced E. coli
-Increased ADG and FCR improvement |
Hu et al. [80] |
Giang and co-authors have isolated LAB from different parts of the intestines of healthy fattening pigs [63]. These bacterial strains included Enterococcus faecium, Lactobacillus acidophilus, Pediococcus pentosaceus and L. plantarum and were used as probiotic complexes to improve the growth of weaned piglets. Iniguez-Palomares et al. [81] reported Lactobacillus strains isolated from the small intestines of piglets; most of these strains were of the L. salivarius species. These strains showed promising probiotic properties, including resistance to a pH of 3 and to conjugated porcine bile, auto-aggregation effects and an ability to strongly inhibit the pathogen E. coli K88.
LAB species have been isolated from silages in hot and humid weather. They can be used as starter cultures in silages for ruminant. These strains were Lactobacillus plantarum, L. pentosus, L. rhamnosus, L. buchneri, L. rapi, Pediococcus pentosaceus and P. lolii [82]. LAB isolated from fecal young calves such as L. murinus, L. johnsonii and L. salivarius had an ability to produce bacteriocin-like activity against pathogens [83].
Fuller reported microbial communities in chicken guts, found amount 29 genera. Each genus is distinguished as being of 3 to 4 species and each species is separated into 3 to 4 subspecies depending on the different mechanisms of the microorganisms. Therefore, the number of types of microorganisms in chicken guts is found to be over 200 species. In addition, wild types of chicken were found to have a greater amount of intestinal microflora than chickens that were commercially raised due to an opportunity for these microbes to receive from hens [2].
Probiotic bacteria widely used in aquatic animals are LAB and Bacillus species. Most these probiotic strains are isolated from aquatic animals from their gastrointestinal tracts. Several studies have reported the probiotic isolates, isolated from both fresh water and sea water animals. Diaz et al. reported that Lactobacillus salivarius from bottlenose dolphin can inhibit the growth of Salmonalla Enteritidis strains that isolated from both marine animals and humans [84]. Leuconostoc mesenteroides strains have been isolated from the intestines of fresh water fishes such as snakehead fish [85] and Nile tilapia fish [86]. They showed an ability to inhibit the growth of fish pathogens. Bacillus pumilus and B. clausii isolated from the guts of fish grouper Epinephelus coioides showed beneficial effects, including an ability to inhibit the fish pathogens, growth performance improvements and an immune stimulation [87].
Recently, Munoz-Atienza et al. [88] used probiotic Leuconostoc cremoris and Weissella cibaria isolated from Atlantic salmon fish and common octopus, respectively to inhibit the marine fish (turbot fish) pathogens successfully and to stimulate a non-specific immune response. These probiotic isolates can be survived in seawater at 18 °C for 7 days and resisted to pH 3 and 10 % (v/v) turbot bile [52]. Sarkono et al. reported that Lactobacillus paracasei isolated from eyes shellfish (abalone) showed a resistance to acidic and bile conditions and an ability to inhibit pathogenic bacteria such as E. coli, Bacillus cereus and Staphylococcus aureus [89].
Selection for probiotics
Selection of probiotics from different sources involves screening for non-pathogenic bacteria, such as LAB, followed by an evaluation of the basic features of these bacteria, including acid and bile tolerance, ability to adhere to gut epithelial cells and ability to combat pathogens in the GI tract. The pathogenic properties of Lactobacillus salivarius isolated from human milk have been tested by an evaluation of oral toxicity in mice [90]. However, novel probiotics must be tested for beneficial properties in both in vitro and in vivo models. The novelty of probiotic strains is considered together with safety requirements, which include a complete genome description and annotation, knowledge regarding the transferability of antibiotic resistance, selection of the proper in vivo model, toxicological studies and designation of the target population, as recently described by Kumar et al. [91].
Selection procedures for probiotics isolated from fermented foods usually include testing for probiotic properties such as a tolerance to heat, acid, bile salt and NaCl. The probiotic strains are also tested for antimicrobial activity against pathogens. In addition, other features have been considered when selecting probiotics. These include the production of bile salt hydrolase [92], the production of exopolysaccharides (EPS) [19, 61, 93], which increase the colonization of probiotics in the gut, an ability to inhibit the harmful fecal enzymes of intestinal microflora, including β-glucosidase, β-glucuronidase, tryptophanase and ureas [51, 72], and safety features. Safety features include being non-pathogenic microorganisms [90] and not being able to transfer any antibiotic resistance genes to other bacteria.
Selection of LAB for use as potential probiotics begins by screening for exopolysaccharide (EPS)-producing bacteria. [19, 61, 93]. Then, the selected bacteria are tested for other properties, including resistance to acid and bile, and an ability to combat pathogens in the GI tract. Microbes that are potential probiotics have been isolated from vegetables and traditional dairy fermented foods; these microbes have the basic features just mentioned and represent approximately 24 % of all EPS-producing strains [19]. EPS-producing bacteria can be found in both fermented dairy and non-dairy foods. The EPS-producing activity of these bacteria is strain specific [93].
Probiotics isolated from intestines in both humans and animals have some probiotic properties that are different from those of the probiotics originating from dairy products. The adhesion of these probiotics is one of the most notable differences. Intestinal isolates usually exhibit higher adhesion activity than the dairy isolates. Bunesova and colleagues reported that Bifidobacterium animalis isolated from feces showed strong auto-aggregation activity, which is related to adhesion in the GI tract. This activity was not observed in the probiotic strains isolated from dairy products [94]. In addition, the intestinal probiotic isolates are more likely to be resistant to low pH levels and high concentrations of bile than are the probiotics isolated from dairy origins [95]. However, Monteagudo-Mera et al. reported that some Lactobacillus strains isolated from cheese were more tolerant to low pH levels and more adherent to CaCo-2 cells than was Lactobacillus spp. isolated from human feces. Therefore, probiotics in dairy products may select for strains that can be isolated from these products for a more beneficial use [83].
