Abstract
Deregulated cellular energetics was one of the cancer hallmarks. Several underlying mechanisms of deregulated cellular energetics are associated with mitochondrial dysfunction caused by mitochondrial DNA mutations, mitochondrial enzyme defects, or altered oncogenes/tumor suppressors. In this review, we summarize the current understanding about the role of mitochondrial dysfunction in cancer progression. Point mutations and copy number changes are the two most common mitochondrial DNA alterations in cancers, and mitochondrial dysfunction induced by chemical depletion of mitochondrial DNA or impairment of mitochondrial respiratory chain in cancer cells promotes cancer progression to a chemoresistance or invasive phenotype. Moreover, defects in mitochondrial enzymes, such as succinate dehydrogenase, fumarate hydratase, and isocitrate dehydrogenase, are associated with both familial and sporadic forms of cancer. Deregulated mitochondrial deacetylase sirtuin 3 might modulate cancer progression by regulating cellular metabolism and oxidative stress. These mitochondrial defects during oncogenesis and tumor progression activate cytosolic signaling pathways that ultimately alter nuclear gene expression, a process called retrograde signaling. Changes in the intracellular level of reactive oxygen species, Ca2+, or oncometabolites are important in the mitochondrial retrograde signaling for neoplastic transformation and cancer progression. In addition, altered oncogenes/tumor suppressors including hypoxia-inducible factor 1 and tumor suppressor p53 regulate mitochondrial respiration and cellular metabolism by modulating the expression of their target genes. We thus suggest that mitochondrial dysfunction plays a critical role in cancer progression and that targeting mitochondrial alterations and mitochondrial retrograde signaling might be a promising strategy for the development of selective anticancer therapy.
Keywords: Cancer, carcinogenesis, medicine/oncology, metabolism, mitochondrial, DNA
Introduction
Based on the increased understanding in the past few decades, deregulated cellular energetics was recently added as one of the cancer hallmarks.1 Otto Warburg first proposed that tumor cells, unlike normal cells, exhibit increased glycolytic activity and reduced mitochondrial respiration even in the presence of oxygen. This phenomenon is known as the “Warburg effect.” Several underlying mechanisms of deregulated cellular energetics are associated with mitochondrial dysfunction caused by mitochondrial DNA (mtDNA) mutations, mitochondrial enzyme defects, or altered oncogenes/tumor suppressors.2–5
Mitochondria are intracellular organelles in eukaryotic cells that participate in bioenergetic metabolism and cellular homeostasis, including the generation of ATP through electron transport and oxidative phosphorylation in conjunction with the oxidation of metabolites by tricarboxylic acid (TCA) cycle and catabolism of fatty acids by β-oxidation, the production of reactive oxygen species (ROS), and the initiation and execution of apoptosis.6,7 Mitochondria contain multiple copies of mtDNA. Human mtDNA is a 16.6-kb double-stranded, circular DNA molecule that encodes 13 respiratory enzyme complex polypeptides, 22 transfer RNAs, and 2 ribosomal RNAs required for mitochondrial protein synthesis.8 Because mtDNA is essential for the maintenance of functionally competent organelles, the accumulation of mtDNA mutations or decreased mtDNA copy number is expected to affect energy production and enhance ROS generation and cell survival, and these processes may be involved in aging, mitochondrial diseases, or cancer.4,9–11
Although mitochondria have their own genome, most of the proteins and enzymes that reside in mitochondria are nuclear gene products. The enzymes of the TCA cycle are encoded by nuclear DNA and are located in the mitochondrial matrix or embedded in the inner mitochondrial membrane. Sirtuin 3 (SIRT3) is a nuclear-encoded mitochondrial protein deacetylase that regulates the function of several mitochondrial proteins involved in oxidative phosphorylation and intermediate metabolism.12 Mitochondrial dysfunction, caused by mtDNA mutations or mitochondrial enzyme defects, not only perturbs cellular bioenergetics, supporting the metabolic reprogramming of cancer cells, but also triggers tumor-promoting changes mediated by the ROS, Ca2+, or small molecule metabolites released by mitochondria. Moreover, some oncogenes or tumor suppressors including hypoxia-inducible factor 1 (HIF-1) and tumor suppressor p53 (TP53) have been shown to regulate mitochondrial respiration and cellular metabolism. Altered oncogenes/tumor suppressors thus provide a direct link between mitochondrial dysfunction and tumorigenesis. In this review, we discuss the role of mitochondrial dysfunction caused by either mtDNA mutations, mitochondrial enzyme defects, or altered oncogenes/tumor suppressors in initiating a complex cellular reprogramming that supports the formation and progression of cancers.
Somatic mutations and decreased copy number of mtDNA in tumor cells may lead to mitochondrial dysfunction and cancer progression
Several types of mtDNA alterations, such as point mutations, large-scale deletions, insertions, and copy number changes, have been identified in human cancers.13 Point mutations and copy number alterations are the two most common mtDNA alterations in cancers.9
Point mutations
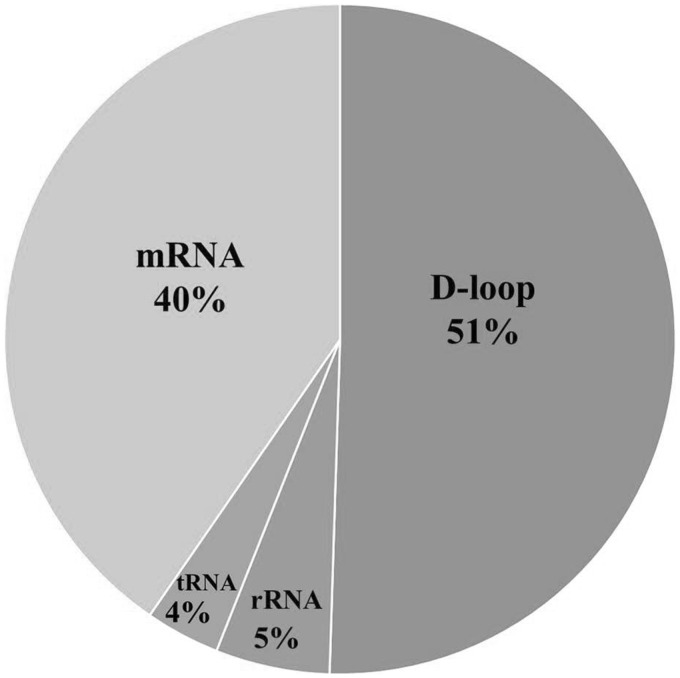
According to an analysis based on a total 859 patients with 20 different types of cancer, 66% of cancers carried at least one somatic point mutation of mtDNA,14 suggesting that somatic point mutation in mtDNA is a common event in human cancer progress. Among these identified mutations, 51% occurred in the D-loop region of mtDNA, 40% were found in the protein-coding region, 5% were located in the rRNA genes, and 4% were observed in the tRNA genes (Figure 1).14
Figure 1.
The location distribution of somatic mutations in mtDNA of human cancers analyzed in a total 859 patients with 20 different types of cancer.
Source: Data adapted from Lee et al.14
The D-loop region of mtDNA is the most frequent site of somatic mutation in cancers. The mononucleotide repeat region of the poly-cytosine (poly-C) sequence at nucleotide positions (np) 303–309 (D310) in mtDNA is a hot spot for somatic mutation.15 Because the D-loop region contains the major regulatory sites for mtDNA replication and transcription, mutations near these sites might affect mtDNA copy number and its transcription in cancer cells.13,16 Importantly, clinical correlation analyses in various cancers showed that cancer patients with mtDNA D-loop mutation in their cancer tissues exhibited poorer prognoses than those who were free of the mtDNA mutations.17–19
Among the identified mutations in the protein-coding regions, 25% were missense, nonsense, or frame-shift mutations that have high potential to cause mitochondrial dysfunction. Several mutations identified in the mtDNA protein-coding region and tRNA genes of cancer tissues were pathogenic or have been reported to be associated with mitochondrial diseases (Table 1). These mutations include frame-shift mutations, e.g. 11032delA in the NADH dehydrogenase subunit 4 (ND4) gene20,28,29 and 12418insA in the ND5 gene,20,21,26,27,30 which result in truncated polypeptides, and tRNA mutations (e.g. T1659C in the tRNAVal gene,20 G5650A in the tRNAAla gene,20 and 7472insC the tRNASer(UCN) gene27 that potentially alter tRNA structure).
Table 1.
Somatic mutations of mitochondrial DNA in human cancers
| Nucleotide position at mtDNA | Cancer type | Mutation | Gene | Amino acid change | Correlated function | References |
|---|---|---|---|---|---|---|
| 956 | Hepatocellular carcinoma | Poly-C | 12s rRNA | 20 | ||
| 1499 | Breast cancer | T → C/T | 12S rRNA | 21 | ||
| 1659 | Hepatocellular carcinoma | T → C/T | tRNAVal | 20 | ||
| 1913 | Breast cancer | G → A | 16S rRNA | 21 | ||
| 3409 | Breast cancer | A3 → A3/2 | ND1 | Frame shift | Potential to cause mitochondrial Complex I dysfunction (truncated ND1) | 21 |
| 3697 | Gastric cancer | G → A | ND1 | Gly (GGC) → Ser (AGC) | Potential to cause mitochondrial Complex I dysfunction (mutation in highly conserved residue) | 22 |
| 3842 | Hepatocellular carcinoma | G → A/G | ND1 | Trp (TGA) → Stop (TAA) | Potential to cause mitochondrial Complex I dysfunction (truncated ND1) | 20 |
| 3894–3960/3901–3967 | Hepatocellular carcinoma | 66 bp del | ND1 | Frame shift | Potential to cause mitochondrial Complex I dysfunction (truncated ND1) | 20 |
| 4561 | Breast cancer | T → TT/T | ND2 | Frame shift | Potential to cause mitochondrial Complex I dysfunction (truncated ND2) | 21 |
| 4605 | Breast cancer | A7 → A7/8 | ND1 | Frame shift | Potential to cause mitochondrial Complex I dysfunction (truncated ND2) | 21 |
| 4996 | Gastric cancer | G → A | ND2 | Arg (CGC) → His (CAC) | Potential to cause mitochondrial Complex I dysfunction (mutation in highly conserved residue) | 22 |
| 5112 | Breast cancer | G → A/G | ND2 | Ala (GCA) → Thr (ACA) | 21 | |
| 5522 | Breast cancer | G → A/G | tRNATrp | 21 | ||
| 5650 | Hepatocellular carcinoma | G → A/G | tRNAAla | Decreased Complex I and IV activity | 20, 23 | |
| 5809 | Breast cancer | G/A → A/G | tRNACys | 21 | ||
| 5895 | Gastric cancer | C19/n → C18/n | Non-coding nucleotides | 22 | ||
| 6384 | Breast cancer | G → A/G | COI | Ala (GCC) → Thr (ACC) | Potential to cause mitochondrial Complex IV dysfunction (mutation in highly conserved residue) | 21 |
| 6768 | Breast cancer | G → A/G | COI | Ala (GCA) → Thr (ACA) | Potential to cause mitochondrial Complex IV dysfunction (mutation in highly conserved residue) | 21 |
| 6787 | Hepatocellular carcinoma | T → C | COI | Val (GTA) → Ala (GCA) | Potential to cause mitochondrial Complex IV dysfunction (mutation in highly conserved residue) | 20 |
| 7293 | Breast cancer | G → A | COI | Ala (GCA) → Thr (ACA) | Potential to cause mitochondrial Complex IV dysfunction (mutation in highly conserved residue) | 21 |
| 7472 | Gastric cancer | insC | tRNASer(UCN) | Decreased Complex I activity, lower oxygen consumption rate, and higher lactic acid production | 22, 24, 25 | |
| 7976 | Hepatocellular carcinoma | G → A | COII | Gly (GGC) → Ser (AGC) | Potential to cause mitochondrial Complex IV dysfunction (mutation in highly conserved residue) | 20 |
| 9263 | Hepatocellular carcinoma | A → G | COIII | Thr (ACA) → Thr (ACG) | 20 | |
| 9267 | Hepatocellular carcinoma | G → A | COIII | Ala (GCC) → Thr (ACC) | Potential to cause mitochondrial Complex IV dysfunction (mutation in highly conserved residue) | 20 |
| 9412 | Breast cancer | G → A/G | COIII | Gly (GGC) → Asp (GAC) | Potential to cause mitochondrial Complex IV dysfunction (mutation in highly conserved residue) | 21 |
| 9545 | Hepatocellular carcinoma | A/G → G | COIII | Gly (GGA) → Gly (GGG) | 20 | |
| 9774 | Breast cancer | G → A/G | COIII | Asp (GAC) → Asn (AAC) | Potential to cause mitochondrial Complex IV dysfunction (mutation in highly conserved residue) | 21 |
| 9901 | Breast cancer | A → C/A | COIII | His (CAC) → Pro (CCC) | Potential to cause mitochondrial Complex IV dysfunction (mutation in highly conserved residue) | 21 |
| 9986 | Gastric cancer | C → A | COIII | Gly (GGG) → Gly (GGA) | 22 | |
| 10599 | Breast cancer | G → A/G | ND4L | Ala (GCT) → Thr (ACT) | 21 | |
| 11032 | Prostate cancer Renal oncocytomas Hepatocellular carcinoma | A7 → A6/7 | ND4 | Frame shift | Loss of Complex I activity | 20, 28, 29 |
| 11708 | Hepatocellular carcinoma | A → G | ND4 | Ile (ATC) → Val (GTC) | 20 | |
| 12405 | Gastric cancer | C → T | ND5 | Leu (CTC) → Leu (CTT) | 22 | |
| 12418 | Colorectal cancer Hepatocellular carcinoma Breast cancer Gastric cancer | A8 → A8/9 | ND5 | Frame shift | Defective mitochondrial respiratory function, higher lactate production and increased tumorigenesis | 20, 21, 22, 26, 27 |
