Abstract
Cancer-associated genes serve a crucial role in carcinogenesis. The present study aimed to investigate the mRNA expression levels of microspherule protein 1 (MCRS1) and MCRS2 in colorectal cancer (CRC) and their association with clinical variables. The mRNA expression levels of MCRS1 and MCRS2 were assessed by semi-quantitative reverse transcription polymerase chain reaction in the tumor and corresponding non-tumor tissues of 54 newly-diagnosed CRC patients, as well as in the normal colonic mucosa tissue of 19 age/gender-matched healthy controls. Immunofluorescence was also employed to identify the expression of MCRS1 in CRC tissues, while the concentration of serum carcinoembryonic antigen (CEA) was determined by an enzyme-linked immunoassay. The results identified a negative correlation between MCRS1 and MCRS2 expression levels (r=−0.3018, P=0.0266). MCRS1 mRNA expression was significantly increased and MCRS2 mRNA expression was decreased in CRC tissues compared with the levels in the corresponding normal tissues (both P<0.001). An increase in MCRS1 expression and a decrease in MCRS2 expression was detected in advanced stage when compared with early stage CRC patients. Immunofluorescence analysis revealed increased expression of MCRS1 in CRC patients. Furthermore, the expression levels of MCRS1 displayed positive correlation, whilst those of MCRS2 displayed negative correlation, with the serum CEA level in patients with CRC. The results suggest that increased MCRS1 and decreased MCRS2 expression appeared to be involved in the pathogenesis of CRC. The present study provides evidence suggesting that MCRS1 and MCRS2 may identify CRC patients at a risk of disease relapse, and thus, may be potential tools for monitoring disease activity and act as novel diagnostic markers in the treatment of CRC.
Keywords: colorectal cancer, microspherule protein 1, microspherule protein 2, carcinoembryonic antigen
Introduction
Colorectal cancer (CRC) is one of the most prevalent malignant tumors, with high incidence rate and mortality. It has been reported that CRC affects >1.2 million individuals globally each year (1). CRC is the third most common cancer and the leading cause of cancer-associated mortality in the United States (1,2) and the incidence of CRC is increasing in China (3–5).
The morphological changes involved in the progression of CRC from a benign adenoma to a malignant carcinoma have a complex biological and molecular underlying process (6). Cancer-associated genes serve an important role in carcinogenesis, including the processes of cell proliferation, transformation, angiogenesis, invasion and metastasis. The increased activity of oncogenes or decreased activity of tumor-suppressor genes have been found to participate in the development of tumors (7). A number of CRC cases exhibit mutations in the KRAS oncogene (8–11), as well as in p53 (12), APC and SMAD4/DPPC4 (13–15) tumor-suppressor genes. Furthermore, a large number of mutated genes have been identified in CRC, including genes that encode proteins with vital roles in CRC carcinogenesis. However, the molecular mechanism of CRC carcinogenesis has yet to be elucidated. The major cause of CRC mortality and morbidity is metastasis, with >30% of patients with CRC eventually developing metastatic disease (16–18). Thus, there is an urgent requirement for specific and sensitive biomarkers to aid the early diagnosis and prognosis of CRC.
Microspherule protein 1 (MCRS1), also known as MSP58, is involved in various pivotal cellular processes, such as DNA repair, regulation of the cell cycle, development of malignancy, transcription and mitosis (19–21). MCRS1 is a 462-amino acid protein which was originally identified as the interaction partner of the p120 nucleolar protein (22), and its overexpression results in the enlargement of nucleoli. Studies have indicated that the interaction of MCRS1 with the transcription factors Daxx (23), STRA13 (24), REP (25), and REP (24) confirmed the function of regulation in transcriptional activity. Increasing evidence has demonstrated that MCRS1 interacts with various proteins that serve critical roles in tumor proliferation; for instance, it is reported that p78, an isoform of MCRS1, has been identified as a centrosomal protein that is required for maintaining centrosome homeostasis (26). Notably, MCRS1 has been identified to be highly expressed in CRC (27), hepatocellular carcinoma (28), glioma (29–31) and non-small-cell lung cancer (32), and to be correlated with poor prognosis. The transformation activity of MCRS1 is inhibited by physical interaction with the PTEN tumor suppressor, with the majority of evidence suggesting that MCRS1 behaves as an oncogene (31,33). By contrast, MCRS2 has been identified as an interacting partner and a potent inhibitor of telomerase, and long-term overexpression of MCRS2 in cancer cell lines leads to telomere shortening (34–36). Drosophila MCRS2 is co-purified with RNA polymerase II complexes and is required for normal levels of cycling gene expression (37). A previous study revealed that MCRS1 and MCRS2 are involved in cell cycle regulation and carcinogenesis (20,32). Carcinoembryonic antigen (CEA) was first identified in 1965 (38) and has now become the most widely used antigen for diagnosing and for monitoring the prognostic significance of CRC (39). However, a lack of sensitivity renders its use limited in clinical diagnosis (40). Thus, it is important to identify novel biomarkers and develop novel treatment strategies for CRC.
Based upon the crucial role of MCRS1 and MCRS2 in tumorigenesis, semi-quantitative reverse transcription polymerase chain reaction (RT-PCR) was used in the present study to detect the mRNA expression levels of MCRS1 and MCRS2 in 54 fresh tissue samples from patients with CRC, along with their corresponding normal tissues. MCRS1 expression was also detected by immunofluorescence. In addition, the association of MCRS1 and MCRS2 expression with the clinicopathological features of CRC was also assessed. MCRS1 and MCRS2 expression and the clinical variables were analyzed in order to evaluate the clinical significance of differences in the gene expression levels.