Safety properties of probiotics
One of the important safety properties of probiotic microbes is an antibiotic resistant feature. Generally, antimicrobial resistances of probiotic microorganisms are two characteristics: (i) natural or intrinsic resistance, in which case resistance is not transferable; (ii) acquired resistance, usually caused from bacterial mutation or may carry plasmid encoding of antibiotic resistance genes and potentially transferable to other commensal or pathogenic bacteria [96]. Studies of antimicrobial resistance of probiotic microbes have reported depending on isolation sources and antibiotic tested groups. LAB is one of the large groups to select as potential probiotics and also to test for antimicrobial susceptibility. Analyses of antibiotic resistance of probiotics are in both phenotypic and molecular methods. Broad spectrum antibiotics such as tetracycline and chloramphenicol have been detected antibiotic resistant genes as a horizontal gene transfer in LAB probiotics or starters mainly lactobacilli, including tet(W), tet(M), tet(S), tet(O),tet(Q), tet(36), tet(Z), tet(O/W/32/O/W/O), tet(W/O), tet(K), tet(L) and cat gene, respectively [97]. LAB mostly lactobacilli isolated from various sources, including GI tracts of animals such as chickens [98], dogs [99] and wild boars [100], human feces [101], fermented food products [29, 102] and fermented milk products. Njage et al. have been reported that they can be resisted to tetracycline that they may acquire the resistance gene from other bacteria [103].
Isolation and identification of probiotics
An initial isolation of probiotic LAB from uncommon sources or non-intestinal sources is cultivation using a high nutritional medium which is different or modification from conventional de Man, Rgosa and Sharp (MRS) medium. For example, LAB isolated from paddy rice silage, crop and silage fermentation have been cultivated using medium consisting mainly glucose, yeast extract and peptone (GYP) [104, 105]. LAB isolated from soil (rhizospheres of fruit trees and soil around animal farms) was cultivated successfully using GYP plus BM medium [17, 18]. BM medium containing the most of tomato juices, peptone, liver extracts and glucose is used to cultivate malolactic-producing LAB such as Oenococcus oeni from red wine- making in Japan [106, 107].
LAB screening from various parts of GI tracts of animals is also used a modified MRS medium. In this case, high producing lactic acid of LAB isolates that are usually found in the GI tracts of animals may require some substrates for suitable growths such as pH conditions and nutrients. LAB has been isolated from GI tracts of animals using a modified MRS medium by adding 0.3–1 % (w/v) CaCO3. This medium has been used to isolate LAB successfully in various animals, including chickens [62, 98], cattle [108] and dogs [99]. The medium has also been used to cultivate LAB from traditional fermented foods which were made from various raw materials such as fish (Pla-chom) [102] and beef (Mum) [29]. LAB was cultivated from fermented foods in acidic conditions such as a traditional pickle food using medium consisting the most of glucose, yeast extract and peptone and 0.5 % (w/v) CaCO3 [105].
LAB species were isolated from an air surrounding sourdough and bakery room productions. Identification of these LAB using molecular methods depends on culture-dependent assays that they were cultivated using MRS-5, a modified version of high nutritional MRS medium [109], for a sample enrichment that these LAB were successfully growths [27]. Thus, screening of probiotic LAB from uncommon sources, such as soil and air can be succeeded using accumulation methods that they are cultivated using high nutrition modified MRS medium such as MRS-5 and GYP plus BM medium under anaerobic conditions.
Screening of some intestinal LAB probiotic and Bifidobacteria are also not easy using the best anaerobic culture methods because a large complex bacterial community inhabiting the GI tract and many species of the microbes have never been cultivated under laboratory environments. Indigenous microbiota in intestinal sources in both humans and animals are identified by two main methods. The first one, a culture-based method, is observed phenotype characteristics including biochemical, physiological and morphological tests in accordance with Bergey’s Manual. The culture-based method associated with biological molecular technique is a way for more effective identifications. The second one, a culture-independent method is considered as alternative techniques to investigate the large proportion of the both cultured and uncultured bacteria in GI tracts. These methods are developed to discriminate between species of bacteria from the difference of their DNA fragments in band profiles, such as the PCR in Denaturing Gradient Gel Electrophoresis (PCR-DGGE) and the PCR in Temperature Gradient Gel Electrophoresis (PCR-TGGE). The fluorescence in situ hybridization (FISH) technique was also used to detect the uncultured bacteria [110]. Biochemical testing of LAB identification is widely used by carbohydrate fermentation patterns such as the API (API system, Biomerieux, France). Biological molecular methods have been increasingly used and there are many methods, including a DNA base composition (mol % of guanine plus cytosine), a DNA homology accompanied with polymerase chain reaction (PCR) technique and DNA sequencing using 16S rRNA gene region. 16S rRNA gene sequencing is developed to simplify sequences using species-specific PCR primers that targeted some regions of lactobacilli such as the 16S – 23S rRNA spacer region [111], the shuttle cloning vector to extract the bacterial plasmids [112], internal transcribed spacer PCR (ITS-PCR) [113] and amplifications with amplified ribosomal DNA restriction analysis (ARDRA) [114]. Fingerprinting techniques are the methods to differentiate the microbial community at strain levels. These methods have been developed to discriminate strains of LAB, especially lactobacilli, including the ribotyping [115], randomly amplified polymorphic DNA (RAPD)-PCR [116], amplified fragment length polymorphism (AFLP) [117], plasmid profiling [118] and pulsed-field gel electrophoresis (PFGE) [112]. A PFGE technique is high discriminatory power using the specific restriction enzyme of the genomic DNA of bacteria. A pyrosequencing technique with specific primers [119] is also useful methods to differentiate these probiotic species.
Conclusions
Alternative sources of probiotics, such as non-dairy fermented food products, present an advantage in the search for new probiotic strains. Increasingly, these probiotic sources are being selected for use in people who are lactose intolerant. The selection of probiotics from different sources involves screening for non-pathogenic microbes followed by an evaluation of basic properties, including acid and bile tolerance, an ability to adhere to gut epithelial cells, an ability to combat against pathogens in the GI tract, and the safety-enhancing property of an inability to transfer any antibiotic resistance genes to other bacteria. Selected probiotics isolated from intestinal sources in both humans and animals are identified using molecular methods by two main methods: culture-based and culture-independent methods. The culture-independent method is considered as alternative techniques to investigate the large proportion of the both cultured and uncultured bacteria in the GI tract.