| 13015 | Gastric cancer | T → C | ND5 | Leu (TTA) → Leu (CTA) | 22 | |
| 13878 | Breast cancer | A → G/A | ND5 | Lys (AAA) → Lys (AAG) | 21 | |
| 13980 | Breast cancer | G → C/G | ND5 | Leu (CTG) → Leu (CTC) | 21 | |
| 15416 | Breast cancer | T → C/T | CytB | Tyr (TAC) → His (CAC) | Potential to cause mitochondrial Complex III dysfunction (mutation in highly conserved residue) | 21 |
The 11032delA mutation was reported to be identified in prostate cancer,28 renal oncocytoma tissues,29 and hepatocellular carcinoma (HCC).20 The 11032delA mutation is an “A” nucleotide deletion in the mononucleotide repeat of a poly-adenosine (poly-A) sequence at np 11032–11038 in mtDNA. The mutation causes a frame-shift and premature termination of the ND4 gene, thereby resulting in a truncated ND4 subunit protein. Moreover, the 11032delA mutation is correlated with a loss of Complex I activity in renal oncocytomas.29
The 12418insA mutation was also identified in several types of cancer, including the rotenone-resistant VA2B cell line,30 colorectal cancer,26 HCC,20 gastric cancer,27 and breast cancer specimens.21 The 12418insA mutation is an “A” nucleotide insertion in the mononucleotide repeat of a poly-A sequence at np 12418–12425 in mtDNA. The mutation causes a frame-shift and premature termination of the ND5 gene, thereby resulting in a truncated ND5 subunit protein. Using a cybrid cell model, a heteroplasmic 12418insA mutation was demonstrated to reduce oxidative phosphorylation and increase ROS production in human cancer cells and promotes tumor growth in nude mice, suggesting that this mtDNA mutation contributes to tumorigenesis.22
The T1659C transition in the tRNAVal gene and the G5650A transition in the tRNAAla gene were identified in two independent HCC tissues.20 Clinically, the two mutations have been previously reported to be associated with distinct mitochondrial disorders.23,31,32 The T1659C transition was found at a very high level of heteroplasmy in the blood, and skeletal muscle from a young girl with learning difficulties, hemiplegia, and a movement disorder, as well as an increased lactate level in the cerebrospinal fluid.32 The G5650A transition was reported at a high level of heteroplasmy in the muscle and blood of a patient with a stereotypic clinical presentation of cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy (CADASIL) and myopathy with ragged-red fibers.31 The mtDNA mutation was also found in a family with a predominantly proximal myopathy and was associated with a large number of cytochrome c oxidase-deficient fibers and a marked decrease in the activities of both Complex I and Complex IV.23
The 7472insC was identified in a gastric cancer patient27 and was a “C” nucleotide insertion in the mononucleotide repeat of a poly-C sequence at np 7466–7472 in mtDNA, which could alter the structure of the TψC loop in the clover leaf secondary structure of tRNASer(UCN). The mutation was reported to be associated with maternally inherited hearing loss, ataxia and myoclonus syndrome,24,25 rapidly progressive neurodegeneration,33 and progressive myoclonus epilepsy with ragged-red fibers (MERRF) and a MERRF-like phenotype.34,35 It has been shown that the cybrids carrying homoplasmic 7472insC mutant mtDNA exhibited decreased Complex I activity, low oxygen consumption rate and high lactic acid production.24,25
Another pathogenic mutation T8993G in the ATP synthase subunit 6 (ATPase 6) gene of mtDNA was introduced into the PC3 prostate cancer cell line by a cybrid transfer technique, and the mtDNA mutation exhibited enhanced tumorigenesis in nude mice.36 Moreover, using cybrids containing the T8993G or T9176C mutation in the ATPase 6 gene in the HeLa cell line, these mtDNA mutations were also shown to confer an advantage in tumor growth in nude mice by preventing apoptosis.37
These findings suggest that most mtDNA point mutations identified in cancer tissues have a high potential to result in mitochondrial dysfunction. Some point mutations were shown to contribute to tumorigenesis. However, it is unclear whether the different mtDNA point mutations play similar role in cancer progression. The cybrid transfer technique provides a strategy to approach this issue for specific mtDNA mutation.
Copy number changes
A decrease in mtDNA copy number was frequently detected in cancer tissues compared with corresponding noncancerous tissues. Alterations in mtDNA copy number of cancers appear to be tissue specific.13,15 A decreased mtDNA copy number is frequently found in the majority of HCC, gastric cancers, and breast cancers.16,19,38
In HCC, a decrease in mtDNA copy number was found to more frequently occur in females than in males, indicating that the change of mtDNA copy number might contribute to the differences in clinical manifestations including tumorigenesis, cancer progression, metastasis, and prognosis.39 HCC patients with lower mtDNA content exhibited poorer 5-year survival than patients with higher mtDNA content.40 In gastric cancer, a decrease in mtDNA copy number was reported to be correlated with patients with ulcerated and infiltrating or diffusely thick types, which were associated with poor prognoses and lower 5-year survival rates after gastric resection.38 An analysis of a different set of patients revealed that increased mtDNA copy number is associated with worse survival in patients with late-stage tumors.41 In breast cancer, a decrease in mtDNA copy number was correlated with higher histological grade, poorer disease-free survival, and lower overall survival.42 In colorectal cancer, patients with lower mtDNA copy number showed higher TNM stages and poorer differentiation.43
On the other hand, in head and neck cancers, an increase in mtDNA copy number was positively correlated with the histopathological grade, and mtDNA copy number in saliva from patients with head and neck squamous cell carcinoma was higher than controls and was associated with advanced tumor stage.44,45 In ovarian cancer, the mtDNA copy number in patients with pathologically high-grade tumors was higher than in patients with low-grade tumors.46 Similarly, in esophageal squamous cell carcinoma, the mtDNA copy number in the cancers of metastatic lymph nodes is higher than that in noncancerous tissue.47,48
The factor involving in the tissue-specific changes of mtDNA copy number in cancers is still unclear. Based on the clinicopathological correlations with changes of mtDNA copy number, these mtDNA copy number changes can potentially be used as a molecular prognostic marker of some types of cancer.
These findings suggest that somatic point mutations or decreased copy number of mtDNA in cancers may result in mitochondrial dysfunction. It needs to further evaluate whether these somatic mtDNA alterations might contribute to cancer progression or might be bystanders during cancer progression.
Mitochondrial dysfunction induced by somatic mtDNA alterations promotes cancer progression
Mitochondrial dysfunction induced by somatic mtDNA alterations in cancers might provide a mechanism to trigger the energy metabolism change of tumor cells from oxidative phosphorylation to glycolysis. It was thus hypothesized that mitochondrial dysfunction might contribute to cancer progression. To examine this hypothesis, several strategies, including chemical depletion of mtDNA, chemical impairment of mitochondrial respiratory chain, and cybrid transfer technique, were adopted to evaluate whether mitochondrial dysfunction promotes cancer progression to an apoptosis-resistant/chemo-resistant and/or invasive phenotype and to dissect the underlying mechanism.
It was demonstrated that depletion of mtDNA in HeLa cells prevents activation of adriamycin in the cells through impairing mitochondrial Complex I activity and subsequently results in resistance to this drug.49 Moreover, depletion of mtDNA in human hepatoma SK-Hep1 cells results in an adaptive increase in the expression of manganese superoxide dismutase (MnSOD) and other antioxidant enzymes that enabled the cancer cells to counteract oxidative stress or chemotherapeutic agents.50 The depletion of mtDNA was also shown to increase the expression of the multidrug resistance 1 (MDR1) gene and hence to exhibit higher tolerance to anti-cancer agents in human colon cancer HCT-8 cells,51 osteosarcoma 143B cells,52 and hepatoma cells.53 Chloramphenicol-induced mitochondrial stress in human hepatoma HepG2 cells and non-small cell lung cancer H1299 cells increases p21 expression and prevents mitomycin-induced apoptosis through a p21-dependent pathway.54 Down-regulation of the α subunit of ATP synthase and low ATP synthase activity in human colon cancer cells exhibits resistance to 5-fluorouracil.55 Mitochondrial respiration defects in cancer cells cause activation of the Akt survival pathway and contribute to drug resistance through a redox mediated mechanism.56 We also found that chemical impairment of the mitochondrial respiratory chain enhances cisplatin-resistance in human HepG2 cells through up-regulated expression and secretion of amphiregulin57 and in human gastric cancer cells through a ROS-mediated regulation.58
In addition, mtDNA copy number might affect hormone dependence in prostate cancer cells59 and breast cancer cells.60 It was demonstrated that the depletion of mtDNA in androgen-dependent LNCaP cells results in a loss of androgen dependence.61 The depletion of mtDNA in MCF-7 cells exhibits resistance to hydroxytamoxifen and ICI182780.62 These findings suggest that mtDNA defects may play an important role in the development of hormone independence, which may contribute to the progression of these cancers.
It was demonstrated that partial chemical depletion of mtDNA and impairment of mitochondrial respiratory chain in human lung cancer A549 cells induce an invasive phenotype.63 It was also reported that the mitochondrial dysfunction-modulated invasive phenotype is induced by transcriptional regulation of extracellular matrix-remodeling genes.61 Moreover, depletion of mtDNA in LNCaP and MCF-7 cells exhibits an invasive phenotype by promoting epithelial-mesenchymal transition.62 Depletion of mtDNA in breast cancer cells by expressing mutant mitochondrial DNA polymerase γ increases Matrigel invasion.64 In addition, chemical impairment of the mitochondrial respiratory chain in HepG2 cells enhances their migratory phenotype through the overexpression and secretion of amphiregulin by an autocrine or paracrine loop,57 and overexpression of peroxisome proliferator-activated receptor-gamma coactivator 1 alpha (PGC-1α) to promote mitochondrial biogenesis in HepG2 cells can markedly repress cell migration through the upregulation of E-cadherin expression.65,66 We also found that chemical impairment of the mitochondrial respiratory chain in human gastric cancer cells enhances cell migration through ROS-induced integrin β5 expression.58
To rule out the toxic effects of chemical treatments, the cybrid transfer technique for replacement of mtDNA in tumor cells was used to demonstrate that mtDNA mutations may promote tumorigenesis and contribute to cancer progression. It was demonstrated that the heteroplasmic 12418insA mutation,22 the T8993G mutation,36,37 and the T9176C mutation37 promotes tumor growth in nude mice. Moreover, ROS-generating mtDNA mutations were found to enhance the metastatic potential of tumor cells.67
These findings together suggest that mitochondrial dysfunction may enhance tumor growth or promote cancer progression to an apoptosis-resistant/chemo-resistant and/or invasive phenotype through various mechanisms.
Defects in mitochondrial enzyme in tumor cells may lead to mitochondrial dysfunction and cancer progression
In addition to somatic mtDNA alterations, deregulated cellular energetics in cancer cells might result from defects in the nuclear-encoded mitochondrial enzymes, including several enzymes of the TCA cycle and mitochondrial deacetylase SIRT3.