Materials and methods
Patients and tissues
A total of 54 newly-diagnosed CRC patients at the inpatient service of the First Hospital of Jilin University (Changchun, China) were recruited into the study between April 2011 and August 2013. In addition, 19 healthy individuals were recruited as controls. Written informed consent was obtained from individual participants. The experimental protocol was established according to the Declaration of Helsinki (sixth revision, 2008) and approved by the Medical Ethics Committee of the First Hospital of Jilin University. The baseline demographic and clinical data of individual participants were collected from hospital records and reviewed by experienced surgeons. The demographic and clinical characteristics of participants were obtained.
Patients were initially screened with fecal occult blood test (Benzidine test) to detect potential bleeding in the digestive tract. A piece of faeces was emulsified and 1 ml saturated benzidine solution (Sangon Biotech, Co., Ltd., Shanghai, China) was added. Thereafter, hydrogen peroxide was slowly added followed by vigorous shaking of the solution. A color change from green to blue was regarded as a positive result, whereas, a change to deep purple without first becoming green or blue was considered to be a negative result.
Furthermore, patients were diagnosed following the histological examination of biopsy [hematoxylin and eosin (HE) staining (Beijing Zhongshan Golden Bridge Biotechnology, Co., Ltd., Beijing, China) or immunohistochemical staining (with Bcl-2, Her-2, EGFR, p53, Ki-67; Fuzhou Maixin Biotech. Co., Ltd., Fuzhou, China) or both]. Tumor tissues were obtained during colonoscopy with the video endoscopy system Olympus Evis Lucera Elite (Olympus, Tokyo, Japan), which was followed by a computed tomography scan (Brilliance CT 64-slice, Philips, Holland). The tumor classification, histological grades and lymph node metastasis status of individual tumor samples were evaluated by pathologists in a blinded manner, and staged according to the tumor-node-metastasis (TNM) classification system of the International Union against Cancer (edition 7). (41). Patients with stage I/II of CRC were classified as early stage patients while those with stage III/IV of CRC were grouped as advanced stage patients. A total of 54 freshly resected surgical tumour tissues and the corresponding non-tumor tissues that were obtained from a location ≥5 cm away from the centre of CRC were assessed by HE staining. Moreover, 19 normal mucosa samples were obtained from patients with hemorrhoids when they underwent a procedure for treatment of hemorrhoids at the same department. These healthy control patients had no other gastrointestinal disease and their tissue samples served as the healthy control tissues (Table I). Individual patients were excluded if they had a history of previous tumor and received radiotherapy or chemotherapy, poor physical condition, undergone treatment with immunosuppressants during the previous three months or were >80-years-old. Normal colonic mucosa, fresh tumor tissues and the corresponding non-tumor control tissues were obtained from patients during surgery and were used for RNA extraction.
Table I.
Demographic and clinical characteristics of subjects.
| Characteristic | Healthy controls (n=19) | Non-tumor controls (n=54) | Early CRC (n=23) | Advanced CRC (n=31) |
|---|---|---|---|---|
| Mean age (range), years | 55 (43–69) | 58 (44–71) | 57 (44–71) | 60 (48–70) |
| Gender (male/female) | 11/8 | 32/22 | 13/10 | 19/12 |
| Tumor location (colon/rectum) | N/A | 21/33 | 8/15 | 13/18 |
| TNM stage (I/II or III/IV) | N/A | N/A | 7/16 (I/II) | 21/10 (III/IV) |
| Differentiation (good/moderate/poor) | N/A | N/A | 7/9/7 | 5/11/15 |
| Serum CEA, ng/ml | 0.36 (0.15–4.58) | 7.24 (0.17–38.34)a | 4.37 (0.17–26.74) | 21.16 (2.7–38.34)a |
Data are presented as the median and range unless otherwise specified.
P<0.05 vs. healthy controls. CRC, colorectal cancer; TNM, Tumor-Node-Metastasis; CEA, carcinoembryonic antigen.
Semi-quantitative RT-PCR
Total RNA was extracted from the normal colonic mucosa, fresh surgical tumor and non-tumor tissue samples using the Eastep Super Total RNA Extraction kit (LS1000, Promega Corp., Shanghai, China), according to the manufacturer's protocol. RNA quantity and quality were assessed using a Synergy HTX Multi-Mode Microplate Reader (BioTek Instruments, Inc., Winooski, VT, USA) and the absorbance ratio of 260/280 nm should have been in the range between 1.8 to 2.0. The integrity of RNA was confirmed using a 2.0% agarose gel electrophoresis and visualized with a Tanon-2500R Gel Imaging system (Tanon Science and Technology Co., Ltd., Shanghai, China). Electrophoresis of RNA on the agarose gel demonstrated the appearance of a 5S band, and clear 28S and 18S bands that suggested the integrity of RNA. The integrity of RNA was confirmed using agarose gel electrophoresis. First strand cDNA was obtained using the SuperScript II Reverse Transcriptase (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) according to the manufacturer's instructions, using an input of 1 µg total RNA. Simultaneously, semi-quantitative RT-PCR for the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was performed as a positive internal control. The presence of MCRS1, MCRS2 and GAPDH in the samples was confirmed using the SYBR Green Mastermix (Invitrogen; Thermo Fisher Scientific, Inc.). Specific primers and optimized annealing temperatures displayed in Table II. The GAPDH primer sequences were designed specifically as previously described (42) and the primers for MCRS1, MCRS2 and GAPDH were designed and synthesized by Sangon Biotech Co., Ltd.