Abbreviations
CFU, colony forming unit; DFM, direct fed microbial; EPS, exopolysaccharides; FISH, fluorescence in situ hybridization; GI, gastrointestinal; GYP, glucose, yeast extract and peptone; LAB, lactic acid bacteria; MRS, de Man, Rogosa and Sharp; PCR-DGGE, PCR in denaturing gradient Gel electrophoresis; PCR-TGGE, PCR in temperature gradient Gel electrophoresis; PFGE, pulsed-field gel electrophoresis; RAPD, randomly amplified polymorphic DNA
Acknowledgements
The authors would like to thank Professor Dr. Min-Tze Liong from School of Industrial Technology, University Sains Malaysia and Associate Professor Dr. Pornthap Thanonkeo, Faculty of Technology, Khon Kaen University, Thailand for helpful comments on the manuscript.
Funding
The authors would like to acknowledge Thailand Research Fund organization (Research and Researcher for Industry 2014) (PHD5710071) for the financial support in writing the manuscript.
Availability of data and materials
Not applicable.
Authors’ contributions
Wrote the manuscript: PS. Both authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Contributor Information
Pairat Sornplang, Email: pirson@kku.ac.th.
Sudthidol Piyadeatsoontorn, Email: sudthidol@gmail.com.
References
- 1.Gilliland SE. Health and nutritional benefits from lactic acid bacteria. FEMS Microbiol Rev. 1990;7:175–86. doi: 10.1111/j.1574-6968.1990.tb04887.x. [DOI] [PubMed] [Google Scholar]
- 2.Fuller R. Probiotics in man and animals. J Appl Bacteriol. 1989;66:365–78. doi: 10.1111/j.1365-2672.1989.tb05105.x. [DOI] [PubMed] [Google Scholar]
- 3.FAO/WHO. Guidelines for the evaluation of probiotics in food. London, Ontario, Canada: Food and Agriculture Organization of the United Nations and World Health Organization Working Group Report; 2002. Available at: ftp://ftp.fao.org/es/esn/food/wgreport2.pdf. Accessed 7 June 2016.
- 4.Fuller R. Probiotic: the scientific basis. London: Chapman and Hall; 1992. [Google Scholar]
- 5.Nousiainen J, Setala J. Lactic acid bacteria as animal probiotics. In: Salminen S, von WA, editors. Lactic acid bacteria. New York: Mercel Dekker Inc; 1998. [Google Scholar]
- 6.Soccol CR, Vandenberghe LPDS, Spier MR, Medeiros ABP, Yamaguishi CT, Lindner JDD, Pandey A, Thomaz-Soccol V. The potential of probiotics: a review. Food Technology and Biotechnology. 2010;48:413–34. [Google Scholar]
- 7.Hyronimus B, Le Marrec C, Sassi AH, Deschamps A. Acid and bile tolerance of spore-forming lactic acid bacteria. Int J Food Microbiol. 2000;61:193–97. doi: 10.1016/S0168-1605(00)00366-4. [DOI] [PubMed] [Google Scholar]
- 8.Hong HA, Cutting SM. The use of bacterial spore formers as probiotics. FEMS Microbiol Rev. 2005;29:813–35. doi: 10.1016/j.femsre.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 9.Bayane A, Diawara B, Dubois RD, Destain J, Roblain D, Thonart P. Isolation and characterization of new spore-forming lactic acid bacteria with prospects of use in food fermentations and probiotic preparations. Afr J Microbiol Res. 2010;4:1016–25. [Google Scholar]
- 10.Siddiqee MH, Sarker H, Shurovi KM. Assessment of probiotic application of lactic acid bacteria (LAB) isolated from different food items. Stamford J Microbiol. 2013;2:10–4. [Google Scholar]
- 11.Ramirez-Chavarin ML, Wacher C, Eslava-Campos CA, Perez-Chabela ML. Probiotic potential of thermotolerant lactic acid bacteria strains isolated from cooked meat products. Int Food Res J. 2013;20:991–1000. [Google Scholar]
- 12.Pundir RK, Rana S, Kashyap N, Kaur A. Probiotic potential of lactic acid bacteria isolated from food samples: an in vitro study. J Appl Pharm Sci. 2013;3:85–93. [Google Scholar]
- 13.Schoster A, Weese JS, Guardabassi L. Probiotic use in horses – What is the evidence for their clinical efficacy. J Vet Intern Med. 2014;28:1640–52. doi: 10.1111/jvim.12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Naeem M, Ilyas M, Haider S, Baig S, Saleem M. Isolation characterization and identification of lactic acid bacteria from fruit juices and their efficacy against antibiotics. Pak J Bot. 2012;44:323–8. [Google Scholar]
- 15.Hamet MF, Londero A, Medrano M, Vercammen E, Van Hoorde K, Garrote GL, Huys G, Vandamme P, Abraham AG. Application of culture-dependent and culture-independent methods for the identification of Lactobacillus kefiranofaciens in microbial consortia present in kefir grains. Food Microbiol. 2013;36:327–34. doi: 10.1016/j.fm.2013.06.022. [DOI] [PubMed] [Google Scholar]
- 16.Tajabadi N, Mardan M, Manap MYA, Mustafa S. Molecular identification of Lactobacillus spp. isolated from the honey comb of the honey bee (Apis dorsata) by 16S rRNA gene sequencing. J Apic Res. 2013;52:235–41. doi: 10.3896/IBRA.1.52.5.10. [DOI] [Google Scholar]
- 17.Chen YS, Yanagida F, Shinohara T. Isolation and identification of lactic acid bacteria from soil using an enrichment procedure. Lett Appl Microbiol. 2005;40:195–200. doi: 10.1111/j.1472-765X.2005.01653.x. [DOI] [PubMed] [Google Scholar]
- 18.Yanagida F, Chen YS, Shinohara T. Searching for bacteriocin-producing lactic acid bacteria in soil. J Gen Appl Microbiol. 2006;52:21–8. doi: 10.2323/jgam.52.21. [DOI] [PubMed] [Google Scholar]