Succinate dehydrogenase (SDH)
The SDH (also known as respiratory Complex II) is a heterotetrameric protein complex that is located on the mitochondrial inner membrane, and contains two catalytic subunits (SDHA and SDHB) and two integral subunits (SDHC and SDHD).68 The four subunits are encoded by nuclear genes and two assembly factors, SDH assembly factor 1 (SDHAF1) and SDHAF2, are essential for the assembly and activity of the SDH protein complex.69,70 In the TCA cycle, the SDH complex converts succinate into fumarate in a reaction that is coupled to the reduction of flavin adenine dinucleotide (FAD) to FADH2, and electrons are transferred to coenzyme Q. Inactivating mutations in the genes of the SDH subunits and assembly factors have been identified in pheochromocytoma (PCC),71 paraganglioma (PGL),71–74 gastrointestinal stromal tumors (GISTs),75 renal carcinoma,76,77 thyroid tumors, testicular seminoma, neuroblastomas, and breast cancer.78
The oncogenic activity of SDH defects has been attributed to succinate accumulation. Defects in SDH have been shown to result in the accumulation of extra-mitochondrial succinate and are linked to the inhibition of prolyl hydroxylases and hence the stabilization and activation of HIF-1α under normoxic conditions.79–81 The SDH defects thus establish a tumorigenic “pseudo-hypoxic” state. In addition, SDH mutations have been reported to trigger ROS production, which might result in HIF activation through the inactivation of prolyl hydroxylases68,82 or might lead to genomic instability.83 Moreover, the accumulation of succinate was found to result in epigenetic changes in gene expression by the inhibition of α-ketoglutarate-dependent histone and DNA demethylases.68,84,85
Fumarate hydratase
Fumarate hydratase (also known as fumarase) is a nuclear-encoding mitochondrial matrix enzyme, which is involved in the TCA cycle for converting fumarate to malate. Loss of functional fumarate hydratase lead to low respiratory rate and increased glucose addition and lactate production in a UOK262 kidney cancer cell line derived from a metastasis in a patient with hereditary leiomyomatosis renal cell carcinoma (HLRCC).86,87 The altered mitochondrial respiration and cellular metabolism in the fumarase-deficient cells might result from ROS-dependent stabilization and activation of HIF-1α and lower expression of TP53.86,88
Fumarate hydratase mutations have been found in uterine leiomyomas and renal cell cancer,89 dominantly inherited uterine fibroids, skin leiomyomata and papillary renal cell cancer,90 clear cell renal cancer,91 and PGL/PCC.92 Similar to SDH, the tumorigenic activity of fumarate hydratase defects is attributed to the abnormal accumulation of fumarate and the stabilization and activation of HIF-1α by inhibition of prolyl hydroxylases.80,93 Moreover, it was found that fumarate covalently binds to cysteine residues in protein, a process called succination, and modulates the enzyme activity. The accumulation of intracellular fumarate can result in the succination of Kelch-like ECH-associated protein 1 (Keap1) and abrogates Keap1-mediated degradation of the nuclear factor erythroid 2-related factor 2 (NRF2).94 The increased NRF2 activates several antioxidant genes and supports tumor formation.94 In addition, the accumulation of fumarate might promote tumorigenesis by inhibiting α-ketoglutarate-dependent genome-wide histones and DNA methylations, hence resulting in epigenetic alterations in gene expression,84 or by increasing ROS-dependent signaling via glutathione succination.95
Isocitrate dehydrogenase (IDH)
There are three isozymes of IDH in mammalian cell: IDH1, IDH2, and IDH3. IDH1 and IDH2 are homodimeric NADP+-dependent enzymes located in the cytosol and mitochondrial matrix, respectively. IDH3 is a heterotetrameric NAD+-dependent enzyme (consist of two alpha subunits, one beta subunit, and one gamma subunit) located in the mitochondrial matrix. The IDHs can oxidatively decarboxylate isocitrate to α-ketoglutarate. Mutations in IDH1 and IDH2 frequently occur in glioma (>75%)96 and secondary glioblastoma (GBM) (70–75%) and are rarely detected in primary glioblastoma (5%).97,98 Mutations in IDH1 and IDH2 have also been found in various human cancers with different frequencies, including acute myeloid leukemia (AML),99 angioimmunoblastic T-cell lymphoma,100 cholangiocarcinoma,101,102 chondrosarcoma,103 colon cancer,104 giant cell tumor of bone,105 melanoma,106 prostate cancer,107 and osteosarcoma.108
Most IDH1 and IDH2 mutations in cancers are often heterozygous with a wild-type allele,96,97,109 and the mutations have been found to result in a neomorphic enzyme activity that converts α-ketoglutarate into the R-enantiomer of 2-hydroxyglutarate.110–113 2-Hydroxyglutarate inhibits the enzymatic activity of Complex IV (cytochrome c oxidase) and Complex V (ATP synthase)114,115 and alters the gene expression of TCA cycle enzymes in cancer cells.116 These findings suggest that the accumulation of 2-hydroxyglutarate contributes to the alterations in energy metabolism in IDH-mutant cancer cells.
2-Hydroxyglutarate is considered a major contributor to the oncogenic activity of IDH mutations and is thus identified as an “onco-metabolite” that promotes tumorigenesis.112 The increased 2-hydroxyglutarate levels were shown to be associated with DNA hypermethylation and a broad epigenetic change, which results in epigenetic alterations of gene expression.113,117,118 The oncogenic activity of 2-hydroxyglutarate has been attributed to its accumulation and the inhibitory effect on various α-ketoglutarate-dependent dioxygenases, including the prolyl hydroxylases, histone demethylase KDM4C, and 5-methylcytosine hydroxylase TET2.113,117,119,120 However, the effects of 2-hydroxyglutarate on HIF-1α and prolyl hydroxylases could be controversial. Some studies reported that 2-HG leads to inhibition of HIF-1α via activation of prolyl hydroxylases, whereas other studies showed that 2-hydroxyglutarate inhibits prolyl hydroxylases and induces HIF-1α expression.100,113,121,122 These findings imply that the effects of 2-hydroxyglutarate on HIF-1α signaling might be cell type dependent and the role of IDH mutants in tumorigenesis by HIF-1α signaling needs further investigation.
SIRT3
SIRT3 is one of the nuclear-encoded mitochondrial protein deacetylases. SIRT3 regulates the function of several mitochondrial proteins involved in oxidative phosphorylation, fatty acid oxidation, the urea cycle, and the antioxidant response system.123–131 SIRT3 is thought to play a tumor suppressor role in various cancers, including breast, colorectal, HCC, lung and gastric cancers.132–137 Loss of SIRT3 was found to increase intracellular ROS levels and result in the stability and activation of HIF1α and the Warburg effect phenotype, which promote tumorigenesis.133
These findings together suggest that mitochondrial dysfunction induced by these defects in several nuclear-encoded enzymes of the TCA cycle, such as SDH, fumarate hydratase, and IDH, and the down-regulation of mitochondrial deacetylase SIRT3 results in mitochondrial dysfunction, and enhances tumor growth, as well as promotes cancer progression.
Altered oncogene/tumor suppressor gene represses mitochondrial respiratory function in cancers
Alterations in oncogene/oncosuppressor including HIF-1 and TP53 have been shown to regulate mitochondrial respiration and cellular metabolism.
HIF-1
HIF-1 is a heterodimeric protein consisting of HIF-1α and HIF-1β subunits, both of which are members of the basic helix-loop-helix family of transcription factors, that modulates the cellular response to hypoxia in normal and tumor cells.138 Although HIF-1β is a constitutively expressed nuclear protein, HIF-1α is tightly regulated by oxygen availability. Under normoxia, HIF-1α subunit is rapidly hydroxylated on the oxygen-dependent degradation (ODD) domain by prolyl hydroxylases and degraded through von Hippel–Lindau tumor suppressor protein (pVHL)-mediated proteasome degradation pathway.139,140 In tumor cells, HIF-1α plays a crucial role in angiogenesis, proliferation, invasion, and metastasis.138,141 Overexpressed HIF-1α is associated with malignancy in many types of tumor including bladder, brain, breast, colon, oral, liver, lung, pancreas, skin, stomach, uterus, and leukemia.142 Several factors have been reported to increase HIF-1α expression, including inhibition of SDH and fumarate hydratase, or activation of phosphoinositide 3-kinase (PI3K) and viral transforming genes.29,143 Moreover, HIF1α plays a critical role in the regulation of mitochondrial respiration and cellular metabolism.144,145 The activation of HIF1α in tumor can alter energy metabolism from oxidative phosphorylation to glycolysis through the expression of pyruvate dehydrogenase kinase 1 (PDK1), which inhibits the conversion of pyruvate to acetyl-CoA as the substrate for the TCA cycle by phosphorylation of pyruvate dehydrogenase. The inhibition of pyruvate dehydrogenase leads to repression of mitochondrial oxidative metabolism. Moreover, HIF-1α increases glycolysis by up-regulation the gene expressions of the glucose transporters, glycolytic enzymes, and lactate dehydrogenase A (LDHA).143 HIF-1α is thought to contribute to reprogramming energy metabolism in tumor cells from oxidative phosphorylation to glycolysis. In addition, HIF-1α induces the expression of mitochondrial LON protease, which is important in the degradation of the COX4-1 subunit of the cytochrome c oxidase complex (COX), and the increased expression of COX4-2, a downstream target gene of HIF-1α, is used to substitute COX4-1. It was found that the replacement of COX4-1 with COX4-2 in cancer cells increases the efficiency of electron transfer to oxygen and continued respiration and decreases the ROS production under limited oxygen availability.146 A recent study also showed that the expression of mitochondrial chaperone TRAP1 is increased in cancer cells, and the chaperone interacts with and inhibits SDH. The interaction between TRAP1 and SDH results in the accumulation of succinate and the stabilization of HIF-1α and hence induces metabolic reprogramming and promotes tumorigenesis.147 These findings suggest that HIF-1α plays a crucial role in the cellular metabolism and tumorigenesis of cancer cells.
TP53
TP53 is well known for the functions in DNA damage response (DDR), cell cycle arrest, and apoptosis. Loss or inactive mutations of p53 were often observed in several types of tumor (approximately 60%) (TP53 website). Moreover, animals lacking TP53 have been shown to exhibit the spontaneous development of tumors.148,149 In addition, TP53 was found to be important for maintenance of the mitochondrial respiration and regulation of cellular metabolism. TP53 defects are thought to contribute to the Warburg effect in cancer cells.
TP53 can repress the transcription of glucose transporter (GLUT) isoform 1 (GLUT1) and GLUT4 and reduce the expression of GLUT3 through an IKK/nuclear factor-κB (NFκB)-dependent manner.150,151 These glucose transporters are important in glucose uptake, and the loss of TP53 can increase glucose consumption in cancer cells. Moreover, TP53 was found to modulate glycolytic enzymes through several mechanisms.149 TP53 can stimulate the expression of the TP53-induced glycolysis and the apoptosis regulator (TIGAR) and repress glycolysis by degrading fructose-2,6-bisphosphate, which is an activator of phosphofructokinase 1.152 In addition, TP53 can promote the degradation of phosphoglycerate mutase and hence can repress glycolysis.153 TP53 defects can result in an increase in glycolytic flux by reduced TIGAR and increased phosphoglycerate mutase.
On the other hand, T53 was found to transcriptionally induce the expression of subunit I of cytochrome c oxidase154 and the apoptosis-inducing factor (AIF, which is important in the maintenance of mitochondrial Complex I)155,156 and the synthesis of cytochrome c oxidase 2 (SCO2, which is essential to the assembly of the cytochrome c oxidase complex)157 and glutaminase 2 (GLS2, which catalyzes glutamine to glutamate for fuel for the TCA cycle).158,159 These findings suggest that TP53 defects can result in down-regulation of mitochondrial respiration and oxidative metabolism.
In addition, TP53 could transcriptionally inactivate the expression of malic enzymes ME1 and ME2, which are involved in recycling of malate to pyruvate, and hence inhibits the intermediates of the TCA cycle entering biosynthetic pathways.160 Moreover, TP53 was reported to increase the expression of ribonucleotide reductase subunit TP53R2 for maintaining mtDNA integrity.161 TP53 was also shown to interact with mitochondrial DNA polymerase γ and play an important role in mtDNA maintenance in response to oxidative damage.162 These findings indicate that TP53 is involved in the regulation of mitochondrial respiration and cellular metabolism, and a loss function of TP53 can contribute to the Warburg effect in tumor cells.
Activation of mitochondrial retrograde signaling contributes to mitochondrial dysfunction-induced tumorigenesis and cancer progression
Because of the roles of HIF-1α and TP53 in both mitochondrial function and tumorigenesis, they might directly link between mitochondrial dysfunction and the formation and progression of cancer. Unlike HIF-1α and TP53, mitochondrial dysfunction caused by mtDNA mutations and the nuclear-encoded mitochondrial enzyme defects might contribute to the formation and progression of cancer by triggering cytosolic signaling pathways that ultimately alter nuclear gene expression, a process called retrograde signaling. Recent studies revealed that changes in the intracellular levels of ROS, Ca2+, and oncometabolites that are released from mitochondria might be important in the mitochondrial retrograde signaling for neoplastic transformation.