Table II.
Primer sequences of MCRS1, MCRS2 and GAPDH genes and optimized annealing temperatures.
| Gene | Primer sequences (5′-3′) | Product size (bp) | Annealing temperatures (°C) |
|---|---|---|---|
| MCRS1 | F: ACGCCCTGCTCTACGAT | 483 | 56.3 |
| R: TCATGCCTGTGATGCTGTC | |||
| MCRS2 | F: CCGGAATTCATGACACGTGGCACCG | 1428 | 58.4 |
| R: CCGCTCGAGTCACTGTGGTGTGATCTTG | |||
| GAPDH | F: AGGTCCACCACTGACACGTT | 310 | 55.0 |
| R: GCCTCAAGATCATCAGCAAT |
MCRS, microspherule protein; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; F, forward; R, reverse.
Optimized PCR cycling parameters for MCRS1 and MCRS2 were as follows: 94°C for 5 min, followed by 35 cycles of 94°C for 30 sec, 58°C for 1 min, and 72°C for 1 min, and then 72°C for 10 min. For GAPDH, 5 min initial denaturation at 95°C, 30 cycles of 30 sec annealing at 72°C and 5 min final extension at 72°C were performed. The PCR reaction mixture (total volume, 25 µl) contained 0.1 µl Takara LA Taq (Takara Biotechnology Co., Ltd., Dalian, China), 12.5 µl 2X mix (Taq PCR Master Mix, Lifefeng Technology, Shanghai, China), 1.6 µl dNTP mixture (2.5 mm each), 2.5 µl template cDNA, 0.25 µl forward primer (10 µm) and reverse primer (10 µm) mixture, and 7.8 µl triple-distilled H2O. Next, the PCR products were subjected to electrophoresis in 2% agarose gel in Tris/borate/EDTA buffer (89 mM Tris-base, pH 7.6; 89 mM boric acid; 2 mM EDTA) and images were captured in a UV light box (UVItec, Ltd., Cambridge, UK). For each semi-quantitative RT-PCR reaction products, electrophoresis images were scanned and evaluated using a Tanon-2500R Automatic Digital gel Imaging Analysis System (Tanon Science and Technology Co., Ltd., Shanghai, China). Next, the original intensities of specific bands were quantified by a Gel Image System Version 4.0 software (Tanon Science and Technology Co., Ltd., Shanghai, China). The final data were obtained after normalization by the intensity of GAPDH and according to a previous study (43) that included the expression amount of every gene in CRC tissues, the corresponding non-tumor tissues and normal mucosa tissues. (37).
Immunofluorescence
Due to technical limitations in obtaining antibodies for MCRS2, the expression of MCRS1 in the colorectal tissue sections was identified by immunofluorescence. Briefly, the colorectal tissue samples were fixed with formalin and embedded in paraffin. The tissue sections (4 µm) were dehydrated and subjected to sodium citrate (pH=6.0, 0.1, mol/l antigen retrieval in a microwave (M3-L233C, Midea Corp., Foshan, Guangdong, China). The slides were placed on a glass holder filled with sodium citrate and the slide holder was placed in 500 ml sodium citrate (pH 6.0, 0.1 mol/l) in a 2 L glass beaker. The beaker was heated in the microwave for 20 min and the evaporated water was replaced every 5 min. The slides were cooled down at room temperature. Subsequent to blocking with 5% BSA (Boster Bio, Wuhan, Hubei, China) donkey sera from healthy animals for 40 min at room temperature, the sections were stained with mouse anti human polyclonal antibody against MCRS1 (polyclonal donkey antibody; 1:100 dilution; ab67393; Abcam, Cambridge, UK) and incubated at 4°C overnight. Next, the bound antibodies were detected using the corresponding donkey anti-mouse Alexa Fluor 488 antibody (1:500; ab150105; Abcam). The sections were mounted with Prolong Gold antifade agent with DAPI (Invitrogen; Thermo Fisher Scientific, Inc.) and were photographed under an Olympus FV1000 confocal microscope (Olympus Corp., Tokyo, Japan). The number of MCRS1 cells in three fields (magnification, ×200) selected randomly in each tumor and corresponding non-tumor samples were counted in a blinded manner and the data were analyzed using ImageJ (National Institutes of Health, Bethesda, MD, USA).
CEA assay
The concentrations of serum CEA in individual serum samples were determined by ELISA. In brief, 3 ml fasting venous blood was collected from patients with CRC (n=54) or hemorrhoids (n=19) before the operation and centrifuged at 1,000xg at room temperature for 15 mins. The separated serum levels of CEA were detected by a human CEA ELISA kit (QiaoDu Biotechnology, Shanghai, China) according to the manufacturer's instructions and the absorbance was measured at 450 nm by an ADVIA Centaur XP immunoassay system (Siemens Healthcare Diagnostics, Tarrytown, NY, USA). Next, the average absorbance values were calculated for each set of reference standards, controls and samples in order to plot a standard curve. In order to determine the corresponding concentration of CEA in ng/ml from the standard curve, the mean absorbance value for each sample was used. The cut-off value for serum CEA was 5 ng/ml.