- 19.Patel A, Prajapati JB, Holst O, Ljungh A. Determining probiotic potential of exopolysaccharide producing lactic acid bacteria isolated from vegetables and traditional Indian fermented food products. Food Biosci. 2014;5:27–33. doi: 10.1016/j.fbio.2013.10.002. [DOI] [Google Scholar]
- 20.Yoon KY, Woodams EE, Hang YD. Probiotication of tomato juice by lactic acid bacteria. The J Microbiol. 2004;42:315–8. [PubMed] [Google Scholar]
- 21.Miranda JM, Samuel A, Nebot CG, Cepeda A, Franco CM, Calo-Mata MP. Technological characterization of lactic acid bacteria isolated from beef stored on Vacuum-Packaged and Advanced Vacuum Skin Packaged System. J Food Process Technol. 2014;5:1–6. [Google Scholar]
- 22.El-Mabrok ASW, Hassan Z, Mokhtar AM, Hussain KMA, Kahar FKSBA. Sceening of lactic acid bacteria as biocontrol against Colletotrichum capsici on chilli Bangi. Res J Appl Sci. 2012;7:466–73. [Google Scholar]
- 23.Sirilun S, Chaiyasut C, Kantachote D, Luxananil P. Characterisation of non human origin probiotic Lactobacillus plantarum with cholesterol-lowering olesterol-lowering property. Afr J Microbiol Res. 2010;4:994–1000. [Google Scholar]
- 24.Zendo T. Screening and characterization of novel bacteriocins from lactic acid bacteria. Biosci Biotechnol Biochem. 2013;77:893–9. doi: 10.1271/bbb.130014. [DOI] [PubMed] [Google Scholar]
- 25.Olofsson TC, Vasquez A. Detection and identification of a novel lactic acid bacterial flora within the honey stomach of the honeybee Apis mellifera. Curr Microbiol. 2008;57:356–63. doi: 10.1007/s00284-008-9202-0. [DOI] [PubMed] [Google Scholar]
- 26.Shiphrah VH, Sahu S, Thakur AR, Chaudhuri SR. Screening of bacteria for lactic acid production from whey water. Am J Biochem Biotechnol. 2013;9:118–23. doi: 10.3844/ajbbsp.2013.118.123. [DOI] [Google Scholar]
- 27.Scheirlinck I, Van der Meulen R, De Vuyst L, Vandamme P, Huys G. Molecular source tracking of predominant lactic acid bacteria in traditional Belgian sourdoughs and their production environments. J Appl Microbiol. 2009;106:1081–92. doi: 10.1111/j.1365-2672.2008.04094.x. [DOI] [PubMed] [Google Scholar]
- 28.Paludan-Müller C, Huss HH, Gram L. Characterization of lactic acid bacteria isolated from a Thai low-salt fermented fish product and the role of garlic as substrate for fermentation. Int J Food Microbiol. 1999;46:219–29. doi: 10.1016/S0168-1605(98)00204-9. [DOI] [PubMed] [Google Scholar]
- 29.Sornplang P, Yowarach S, Leelavatcharamas V. Isolation and susceptibility to antibiotics of lactic acid bacteria from fermented beef product, Mum. Paper presented at the In: Proceeding of the 22nd Annual Meeting of the Thai Society for Biotechnology. TSB 2010 International Conference on Biotechnology for Healthy Living., Prince of Songkla University, Trang Campus, Thailand., 20–22 October 2010; 2010
- 30.Sornplang P, Leelavatcharamas V. Antimicrobial susceptibility of probiotic lactobacilli isolated from chicken feces. KKU Res J. 2010;15:693–701. [Google Scholar]
- 31.Siripornadulsil W, Tasaku S, Buahorm J, Siripornadulsil S. Probiotic properties of lactic acid bacteria isolated from fermented food. Intl J Biol food Vet Agri Eng. 2014;8:364–6. [Google Scholar]
- 32.Senthong R, Chanthachum S, Sumpavapol P. Screening and identification of probiotic lactic acid bacteria isolated from Poo-Khem, A traditional salted crab. Paper presented at the International Conference on Nutrition and Food Sciences; 2012
- 33.Nanasombat S, Phunpruch S, Jaichalad T. Screening and identification of lactic acid bacteria from raw seafood and Thai fermented seafood products for their potential use as starter cultures. Songklanakarin J Sci Technol. 2012;34:255–62. [Google Scholar]
- 34.Cho MJ, Lee JY, Kim JH. Microbial and physiochemical properties of Cheonggukjang fermented using Bacillus strains with antibacterial or antifungal activities. Food Sci Biotechnol. 2014;23:1525–32. doi: 10.1007/s10068-014-0208-z. [DOI] [Google Scholar]
- 35.Miyashita M, Yukphan P, Chaipitakchonlatarn W, Malimas T, Sugimoto M, Yoshino M, Potacharoen W, Tanasupawat S, Nakagawa Y, Kirtikara K, Tanticharoen M. 16S rRNA gene sequence analysis of lactic acid bacteria isolated from fermented foods in Thailand. Microbiol Cult Coll. 2012;28:1–9. [Google Scholar]
- 36.Kanpiengjai A, Rieantrakoonchai W, Pratanaphon R, Pathom-aree W, Lumyong S, Khanongnuch C. High efficacy bioconversion of starch to lactic acid using an amylolytic lactic acid bacterium isolated from Thai indigenous fermented rice noodles. Food Sci Biotechnol. 2014;23:1541–50. doi: 10.1007/s10068-014-0210-5. [DOI] [Google Scholar]
- 37.Bacha K, Mehari T, Ashenafi M. In-vitro probiotic potential of lactic acid bacteria isolated from ‘Wakalim’, a traditional Ethiopian fermented beef sausage. Ethiop J Health Sci. 2009;19:21–9. [Google Scholar]
- 38.Sim KY, Chye FY, Anton A. Probiotic potential and antimicrobial activities of micro- organisms isolated from an indigenous fish sauce. Borneo Science. 2012;31:57–63. [Google Scholar]