Increased low levels of mitochondria-derived ROS can function as signaling messengers by reversibly oxidizing protein thiol groups, thereby modifying protein structure and function. Higher levels of ROS can nonspecifically damaging DNA, proteins, and lipids, which could result in disruption of mitochondrial electron transfer chain and consequently collapse of mitochondrial function and threaten cell survival. Accumulating evidence has shown that hypoxia, activation of oncogenes, loss of tumor suppressors, and mitochondrial dysfunction induced by mtDNA mutations or mitochondrial enzyme defects increase production of mitochondrial ROS.163 Recent evidence revealed that tumor cells can enhance glutathione and thioredoxin antioxidant systems to drive cancer initiation and progression by preventing increased ROS from reaching cytotoxic levels.164 This range of ROS might be capable of increasing tumorigenesis and/or promoting cancer progression by activating signaling pathways that regulate cellular proliferation, metabolic adaptation, antioxidant systems, apoptosis-resistance, chemoresistance, and cellular migration/invasion.163
In cytoplasmic hybrids for replacement of mtDNA in tumor cells, ROS-generating mtDNA mutations have been found to increase tumorigenesis22 and to enhance the metastatic potential of tumor cells.67 Loss of SDHB81 and mutations in SDHC165 were also shown to increase tumorigenesis by enhancing mitochondrial ROS levels. Fumarase-deficient cancer cells were found to increase mitochondrial ROS and HIF-1a stabilization.86 Moreover, mitochondrial dysfunctions caused by various respiration inhibitors have been shown to promote cell migration via ROS-enhanced β5-integrin expression in gastric cancer SC-M1 cells58 or ROS-mediated upregulation of amphiregulin in human hepatoma HepG2 cells.57
In addition to ROS, Ca2+ was found to up-regulate amphiregulin and induce chemo-resistance and migration of human hepatoma HepG2 cells.57 Moreover, elevated cytosolic Ca2+ was found in cancer cells with mtDNA depletion by ethidium bromide166 and was involved in the mtDNA depletion-induced expression of the invasive markers, cathepsin L and TGFβ1 as well as the invasive phenotype.61 Activation of protein kinase C (PKC) and calcineurin-induced signaling might mediate the mtDNA depletion-induced invasive phenotype.166 It was further demonstrated that calcineurin can stimulate IκBβ-dependent NFκB and nuclear translocation of cRel-p50, which results in the changes in the malignant characteristics including metabolic reprogramming, invasive behavior, and resistance to apoptosis.167 These findings suggest that increased cytosolic Ca2+ induced by mitochondrial dysfunction can activate signaling pathways that regulate metabolic alterations, cell invasion, and apoptosis-resistance to promote cancer progression.
As mentioned above, defects in SDH, fumarate hydratase, and IDH can result in the accumulation of metabolites that are released from mitochondria; these metabolites, such as succinate, fumarate, and 2-hydroxyglutarate, have been found to be important for tumorigenesis activity of these mitochondrial enzyme defects. These metabolites are thus considered oncometabolites. The accumulation of oncometabolites can promote tumorigenesis by inhibiting prolyl hydroxylases and stabilizing HIF-1α.92,168 Moreover, the accumulation of these oncometabolites might affect α-ketoglutarate-dependent genome-wide histone and DNA methylations and hence might result in epigenetic alterations of gene expression, which contribute to tumorigenesis.83
In addition, mitochondrial dysfunctions were found to repress the protein synthesis, trans-activation activity, and targeting gene expressions of HIF-1α through an energy sensor, the AMP-activated protein kinase (AMPK)-dependent pathway.169 This provides another mechanism for the communication between the mitochondria and the nucleus. We further found that loss of this communication renders HCC cancer cells to exhibit drug resistance.170 These findings suggest that the mitochondrial retrograde signaling pathway might regulate the sensitivity of cancer cells to anti-cancer agents.
Interestingly, a recent study found that pyruvate dehydrogenase complex (PDC), which is originally found to be located in mitochondria and catalyzes the pyruvate to acetyl-CoA reaction, can be translocated from the mitochondria to the nucleus and can generate acetyl-CoA in the nucleus to be used for histone acetylation under conditions of mitochondrial dysfunction. The nucleus translocation of PDC was found to be important for cell cycle G1-S phase progression in response to exposure of serum and growth factor (such as EGF), suggesting a potential role in cancer with proliferative signals or mitochondrial dysfunction.171 These findings suggest that the communication between the nucleus and the mitochondria might play an important role in tumorigenesis.
Summary
We have shown several lines of evidence that suggest that the underlying mechanisms of deregulated cellular energetics are associated with mitochondrial dysfunction caused by mtDNA mutations, mitochondrial enzyme defects, or altered oncogenes/tumor suppressors. Mitochondrial dysfunction can promote cancer progression to an apoptosis-resistant/chemo-resistant and/or invasive phenotype through various mechanisms. These mitochondrial alterations during oncogenesis and tumor progression can activate cytosolic signaling pathways from mitochondria to the nucleus and ultimately alter nuclear gene expression for neoplastic transformation (Figure 2). However, it is still largely unclear how specific mtDNA mutations regulate the formation and progression of cancer. Elucidation of the detailed molecular mechanisms for the mitochondrial retrograde signaling still requires further investigation. Based on these findings that mitochondrial dysfunction may contribute to cancer progression, we suggest that targeting mitochondrial alterations and mitochondrial retrograde signaling might be a promising strategy for the development of selective anticancer therapy.
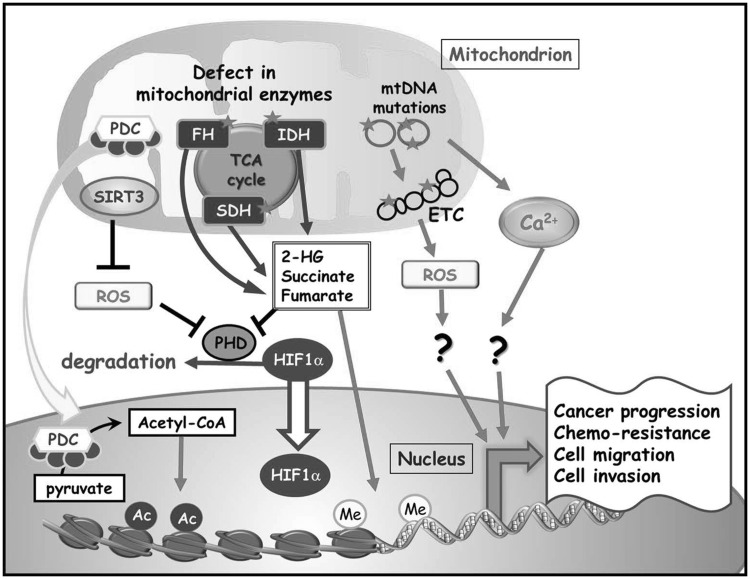
Figure 2.
Mitochondrial dysfunction caused by mitochondrial DNA (mtDNA) mutations, mitochondrial enzyme defects, or altered oncogenes/tumor suppressors might contribute to the formation and progression of cancer. Somatic alterations in mtDNA and defects in mitochondrial enzymes, such as succinate dehydrogenase (SDH), fumarate hydratase (FH), and isocitrate dehydrogenase (IDH) were found to be associated with both familial and sporadic forms of cancer. Deregulated mitochondrial deacetylase sirtuin 3 (SIRT3) might modulate cancer progression by regulating cellular metabolism and oxidative stress. These mitochondrial alterations during oncogenesis and tumor progression activate cytosolic signaling pathways that ultimately alter nuclear gene expression, a process called retrograde signaling. Changes in reactive oxygen species (ROS), Ca2+, or oncometabolites are involved in the mitochondrial retrograde signaling for neoplastic transformation by inhibiting prolyl hydroxylases and stabilizing hypoxia-inducible factor 1α (HIF-1α) or by modulating α-ketoglutarate-dependent genome-wide histone and DNA methylations, resulting in epigenetic alterations of gene expression. In addition, altered oncogene/oncosuppressor including HIF-1 and tumor suppressor p53 (TP53) not only regulates mitochondrial respiration and cellular metabolism but also promote the formation and progression of cancer
Acknowledgements
This work was partly supported by the grants from the Center of Excellence for Cancer Research at Taipei Veterans General, the Ministry of Health and Welfare (MOHW103-TD-B-111-02; MOHW104-TDU-B-211-124-001; MOHW105-TDU-B-211-134003), Executive Yuan; the grant from the Ministry of Education, Aim for the Top University Plan; and the grants MOST101-2320-B-010-068-MY3, MOST101-2314-B-075-015-MY3, and MOST 104-2320-B-010-031 from the Ministry of Science and Technology, Taiwan.
Authors' contributions
CCH reviewed the literature and wrote the first draft; LMT and HCL edited the final draft. All authors participated in the writing of this manuscript.
