Abstract
Neoplasms of the ovary are the second most common tumor of the female reproductive system, and the most lethal of the gynecological malignancies. Ovarian tumors are divided into a copious number of different groups reflecting their different features. The present study analyzed 187 ovarian tumors (39 sex-cord stromal tumors, 22 borderline tumors and 126 carcinomas) for the expression of the high-mobility group AT-hook 2 (HMGA2) gene, for mutations in the isocitrate dehydrogenase (NADP(+)) 1, cytosolic (IDH1), isocitrate dehydrogenase (NADP(+)) 2, mitochondrial (IDH2) and telomerase reverse transcriptase (TERT) genes, and for methylation of the O6-methylguanine-DNA methyltransferase (MGMT) promoter. Reverse transcription-polymerase chain reaction analysis showed that HMGA2 was expressed in 74.5% of the samples (120/161). A truncated transcript of HMGA2 was identified in 11 cases. A novel truncated form of HMGA2 was found in 4 serous high-grade carcinomas. Only 4 tumors (4/185) showed the TERT C228T mutation. No IDH1 or IDH2 mutations were found. Methylation of the promoter of MGMT was found in 2 borderline tumors (2/185). HMGA2 was expressed, in its truncated and native form, in different ovarian tumors, even the less aggressive types, underscoring the general importance of this gene in ovarian tumorigenesis. Mutations involving TERT, as well as MGMT promoter methylation, are rare events in ovarian tumors.
Keywords: HMGA2, IDH1, IDH2, TERT, MGMT, ovarian tumors
Introduction
Tumors of the ovaries account for 30% of all cancers of the female genital system; they are a heterogeneous group of neoplasms, divided into a number of different subgroups, depending largely on histological and cytological features (1).
The majority (90%) of ovarian tumors are epithelial in nature. Malignant epithelial ovarian tumors are currently divided into high-grade serous, low-grade serous, endometrioid, mucinous and clear cell carcinomas (2).
Borderline tumors of the ovary are neoplasms of low malignant potential. These tumors present with cellular atypia, but are not invasive. A few of these tumors share genomic and molecular features with carcinomas (3), but it remains unclear as to whether this is a feature of only a subset or of borderline tumors in general.
Sex-cord stromal tumors account for 8% of all ovarian tumors and are further classified based on their predominant cell content; for example, granulosa cell tumors contain ≥10% granulosa cells. The thecoma-fibroma group of tumors is dominated by theca cells (thecoma), stromal fibroblasts (fibroma) or the two cell types in different proportions (thecofibroma) (4).
Heterogeneity among ovarian tumors biologically and clinically represents a considerable challenge. The molecular and genetic profiles of the tumors could aid in predicting their inherent aggressiveness and could also provide keys to more specific treatments. As a contribution toward this goal, the present study analyzed 187 samples of different types of ovarian tumors, namely, granulosa cell tumors, thecofibromas, fibromas, teratomas, borderline tumors and infiltrating carcinomas of different histologies, for their expression of the high-mobility group AT-hook 2 (HMGA2) gene, for mutations of the isocitrate dehydrogenase (NADP(+)) 1, cytosolic (IDH1), isocitrate dehydrogenase (NADP(+)) 2, mitochondrial (IDH2) and telomerase reverse transcriptase (TERT) genes, and for methylation of the promoter of the O6-methylguanine-DNA methyltransferase (MGMT) gene.
Materials and methods
Tumor material
The material consisted of fresh frozen samples from 187 ovarian tumors surgically resected at The Norwegian Radium Hospital (Oslo, Norway) between December 1999 and January 2010. The tumors have been previously characterized for chromosomal aberrations and genomic imbalances (3,5,6). The present series consisted of 39 sex-cord stromal tumors (19 thecofibromas, 16 fibromas, 2 granulosa cell tumors and 2 teratomas), 22 borderline tumors and 126 carcinomas (56 serous high-grade carcinomas, 30 endometrioid carcinomas, 18 mucinous carcinomas, 12 clear cell carcinomas, 4 serous low-grade carcinomas, 1 undifferentiated carcinoma, and 5 mixed-type carcinomas). This study was approved by the regional ethics committee (Regional Komité For Medisinsk Forskning-Setikk Sør-Øst, Norge; http://helseforskning.etikkom.no) and written informed consent was obtained from the patients.