- 39.Agaliya PJ, Jeevaratnam K. Screening of Lactobacillus plantarum isolated from fermented idli batter for probiotic properties. Afr J Biotechnol. 2012;11:12856–64. [Google Scholar]
- 40.Thokchom S, Joshi SR. Antibiotic resistance and probiotic properties of dominant lactic microflora from Tungrymbai, an ethnic fermented soybean food of Indian. J Microbiol. 2012;50:535–9. doi: 10.1007/s12275-012-1409-x. [DOI] [PubMed] [Google Scholar]
- 41.De Bellis P, Valerio F, Sisto A, Lonigro SL, Lavermicocca P. Probiotic table olives: microbial populations adhering on olive surface in fermentation sets inoculated with the probiotic strain Lactobacillus paracasei IMPC2.1 in an industrial plant. Int J Food Microbiol. 2010;140:6–13. doi: 10.1016/j.ijfoodmicro.2010.02.024. [DOI] [PubMed] [Google Scholar]
- 42.Argyri AA, Zoumpopoulou G, Karatzas KAG, Tsakalidou E, Nychas GJE, Panagou E, Tassou CC. Selection of potential probiotic lactic acid bacteria from fermented olives by in vitro tests. Food microbial. 2013;33:282–91. doi: 10.1016/j.fm.2012.10.005. [DOI] [PubMed] [Google Scholar]
- 43.Doulgeraki AI, Pramateftaki P, Argyri AA, Nychas GJE, Tassou CC, Panagou EZ. Molecular characterization of lactic acid bacteria isolated from industrially fermented Greek table olives. LWT- Food Sci Technol. 2013;50:353–6. doi: 10.1016/j.lwt.2012.07.003. [DOI] [Google Scholar]
- 44.Rauta PR, Dhupal M, Nayak B. Screening and characterization of potential probiotic lactic acid bacteria isolated from vegetable waste and fish intestine. Int J Curr Microbiol App Sci. 2013;2:234–44. [Google Scholar]
- 45.Fleck ZC, Savic V, Kozacins L, Njari B, Zdolec N, Filipovic I. Identification of lactic acid bacteria isolated from dry fermented sausages. Vet Arhiv. 2012;82:265–72. [Google Scholar]
- 46.Mahasneh AM, Abbas MM. Probiotics and traditional fermented foods: The eternal connection (Mini-Review) J J Biol Sci. 2010;3:133–40. [Google Scholar]
- 47.Krishnamoorthy M, Arjun P. Probiotic and antimicrobial activity of bacteria from fermented toddy of Cocus nucifera. J Acad Indus Res. 2012;1:127–31. [Google Scholar]
- 48.Vasudha S, Mishra HN. Non dairy probiotic beverages. Int Food Res J. 2013;20:7–15. [Google Scholar]
- 49.Kivanc M, Yilmaz M, Cakir E. Isolation and identification of lactic acid bacteria from Boza, and their microbial activity against several reporter strains. Turk J Biol. 2011;35:313–24. [Google Scholar]
- 50.Oluwajoba SO, Akinyosoye FA, Oyetayo VO. In vitro screening and selection of probiotic lactic acid bacteria isolated from spontaneously fermenting Kunu-Zaki. Adv Microbiol. 2013;3:309–16. doi: 10.4236/aim.2013.34044. [DOI] [Google Scholar]
- 51.Choi SS, Kang BY, Chung MJ, Kim SD, Park SH, Kim JS, Ha NJ. Safety assessment of potential lactic acid bacteria Bifidobacterium longum SPM1205 isolated from healthy Koreans. J Microbiol. 2005;43:493–8. [PubMed] [Google Scholar]
- 52.Munoz JAM, Chenoll E, Casinos B, Bataller E, Ramon D, Genoves S, Montava R, Ribes JM, Buesa J, Fàbrega J, Rivero M. Novel probiotic Bifidobacterium longum subsp.infantis CECT 7210 strain active against rotavirus infections. Appl Environ Microbiol. 2011;7724:8775–83. doi: 10.1128/AEM.05548-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martin R, Jimenez E, Olivares M, Marin ML, Fernandez L, Xaus J, Rodríguez JM. Lactobacillus salivarius CECT 5713, a potential probiotic strain isolated from infant feces and breast milk of a mother–child pair. Int J Food Microbiol. 2006;112:35–43. doi: 10.1016/j.ijfoodmicro.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 54.Kavitha JR, Devasena T. Isolation, characterization, determination of probiotic properties of lactic acid bacteria from human milk. OSR J Pharm Biol Sci. 2013;7:1–7. [Google Scholar]
- 55.Shokryazdan P, Sieo CC, Kalavathy R, Liang JB, Alitheen NB, Faseleh Jahromi M, Ho YW. Probiotic potential of Lactobacillus strains with antimicrobial activity against some human pathogenic strains. Biomed Res Int. 2014;2014:1–16. doi: 10.1155/2014/927268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Verdenelli MC, Ghelfi F, Silvi S, Orpianesi C, Cecchini C, Cresci A. Probiotic properties of Lactobacillus rhamnosus and Lactobacillus paracasei isolated from human faeces. Eur J Nutr. 2009;48:355–63. doi: 10.1007/s00394-009-0021-2. [DOI] [PubMed] [Google Scholar]
- 57.dos Santos Pozza MS, da Silva Miglioranza LH, Garcia JE, Garcia S, Pozza PC. Human gastrointestinal tract resistance of Lactobacillus strains isolated from infant faeces. Semina: Ciencias Agrarias. 2011;32:1021–32. [Google Scholar]
- 58.Nueno-Palop C, Narbad A. Probiotic assessment of Enterococcus faecalis CP58 isolated from human gut. Int J Food Microbiol. 2011;145:390–4. doi: 10.1016/j.ijfoodmicro.2010.12.029. [DOI] [PubMed] [Google Scholar]
- 59.Jimenez E, Martin R, Maldonado R, Martín A, de Segura V, Fernandez AGL, Rodriguez JM. Complete genome sequence of Lactobacillus salivarius CECT 5713, a probiotic strain isolated from human milk and infant feces. J bacterial. 2010;92:5266–7. doi: 10.1128/JB.00703-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Osmanagaoglu O, Kiran F, Nes IF. A probiotic bacterium, Pediococcus pentosaceus OZF, isolated from human breast milk produces pediocin AcH/PA-1. Afr J Biotechnol. 2013;10:2070–9. [Google Scholar]