Declaration of conflicting interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011; 144: 646–74. [DOI] [PubMed] [Google Scholar]
- 2.Chen Z, Lu W, Garcia-Prieto C, Huang P. The Warburg effect and its cancer therapeutic implications. J Bioenerg Biomembr 2007; 39: 267–74. [DOI] [PubMed] [Google Scholar]
- 3.Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer 2011; 11: 85–95. [DOI] [PubMed] [Google Scholar]
- 4.Wallace DC. Mitochondria and cancer. Nat Rev Cancer 2012; 12: 685–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Upadhyay M, Samal J, Kandpal M, Singh OV, Vivekanandan P. The Warburg effect: insights from the past decade. Pharmacol Ther 2013; 137: 318–30. [DOI] [PubMed] [Google Scholar]
- 6.Wallace DC, Fan W, Procaccio V. Mitochondrial energetics and therapeutics. Annu Rev Pathol 2010; 5: 297–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galluzzi L, Kepp O, Kroemer G. Mitochondria: master regulators of danger signalling. Nat Rev Mol Cell Biol 2012; 13: 780–8. [DOI] [PubMed] [Google Scholar]
- 8.Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJ, Staden R, Young IG. Sequence and organization of the human mitochondrial genome. Nature 1981; 290: 457–65. [DOI] [PubMed] [Google Scholar]
- 9.Lee HC, Chang CM, Chi CW. Somatic mutations of mitochondrial DNA in aging and cancer progression. Ageing Res Rev 2010; 9 Suppl 1: S47–58. [DOI] [PubMed] [Google Scholar]
- 10.Schon EA, DiMauro S, Hirano M. Human mitochondrial DNA: roles of inherited and somatic mutations. Nat Rev Genet 2012; 13: 878–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wallace DC. A mitochondrial bioenergetic etiology of disease. J Clin Invest 2013; 123: 1405–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Y, Fu LL, Wen X, Wang XY, Liu J, Cheng Y, Huang J. Sirtuin-3 (SIRT3), a therapeutic target with oncogenic and tumor-suppressive function in cancer. Cell Death Dis 2014; 5: e1047–e1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee HC, Yin PH, Lin JC, Wu CC, Chen CY, Wu CW, Chi CW, Tam TN, Wei YH. Mitochondrial genome instability and mtDNA depletion in human cancers. Ann NY Acad Sci 2005; 1042: 109–22. [DOI] [PubMed] [Google Scholar]
- 14.Lee HC, Huang KH, Yeh TS, Chi CW. Somatic alterations in mitochondrial DNA and mitochondrial dysfunction in gastric cancer progression. World J Gastroenterol 2014; 20: 3950–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee HC, Wei YH. Mitochondrial DNA instability and metabolic shift in human cancers. Int J Mol Sci 2009; 10: 674–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee HC, Li SH, Lin JC, Wu CC, Yeh DC, Wei YH. Somatic mutations in the D-loop and decrease in the copy number of mitochondrial DNA in human hepatocellular carcinoma. Mutat Res 2004; 547: 71–8. [DOI] [PubMed] [Google Scholar]
- 17.Matsuyama W, Nakagawa M, Wakimoto J, Hirotsu Y, Kawabata M, Osame M. Mitochondrial DNA mutation correlates with stage progression and prognosis in non-small cell lung cancer. Hum Mutat 2003; 21: 441–3. [DOI] [PubMed] [Google Scholar]
- 18.Lievre A, Chapusot C, Bouvier AM, Zinzindohoue F, Piard F, Roignot P, Arnould L, Beaune P, Faivre J, Laurent-Puig P. Clinical value of mitochondrial mutations in colorectal cancer. J Clin Oncol 2005; 23: 3517–25. [DOI] [PubMed] [Google Scholar]
- 19.Tseng LM, Yin PH, Chi CW, Hsu CY, Wu CW, Lee LM, Wei YH, Lee HC. Mitochondrial DNA mutations and mitochondrial DNA depletion in breast cancer. Genes Chromosomes Cancer 2006; 45: 629–38. [DOI] [PubMed] [Google Scholar]
- 20.Yin PH, Wu CC, Lin JC, Chi CW, Wei YH, Lee HC. Somatic mutations of mitochondrial genome in hepatocellular carcinoma. Mitochondrion 2010; 10: 174–82. [DOI] [PubMed] [Google Scholar]
- 21.Tseng LM, Yin PH, Yang CW, Tsai YF, Hsu CY, Chi CW, Lee HC. Somatic mutations of the mitochondrial genome in human breast cancers. Genes Chromosomes Cancer 2011; 50: 800–11. [DOI] [PubMed] [Google Scholar]
- 22.Park JS, Sharma LK, Li H, Xiang R, Holstein D, Wu J, Lechleiter J, Naylor SL, Deng JJ, Lu J, Bai Y. A heteroplasmic, not homoplasmic, mitochondrial DNA mutation promotes tumorigenesis via alteration in reactive oxygen species generation and apoptosis. Hum Mol Genet 2009; 18: 1578–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McFarland R, Swalwell H, Blakely EL, He L, Groen EJ, Turnbull DM, Bushby KM, Taylor RW. The m.5650G>A mitochondrial tRNAAla mutation is pathogenic and causes a phenotype of pure myopathy. Neuromuscul Disord 2008; 18: 63–7. [DOI] [PubMed] [Google Scholar]
- 24.Tiranti V, Chariot P, Carella F, Toscano A, Soliveri P, Girlanda P, Carrara F, Fratta GM, Reid FM, Mariotti C, Zeviani M. Maternally inherited hearing loss, ataxia and myoclonus associated with a novel point mutation in mitochondrial tRNASer(UCN) gene. Hum Mol Genet 1995; 4: 1421–7. [DOI] [PubMed] [Google Scholar]
- 25.Toompuu M, Tiranti V, Zeviani M, Jacobs HT. Molecular phenotype of the np 7472 deafness-associated mitochondrial mutation in osteosarcoma cell cybrids. Hum Mol Genet 1999; 8: 2275–83. [DOI] [PubMed] [Google Scholar]
- 26.Polyak K, Li Y, Zhu H, Lengauer C, Willson JK, Markowitz SD, Trush MA, Kinzler KW, Vogelstein B. Somatic mutations of the mitochondrial genome in human colorectal tumours. Nat Genet 1998; 20: 291–3. [DOI] [PubMed] [Google Scholar]
- 27.Hung WY, Wu CW, Yin PH, Chang CJ, Li AF, Chi CW, Wei YH, Lee HC. Somatic mutations in mitochondrial genome and their potential roles in the progression of human gastric cancer. Biochim Biophys Acta 2010; 1800: 264–70. [DOI] [PubMed] [Google Scholar]
- 28.Jeronimo C, Nomoto S, Caballero OL, Usadel H, Henrique R, Varzim G, Oliveira J, Lopes C, Fliss MS, Sidransky D. Mitochondrial mutations in early stage prostate cancer and bodily fluids. Oncogene 2001; 20: 5195–8. [DOI] [PubMed] [Google Scholar]
- 29.Mayr JA, Meierhofer D, Zimmermann F, Feichtinger R, Kogler C, Ratschek M, Schmeller N, Sperl W, Kofler B. Loss of complex I due to mitochondrial DNA mutations in renal oncocytoma. Clin Cancer Res 2008; 14: 2270–5. [DOI] [PubMed] [Google Scholar]
- 30.Hofhaus G, Attardi G. Efficient selection and characterization of mutants of a human cell line which are defective in mitochondrial DNA-encoded subunits of respiratory NADH dehydrogenase. Mol Cell Biol 1995; 15: 964–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Finnila S, Tuisku S, Herva R, Majamaa K. A novel mitochondrial DNA mutation and a mutation in the Notch3 gene in a patient with myopathy and CADASIL. J Mol Med (Berl) 2001; 79: 641–7. [DOI] [PubMed] [Google Scholar]
- 32.Blakely EL, Poulton J, Pike M, Wojnarowska F, Turnbull DM, McFarland R, Taylor RW. Childhood neurological presentation of a novel mitochondrial tRNA(Val) gene mutation. J Neurol Sci 2004; 225: 99–103. [DOI] [PubMed] [Google Scholar]
- 33.Cardaioli E, Da Pozzo P, Cerase A, Sicurelli F, Malandrini A, De Stefano N, Stromillo ML, Battisti C, Dotti MT, Federico A. Rapidly progressive neurodegeneration in a case with the 7472insC mutation and the A7472C polymorphism in the mtDNA tRNA ser(UCN) gene. Neuromuscul Disord 2006; 16: 26–31. [DOI] [PubMed] [Google Scholar]
- 34.Jaksch M, Klopstock T, Kurlemann G, Dorner M, Hofmann S, Kleinle S, Hegemann S, Weissert M, Muller-Hocker J, Pongratz D, Gerbitz KD. Progressive myoclonus epilepsy and mitochondrial myopathy associated with mutations in the tRNA(Ser(UCN)) gene. Ann Neurol 1998; 44: 635–40. [DOI] [PubMed] [Google Scholar]
- 35.Pulkes T, Liolitsa D, Eunson LH, Rose M, Nelson IP, Rahman S, Poulton J, Marchington DR, Landon DN, Debono AG, Morgan-Hughes JA, Hanna MG. New phenotypic diversity associated with the mitochondrial tRNA(SerUCN) gene mutation. Neuromuscul Disord 2005; 15: 364–71. [DOI] [PubMed] [Google Scholar]
- 36.Petros JA, Baumann AK, Ruiz-Pesini E, Amin MB, Sun CQ, Hall J, Lim S, Issa MM, Flanders WD, Hosseini SH, Marshall FF, Wallace DC. mtDNA mutations increase tumorigenicity in prostate cancer. Proc Natl Acad Sci USA 2005; 102: 719–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shidara Y, Yamagata K, Kanamori T, Nakano K, Kwong JQ, Manfredi G, Oda H, Ohta S. Positive contribution of pathogenic mutations in the mitochondrial genome to the promotion of cancer by prevention from apoptosis. Cancer Res 2005; 65: 1655–63. [DOI] [PubMed] [Google Scholar]
- 38.Wu CW, Yin PH, Hung WY, Li AF, Li SH, Chi CW, Wei YH, Lee HC. Mitochondrial DNA mutations and mitochondrial DNA depletion in gastric cancer. Genes Chromosomes Cancer 2005; 44: 19–28. [DOI] [PubMed] [Google Scholar]
- 39.Yin PH, Lee HC, Chau GY, Wu YT, Li SH, Lui WY, Wei YH, Liu TY, Chi CW. Alteration of the copy number and deletion of mitochondrial DNA in human hepatocellular carcinoma. Br J Cancer 2004; 90: 2390–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamada S, Nomoto S, Fujii T, Kaneko T, Takeda S, Inoue S, Kanazumi N, Nakao A. Correlation between copy number of mitochondrial DNA and clinico-pathologic parameters of hepatocellular carcinoma. Eur J Surg Oncol 2006; 32: 303–7. [DOI] [PubMed] [Google Scholar]
- 41.Zhang G, Qu Y, Dang S, Yang Q, Shi B, Hou P. Variable copy number of mitochondrial DNA (mtDNA) predicts worse prognosis in advanced gastric cancer patients. Diagn Pathol 2013; 8: 173–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu M, Zhou Y, Shi Y, Ning L, Yang Y, Wei X, Zhang N, Hao X, Niu R. Reduced mitochondrial DNA copy number is correlated with tumor progression and prognosis in Chinese breast cancer patients. IUBMB Life 2007; 59: 450–7. [DOI] [PubMed] [Google Scholar]
- 43.Lin PC, Lin JK, Yang SH, Wang HS, Li AF, Chang SC. Expression of beta-F1-ATPase and mitochondrial transcription factor A and the change in mitochondrial DNA content in colorectal cancer: clinical data analysis and evidence from an in vitro study. Int J Colorectal Dis 2008; 23: 1223–32. [DOI] [PubMed] [Google Scholar]
- 44.Kim MM, Clinger JD, Masayesva BG, Ha PK, Zahurak ML, Westra WH, Califano JA. Mitochondrial DNA quantity increases with histopathologic grade in premalignant and malignant head and neck lesions. Clin Cancer Res 2004; 10: 8512–5. [DOI] [PubMed] [Google Scholar]
- 45.Jiang WW, Masayesva B, Zahurak M, Carvalho AL, Rosenbaum E, Mambo E, Zhou S, Minhas K, Benoit N, Westra WH, Alberg A, Sidransky D, Koch W, Califano J. Increased mitochondrial DNA content in saliva associated with head and neck cancer. Clin Cancer Res 2005; 11: 2486–91. [DOI] [PubMed] [Google Scholar]
- 46.Wang Y, Liu VW, Xue WC, Cheung AN, Ngan HY. Association of decreased mitochondrial DNA content with ovarian cancer progression. Br J Cancer 2006; 95: 1087–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin CS, Chang SC, Wang LS, Chou TY, Hsu WH, Wu YC, Wei YH. The role of mitochondrial DNA alterations in esophageal squamous cell carcinomas. J Thorac Cardiovasc Surg 2010; 139: 189–97 e4. [DOI] [PubMed] [Google Scholar]
- 48.Lin CS, Lee HT, Lee SY, Shen YA, Wang LS, Chen YJ, Wei YH. High mitochondrial DNA copy number and bioenergetic function are associated with tumor invasion of esophageal squamous cell carcinoma cell lines. Int J Mol Sci 2012; 13: 11228–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Singh KK, Russell J, Sigala B, Zhang Y, Williams J, Keshav KF. Mitochondrial DNA determines the cellular response to cancer therapeutic agents. Oncogene 1999; 18: 6641–6. [DOI] [PubMed] [Google Scholar]
- 50.Park SY, Chang I, Kim JY, Kang SW, Park SH, Singh K, Lee MS. Resistance of mitochondrial DNA-depleted cells against cell death: role of mitochondrial superoxide dismutase. J Biol Chem 2004; 279: 7512–20. [DOI] [PubMed] [Google Scholar]