DNA and RNA extraction, and cDNA synthesis
DNA extraction was performed using the Maxwell 16 Extractor (Promega, Madison, WI, USA) and the Maxwell 16 Tissue DNA Purification kit (Promega) according to the manufacturer's instructions. RNA was extracted using the miRNeasy kit (Qiagen GmbH, Hilden, Germany) and QIAcube (Qiagen GmbH). The concentration and purity of the DNA and RNA were measured with the Nanovue Spectrophotometer (GE Healthcare, Pittsburgh, PA, USA). Extracted RNA (1 µg) was reverse-transcribed in a 20-µl reaction volume using the iScript Advanced cDNA Synthesis kit according to the manufacturer's instructions (Bio-Rad Laboratories, Oslo, Norway).
Molecular analyses
Molecular analyses of IDH1, IDH2, TERT and MGMT were performed according to previously described protocols (7).
HMGA2
Molecular analyses of HMGA2 were performed using a previously described protocol (7), but with minor variations. In cases 7, 8, 9, 10 and 11, the nested polymerase chain reaction (PCR) of the 3′ rapid amplification of cDNA ends (3′RACE) was performed using the Touchdown-PCR conditions described by Korbie et al (8) to increase the specificity of the PCR and improve the quality of the products. The 3′RACE results for cases 7, 8, 9 and 11 were validated through additional PCRs using primers obtained by the sequence of the 3′RACE products (Table I). More specifically, HMGA2-928-F1 and HMGA2R169 were used in case 7, HMGA2F233 and HMGA2R233 were used in case 8, HMGA2-928F and HMGA2R263 were used in case 9, and HMGA2-928F and HMGA2R175 were used in case 11 (Table II). The products of these PCRs were sequenced and analyzed by BLAT (http://genome.ucsc.edu/cgi-bin/hgBlat).
Table I.
Specific primers for the 3′ rapid amplification of cDNA ends product amplification.
| Primer name | Primer sequence |
|---|---|
| HMGA2-982-F1 | 5′-CAAGAGTCCCTCTAAAGCAGCTCA-3′ |
| HMGA2R169 | 5′-TGGGATGCAGACTTCAGTTGGAA-3′ |
| HMGA2F323 | 5′-CCCTATCACCTCATCTCCCG-3′ |
| HMGA2R323 | 5′-TTGTCCACTCATTCAGCAGATC-3′ |
| HMGA2R363 | 5′-CAGGCATGGCTCTGCATGTG-3′ |
| HMGA2R175 | 5′-TGACCACTGAATTCTGGCCTCA-3′ |
Table II.
Truncated forms of HMGA2 found in ovarian tumors.
| Case/lab no. | Diagnosis | RT-PCR | Sequence | Karyotype |
|---|---|---|---|---|
| 1/05–1270 | Mucinous carcinoma | Truncated transcript | U29117 | Culture failure |
| 2/07–1630 | Mucinous carcinoma | Truncated transcript | Not available | 46,XX[69] |
| 3/08–1650 | Mucinous carcinoma | Truncated transcript | U29117 | 46, XX, del(1)(q21)[2] / 46, XX[88] |
| 4/01–196 | Endometrioid carcinoma | Truncated transcript | U29117 | 46,XX[16] |
| 5/07–0449 | Endometrioid carcinoma | Truncated transcript | U29117 | 46,XX[12] |
| 6/03–481 | Borderline | Truncated transcript | AK31139 | 46,XX[32] |
| 7/01–169 | Serous high-grade | Truncated transcript | Novel | 46,XX[48] |
| 8/02–333 | Serous high-grade | Truncated transcript | Novel | 47~49,XX,+8,+9[2]/49,idem,+5,-6,+7[4]/54, idem,+3,+5,+6,+7,+14,+17,+19[5] |
| 9/02–363 | Serous high-grade | Truncated transcript | Not available | 65~68,XX,-X,+1,+2,del(3)(p13),+4, add(4)(p12) ×2, add(5)(p15),-6,+7, add(7)(p15),-8,-9,-12,-13,-14,-15,-17,-18,der(19)add(19)(p13)add(19)(q13)+20,+20,-21,-22,+2mar[9] |
| 10/04–499 | Serous high-grade | Truncated transcript | Not available | 46,XX[5] |
| 11/04–715 | Serous high-grade | Truncated transcript | Novel | Culture failure |
HMGA2, high-mobility group AT-hook 2; RT-PCR, reverse transcription-polymerase chain reaction.