- 61.Tulumoglu S, Yuksekdag ZN, Beyatli Y, Simsek O, Cinar B, Yaşar E. Probiotic properties of lactobacilli species isolated from children’s feces. Anaerobe. 2013;24:36–42. doi: 10.1016/j.anaerobe.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 62.Ibourahema C, Dauphin RD, Jacqueline D, Thonart P. Characterization of lactic acid bacteria isolated from poultry farms in Senegal. Afr J Biotechnol. 2008;7:2006–12. doi: 10.5897/AJB2008.000-5048. [DOI] [Google Scholar]
- 63.Giang HH, Viet TQ, Ogle B, Lindberg JE. Effects of different probiotic complexes of lactic acid bacteria on growth performance and gut environment of weaned piglets. Livest Sci. 2010;133:182–4. doi: 10.1016/j.livsci.2010.06.059. [DOI] [Google Scholar]
- 64.Ghorbani GR, Morgavi DP, Beauchemin KA, Leedle JAZ. Effects of bacterial direct-fed microbials on ruminal fermentation, blood variables, and the microbial populations of feedlot cattle. J Anim Sci. 2002;80:1977–86. doi: 10.2527/2002.8071977x. [DOI] [PubMed] [Google Scholar]
- 65.Belkacem B, Meriem M, Mebrouk K. Probiotic potential of thermotolerants lactobacilli isolated from chicken gastrointestinal digestive and their use as poultry feed. World Appl Sci J. 2009;7:951–7. [Google Scholar]
- 66.Petsuriyawong B, Khunajakr N. Screening of probiotic lactic acid bacteria from piglet feces. Kasetsart J. 2011;45:245–53. [Google Scholar]
- 67.Stein DR, Allen DT, Perry EB, Bruner JC, Gates KW, Rehberger T, Mertz K, Jones D, Spicer LJ. Effects of feeding propionicbacteria to dairy cows on milk yield, milk components, and reproduction. J Dairy Sci. 2006;89:111–25. doi: 10.3168/jds.S0022-0302(06)72074-4. [DOI] [PubMed] [Google Scholar]
- 68.Prieto ML, O’Sullivan L, Tan SP, McLoughlin P, Hughes H, Gutierrez M, Lane JA, Hickey RM, Lawlor PG, Gardiner GE. In vitro assessment of marine bacillus for use as livestock probiotics. Mar Drugs. 2014;12:2422–45. doi: 10.3390/md12052422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cousin FJ, Jouan-Lanhouet S, Dimanche-Boitrel MT, Corcos L, Jan G. Milk fermented by Propionibacterium freudenreichii induces apoptosis of HGT-1 human gastric cancer cells. PLoS One. 2012;7:31892. doi: 10.1371/journal.pone.0031892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Latha S, Vinothini G, John Dickson Calvin D, Dhanasekaran D. In vitro probiotic profile based selection of indigenous actinobacterial probiont Streptomyces sp. JD9 for enhanced broiler production. J Biosci Bioeng. 2015;121:124–31. doi: 10.1016/j.jbiosc.2015.04.019. [DOI] [PubMed] [Google Scholar]
- 71.García-Hernández Y, Rodríguez Z, Brandão LR, Rosa CA, Nicoli JR, Elías Iglesias A, Peréz-Sanchez T, Salabarría RB, Halaihel N. Identification and in vitro screening of avian yeasts for use as probiotic. Res Vet Sci. 2012;93:798–802. doi: 10.1016/j.rvsc.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 72.Bujnakova D, Strakova E, Kmet V. In vitro evaluation of the safety and probiotic properties of lactobacilli isolated from chicken and calves. Anaerobe. 2014;29:118–27. doi: 10.1016/j.anaerobe.2013.10.009. [DOI] [PubMed] [Google Scholar]
- 73.Ben Salah R, Trabelsi I, Ben Mansour R, Lassoued S, Chouayekh H, Bejar S. A new Lactobacillus plantarum strain, TN8, from the gastro intestinal tract of poultry induces high cytokine production. Anaerobe. 2012;18:436–44. doi: 10.1016/j.anaerobe.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 74.Argañaraz-Martínez E, Babot JD, Apella MC, Perez CA. Physiological and functional characteristics of Propionibacterium strains of the poultry microbiota and relevance for the development of probiotic products. Anaerobe. 2013;23:27–37. doi: 10.1016/j.anaerobe.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 75.Wang L, Liu C, Chen M, Ya T, Huang W, Gao P, Zhang H. A novel Lactobacillus plantarum strain P-8 activates beneficial immune response of broiler chickens. Int Immunopharmacol. 2015;29:901–7. doi: 10.1016/j.intimp.2015.07.024. [DOI] [PubMed] [Google Scholar]
- 76.Blajman J, Gaziano C, Zbrun MV, Soto L, Astesana D, Berisvil A, Scharpen AR, Signorini M, Frizzo L. In vitro and in vivo screening of native lactic acid bacteria toward their selection as a probiotic in broiler chickens. Res Vet Sci. 2015;101:50–6. doi: 10.1016/j.rvsc.2015.05.017. [DOI] [PubMed] [Google Scholar]
- 77.Pringsulaka O, Rueangyotchanthana K, Suwannasai N, Watanapokasin R, Amnueysit P, Sunthornthummas S, Sukkhum S, Sarawaneeyaruk S, Rangsiruji A. In vitro screening of lactic acid bacteria for multi-strain probiotics. Livest Sci. 2015;174:66–73. doi: 10.1016/j.livsci.2015.01.016. [DOI] [Google Scholar]
- 78.Maldonado NC, de Ruiz CS, Otero MC, Sesma F, Nader-Macías ME. Lactic acid bacteria isolated from young calves--characterization and potential as probiotics. Res Vet Sci. 2012;92:342–9. doi: 10.1016/j.rvsc.2011.03.017. [DOI] [PubMed] [Google Scholar]