Statistical analysis
The difference between the groups was analyzed using the Mann-Whitney U test. The associations of the MCRS1 and MCRS2 expression levels with each clinicopathological parameter were examined using the χ2 test or Fisher's exact test (two sided). Student's t-tests (independent samples t-test) or a u-test were adopted to determine the difference between two sample means. One-way analysis of variance was used to assess the difference among three or four sample means, and Spearman's rank test was used to assess the correlation between MCRS1 and MCRS2 expression levels. All statistical tests were performed using SPSS for Windows (version 19.0; IBM SPSS, Chicago, IL, USA). A two-sided P-value of <0.05 was considered to indicate a statistically significant difference.
Results
Demographic and clinical characteristics of CRC patients
A total of 54 newly-diagnosed CRC patients and 19 healthy individuals were recruited to determine the expression status of MCRS1 and MCRS2 in CRC patients. The demographic and clinical characteristics of the patients and healthy individuals are displayed in Table I. There was no significant difference in the distribution of age and gender between the patients and healthy controls. Moreover, there was no significant difference in the tumor location (colon/rectum), TNM stage (I/II or III/IV) and differentiation (good/moderate/poor) between these two groups of CRC. In addition, it was observed that the serum CEA levels in the CRC patients were significantly higher compared with those in the healthy controls (P<0.05).
mRNA expression levels of MCRS1 and MCRS2 in CRC tissues
Semi-quantitative RT-PCR was performed to detect the mRNA expression levels of MCRS1 and MCRS2 in 54 newly-diagnosed CRC tissues, their corresponding normal tissues and 19 healthy controls. The results of the agarose gel electrophoresis for the tumor and corresponding non-tumor tissues of 5 patients selected at random are displayed in Fig. 1A. The relative MCRS1 expression was significantly increased in CRC tissues compared with the corresponding normal tissues (0.255 vs. 0.1026, P<0.0001) and healthy controls (0.2555 vs. 0.08523, P<0.0001; Fig. 1B). Notably, MCRS2 expression was significantly lower in the CRC tissues compared with that in the corresponding normal tissues (0.15248 vs. 0.2763, P<0.0001) and healthy controls (0.15248 vs. 0.32977, P<0.0001; Fig. 1C). There was no significant difference in MCRS1 and MCRS2 mRNA expression with regard to age, gender of patients, tumor location or histology (data not shown).
Figure 1.
Relative mRNA expression levels of MCRS1 and MCRS2 in CRC tissues, corresponding normal tissues and healthy control tissues. (A) Agarose gel images showing the expression levels of MCRS1, MCRS2 and internal control GAPDH detected by reverse transcription-quantitative polymerase chain in tumor tissues and corresponding normal tissues of 5 randomly-selected CRC patients. Relative mRNA expression levels of (B) MCRS1 and (C) MCRS2 in CRC tissues, corresponding normal tissues and healthy control tissues (normal colonic mucosa). Data are expressed as the mean ± standard deviation of individual groups, as well as the mean values of individual subjects. MCRS, microspherule protein; CRC, colorectal cancer; GAPDH, glyceraldehyde-3-phosphate dehydrogenase. T, tumor tissue; N, normal tissue; H, healthy control mucosa tissue.
Correlation between MCRS1 and MCRS2 expression in CRC
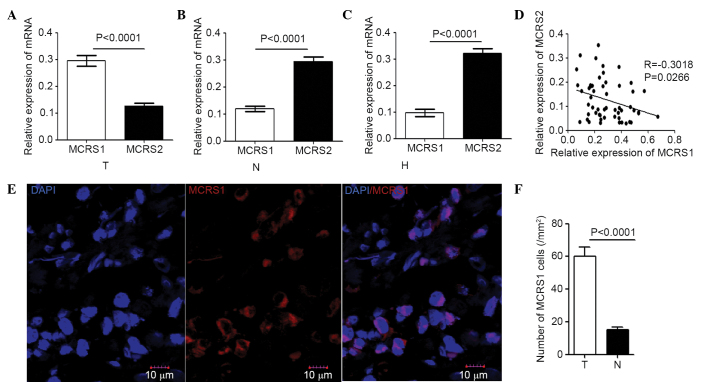
The correlation between mRNA levels of MCRS1 and MCRS2 expression in the CRC tissues, corresponding normal tissues and healthy controls were analyzed. In CRC tissues, the expression of MCRS1 was significantly higher compared with that of MCRS2 (0.255 vs. 0.15248, P<0.0001; Fig. 2A). By contrast, the expression of MCRS2 was significantly higher compared with that of MCRS1 in the corresponding normal tissues (0.2763 vs. 0.1026, P<0.0001; Fig. 2B) and the healthy control colonic mucosa (0.32977 vs. 0.08523, P<0.0001; Fig. 2C). A significant negative correlation was observed between the MCRS1 and MCRS2 expression levels in the CRC samples (r=−0.3018, P=0.0266; Fig. 2D). In addition, immunofluorescent assays were performed and the expression status of MCRS1 was predominately localized in the nucleus (Fig 2E). The results indicated that the number of MCRS1 cells/mm2 in the tumor sections (59 cells/mm2; P<0.0001) was significantly greater compared with that in the non-tumor tissue sections (16 cells/mm2; P<0.0001; Fig. 2E and F), and the healthy control colonic mucosa (14 cells/mm2; P<0.0001; Fig. 2F). Thus, increased expression of MCRS1 was present in the colorectal neoplasms.