Methylation-specific quantitative-polymerase chain reaction (MSP-qPCR)
Unmethylated cytosine residues were converted to uracil by bisulfite treatment of 1.3 µg DNA using the EpiTect Bisulfite Kit (Qiagen GmbH) according to the manufacturers' protocol. Following conversion, DNA was eluted in buffer (Qiagen GmbH) to a final concentration of 30 ng/µl. Bisulfite-treated DNA (2 µl) was amplified with a qPCR in a 20-µl reaction mixture containing 10 µl of Precision Mix (Bio-Rad Laboratories, Oslo, Norway), 0.4 µl of forward and reverse primers, and 7.2 µl of H2O. Two different mixes were used, one containing the primer sequences of MGMT for the unmethylated reaction [5′-TTTGTGTTTTGATGTTTGTAGGTTTTTGT-3′ (forward) and 5′-AACTCCACACTCTTCCAAAAACAAAACA-3′ (reverse)] and one mix containing the primers for the methylated reaction [5′-TTTCGACGTTCGTAGGTTTTCGC-3′ (forward) and 5′-GCACTCTTCCGAAAACGAAACG-3′ (reverse)]. All qPCR analyses were performed in triplicate with the CFX96 Touch Real-Time PCR detection system (Bio-Rad Laboratories) and methylated and unmethylated standards (Epitect PCR Control DNA Set; Qiagen GmbH) were used as controls. The thermal cycling included 2 min at 90°C, followed by 40 cycles of 50 sec at 95°C, 50 sec at 60°C, 50 sec at 72°C, and 10 sec at 95°C, and a final process in which the temperature increased by 0.5°C in steps, starting from 65°C and ending at 95°C. Data were analyzed with the CFX Manager software (Bio-Rad Laboratories), and all the melting curves of the samples were referenced to the methylated and unmethylated controls (9).
Results
HMGA2
Informative results for HMGA2 expression were obtained for all 161 tumors from which RNA was available. The gene was found to be expressed in 74.5% of the samples (120/161 tumors). The frequency varied among the different tumor types and histological subgroups, with the thecofibromas showing the highest frequency (100%; 16/16 tumors), followed by the high-grade serous carcinomas (90.2%; 37/41 tumors), the fibromas (73.3%; 11/15), the borderline tumors (66.7%; 14/21 tumors), the mucinous carcinomas (61.1%; 11/18 tumors), the endometrioid carcinomas (60.0%; 18/30 tumors) and the clear cell carcinomas with only 18.2% (2/11 tumors).
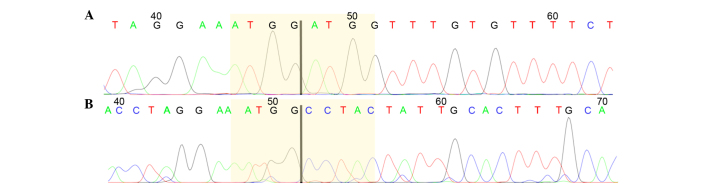
The samples were run for two parallel PCRs, which amplified exons 1–3 and exons 1–5 of HMGA2, respectively. Out of the 161 samples analyzed, 109 showed expression of full-length HMGA2, whereas 41 tumors showed no HMGA2 expression. The remaining 11 tumors showed expression of a truncated HMGA2, i.e., exons 1–3 (Table II). In these cases, 3′RACE PCR was performed in search of possible fusion transcripts. In 9 cases, the HMGA2 transcript could be identified by Sanger sequencing, whereas in cases 2 and 10, sequencing was not informative. In 4 out of the 9 tumors showing a truncated form of HMGA2 (cases 1, 3, 4 and 5), an ectopic sequence was found fused with HMGA2. This sequence has been previously found in human lipomas (accession number U29117). A single tumor (case 6) displayed a truncated transcript of HMGA2 with a sequence similar to the previously reported Homo sapiens cDNA FLJ18441 (accession number AK311399). Another 4 serous high-grade tumors (cases 7, 8, 9 and 11) showed a truncated form of HGMA2 not reported previously. 3′RACE results for these cases were validated through an additional PCR using the primers obtained by the sequence of the 3′RACE products. This was informative for 3 out of 4 cases, but gave no informative result for case 9. The transcripts contained the normal mRNA sequence until exon 3, with different regions of the third intron varying in length from case to case (Fig. 1).
Figure 1.