- 51.Ferraresi R, Troiano L, Pinti M, Roat E, Lugli E, Quaglino D, Taverna D, Bellizzi D, Passarino G, Cossarizza A. Resistance of mtDNA-depleted cells to apoptosis. Cytometry A 2008; 73: 528–37. [DOI] [PubMed] [Google Scholar]
- 52.Lee W, Choi HI, Kim MJ, Park SY. Depletion of mitochondrial DNA up-regulates the expression of MDR1 gene via an increase in mRNA stability. Exp Mol Med 2008; 40: 109–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gonzalez-Sanchez E, Marin JJ, Perez MJ. The expression of genes involved in hepatocellular carcinoma chemoresistance is affected by mitochondrial genome depletion. Mol Pharm 2014; 11: 1856–68. [DOI] [PubMed] [Google Scholar]
- 54.Li CH, Tzeng SL, Cheng YW, Kang JJ. Chloramphenicol-induced mitochondrial stress increases p21 expression and prevents cell apoptosis through a p21-dependent pathway. J Biol Chem 2005; 280: 26193–9. [DOI] [PubMed] [Google Scholar]
- 55.Shin YK, Yoo BC, Chang HJ, Jeon E, Hong SH, Jung MS, Lim SJ, Park JG. Down-regulation of mitochondrial F1F0-ATP synthase in human colon cancer cells with induced 5-fluorouracil resistance. Cancer Res 2005; 65: 3162–70. [DOI] [PubMed] [Google Scholar]
- 56.Pelicano H, Xu RH, Du M, Feng L, Sasaki R, Carew JS, Hu Y, Ramdas L, Hu L, Keating MJ, Zhang W, Plunkett W, Huang P. Mitochondrial respiration defects in cancer cells cause activation of Akt survival pathway through a redox-mediated mechanism. J Cell Biol 2006; 175: 913–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chang CJ, Yin PH, Yang DM, Wang CH, Hung WY, Chi CW, Wei YH, Lee HC. Mitochondrial dysfunction-induced amphiregulin upregulation mediates chemo-resistance and cell migration in HepG2 cells. Cell Mol Life Sci 2009; 66: 1755–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hung WY, Huang KH, Wu CW, Chi CW, Kao HL, Li AF, Yin PH, Lee HC. Mitochondrial dysfunction promotes cell migration via reactive oxygen species-enhanced beta5-integrin expression in human gastric cancer SC-M1 cells. Biochim Biophys Acta 2012; 1820: 1102–10. [DOI] [PubMed] [Google Scholar]
- 59.Higuchi M, Kudo T, Suzuki S, Evans TT, Sasaki R, Wada Y, Shirakawa T, Sawyer JR, Gotoh A. Mitochondrial DNA determines androgen dependence in prostate cancer cell lines. Oncogene 2006; 25: 1437–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Naito A, Carcel-Trullols J, Xie CH, Evans TT, Mizumachi T, Higuchi M. Induction of acquired resistance to antiestrogen by reversible mitochondrial DNA depletion in breast cancer cell line. Int J Cancer 2008; 122: 1506–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van Waveren C, Sun Y, Cheung HS, Moraes CT. Oxidative phosphorylation dysfunction modulates expression of extracellular matrix–remodeling genes and invasion. Carcinogenesis 2006; 27: 409–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Naito A, Cook CC, Mizumachi T, Wang M, Xie CH, Evans TT, Kelly T, Higuchi M. Progressive tumor features accompany epithelial-mesenchymal transition induced in mitochondrial DNA-depleted cells. Cancer Sci 2008; 99: 1584–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Amuthan G, Biswas G, Ananadatheerthavarada HK, Vijayasarathy C, Shephard HM, Avadhani NG. Mitochondrial stress-induced calcium signaling, phenotypic changes and invasive behavior in human lung carcinoma A549 cells. Oncogene 2002; 21: 7839–49. [DOI] [PubMed] [Google Scholar]
- 64.Singh KK, Ayyasamy V, Owens KM, Koul MS, Vujcic M. Mutations in mitochondrial DNA polymerase-gamma promote breast tumorigenesis. J Hum Genet 2009; 54: 516–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee HJ, Su Y, Lui WY, Chau GY, Yin PH, Lee HC, Chi CW. Peroxisome proliferator-activated receptor gamma coactivator-1 alpha (PGC-1alpha) upregulated E-cadherin expression in HepG2 cells. FEBS Lett 2008; 582: 627–34. [DOI] [PubMed] [Google Scholar]
- 66.Lee HJ, Su Y, Yin PH, Lee HC, Chi CW. PPAR(gamma)/PGC-1(alpha) pathway in E-cadherin expression and motility of HepG2 cells. Anticancer Res 2009; 29: 5057–63. [PubMed] [Google Scholar]
- 67.Ishikawa K, Takenaga K, Akimoto M, Koshikawa N, Yamaguchi A, Imanishi H, Nakada K, Honma Y, Hayashi J. ROS-generating mitochondrial DNA mutations can regulate tumor cell metastasis. Science 2008; 320: 661–4. [DOI] [PubMed] [Google Scholar]
- 68.Bardella C, Pollard PJ, Tomlinson I. SDH mutations in cancer. Biochim Biophys Acta 2011; 1807: 1432–43. [DOI] [PubMed] [Google Scholar]
- 69.Ghezzi D, Goffrini P, Uziel G, Horvath R, Klopstock T, Lochmuller H, D'Adamo P, Gasparini P, Strom TM, Prokisch H, Invernizzi F, Ferrero I, Zeviani M. SDHAF1, encoding a LYR complex-II specific assembly factor, is mutated in SDH-defective infantile leukoencephalopathy. Nat Genet 2009; 41: 654–6. [DOI] [PubMed] [Google Scholar]
- 70.Hao HX, Khalimonchuk O, Schraders M, Dephoure N, Bayley JP, Kunst H, Devilee P, Cremers CW, Schiffman JD, Bentz BG, Gygi SP, Winge DR, Kremer H, Rutter J. SDH5, a gene required for flavination of succinate dehydrogenase, is mutated in paraganglioma. Science 2009; 325: 1139–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Astuti D, Latif F, Dallol A, Dahia PL, Douglas F, George E, Skoldberg F, Husebye ES, Eng C, Maher ER. Gene mutations in the succinate dehydrogenase subunit SDHB cause susceptibility to familial pheochromocytoma and to familial paraganglioma. Am J Hum Genet 2001; 69: 49–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Baysal BE, Ferrell RE, Willett-Brozick JE, Lawrence EC, Myssiorek D, Bosch A, van der Mey A, Taschner PE, Rubinstein WS, Myers EN, Richard CW, 3rd, Cornelisse CJ, Devilee P, Devlin B. Mutations in SDHD, a mitochondrial complex II gene, in hereditary paraganglioma. Science 2000; 287: 848–51. [DOI] [PubMed] [Google Scholar]
- 73.Niemann S, Muller U. Mutations in SDHC cause autosomal dominant paraganglioma, type 3. Nat Genet 2000; 26: 268–70. [DOI] [PubMed] [Google Scholar]
- 74.Hensen EF, Siemers MD, Jansen JC, Corssmit EP, Romijn JA, Tops CM, van der Mey AG, Devilee P, Cornelisse CJ, Bayley JP, Vriends AH. Mutations in SDHD are the major determinants of the clinical characteristics of Dutch head and neck paraganglioma patients. Clin Endocrinol (Oxf) 2011; 75: 650–5. [DOI] [PubMed] [Google Scholar]
- 75.Janeway KA, Kim SY, Lodish M, Nose V, Rustin P, Gaal J, Dahia PL, Liegl B, Ball ER, Raygada M, Lai AH, Kelly L, Hornick JL, Pediatric NIH, Wild-Type GC, O'Sullivan M, de Krijger RR, Dinjens WN, Demetri GD, Antonescu CR, Fletcher JA, Helman L, Stratakis CA. Defects in succinate dehydrogenase in gastrointestinal stromal tumors lacking KIT and PDGFRA mutations. Proc Natl Acad Sci USA 2011; 108: 314–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ricketts C, Woodward ER, Killick P, Morris MR, Astuti D, Latif F, Maher ER. Germline SDHB mutations and familial renal cell carcinoma. J Natl Cancer Inst 2008; 100: 1260–2. [DOI] [PubMed] [Google Scholar]
- 77.Paik JY, Toon CW, Benn DE, High H, Hasovitz C, Pavlakis N, Clifton-Bligh RJ, Gill AJ. Renal carcinoma associated with succinate dehydrogenase B mutation: a new and unique subtype of renal carcinoma. J Clin Oncol 2014; 32: e10–3. [DOI] [PubMed] [Google Scholar]
- 78.Kim S, Kim do H, Jung WH, Koo JS. Succinate dehydrogenase expression in breast cancer. Springerplus 2013; 2: 299–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Briere JJ, Favier J, Benit P, El Ghouzzi V, Lorenzato A, Rabier D, Di Renzo MF, Gimenez-Roqueplo AP, Rustin P. Mitochondrial succinate is instrumental for HIF1alpha nuclear translocation in SDHA-mutant fibroblasts under normoxic conditions. Hum Mol Genet 2005; 14: 3263–9. [DOI] [PubMed] [Google Scholar]
- 80.Pollard PJ, Briere JJ, Alam NA, Barwell J, Barclay E, Wortham NC, Hunt T, Mitchell M, Olpin S, Moat SJ, Hargreaves IP, Heales SJ, Chung YL, Griffiths JR, Dalgleish A, McGrath JA, Gleeson MJ, Hodgson SV, Poulsom R, Rustin P, Tomlinson IP. Accumulation of Krebs cycle intermediates and over-expression of HIF1alpha in tumours which result from germline FH and SDH mutations. Hum Mol Genet 2005; 14: 2231–9. [DOI] [PubMed] [Google Scholar]
- 81.Selak MA, Armour SM, MacKenzie ED, Boulahbel H, Watson DG, Mansfield KD, Pan Y, Simon MC, Thompson CB, Gottlieb E. Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-alpha prolyl hydroxylase. Cancer Cell 2005; 7: 77–85. [DOI] [PubMed] [Google Scholar]
- 82.Guzy RD, Sharma B, Bell E, Chandel NS, Schumacker PT. Loss of the SdhB, but Not the SdhA, subunit of complex II triggers reactive oxygen species-dependent hypoxia-inducible factor activation and tumorigenesis. Mol Cell Biol 2008; 28: 718–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Owens KM, Aykin-Burns N, Dayal D, Coleman MC, Domann FE, Spitz DR. Genomic instability induced by mutant succinate dehydrogenase subunit D (SDHD) is mediated by O2(-*) and H2O2. Free Radic Biol Med 2012; 52: 160–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Xiao M, Yang H, Xu W, Ma S, Lin H, Zhu H, Liu L, Liu Y, Yang C, Xu Y, Zhao S, Ye D, Xiong Y, Guan KL. Inhibition of alpha-KG-dependent histone and DNA demethylases by fumarate and succinate that are accumulated in mutations of FH and SDH tumor suppressors. Genes Dev 2012; 26: 1326–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Letouze E, Martinelli C, Loriot C, Burnichon N, Abermil N, Ottolenghi C, Janin M, Menara M, Nguyen AT, Benit P, Buffet A, Marcaillou C, Bertherat J, Amar L, Rustin P, De Reynies A, Gimenez-Roqueplo AP, Favier J. SDH mutations establish a hypermethylator phenotype in paraganglioma. Cancer Cell 2013; 23: 739–52. [DOI] [PubMed] [Google Scholar]
- 86.Sudarshan S, Sourbier C, Kong HS, Block K, Valera Romero VA, Yang Y, Galindo C, Mollapour M, Scroggins B, Goode N, Lee MJ, Gourlay CW, Trepel J, Linehan WM, Neckers L. Fumarate hydratase deficiency in renal cancer induces glycolytic addiction and hypoxia-inducible transcription factor 1 alpha stabilization by glucose-dependent generation of reactive oxygen species. Mol Cell Biol 2009; 29: 4080–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yang Y, Valera VA, Padilla-Nash HM, Sourbier C, Vocke CD, Vira MA, Abu-Asab MS, Bratslavsky G, Tsokos M, Merino MJ, Pinto PA, Srinivasan R, Ried T, Neckers L, Linehan WM. UOK 262 cell line, fumarate hydratase deficient (FH-/FH-) hereditary leiomyomatosis renal cell carcinoma: in vitro and in vivo model of an aberrant energy metabolic pathway in human cancer. Cancer Genet Cytogenet 2010; 196: 45–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tong WH, Sourbier C, Kovtunovych G, Jeong SY, Vira M, Ghosh M, Romero VV, Sougrat R, Vaulont S, Viollet B, Kim YS, Lee S, Trepel J, Srinivasan R, Bratslavsky G, Yang Y, Linehan WM, Rouault TA. The glycolytic shift in fumarate-hydratase-deficient kidney cancer lowers AMPK levels, increases anabolic propensities and lowers cellular iron levels. Cancer Cell 2011; 20: 315–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Launonen V, Vierimaa O, Kiuru M, Isola J, Roth S, Pukkala E, Sistonen P, Herva R, Aaltonen LA. Inherited susceptibility to uterine leiomyomas and renal cell cancer. Proc Natl Acad Sci USA 2001; 98: 3387–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tomlinson IP, Alam NA, Rowan AJ, Barclay E, Jaeger EE, Kelsell D, Leigh I, Gorman P, Lamlum H, Rahman S, Roylance RR, Olpin S, Bevan S, Barker K, Hearle N, Houlston RS, Kiuru M, Lehtonen R, Karhu A, Vilkki S, Laiho P, Eklund C, Vierimaa O, Aittomaki K, Hietala M, Sistonen P, Paetau A, Salovaara R, Herva R, Launonen V, Aaltonen LA, Multiple Leiomyoma C. Germline mutations in FH predispose to dominantly inherited uterine fibroids, skin leiomyomata and papillary renal cell cancer. Nat Genet 2002; 30: 406–10. [DOI] [PubMed] [Google Scholar]