Figure 2.
Correlation between MCRS1 and MCRS2 expression levels in (A) tumor tissues, (B) corresponding non-tumor tissues and (C) healthy control tissues. (D) Correlation between mRNA relative expression levels of MCRS1 and MCRS2 in tumor tissues of CRC patients. (E) Immunofluorescence analysis of MCRS1 in tumor tissue samples. (F) Quantitative analysis performed to determine the number of MCRS1 cells/mm2 in tumor tissue sections. MCRS, microspherule protein; CRC, colorectal cancer; T, tumor tissue; N, normal tissue; H, healthy control mucosa tissue.
Association of MCRS1 and MCRS2 expression with clinicopathological factors in CRC patients
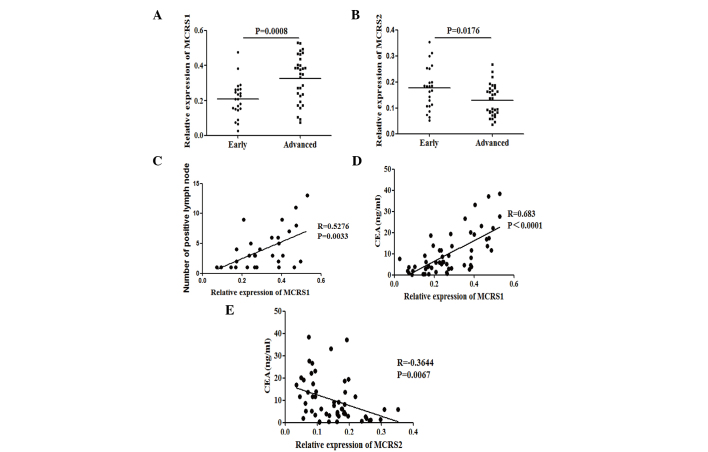
The present study examined whether the expression levels of MCRS1 and MCRS2 were associated with pathological stages in CRC. The patients were stratified according to the pathological TNM stages. Significantly increased MCRS1 mRNA expression was observed in patients with advanced stage CRC compared with early stage CRC (0.3543 vs. 0.2091, P=0.0008; Fig. 3A). Conversely, the mRNA expression of MCRS2 was significant decreased in advanced stage compared with early stage CRC (0.1368 vs. 0.1836, P=0.0176; Fig. 3B). Furthermore, HE stain was performed in resected lymph nodes and the number of positive lymph nodes was counted. The correlation between the number of positive lymph nodes and the expression of MCRS1 or MCRS2 was accessed. The expression levels of MCRS1 correlate positively with the number of positive lymph nodes (r=0.5276, P=0.0033; Fig. 3C), whereas the expression of MCRS2 was negatively correlated with the number of positive lymph nodes.
Figure 3.
Stratification analysis of mRNA expression status of MCRS1 and MCRS2. Correlation of relative mRNA expression levels of (A) MCRS1 and (B) MCRS2 with disease stage. Patients were classified as suffering from early (TNM stage I/II, n=23) or advanced stage CRC (TNM stage III/IV, n=31). In addition, the correlation of serum CEA with the expression levels of MCRS1 with the number of (C) positive lymph nodes was analyzed. In addition, the correlation of serum CEA with the expression levels of (D) MCRS1 and (E) MCRS2 expression was also determined. MCRS, microspherule protein; CRC, colorectal cancer; CEA, carcinoembryonic antigen.
The potential association of the expression levels of MCRS1 and MCRS2 with the serum CEA in this population was also assessed. It was determined that the expression levels of MCRS1 were positively correlated with the serum CEA levels in patients with CRC (r=0.683, P<0.0001; Fig. 3D). By contrast, the expression levels of MCRS2 were negatively correlated with the serum CEA levels in CRC patients (r=−0.3644, P=0.0067; Fig. 3E).
Discussion
The results of the present study indicated that the expression levels of MCRS1 were increased in CRC tissues and were positively correlated with the node metastasis, distant metastasis and the TNM stage of tumors. Notably, a statistically significant association was observed between the MCRS1 expression and serum CEA levels. In a study conducted by Shi et al (27), the expression of MCRS1 was positively correlated with the depth of invasion, local recurrence, tumor grade and TNM stage. Notably, decreased expression levels of MCRS2 in CRC tissues were confirmed for the first time, and the expression of MCRS2 was found to be negatively correlated with the node metastasis, distant metastasis, the TNM stage of tumors and serum CEA levels. The significantly different outcomes between patients expressing high and low levels of MCRS1 and MCRS2 suggested that MCRS1 and MCRS2 may be used to predict the clinical outcome of CRC. The aforementioned findings are valuable in providing potential therapeutic targets for the future treatment of CRC.
Accumulating evidence indicated that the expression of MCRS1 was associated with several types of human malignant solid tumors, including those of CRC (27), hepatocellular carcinoma (28), glioma (29–31) and non small-cell lung cancer (32), and poor prognosis. It has been demonstrated that MCRS1 appears to function as an oncogene and enhance tumor cell survival. In addition, it promotes tumor growth and metastasis in certain tumors via regulation of the transcription factors Daxx (23), STRA13 (24), Mi-2β and REP (25), indicating that MCRS1 is likely to be involved in tumor invasion and metastasis. The present study established that increased MCRS1 expression was correlated with node metastasis, distant metastasis and the advanced TNM stage of tumors. These results have indicated that MCRS1 is involved in the invasion and metastasis of CRC, which is consistent with previous findings of other authors (27,31). The present study also revealed that the expression of MCRS1 was predominately localized in the nucleus, which confirmed that MCRS1 has a positive influence on the cell proliferation capacity.