HMGA2 truncated transcript found in serous high-grade carcinomas. Chromatograms of the HMGA2 transcripts found in (A) cases 7 and 11, and (B) case 8. The junctions between exon 3 (left) and the intronic regions (right) are highlighted in yellow.
TERT
All 185 cases from which DNA was available were analyzed for mutations in the promoter region of TERT. The most frequent mutations, i.e., C228T and C250T, were focused on since they have been observed in a large number of tumors of different types (10). Only 1 borderline tumor and 3 fibromas showed the C228T mutation. All other cases showed no mutation.
IDH1 and IDH2 mutations
A total of 184 samples were analyzed for mutations in IDH1 and IDH2. The following mutation sites were investigated: IDH1R100, IDH1R109, and IDH1R132 of IDH1, and IDH2R140, IDH2R149 and IDH2R172 of IDH2. All analyses gave informative results. No mutations were found in these tumors. In 8 tumors, the single nucleotide polymorphism (SNP) IDH1G105 was identified.
MGMT
The MGMT promoter methylation status of the 185 samples from which DNA was extracted was assessed using MSP-qPCR. All analyses gave informative results. Only 2 borderline tumors were found to have MGMT promoter methylation.
Discussion
The high-mobility group AT-hook proteins, HMGA1 and HMGA2, are non-histone proteins that are involved in a range of nuclear processes from chromatin dynamics to gene regulation. HMGA gene family expression is observed during embryonic development (11), while expression is largely absent in adult normal tissues (11). However, high HMGA2 expression levels have been found in a variety of benign and malignant tumors (12,13). HMGA2 is involved in a number of different processes, from cellular proliferation to epithelial-mesenchymal transition (14). Recent studies have highlighted a pivotal role for HMGA2 in tumor metastasis (15). HMGA2 expression, in its full and/or truncated length, has not been investigated extensively in ovarian tumors, although a study by Hetland et al (16) showed the expression of HGMA2 by immunohistochemistry in primary solid tumors (96%), metastases (90%) and effusions (94%) of serous carcinomas. The present results showed that HMGA2 is expressed in a number of ovarian tumors (74.5%; 120/161 tumors). High-level expression was noted throughout the entire spectrum of malignancy, i.e., from sex-cord stromal neoplasms to borderline tumors to aggressive carcinomas. The only quantitative exception was the clear cell carcinomas, among which only 18.2% showed HMGA2 expression (2/11 tumors). Whether neoplasms of this subtype really express HMGA2 more rarely is a conclusion that must await examination of more tumors.
A truncated gene was found in 11 ovarian tumors of the present cohort. HMGA2 has previously been found disrupted, due to rearrangement of chromosomal band 12q15, in different benign connective tissue tumors, including lipomas (17), pleomorphic salivary gland adenomas (18), uterine leiomyomas (19) and lung hamartomas (20). The alterations involve exon 3 and cause the deletion of downstream regions, resulting in a truncated transcript that is able to evade miRNA-dependent gene silencing. Another alteration involves the chromosomal rearrangement of 12q13-15 to form a fusion gene. In the present study, karyotypic information was available for 9 out of the 11 tumors with a truncated HMGA2 (Table II). The karyotype was normal in 6 cases, whereas case 3 showed a del(1)(q21) as a sole rearrangement, case 8 exhibited a karyotype described as 47~49,XX,+8,+9[2]/49,idem,+5,-6,+7[4]/54,idem,+3,+5,+6,+7,+14,+17,+19[5], and the karyotype of case 9 was interpreted as 65~68,XX,-X,+1,+2, del(3)(p13),+4, add(4) (p12)x2,add(5)(p15),-6,+7,add(7) (p15),-8,-9,-12,-13,-14,-15,-17, −18, der(19) add (19)(p13) add(19)(q13), +20,+20,-21,-22,+2mar[9]. Notably, none of these cases showed a structural rearrangement involving 12q15 despite the fact that a truncated form of the HMGA2 gene was found in all of them. Possibly, this gene-level change may be due to a small deletion not visible at the chromosomal level.