- 79.Apás AL, González SN, Arena ME. Potential of goat probiotic to bind mutagens. Anaerobe. 2014;28:8–12. doi: 10.1016/j.anaerobe.2014.04.004. [DOI] [PubMed] [Google Scholar]
- 80.Hu Y, Dun Y, Li S, Zhao S, Peng N, Liang Y. Effects of Bacillus subtilis KN-42 on growth performance, diarrhea and faecal bacterial flora of weaned piglets. Asian Australasian J Anim Sci. 2014;27:1131–40. doi: 10.5713/ajas.2013.13737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Iniguez-Palomares C, Perez-Morales R, Acedo-Felix E. Evaluation of probiotic properties in Lactobacillus isolated from small intestine of piglets. Rev Latinoam Microbiol. 2007;49:46–54. [Google Scholar]
- 82.Doi K, Nishizaki Y, Kimura H, Kitahara M, Fujino Y, Ohmomo S, Ohshima T, Ogata S. Identification of thermotolerant lactic acid bacteria isolated from silage prepared in the hot and humid climate of Southwestern Japan. Springerplus. 2013;2:485. doi: 10.1186/2193-1801-2-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Monteagudo-Mera A, Rodriguez-Aparicio L, Rua J, Martinez-Blanco H, Navasa N, Garcia-Armesto MR, Ferrero MÁ. In vitro evaluation of physiological probiotic properties of different lactic acid bacteria strains of dairy and human origin. J Funct Foods. 2012;4:531–41. doi: 10.1016/j.jff.2012.02.014. [DOI] [Google Scholar]
- 84.Diaz MA, Bik EM, Carlin KP, Venn‐Watson SK, Jensen ED, Jones SE, Gaston EP, Relman DA, Versalovic J. Identification of Lactobacillus strains with probiotic features from the bottlenose dolphin (Tursiops truncatus) J Appl Microbiol. 2013;115:1037–51. doi: 10.1111/jam.12305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Allameh SK, Daud H, Yusoff FM, Saad CR, Ideris A. Isolation, identification and characterization of Leuconostoc mesenteroides as a new probiotic from intestine of snakehead fish (Channa striatus) Afr J Biotechnol. 2012;11:3810–16. [Google Scholar]
- 86.Zapata AA, Lara-Flores M. Antimicrobial activities of lactic acid bacteria strains isolated from Nile Tilapia intestine (Oreochromis niloticus) J Biol Life Sci. 2012;4:164–71. doi: 10.5296/jbls.v4i1.2408. [DOI] [Google Scholar]
- 87.Sun YZ, Yang HL, Ma RL, Song K, Lin WY. Probiotic applications of two dominant gut Bacillus strains with antagonistic activity improved the growth performance and immune responses of grouper Epinephelus coioides. Fish & Shellfish Immunol. 2010;29:803–9. doi: 10.1016/j.fsi.2010.07.018. [DOI] [PubMed] [Google Scholar]
- 88.Munoz-Atienza E, Araujo C, Magadan S, Hernandez PE, Herranz C, Santos Y, Cintas LM. In vitro and in vivo evaluation of lactic acid bacteria of aquatic origin as probiotics for turbot (Scophthalmus maximus L.) farming. Fish Shellfish Immunol. 2014;41:570–80. doi: 10.1016/j.fsi.2014.10.007. [DOI] [PubMed] [Google Scholar]
- 89.Sarkono F, Sofyan Y. Isolation and identification of lactic acid bacteria from abalone (Haliotis asinina) as a potential candidate of probiotic. Nusantara Biosci. 2010;2:38–42. [Google Scholar]
- 90.Lara-Villoslada F, Sierra S, Díaz-Ropero MP, Olivares M, Xaus J. Safety assessment of the human milk-isolated probiotic Lactobacillus salivarius CECT5713. J Dairy Sci. 2007;90:3583–9. doi: 10.3168/jds.2006-685. [DOI] [PubMed] [Google Scholar]
- 91.Kumar H, Salminen S, Verhagen H, Rowland I, Heimbach J, Banares S, Young T, Nomoto K, Lalonde M. Novel probiotics and prebiotics: road to the market. Curr Opin Biotechnol. 2015;32:99–103. doi: 10.1016/j.copbio.2014.11.021. [DOI] [PubMed] [Google Scholar]
- 92.Liong MT, Shah NP. Bile salt deconjugation ability, bile salt hydrolase activity and cholesterol co - precipitation ability of lactobacilli strains. Int Dairy J. 2005;15:391–8. doi: 10.1016/j.idairyj.2004.08.007. [DOI] [Google Scholar]
- 93.Adebayo-tayo BC, Onilude AA. Screening of lactic acid bacteria strains isolated from some Nigerian fermented foods for EPS production. World Appl Sci J. 2008;4:741–7. [Google Scholar]
- 94.Bunesova V, Vlkova E, Rada V, Rockova S, Svobodova I, Jebavy L, Kmet V. Bifidobacterium animalis subsp. lactis strains isolated from dog faeces. Vet Microbiol. 2012;160:501–5. doi: 10.1016/j.vetmic.2012.06.005. [DOI] [PubMed] [Google Scholar]
- 95.Prasada J, Gilla H, Smarta J, Gopala PK. Selection and characterisation of Lactobacillus and Bifidobacterium strains for use as probiotics. Int Dairy J. 1998;8:993–1002. doi: 10.1016/S0958-6946(99)00024-2. [DOI] [Google Scholar]
- 96.Courvalin P. Antibiotic resistance: the pros and cons of probiotics. Dig Liver Dis. 2006;38(Suppl 2):261–5. doi: 10.1016/S1590-8658(07)60006-1. [DOI] [PubMed] [Google Scholar]
- 97.Sharma P, Tomar SK, Goswami P, Sangwan V, Singh R. Antibiotic resistance among commercially available probiotics. Food Res Int. 2014;57:176–95. doi: 10.1016/j.foodres.2014.01.025. [DOI] [Google Scholar]
- 98.Sornplang P, Uriyapongson S, Poonsuk K, Mahakhan P. Lactic acid bacteria isolated from native chicken feces. KKUVet J. 2007;17:33–42. [Google Scholar]
- 99.Sornplang P, Jitjak T, Prachachit J, Piyadeatsoontorn S. Proceedings of international conference on beneficial microbes ICOBM 2014. Penang: Universiti Sains Malaysia; 2014. Screening and identification of lactic acid bacteria isolated from dogs feces as potential probiotic in dogs. [Google Scholar]