Identification of cancer-specific gene expression and their targets is critical for understanding their roles in tumorigenesis, and may be important in establishing novel prognostic and therapeutic targets. MCRS2 is an isoform of MCRS1 that is 13 amino acids longer than MCRS1 at the N-terminus. It was first identified as an interacting partner of LPTS/PinX1, which is a potent inhibitor of telomerase, and overexpression of MCRS2 in cancer cell lines suppressed telomere elongation (34). Based upon the pivotal role of MCRS2 in carcinogenesis, the present study identified for the first time that low MCRS2 expression correlated negatively with poor differentiation status, node metastasis, distant metastasis and advanced TNM stage in CRC. Therefore, a loss of MCRS2 is a typical feature during CRC progression, indicating a potentially conflicting role of MCRS1 and MCRS2 in CRC. Further studies are required to disclose the possible convergent points of these two genes in order to fully elucidate the association between MCRS1 and MCRS2 in tumorigenesis.
In the present study, MCRS1 and MCRS2 expression levels were examined in 54 CRC patients stratified according to their TNM stage. Initially, the present study demonstrated direct evidence of a negative correlation between MCRS1 and MCRS2 expression, as well as an increase in MCRS1 expression and decrease in MCRS2 expression with regard to malignant CRC transformation. From the present results it may be concluded that MCRS1 has an oncogenic and MCRS2 a tumor suppressive role during the development of CRC, which is consistent with the results of previous studies (44). However, the possible reasons for the functional diversity of MCRS1 and MCRS2 in tumorigenesis still require further assessment.
Serum CEA is a highly specific tumor-associated antigen and is particularly observed in patients with colorectal cancer; however, a lack of sensitivity renders its use limited in clinical diagnosis (40). Thus, it is imperative to identify novel biomarkers and develop novel treatment strategies for CRC. An investigation to determine the association between MCRS1, MCRS2 and serum CEA indicated that the expression levels of MCRS1 displayed positive correlation with the serum CEA levels in patients with CRC. Conversely, the expression levels of MCRS2 exhibited a negative correlation with the serum CEA levels in patients with CRC. These findings suggest that a combination of MCRS1 and MCRS2 expression may be an effective molecular biomarker for the diagnosis of CRC. To the best of our knowledge, this is the first study to demonstrate such findings.
In conclusion, the present data demonstrated the association of MCRS1 and MCRS2 expression with certain clinical characteristics of CRC and investigated their potential roles as biomarkers for diagnosing CRC. A combination of the expression levels of MCRS1 and MCRS2 may potentially be used as an efficient molecular biomarker for diagnosing CRC. Based on the findings of the current study, it is possible to conclude that increased MCRS1 and decreased MCRS2 expression may be involved in the pathogenesis of CRC. The present study also indicated that MCRS1 and MCRS2 may identify CRC patients at a risk of disease relapse, rendering them potential tools for the monitoring of disease activity, and new diagnostic targets for CRC.
References
- 1.Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin. 2014;64:104–117. doi: 10.3322/caac.21220. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, Naishadham D, Jemal A. Cancer statistics for Hispanics/Latinos, 2012. CA Cancer J Clin. 2012;62:283–298. doi: 10.3322/caac.21153. [DOI] [PubMed] [Google Scholar]
- 3.Weitz J, Koch M, Debus J, Höhler T, Galle PR, Büchler MW. Colorectal cancer. Lancet. 2005;365:153–165. doi: 10.1016/S0140-6736(05)17706-X. [DOI] [PubMed] [Google Scholar]
- 4.Lei T, Chen WQ, Zhang SW, Lei TH, Ying Q, He ZY, Wang XH. Prevalence trend of colorectal cancer in 10 cities and counties in China from 1988 to 2002. Zhonghua zhong Liu Za Zhi. 2009;31:428–433. (In Chinese) [PubMed] [Google Scholar]
- 5.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 6.Lech G, Slotwinski R, Krasnodebski IW. The role of tumor markers and biomarkers in colorectal cancer. Neoplasma. 2014;61:1–8. doi: 10.4149/neo_2014_003. [DOI] [PubMed] [Google Scholar]
- 7.Markowitz SD, Bertagnolli MM. Molecular origins of cancer: Molecular basis of colorectal cancer. N Engl J Med. 2009;361:2449–2460. doi: 10.1056/NEJMra0804588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andreyev HJ, Norman AR, Cunningham D, Oates JR, Clarke PA. Kirsten ras mutations in patients with colorectal cancer: The multicenter ‘RASCAL’ study. J Natl Cancer Inst. 1998;90:675–684. doi: 10.1093/jnci/90.9.675. [DOI] [PubMed] [Google Scholar]