The HMGA2 truncated transcripts were further characterized by 3′RACE-PCR, searching for possible fusion partners. However, it emerged that in 9 tumors (2 cases were not informative), the HMGA2 transcript was disrupted in the third exon. Notably, in 4 of them (cases 1, 3, 4 and 5), a sequence previously found in human lipomas was identified. Moreover, a novel truncated transcript was detected for HMGA2 in 4 high-grade serous carcinomas. The sequence of these transcripts contains the normal mRNA sequence until exon 3, followed by different regions of the third intron, which vary in length from case to case. It therefore appears that HMGA2 breakage leads to a truncated transcript that possesses exonic and intronic sequences. He et al (21) suggested that HMGA2 transcript shortening in serous ovarian cancer is the result of alternative polyadenylation that leads to a novel 3′UTR formation. In the present study, the finding of HMGA2 expression in different ovarian tumors, even the less aggressive types such as fibroma, thecofibroma and borderline tumors, highlights once more the importance of HMGA2 for the development of ovarian tumors.
TERT encodes the telomerase reverse transcriptase. It is well known that the gene is involved in cancer, and numerous studies have shown that mutations in the promoter region of the gene can increase telomerase expression (22–24). The present study focused on the most frequently occurring TERT mutations, i.e., C228T and C250T. These mutations introduce a novel binding site (TTCCGG) for members of the E-twenty-six/ternary complex factor transcription factor family (10). The C228T mutation was found in 4 out of the 184 tumors analyzed, 3 of which were fibromas, whereas the fourth was a borderline tumor. The low percentage of TERT mutations found in the various ovarian tumor samples analyzed leads us to hypothesize that it is neither a primary event nor even an important step in the majority of types of ovarian tumors. Notably, TERT C228T appears to be recurrent in ovarian fibromas, but further studies of a larger cohort are necessary to more reliably evaluate the frequency of this mutation. The present study did not find any TERT mutations in the clear cell samples, however, Huang et al (25) found the mutation in 16% of tumors (9/56 tumors) of this carcinoma subtype. The discrepancy between the present data and previous data may be due to the low number of clear cell carcinomas analyzed in the present cohort (n=12).
The IDH1 and IDH2 genes encode two types of isocitrate dehydrogenase. Mutations in either of the genes can result in an enzyme that produces 2-hydroxyglutarate. This metabolite is an inhibitor of α-ketoglutarate-dependent oxygenases, which can cause genome-wide methylations that exhibit an effect on gene expression when impaired. IDH1 and/or IDH2 mutations have been found in gliomas (26) and hematological malignancies (27). SNP IDH1G105, which is an adverse prognostic factor in cytogenetically normal acute myeloid leukemia (28), was identified in 8 out of the 184 tumors analyzed in the present series. This indicates that IDH1 and IDH2 are only rarely involved in ovarian tumorigenesis.
MGMT encodes O6-methylguanine DNA methyltransferase, a DNA repair enzyme that removes alkyl adducts from the O6-position of guanine. Expression of MGMT can result in resistance to alkylating cytostatics. MGMT promoter methylation increases the sensitivity of cells to alkylating drugs, as has been shown in a number of cancer types, particularly gliomas (29). Due to the efficacy of MGMT promoter methylation status as a prognostic and predictive tumor marker, this assessment has become one of the most commonly requested analyses for gliomas (30). The present study found this gene to be altered in only 2 borderline tumors out of 184 analyzed tumors of different types, indicating that MGMT promoter methylation is not a common event in ovarian tumorigenesis. In conclusion, the present study contributes to elucidating the genetic features of the HMGA2 gene in ovarian neoplasms in that it has been found expressed in benign and malignant tumors. Furthermore, a novel truncated form of HMGA2 has been identified.
Acknowledgements
This study was supported by grants from the Norwegian Radium Hospital Foundation, the Norwegian Cancer Society, the Inger and John Fredriksen Foundation for Ovarian Cancer Research, and the Research Council of Norway through its Centers of Excellence funding scheme, project number 179571.