- 100.Klose V, Bayer K, Kern C, Goel F, Fibi S, Wegl G. Antibiotic resistances of intestinal lactobacilli isolated from wild boars. Vet Microbiol. 2014;168:240–4. doi: 10.1016/j.vetmic.2013.11.014. [DOI] [PubMed] [Google Scholar]
- 101.Ammor MS, Florez AB, Mayo B. Antibiotic resistance in non-enterococcal lactic acid bacteria and bifidobacteria. Food Microbiol. 2007;24:559–70. doi: 10.1016/j.fm.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 102.Sornplang P, Leelavatcharamas V, Sukon P, Yowarach S. Antibiotic resistance of lactic acid bacteria isolated from a fermented fish product. Pla-chom. Res J Microbiol. 2011;6:898–903. doi: 10.3923/jm.2011.898.903. [DOI] [Google Scholar]
- 103.Njage PMK, Dolci S, Jans C, Wangoh J, Lacroix C, Meile L. Phenotypic and genotypic antibiotic resistance patterns of Staphylococcus aureus from raw and spontaneously fermented camel milk. Br J Appl Sci Technol. 2013;3:87–98. [Google Scholar]
- 104.Ennahar S, Cai Y, Fujita Y. Phylogenetic diversity of lactic acid bacteria associated with paddy rice silage as determined by 16S ribosomal DNA analysis. Appl Environ Microbiol. 2003;69:444–51. doi: 10.1128/AEM.69.1.444-451.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kawahara T, Otani H. Stimulatory effect of lactic acid bacteria from commercially available Nozawana-zuke pickle on cytokine expression by mouse spleen cells. Biosci Biotechnol Biochem. 2006;70:411–7. doi: 10.1271/bbb.70.411. [DOI] [PubMed] [Google Scholar]
- 106.Sato H, Yanagida F, Shinohara T, Suzuki M, Suzuki KI, Yokotsuka K. Intraspecific diversity of Oenococcus oeni isolated during red wine-making in Japan. FEMS Microbiol Lett. 2001;202:109–14. doi: 10.1111/j.1574-6968.2001.tb10788.x. [DOI] [PubMed] [Google Scholar]
- 107.Yanagida F, Srionnual S, Chen YS. Isolation and characteristics of lactic acid bacteria from koshu vineyards in Japan. Lett Appl Microbiol. 2008;47:134–9. doi: 10.1111/j.1472-765X.2008.02398.x. [DOI] [PubMed] [Google Scholar]
- 108.Puphan K, Sornplang P, Uriyapongson S. Chainarong navanukraw screening of lactic acid bacteria as potential probiotics in beef cattle. Pakist J Nutri. 2015;14:474–9. doi: 10.3923/pjn.2015.474.479. [DOI] [Google Scholar]
- 109.Meroth CB, Walter J, Hertel C, Brandt MJ, Hammes WP. Monitoring the bacterial population dynamics in sourdough fermentation processes by using PCR-denaturing gradient gel electrophoresis. Appl Environ Microbiol. 2003;69:475–82. doi: 10.1128/AEM.69.1.475-482.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Coeuret V, Dubernet S, Bernardeau M, Gueguen M, Vernoux JP. Isolation, haracterisation and identification of lactobacilli focusing mainly on cheeses and other dairy products. Le Lait. 2003;83:269–306. doi: 10.1051/lait:2003019. [DOI] [Google Scholar]
- 111.Tannock GW. Identification of lactobacilli and bifidobacteria. Curr Issues Mol Biol. 1999;1:53–64. [PubMed] [Google Scholar]
- 112.Karen EH, Guan LL, Tannock GW, Korver DR, Allison GE. Detection, characterization, and in vitro and in vivo expression of genes encoding S-proteins in Lactobacillus gallinarum strains isolated from chicken crops. Appl Environ Microbiol. 2005;71:6633–44. doi: 10.1128/AEM.71.11.6633-6643.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kizerwetter-Swida M, Binek M. Selection of potentially probiotic Lactobacillus strains towards their inhibitory activity against poultry enteropathogenic bacteria. Pol J Microbiol. 2005;54:287–94. doi: 10.1099/jmm.0.45888-0. [DOI] [PubMed] [Google Scholar]
- 114.Roy D, SSirois S, Vincent D. Molecular discrimination of lactobacilli used as starter and probiotic cultures by amplified ribosomal DNA restriction analysis. Curr Microbiol. 2001;42:282–9. doi: 10.1007/s002840110218. [DOI] [PubMed] [Google Scholar]
- 115.Rodtong S, Tannock GW. Differentiation of Lactobacillus strains by ribotyping. Appl Environ Microbiol. 1993;59:3480–4. doi: 10.1128/aem.59.10.3480-3484.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Weiss A, Lettner HP, Kramer W, Mayer HK, Knifel W. Molecular methods use for the identification of potentially probiotic Lactobacillus reuteri strains. Food Technol Biotechnol. 2005;43:295–300. [Google Scholar]
- 117.Giraffa G, Lazzi C, Gatti M, Rossetti L, Mora D, Neviani E. Molecular typing of Lactobacillus delbrueckii of dairy origin by PCR-RFLP of protein-coding genes. Int J Food Microbiol. 2003;82:163–72. doi: 10.1016/S0168-1605(02)00262-3. [DOI] [PubMed] [Google Scholar]
- 118.Tannock GW, Fuller R, Pedersen K. Lactobacillus succession in piglet digestive tract demonstrated by plasmid profiling. Appl Environ Microbiol. 2003;56:1310–16. doi: 10.1128/aem.56.5.1310-1316.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Leite AMO, Mayo B, Rachid CTCC, Peixoto RS, Silva JT, Paschoalin VMF, Delgado S. Assessment of the microbial diversity of Brazilian kefir grains by PCR-DGGE and pyrosequencing analysis. Food Microbiol. 2012;31:215–21. doi: 10.1016/j.fm.2012.03.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.