- 9.Andreyev HJ, Norman AR, Cunningham D, Oates J, Dix BR, Iacopetta BJ, Young J, Walsh T, Ward R, Hawkins N, et al. Kirsten ras mutations in patients with colorectal cancer: The ‘RASCAL II’ study. Br J Cancer. 2001;85:692–696. doi: 10.1054/bjoc.2001.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Er TK, Chen CC, Bujanda L, Herreros-Villanueva M. Clinical relevance of KRAS mutations in codon 13: Where are we? Cancer Lett. 2014;343:1–5. doi: 10.1016/j.canlet.2013.09.012. [DOI] [PubMed] [Google Scholar]
- 11.Malapelle U, Carlomagno C, de Luca C, Bellevicine C, Troncone G. KRAS testing in metastatic colorectal carcinoma: Challenges, controversies, breakthroughs and beyond. J Clin Pathol. 2014;67:1–9. doi: 10.1136/jclinpath-2013-201835. [DOI] [PubMed] [Google Scholar]
- 12.Suppiah A, Greenman J. Clinical utility of anti-p53 auto-antibody: Systematic review and focus on colorectal cancer. World J Gastroenterol. 2013;19:4651–4670. doi: 10.3748/wjg.v19.i29.4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Colussi D, Brandi G, Bazzoli F, Ricciardiello L. Molecular pathways involved in colorectal cancer: Implications for disease behavior and prevention. Int J Mol Sci. 2013;14:16365–16385. doi: 10.3390/ijms140816365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hutchison J, Cohen Z, Onyeagucha BC, Funk J, Nelson MA. How microRNAs influence both hereditary and inflammatory-mediated colon cancers. Cancer Genet. 2013;206:309–316. doi: 10.1016/j.cancergen.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Temraz S, Mukherji D, Shamseddine A. Potential targets for colorectal cancer prevention. Int J Mol Sci. 2013;14:17279–17303. doi: 10.3390/ijms140917279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fearon ER. Molecular genetics of colorectal cancer. Annu Rev Pathol. 2011;6:479–507. doi: 10.1146/annurev-pathol-011110-130235. [DOI] [PubMed] [Google Scholar]
- 17.Kelleher FC, Rao A, Maguire A. Circadian molecular clocks and cancer. Cancer Lett. 2014;342:9–18. doi: 10.1016/j.canlet.2013.09.040. [DOI] [PubMed] [Google Scholar]
- 18.Mazzoccoli G, Vinciguerra M, Papa G, Piepoli A. Circadian clock circuitry in colorectal cancer. World J Gastroenterol. 2014;20:4197–4207. doi: 10.3748/wjg.v20.i15.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsu CC, Lee YC, Yeh SH, Chen CH, Wu CC, Wang TY, Chen YN, Hung LY, Liu YW, Chen HK, et al. 58-kDa microspherule protein (MSP58) is novel Brahma-related gene 1 (BRG1)-associated protein that modulates p53/p21 senescence pathway. J Biol Chem. 2012;287:22533–22548. doi: 10.1074/jbc.M111.335331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu CS, Zheng JY, Zhang HL, Zhao HD, Zhang J, Wu GQ, Wu L, Wang Q, Wang WZ, Zhang J. MSP58 knockdown inhibits the proliferation of esophageal squamous cell carcinoma in vitro and in vivo. Asian Pac J Cancer Prev. 2012;13:3233–3238. doi: 10.7314/APJCP.2012.13.7.3233. [DOI] [PubMed] [Google Scholar]
- 21.Hsu CC, Chen CH, Hsu TI, Hung JJ, Ko JL, Zhang B, Lee YC, Chen HK, Chang WC, Lin DY. The 58-kda microspherule protein (MSP58) represses human telomerase reverse transcriptase (hTERT) gene expression and cell proliferation by interacting with telomerase transcriptional element-interacting factor (TEIF) Biochim Biophys Acta. 2014;1843:565–579. doi: 10.1016/j.bbamcr.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 22.Ren Y, Busch RK, Perlaky L, Busch H. The 58-kDa microspherule protein (MSP58), a nucleolar protein, interacts with nucleolar protein p120. Eur J Biochem. 1998;253:734–742. doi: 10.1046/j.1432-1327.1998.2530734.x. [DOI] [PubMed] [Google Scholar]
- 23.Lin DY, Shih HM. Essential role of the 58-kDa microspherule protein in the modulation of Daxx-dependent transcriptional repression as revealed by nucleolar sequestration. J Biol Chem. 2002;277:25446–25456. doi: 10.1074/jbc.M200633200. [DOI] [PubMed] [Google Scholar]
- 24.Ivanova AV, Ivanov SV, Lerman ML. Association, mutual stabilization and transcriptional activity of the STRA13 and MSP58 proteins. Cell Mol Life Sci. 2005;62:471–484. doi: 10.1007/s00018-004-4423-2. [DOI] [PubMed] [Google Scholar]
- 25.Shimono K, Shimono Y, Shimokata K, Ishiguro N, Takahashi M. Microspherule protein 1, Mi-2beta and RET finger protein associate in the nucleolus and up-regulate ribosomal gene transcription. J Biol Chem. 2005;280:39436–39447. doi: 10.1074/jbc.M507356200. [DOI] [PubMed] [Google Scholar]