References
- 1.Tavassoli FA, Devilee P, editors. Tumours of the Breast and Female Genital organs. IARC Press; Lyon: 2003. Pathology and Genetics. [Google Scholar]
- 2.Prat J. Ovarian carcinomas: Five distinct diseases with different origins, genetic alterations, and clinicopathological features. Virchows Arch. 2012;460:237–249. doi: 10.1007/s00428-012-1203-5. [DOI] [PubMed] [Google Scholar]
- 3.Micci F, Haugom L, Ahlquist T, Andersen HK, Abeler VM, Davidson B, Trope CG, Lothe RA, Heim S. Genomic aberrations in borderline ovarian tumors. J Transl Med. 2010;8:21. doi: 10.1186/1479-5876-8-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buy JN, Ghossain M, editors. Gynecological Imaging: A Reference Guide to Diagnosis. Springer-Verlag; Berlin, Heidelberg: 2013. Sex cord-stromal tumors; pp. 329–375. [DOI] [Google Scholar]
- 5.Micci F, Haugom L, Abeler VM, Tropé CG, Danielsen HE, Heim S. Consistent numerical chromosome aberrations in thecofibromas of the ovary. Virchows Arch. 2008;452:269–276. doi: 10.1007/s00428-007-0561-x. [DOI] [PubMed] [Google Scholar]
- 6.Micci F, Haugom L, Abeler VM, Davidson B, Tropé CG, Heim S. Genomic profile of ovarian carcinomas. BMC Cancer. 2014;14:315. doi: 10.1186/1471-2407-14-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Agostini A, Panagopoulos I, Andersen HK, Johannesen LE, Davidson B, Tropé CG, Heim S, Micci F. HMGA2 expression pattern and TERT mutations in tumors of the vulva. Oncol Rep. 2015;33:2675–2680. doi: 10.3892/or.2015.3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Korbie DJ, Mattick JS. Touchdown PCR for increased specificity and sensitivity in PCR amplification. Nat Protoc. 2008;3:1452–1456. doi: 10.1038/nprot.2008.133. [DOI] [PubMed] [Google Scholar]
- 9.Smith E, Jones ME, Drew PA. Quantitation of DNA methylation by melt curve analysis. BMC Cancer. 2009;9:1–12. doi: 10.1186/1471-2407-9-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heidenreich B, Rachakonda PS, Hemminki K, Kumar R. TERT promoter mutations in cancer development. Curr Opin Genet Dev. 2014;24:30–37. doi: 10.1016/j.gde.2013.11.005. [DOI] [PubMed] [Google Scholar]
- 11.Chiappetta G, Avantaggiato V, Visconti R, Fedele M, Battista S, Trapasso F, Merciai BM, Fidanza V, Giancotti V, Santoro M, et al. High level expression of the HMGI (Y) gene during embryonic development. Oncogene. 1996;13:2439–2446. [PubMed] [Google Scholar]
- 12.Rogalla P, Drechsler K, Frey G, Hennig Y, Helmke B, Bonk U, Bullerdiek J. HMGI-C expression patterns in human tissues. Implications for the genesis of frequent mesenchymal tumors. Am J Pathol. 1996;149:775–779. [PMC free article] [PubMed] [Google Scholar]
- 13.Pallante P, Sepe R, Puca F, Fusco A. High mobility group A (HMGA) proteins as tumor markers. Front Med (Lausanne) 2015;2:15. doi: 10.3389/fmed.2015.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu J, Wei JJ. HMGA2 and high-grade serous ovarian carcinoma. J Mol Med (Berl) 2013;91:1155–1165. doi: 10.1007/s00109-013-1055-8. [DOI] [PubMed] [Google Scholar]
- 15.Morishita A, Zaidi MR, Mitoro A, Sankarasharma D, Szabolcs M, Okada Y, D'Armiento J, Chada K. HMGA2 is a driver of tumor metastasis. Cancer Res. 2013;73:4289–4299. doi: 10.1158/0008-5472.CAN-12-3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hetland TE, Holth A, Kærn J, Flørenes VA, Tropé CG, Davidson B. HMGA2 protein expression in ovarian serous carcinoma effusions, primary tumors, and solid metastases. Virchows Arch. 2012;460:505–513. doi: 10.1007/s00428-012-1228-9. [DOI] [PubMed] [Google Scholar]
- 17.Schoenmakers EF, Wanschura S, Mols R, Bullerdiek J, Van den Berghe H, Van de Ven WJ. Recurrent rearrangements in the high mobility group protein gene, HMGI-C, in benign mesenchymal tumours. Nat Genet. 1995;10:436–444. doi: 10.1038/ng0895-436. [DOI] [PubMed] [Google Scholar]