- 91.Sudarshan S, Shanmugasundaram K, Naylor SL, Lin S, Livi CB, O'Neill CF, Parekh DJ, Yeh IT, Sun LZ, Block K. Reduced expression of fumarate hydratase in clear cell renal cancer mediates HIF-2alpha accumulation and promotes migration and invasion. PLoS One 2011; 6: e21037–e21037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Castro-Vega LJ, Buffet A, De Cubas AA, Cascon A, Menara M, Khalifa E, Amar L, Azriel S, Bourdeau I, Chabre O, Curras-Freixes M, Franco-Vidal V, Guillaud-Bataille M, Simian C, Morin A, Leton R, Gomez-Grana A, Pollard PJ, Rustin P, Robledo M, Favier J, Gimenez-Roqueplo AP. Germline mutations in FH confer predisposition to malignant pheochromocytomas and paragangliomas. Hum Mol Genet 2014; 23: 2440–6. [DOI] [PubMed] [Google Scholar]
- 93.Isaacs JS, Jung YJ, Mole DR, Lee S, Torres-Cabala C, Chung YL, Merino M, Trepel J, Zbar B, Toro J, Ratcliffe PJ, Linehan WM, Neckers L. HIF overexpression correlates with biallelic loss of fumarate hydratase in renal cancer: novel role of fumarate in regulation of HIF stability. Cancer Cell 2005; 8: 143–53. [DOI] [PubMed] [Google Scholar]
- 94.Adam J, Hatipoglu E, O'Flaherty L, Ternette N, Sahgal N, Lockstone H, Baban D, Nye E, Stamp GW, Wolhuter K, Stevens M, Fischer R, Carmeliet P, Maxwell PH, Pugh CW, Frizzell N, Soga T, Kessler BM, El-Bahrawy M, Ratcliffe PJ, Pollard PJ. Renal cyst formation in Fh1-deficient mice is independent of the Hif/Phd pathway: roles for fumarate in KEAP1 succination and Nrf2 signaling. Cancer Cell 2011; 20: 524–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sullivan LB, Martinez-Garcia E, Nguyen H, Mullen AR, Dufour E, Sudarshan S, Licht JD, Deberardinis RJ, Chandel NS. The proto-oncometabolite fumarate binds glutathione to amplify ROS-dependent signaling. Mol Cell 2013; 51: 236–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, Friedman H, Friedman A, Reardon D, Herndon J, Kinzler KW, Velculescu VE, Vogelstein B, Bigner DD. IDH1 and IDH2 mutations in gliomas. N Engl J Med 2009; 360: 765–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, Olivi A, McLendon R, Rasheed BA, Keir S, Nikolskaya T, Nikolsky Y, Busam DA, Tekleab H, Diaz LA, Jr., Hartigan J, Smith DR, Strausberg RL, Marie SK, Shinjo SM, Yan H, Riggins GJ, Bigner DD, Karchin R, Papadopoulos N, Parmigiani G, Vogelstein B, Velculescu VE, Kinzler KW. An integrated genomic analysis of human glioblastoma multiforme. Science 2008; 321: 1807–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kloosterhof NK, Bralten LB, Dubbink HJ, French PJ, van den Bent MJ. Isocitrate dehydrogenase-1 mutations: a fundamentally new understanding of diffuse glioma? Lancet Oncol 2011; 12: 83–91. [DOI] [PubMed] [Google Scholar]
- 99.Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, Koboldt DC, Fulton RS, Delehaunty KD, McGrath SD, Fulton LA, Locke DP, Magrini VJ, Abbott RM, Vickery TL, Reed JS, Robinson JS, Wylie T, Smith SM, Carmichael L, Eldred JM, Harris CC, Walker J, Peck JB, Du F, Dukes AF, Sanderson GE, Brummett AM, Clark E, McMichael JF, Meyer RJ, Schindler JK, Pohl CS, Wallis JW, Shi X, Lin L, Schmidt H, Tang Y, Haipek C, Wiechert ME, Ivy JV, Kalicki J, Elliott G, Ries RE, Payton JE, Westervelt P, Tomasson MH, Watson MA, Baty J, Heath S, Shannon WD, Nagarajan R, Link DC, Walter MJ, Graubert TA, DiPersio JF, Wilson RK, Ley TJ. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med 2009; 361: 1058–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cairns RA, Iqbal J, Lemonnier F, Kucuk C, de Leval L, Jais JP, Parrens M, Martin A, Xerri L, Brousset P, Chan LC, Chan WC, Gaulard P, Mak TW. IDH2 mutations are frequent in angioimmunoblastic T-cell lymphoma. Blood 2012; 119: 1901–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Borger DR, Tanabe KK, Fan KC, Lopez HU, Fantin VR, Straley KS, Schenkein DP, Hezel AF, Ancukiewicz M, Liebman HM, Kwak EL, Clark JW, Ryan DP, Deshpande V, Dias-Santagata D, Ellisen LW, Zhu AX, Iafrate AJ. Frequent mutation of isocitrate dehydrogenase (IDH)1 and IDH2 in cholangiocarcinoma identified through broad-based tumor genotyping. Oncologist 2012; 17: 72–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Borger DR, Goyal L, Yau T, Poon RT, Ancukiewicz M, Deshpande V, Christiani DC, Liebman HM, Yang H, Kim H, Yen K, Faris JE, Iafrate AJ, Kwak EL, Clark JW, Allen JN, Blaszkowsky LS, Murphy JE, Saha SK, Hong TS, Wo JY, Ferrone CR, Tanabe KK, Bardeesy N, Straley KS, Agresta S, Schenkein DP, Ellisen LW, Ryan DP, Zhu AX. Circulating oncometabolite 2-hydroxyglutarate is a potential surrogate biomarker in patients with isocitrate dehydrogenase-mutant intrahepatic cholangiocarcinoma. Clin Cancer Res 2014; 20: 1884–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Amary MF, Bacsi K, Maggiani F, Damato S, Halai D, Berisha F, Pollock R, O'Donnell P, Grigoriadis A, Diss T, Eskandarpour M, Presneau N, Hogendoorn PC, Futreal A, Tirabosco R, Flanagan AM. IDH1 and IDH2 mutations are frequent events in central chondrosarcoma and central and periosteal chondromas but not in other mesenchymal tumours. J Pathol 2011; 224: 334–43. [DOI] [PubMed] [Google Scholar]
- 104.Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, Szabo S, Buckhaults P, Farrell C, Meeh P, Markowitz SD, Willis J, Dawson D, Willson JK, Gazdar AF, Hartigan J, Wu L, Liu C, Parmigiani G, Park BH, Bachman KE, Papadopoulos N, Vogelstein B, Kinzler KW, Velculescu VE. The consensus coding sequences of human breast and colorectal cancers. Science 2006; 314: 268–74. [DOI] [PubMed] [Google Scholar]
- 105.Kato Kaneko M, Liu X, Oki H, Ogasawara S, Nakamura T, Saidoh N, Tsujimoto Y, Matsuyama Y, Uruno A, Sugawara M, Tsuchiya T, Yamakawa M, Yamamoto M, Takagi M, Kato Y. Isocitrate dehydrogenase mutation is frequently observed in giant cell tumor of bone. Cancer Sci 2014; 105: 744–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Shibata T, Kokubu A, Miyamoto M, Sasajima Y, Yamazaki N. Mutant IDH1 confers an in vivo growth in a melanoma cell line with BRAF mutation. Am J Pathol 2011; 178: 1395–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kang MR, Kim MS, Oh JE, Kim YR, Song SY, Seo SI, Lee JY, Yoo NJ, Lee SH. Mutational analysis of IDH1 codon 132 in glioblastomas and other common cancers. Int J Cancer 2009; 125: 353–5. [DOI] [PubMed] [Google Scholar]
- 108.Liu X, Kato Y, Kaneko MK, Sugawara M, Ogasawara S, Tsujimoto Y, Naganuma Y, Yamakawa M, Tsuchiya T, Takagi M. Isocitrate dehydrogenase 2 mutation is a frequent event in osteosarcoma detected by a multi-specific monoclonal antibody MsMab-1. Cancer Med 2013; 2: 803–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Watanabe T, Nobusawa S, Kleihues P, Ohgaki H. IDH1 mutations are early events in the development of astrocytomas and oligodendrogliomas. Am J Pathol 2009; 174: 1149–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, Marks KM, Prins RM, Ward PS, Yen KE, Liau LM, Rabinowitz JD, Cantley LC, Thompson CB, Vander Heiden MG, Su SM. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 2009; 462: 739–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gross S, Cairns RA, Minden MD, Driggers EM, Bittinger MA, Jang HG, Sasaki M, Jin S, Schenkein DP, Su SM, Dang L, Fantin VR, Mak TW. Cancer-associated metabolite 2-hydroxyglutarate accumulates in acute myelogenous leukemia with isocitrate dehydrogenase 1 and 2 mutations. J Exp Med 2010; 207: 339–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, Coller HA, Cross JR, Fantin VR, Hedvat CV, Perl AE, Rabinowitz JD, Carroll M, Su SM, Sharp KA, Levine RL, Thompson CB. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell 2010; 17: 225–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, Ito S, Yang C, Wang P, Xiao MT, Liu LX, Jiang WQ, Liu J, Zhang JY, Wang B, Frye S, Zhang Y, Xu YH, Lei QY, Guan KL, Zhao SM, Xiong Y. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell 2011; 19: 17–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kolker S, Pawlak V, Ahlemeyer B, Okun JG, Horster F, Mayatepek E, Krieglstein J, Hoffmann GF, Kohr G. NMDA receptor activation and respiratory chain complex V inhibition contribute to neurodegeneration in d-2-hydroxyglutaric aciduria. Eur J Neurosci 2002; 16: 21–8. [DOI] [PubMed] [Google Scholar]
- 115.Latini A, da Silva CG, Ferreira GC, Schuck PF, Scussiato K, Sarkis JJ, Dutra Filho CS, Wyse AT, Wannmacher CM, Wajner M. Mitochondrial energy metabolism is markedly impaired by D-2-hydroxyglutaric acid in rat tissues. Mol Genet Metab 2005; 86: 188–99. [DOI] [PubMed] [Google Scholar]
- 116.Reitman ZJ, Jin G, Karoly ED, Spasojevic I, Yang J, Kinzler KW, He Y, Bigner DD, Vogelstein B, Yan H. Profiling the effects of isocitrate dehydrogenase 1 and 2 mutations on the cellular metabolome. Proc Natl Acad Sci USA 2011; 108: 3270–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, Li Y, Bhagwat N, Vasanthakumar A, Fernandez HF, Tallman MS, Sun Z, Wolniak K, Peeters JK, Liu W, Choe SE, Fantin VR, Paietta E, Lowenberg B, Licht JD, Godley LA, Delwel R, Valk PJ, Thompson CB, Levine RL, Melnick A. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell 2010; 18: 553–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Chowdhury R, Yeoh KK, Tian YM, Hillringhaus L, Bagg EA, Rose NR, Leung IK, Li XS, Woon EC, Yang M, McDonough MA, King ON, Clifton IJ, Klose RJ, Claridge TD, Ratcliffe PJ, Schofield CJ, Kawamura A. The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO Rep 2011; 12: 463–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, Edwards CR, Khanin R, Figueroa ME, Melnick A, Wellen KE, O'Rourke DM, Berger SL, Chan TA, Levine RL, Mellinghoff IK, Thompson CB. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature 2012; 483: 474–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, Campos C, Fabius AW, Lu C, Ward PS, Thompson CB, Kaufman A, Guryanova O, Levine R, Heguy A, Viale A, Morris LG, Huse JT, Mellinghoff IK, Chan TA. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature 2012; 483: 479–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Koivunen P, Lee S, Duncan CG, Lopez G, Lu G, Ramkissoon S, Losman JA, Joensuu P, Bergmann U, Gross S, Travins J, Weiss S, Looper R, Ligon KL, Verhaak RG, Yan H, Kaelin WG., Jr Transformation by the (R)-enantiomer of 2-hydroxyglutarate linked to EGLN activation. Nature 2012; 483: 484–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Losman JA, Looper RE, Koivunen P, Lee S, Schneider RK, McMahon C, Cowley GS, Root DE, Ebert BL, Kaelin WG., Jr (R)-2-hydroxyglutarate is sufficient to promote leukemogenesis and its effects are reversible. Science 2013; 339: 1621–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hallows WC, Lee S, Denu JM. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc Natl Acad Sci USA 2006; 103: 10230–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hallows WC, Yu W, Smith BC, Devries MK, Ellinger JJ, Someya S, Shortreed MR, Prolla T, Markley JL, Smith LM, Zhao S, Guan KL, Denu JM. Sirt3 promotes the urea cycle and fatty acid oxidation during dietary restriction. Mol Cell 2011; 41: 139–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hirschey MD, Shimazu T, Goetzman E, Jing E, Schwer B, Lombard DB, Grueter CA, Harris C, Biddinger S, Ilkayeva OR, Stevens RD, Li Y, Saha AK, Ruderman NB, Bain JR, Newgard CB, Farese RV, Jr., Alt FW, Kahn CR, Verdin E. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature 2010; 464: 121–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lombard DB, Alt FW, Cheng HL, Bunkenborg J, Streeper RS, Mostoslavsky R, Kim J, Yancopoulos G, Valenzuela D, Murphy A, Yang Y, Chen Y, Hirschey MD, Bronson RT, Haigis M, Guarente LP, Farese RV, Jr, Weissman S, Verdin E, Schwer B. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol Cell Biol 2007; 27: 8807–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Qiu X, Brown K, Hirschey MD, Verdin E, Chen D. Calorie restriction reduces oxidative stress by SIRT3-mediated SOD2 activation. Cell Metab 2010; 12: 662–7. [DOI] [PubMed] [Google Scholar]