- 26.Hirohashi Y, Wang Q, Liu Q, Du X, Zhang H, Sato N, Greene MI. p78/MCRS1 forms a complex with centrosomal protein Nde1 and is essential for cell viability. Oncogene. 2006;25:4937–4946. doi: 10.1038/sj.onc.1209500. [DOI] [PubMed] [Google Scholar]
- 27.Shi H, Li SJ, Zhang B, Liu HL, Chen CS. Expression of MSP58 in human colorectal cancer and its correlation with prognosis. Med Oncol. 2012;29:3136–3142. doi: 10.1007/s12032-012-0284-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhong M, Zhang X, Li B, Chen CS, Ji GL, Li SX, Bi DQ, Zhao QC, Shi H. Expression of MSP58 in hepatocellular carcinoma. Med Oncol. 2013;30:539. doi: 10.1007/s12032-013-0539-2. [DOI] [PubMed] [Google Scholar]
- 29.Lin W, Zhang J, Zhang J, Liu X, Fei Z, Li X, Davidovic L, Tang Z, Shen L, Deng Y, Yang A, et al. RNAi-mediated inhibition of MSP58 decreases tumour growth, migration and invasion in a human glioma cell line. J Cell Mol Med. 2009;13:4608–4622. doi: 10.1111/j.1582-4934.2008.00499.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu L, Zhang ZG, Qin HZ, Zhang J, Gao GD, Lin W, Wang J, Zhang J. Downregulation of MSP58 suppresses cell proliferation in neuroblastoma cell lines. Neuroreport. 2012;23:932–936. doi: 10.1097/WNR.0b013e328359566e. [DOI] [PubMed] [Google Scholar]
- 31.Lin W, Li XM, Zhang J, Huang Y, Wang J, Zhang J, Jiang XF, Fei Z. Increased expression of the 58-kD microspherule protein (MSP58) is correlated with poor prognosis in glioma patients. Med Oncol. 2013;30:677. doi: 10.1007/s12032-013-0677-6. [DOI] [PubMed] [Google Scholar]
- 32.Liang Y, Liu M, Wang P, Ding X, Cao Y. Analysis of 20 genes at chromosome band 12q13: RACGAP1 and MCRS1 overexpression in nonsmall-cell lung cancer. Genes Chromosomes Cancer. 2013;52:305–315. doi: 10.1002/gcc.22030. [DOI] [PubMed] [Google Scholar]
- 33.Okumura K, Zhao M, DePinho RA, Furnari FB, Cavenee WK. PTEN: a novel anti-oncogenic function independent of phosphatase activity. Cell Cycle. 2005;4:540–542. doi: 10.4161/cc.4.4.1614. [DOI] [PubMed] [Google Scholar]
- 34.Song H, Li Y, Chen G, Xing Z, Zhao J, Yokoyama KK, Li T, Zhao M. Human MCRS2, a cell-cycle-dependent protein, associates with LPTS/PinX1 and reduces the telomere length. Biochem Biophys Res Commun. 2004;316:1116–1123. doi: 10.1016/j.bbrc.2004.02.166. [DOI] [PubMed] [Google Scholar]
- 35.Kuo CW, Wang WH, Liu ST. Mapping signals that are important for nuclear and nucleolar localization in MCRS2. Mol Cells. 2011;31:547–552. doi: 10.1007/s10059-011-1033-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu JL, Lin YS, Yang CC, Lin YJ, Wu SF, Lin YT, Huang CF. MCRS2 represses the transactivation activities of Nrf1. BMC Cell Biol. 2009;10:9. doi: 10.1186/1471-2121-10-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Andersen DS, Raja SJ, Colombani J, Shaw RL, Langton PF, Akhtar A, Tapon N. Drosophila MCRS2 associates with RNA polymerase II complexes to regulate transcription. Mol Cell Biol. 2010;30:4744–4755. doi: 10.1128/MCB.01586-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gold P, Freedman SO. Specific carcinoembryonic antigens of the human digestive system. J Exp Med. 1965;122:467–481. doi: 10.1084/jem.122.3.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wiggers T, Arends JW, Volovics A. Regression analysis of prognostic factors in colorectal cancer after curative resections. Diseases of the colon and rectum. 1988;31:33–41. doi: 10.1007/BF02552567. [DOI] [PubMed] [Google Scholar]
- 40.Tan E, Gouvas N, Nicholls RJ, Ziprin P, Xynos E, Tekkis PP. Diagnostic precision of carcinoembryonic antigen in the detection of recurrence of colorectal cancer. Surg Oncol. 2009;18:15–24. doi: 10.1016/j.suronc.2008.05.008. [DOI] [PubMed] [Google Scholar]
- 41.Obrocea FL, Sajin M, Marinescu EC, Stoica D. Colorectal cancer and the 7th revision of the TNM staging system: Review of changes and suggestions for uniform pathologic reporting. Romanian journal of morphology and embryology = Revue roumaine de morphologie et embryologie. 2011;52:537–544. [PubMed] [Google Scholar]
- 42.Tristan C, Shahani N, Sedlak TW, Sawa A. The diverse functions of GAPDH: views from different subcellular compartments. Cell Signal. 2011;23:317–323. doi: 10.1016/j.cellsig.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ji J, Zhao L, Wang X, Zhou C, Ding F, Su L, Zhang C, Mao X, Wu M, Liu Z. Differential expression of S100 gene family in human esophageal squamous cell carcinoma. J Cancer Res Clin Oncol. 2004;130:480–486. doi: 10.1007/s00432-004-0555-x. [DOI] [PubMed] [Google Scholar]
- 44.Zhong M, Zhang X, Li B, Chen CS, Ji GL, Li SX, Bi DQ, Zhao QC, Shi H. Expression of MSP58 in hepatocellular carcinoma. Med Oncol. 2013;30:539. doi: 10.1007/s12032-013-0539-2. [DOI] [PubMed] [Google Scholar]