- 18.Geurts JM, Schoenmakers EF, Van de Ven WJ. Molecular characterization of a complex chromosomal rearrangement in a pleomorphic salivary gland adenoma involving the 3′-UTR of HMGIC. Cancer Genet Cytogenet. 1997;95:198–205. doi: 10.1016/S0165-4608(96)00411-6. [DOI] [PubMed] [Google Scholar]
- 19.Mine N, Kurose K, Nagai H, Doi D, Ota Y, Yoneyama K, Konishi H, Araki T, Emi M. Gene fusion involving HMGIC is a frequent aberration in uterine leiomyomas. J Hum Genet. 2001;46:408–412. doi: 10.1007/s100380170059. [DOI] [PubMed] [Google Scholar]
- 20.Kazmierczak B, Meyer-Bolte K, Tran KH, Wöckel W, Breightman I, Rosigkeit J, Bartnitzke S, Bullerdiek J. A high frequency of tumors with rearrangements of genes of the HMGI(Y) family in a series of 191 pulmonary chondroid hamartomas. Genes Chromosomes Cancer. 1999;26:125–133. doi: 10.1002/(SICI)1098-2264(199910)26:2<125::AID-GCC4>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 21.He X, Yang J, Zhang Q, Cui H, Zhang Y. Shortening of the 3′ untranslated region: An important mechanism leading to overexpression of HMGA2 in serous ovarian cancer. Chin Med J (Engl) 2014;127:494–499. [PubMed] [Google Scholar]
- 22.Killela PJ, Reitman ZJ, Jiao Y, Bettegowda C, Agrawal N, Diaz LA, Jr, Friedman AH, Friedman H, Gallia GL, Giovanella BC, et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc Natl Acad Sci USA. 2013;110:6021–6026. doi: 10.1073/pnas.1303607110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang DS, Wang Z, He XJ, Diplas BH, Yang R, Killela PJ, Meng Q, Ye ZY, Wang W, Jiang XT, et al. Recurrent TERT promoter mutations identified in a large-scale study of multiple tumour types are associated with increased TERT expression and telomerase activation. Eur J Cancer. 2015;51:969–976. doi: 10.1016/j.ejca.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vinagre J, Almeida A, Pópulo H, Batista R, Lyra J, Pinto V, Coelho R, Celestino R, Prazeres H, Lima L, et al. Frequency of TERT promoter mutations in human cancers. Nat Commun. 2013;4:2185. doi: 10.1038/ncomms3185. [DOI] [PubMed] [Google Scholar]
- 25.Huang HN, Chiang YC, Cheng WF, Chen CA, Lin MC, Kuo KT. Molecular alterations in endometrial and ovarian clear cell carcinomas: Clinical impacts of telomerase reverse transcriptase promoter mutation. Mod Pathol. 2015;28:303–311. doi: 10.1038/modpathol.2014.93. [DOI] [PubMed] [Google Scholar]
- 26.Havik AB, Lind GE, Honne H, Meling TR, Scheie D, Hall KS, van den Berg E, Mertens F, Picci P, Lothe RA, et al. Sequencing IDH1/2 glioma mutation hotspots in gliomas and malignant peripheral nerve sheath tumors. Neuro Oncol. 2014;16:320–322. doi: 10.1093/neuonc/not230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patel KP, Barkoh BA, Chen Z, Ma D, Reddy N, Medeiros LJ, Luthra R. Diagnostic testing for IDH1 and IDH2 variants in acute myeloid leukemia an algorithmic approach using high-resolution melting curve analysis. J Mol Diagn. 2011;13:678–686. doi: 10.1016/j.jmoldx.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wagner K, Damm F, Göhring G, Görlich K, Heuser M, Schäfer I, Ottmann O, Lübbert M, Heit W, Kanz L, et al. Impact of IDH1 R132 mutations and an IDH1 single nucleotide polymorphism in cytogenetically normal acute myeloid leukemia: SNP rs11554137 is an adverse prognostic factor. J Clin Oncol. 2010;28:2356–2364. doi: 10.1200/JCO.2009.27.6899. [DOI] [PubMed] [Google Scholar]
- 29.Margison GP, Santibáñez-Koref MF. O6-alkylguanine-DNA alkyltransferase: Role in carcinogenesis and chemotherapy. Bioessays. 2002;24:255–266. doi: 10.1002/bies.10063. [DOI] [PubMed] [Google Scholar]
- 30.Håvik AB, Brandal P, Honne H, Dahlback HS, Scheie D, Hektoen M, Meling TR, Helseth E, Heim S, Lothe RA, Lind GE. MGMT promoter methylation in gliomas-assessment by pyrosequencing and quantitative methylation-specific PCR. J Transl Med. 2012;10:36. doi: 10.1186/1479-5876-10-36. [DOI] [PMC free article] [PubMed] [Google Scholar]