- 128.Schlicker C, Gertz M, Papatheodorou P, Kachholz B, Becker CF, Steegborn C. Substrates and regulation mechanisms for the human mitochondrial sirtuins Sirt3 and Sirt5. J Mol Biol 2008; 382: 790–801. [DOI] [PubMed] [Google Scholar]
- 129.Schwer B, Bunkenborg J, Verdin RO, Andersen JS, Verdin E. Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc Natl Acad Sci USA 2006; 103: 10224–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Shimazu T, Hirschey MD, Hua L, Dittenhafer-Reed KE, Schwer B, Lombard DB, Li Y, Bunkenborg J, Alt FW, Denu JM, Jacobson MP, Verdin E. SIRT3 deacetylates mitochondrial 3-hydroxy-3-methylglutaryl CoA synthase 2 and regulates ketone body production. Cell Metab 2010; 12: 654–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yu W, Dittenhafer-Reed KE, Denu JM. SIRT3 protein deacetylates isocitrate dehydrogenase 2 (IDH2) and regulates mitochondrial redox status. J Biol Chem 2012; 287: 14078–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kim HS, Patel K, Muldoon-Jacobs K, Bisht KS, Aykin-Burns N, Pennington JD, van der Meer R, Nguyen P, Savage J, Owens KM, Vassilopoulos A, Ozden O, Park SH, Singh KK, Abdulkadir SA, Spitz DR, Deng CX, Gius D. SIRT3 is a mitochondria-localized tumor suppressor required for maintenance of mitochondrial integrity and metabolism during stress. Cancer Cell 2010; 17: 41–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Finley LW, Carracedo A, Lee J, Souza A, Egia A, Zhang J, Teruya-Feldstein J, Moreira PI, Cardoso SM, Clish CB, Pandolfi PP, Haigis MC. SIRT3 opposes reprogramming of cancer cell metabolism through HIF1alpha destabilization. Cancer Cell 2011; 19: 416–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bell EL, Emerling BM, Ricoult SJ, Guarente L. SirT3 suppresses hypoxia inducible factor 1alpha and tumor growth by inhibiting mitochondrial ROS production. Oncogene 2011; 30: 2986–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zhang YY, Zhou LM. Sirt3 inhibits hepatocellular carcinoma cell growth through reducing Mdm2-mediated p53 degradation. Biochem Biophys Res Commun 2012; 423: 26–31. [DOI] [PubMed] [Google Scholar]
- 136.Xiao K, Jiang J, Wang W, Cao S, Zhu L, Zeng H, Ouyang R, Zhou R, Chen P. Sirt3 is a tumor suppressor in lung adenocarcinoma cells. Oncol Rep 2013; 30: 1323–8. [DOI] [PubMed] [Google Scholar]
- 137.Huang KH, Hsu CC, Fang WL, Chi CW, Sung MT, Kao HL, Li AF, Yin PH, Yang MH, Lee HC. SIRT3 expression as a biomarker for better prognosis in gastric cancer. World J Surg 2014; 38: 910–7. [DOI] [PubMed] [Google Scholar]
- 138.Mucaj V, Shay JE, Simon MC. Effects of hypoxia and HIFs on cancer metabolism. Int J Hematol 2012; 95: 464–70. [DOI] [PubMed] [Google Scholar]
- 139.Ivan M, Kondo K, Yang H, Kim W, Valiando J, Ohh M, Salic A, Asara JM, Lane WS, Kaelin WG., Jr HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 2001; 292: 464–8. [DOI] [PubMed] [Google Scholar]
- 140.Jaakkola P, Mole DR, Tian YM, Wilson MI, Gielbert J, Gaskell SJ, von Kriegsheim A, Hebestreit HF, Mukherji M, Schofield CJ, Maxwell PH, Pugh CW, Ratcliffe PJ. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 2001; 292: 468–72. [DOI] [PubMed] [Google Scholar]
- 141.Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer 2003; 3: 721–32. [DOI] [PubMed] [Google Scholar]
- 142.Semenza GL. Defining the role of hypoxia-inducible factor 1 in cancer biology and therapeutics. Oncogene 2010; 29: 625–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Semenza GL. Hypoxia-inducible factors: mediators of cancer progression and targets for cancer therapy. Trends Pharmacol Sci 2012; 33: 207–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Zhang H, Gao P, Fukuda R, Kumar G, Krishnamachary B, Zeller KI, Dang CV, Semenza GL. HIF-1 inhibits mitochondrial biogenesis and cellular respiration in VHL-deficient renal cell carcinoma by repression of C-MYC activity. Cancer Cell 2007; 11: 407–20. [DOI] [PubMed] [Google Scholar]
- 145.Yeung SJ, Pan J, Lee MH. Roles of p53, MYC and HIF-1 in regulating glycolysis - the seventh hallmark of cancer. Cell Mol Life Sci 2008; 65: 3981–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Fukuda R, Zhang H, Kim JW, Shimoda L, Dang CV, Semenza GL. HIF-1 regulates cytochrome oxidase subunits to optimize efficiency of respiration in hypoxic cells. Cell 2007; 129: 111–22. [DOI] [PubMed] [Google Scholar]
- 147.Sciacovelli M, Guzzo G, Morello V, Frezza C, Zheng L, Nannini N, Calabrese F, Laudiero G, Esposito F, Landriscina M, Defilippi P, Bernardi P, Rasola A. The mitochondrial chaperone TRAP1 promotes neoplastic growth by inhibiting succinate dehydrogenase. Cell Metab 2013; 17: 988–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Vousden KH, Ryan KM. p53 and metabolism. Nat Rev Cancer 2009; 9: 691–700. [DOI] [PubMed] [Google Scholar]
- 149.Berkers CR, Maddocks OD, Cheung EC, Mor I, Vousden KH. Metabolic regulation by p53 family members. Cell Metab 2013; 18: 617–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Schwartzenberg-Bar-Yoseph F, Armoni M, Karnieli E. The tumor suppressor p53 down-regulates glucose transporters GLUT1 and GLUT4 gene expression. Cancer Res 2004; 64: 2627–33. [DOI] [PubMed] [Google Scholar]
- 151.Kawauchi K, Araki K, Tobiume K, Tanaka N. p53 regulates glucose metabolism through an IKK-NF-kappaB pathway and inhibits cell transformation. Nat Cell Biol 2008; 10: 611–8. [DOI] [PubMed] [Google Scholar]
- 152.Bensaad K, Tsuruta A, Selak MA, Vidal MN, Nakano K, Bartrons R, Gottlieb E, Vousden KH. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell 2006; 126: 107–20. [DOI] [PubMed] [Google Scholar]
- 153.Kondoh H, Lleonart ME, Gil J, Wang J, Degan P, Peters G, Martinez D, Carnero A, Beach D. Glycolytic enzymes can modulate cellular life span. Cancer Res 2005; 65: 177–85. [PubMed] [Google Scholar]
- 154.Okamura S, Ng CC, Koyama K, Takei Y, Arakawa H, Monden M, Nakamura Y. Identification of seven genes regulated by wild-type p53 in a colon cancer cell line carrying a well-controlled wild-type p53 expression system. Oncol Res 1999; 11: 281–5. [PubMed] [Google Scholar]
- 155.Vahsen N, Cande C, Briere JJ, Benit P, Joza N, Larochette N, Mastroberardino PG, Pequignot MO, Casares N, Lazar V, Feraud O, Debili N, Wissing S, Engelhardt S, Madeo F, Piacentini M, Penninger JM, Schagger H, Rustin P, Kroemer G. AIF deficiency compromises oxidative phosphorylation. EMBO J 2004; 23: 4679–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Stambolsky P, Weisz L, Shats I, Klein Y, Goldfinger N, Oren M, Rotter V. Regulation of AIF expression by p53. Cell Death Differ 2006; 13: 2140–9. [DOI] [PubMed] [Google Scholar]
- 157.Matoba S, Kang JG, Patino WD, Wragg A, Boehm M, Gavrilova O, Hurley PJ, Bunz F, Hwang PM. p53 regulates mitochondrial respiration. Science 2006; 312: 1650–3. [DOI] [PubMed] [Google Scholar]
- 158.Hu W, Zhang C, Wu R, Sun Y, Levine A, Feng Z. Glutaminase 2, a novel p53 target gene regulating energy metabolism and antioxidant function. Proc Natl Acad Sci USA 2010; 107: 7455–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Suzuki S, Tanaka T, Poyurovsky MV, Nagano H, Mayama T, Ohkubo S, Lokshin M, Hosokawa H, Nakayama T, Suzuki Y, Sugano S, Sato E, Nagao T, Yokote K, Tatsuno I, Prives C. Phosphate-activated glutaminase (GLS2), a p53-inducible regulator of glutamine metabolism and reactive oxygen species. Proc Natl Acad Sci USA 2010; 107: 7461–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Jiang P, Du W, Mancuso A, Wellen KE, Yang X. Reciprocal regulation of p53 and malic enzymes modulates metabolism and senescence. Nature 2013; 493: 689–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Bourdon A, Minai L, Serre V, Jais JP, Sarzi E, Aubert S, Chretien D, de Lonlay P, Paquis-Flucklinger V, Arakawa H, Nakamura Y, Munnich A, Rotig A. Mutation of RRM2B, encoding p53-controlled ribonucleotide reductase (p53R2), causes severe mitochondrial DNA depletion. Nat Genet 2007; 39: 776–80. [DOI] [PubMed] [Google Scholar]
- 162.Achanta G, Sasaki R, Feng L, Carew JS, Lu W, Pelicano H, Keating MJ, Huang P. Novel role of p53 in maintaining mitochondrial genetic stability through interaction with DNA Pol gamma. EMBO J 2005; 24: 3482–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Sullivan LB, Chandel NS. Mitochondrial reactive oxygen species and cancer. Cancer Metab 2014; 2: 17–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Harris IS, Treloar AE, Inoue S, Sasaki M, Gorrini C, Lee KC, Yung KY, Brenner D, Knobbe-Thomsen CB, Cox MA, Elia A, Berger T, Cescon DW, Adeoye A, Brüstle A, Molyneux SD, Mason JM, Li WY, Yamamoto K, Wakeham A, Berman HK, Khokha R, Done SJ, Kavanagh TJ, Lam CW, Mak TW. Glutathione and thioredoxin antioxidant pathways synergize to drive cancer initiation and progression. Cancer Cell 2015; 27: 211–22. [DOI] [PubMed] [Google Scholar]
- 165.Ishii T, Yasuda K, Akatsuka A, Hino O, Hartman PS, Ishii N. A mutation in the SDHC gene of complex II increases oxidative stress, resulting in apoptosis and tumorigenesis. Cancer Res 2005; 65: 203–9. [PubMed] [Google Scholar]
- 166.Amuthan G, Biswas G, Zhang SY, Klein-Szanto A, Vijayasarathy C, Avadhani NG. Mitochondria-to-nucleus stress signaling induces phenotypic changes, tumor progression and cell invasion. EMBO J 2001; 20: 1910–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Biswas G, Tang W, Sondheimer N, Guha M, Bansal S, Avadhani NG. A distinctive physiological role for IkappaBbeta in the propagation of mitochondrial respiratory stress signaling. J Biol Chem 2008; 283: 12586–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Kurelac I, Romeo G, Gasparre G. Mitochondrial metabolism and cancer. Mitochondrion 2011; 11: 635–7. [DOI] [PubMed] [Google Scholar]
- 169.Hsu CC, Wang CH, Wu LC, Hsia CY, Chi CW, Yin PH, Chang CJ, Sung MT, Wei YH, Lu SH, Lee HC. Mitochondrial dysfunction represses HIF-1alpha protein synthesis through AMPK activation in human hepatoma HepG2 cells. Biochim Biophys Acta 2013; 1830: 4743–51. [DOI] [PubMed] [Google Scholar]
- 170.Hsu CC, Wu LC, Hsia CY, Yin PH, Chi CW, Yeh TS, Lee HC. Energy metabolism determines the sensitivity of human hepatocellular carcinoma cells to mitochondrial inhibitors and biguanide drugs. Oncol Rep 2015; 34: 1620–8. [DOI] [PubMed] [Google Scholar]
- 171.Sutendra G, Kinnaird A, Dromparis P, Paulin R, Stenson TH, Haromy A, Hashimoto K, Zhang N, Flaim E, Michelakis ED. A nuclear pyruvate dehydrogenase complex is important for the generation of acetyl-CoA and histone acetylation. Cell 2014; 158: 84–97. [DOI] [PubMed] [Google Scholar]