Abstract
In this review, we provide a description of those candidate biomarkers which have been demonstrated by multiple-omics approaches to vary in correlation with specific clinical manifestations of sickle cell severity. We believe that future clinical analyses of severity phenotype will require a multiomic analysis, or an omics stack approach, which includes integrated interactomics. It will also require the analysis of big data sets. These candidate biomarkers, whether they are individual or panels of functionally linked markers, will require future validation in large prospective and retrospective clinical studies. Once validated, the hope is that informative biomarkers will be used for the identification of individuals most likely to experience severe complications, and thereby be applied for the design of patient-specific therapeutic approaches and response to treatment. This would be the beginning of precision medicine for sickle cell disease.
Keywords: Genomics, interactomics, metabolomics, precision medicine, proteomics, sickle cell disease
Introduction
Sickle cell anemia (SCA) is a monogenic hemoglobinopathy, yet it has great variation in the clinical severity and outcome of the disease. Our focus in this minireview will be primarily on SCA, which is the major form of several related hemoglobinopathies referred to as SCD. Many SCA patients have a mild clinical course experiencing few vasoocclusive events (crises) and sometimes living into their sixth and seventh decade. Other patients with SCA have a much more severe clinical course with multiple crises per year, significant organ damage and a shortened life span.1–6
SCA morbidity and mortality are significant, but patients demonstrate great variance in the manner in which they are affected by the disease. Although the Mendelian genetics of SCA are straightforward, the correlation between genotypes and phenotypes is far less clear. There is substantial phenotypic heterogeneity among individuals with identical alleles at the beta globin gene.7–10
The vasoocclusive crisis (VOC) rate is one measure of clinical severity that correlates with premature death in SCD patients over 20 years of age.1 Platt et al. reported that 39% of subjects had no painful crises during the period of study, while 1% had more than six episodes per year. The 5% of individuals who had between 3 and 10 crises per year accounted for 33% of all episodes of pain in this cohort.1 A subsequent study demonstrated an increased risk of early death in those sickle cell patients who experienced acute chest syndrome (ACS).2,11 In searching for biomarkers that predict variations in disease severity, a good starting point is identifying those which correlate with VOC and ACS rates.4–6
The lack of validated biomarkers for SCA severity represents a void in the state of knowledge of SCD that creates a critical roadblock in the design of clinical trials, the development of novel therapies and the emergence of precision medicine for SCD patients. The discovery of validated biomarkers will require teams of researchers with expertise in multiomic platforms (omics stacks) and sufficient numbers of subjects to perform retrospective and prospective clinical studies. This review provides candidate omic biomarkers, which require future verification and validation.
Fetal hemoglobin and sickle cell severity
Although SCA is a monogenic disorder due to a single nucleotide polymorphism (SNP) in the sixth codon of β-globin (HBB), genetic and environmental factors contribute to variations in the severity of clinical complications. The sequential expression of the developmentally regulated β-like globin genes (ɛ, Gγ, Aγ, δ, and β) located in the HBB locus on chromosome 11 has been investigated for many decades. During the first year of life, the γ-globin gene is silenced and β-globin is activated, a process known as hemoglobin switching.12 However in children with SCA, this process is attenuated leading to sustained fetal hemoglobin (HbF; α2γ2) synthesis with a mean HbF level of 28.1% at 6 months13 compared with 7.3% in babies with normal hemoglobin A production. HbF is a potent modifier of clinical severity in SCA because of its ability to directly inhibit sickle hemoglobin polymerization under deoxygenated conditions.14 Shortly after birth when HbF levels are high, infants are protected against the VOC complications of SCA however as HbF drops, clinical complications occur such as splenic sequestration and high risk for serious infection among others.
After many years of clinical research to develop specific therapies, hydroxyurea is the only FDA approved drug for treating adults with SCA primarily through HbF induction.15,16 Subsequent clinical trials in children culminating with the BABY HUG study showed hydroxyurea to be effective in young children,17,18 but this agent did not reverse abnormal spleen and kidney function observed in the first year of life. Recently, evidence-based guidelines were released that recommend offering hydroxyurea at 9 months of age to children with SCA regardless of clinical symptoms.19 However, clinical providers remain concerned about the optimal age to safely initiate this therapy in young children. Therefore, other biomarkers of SCA clinical severity that can be targeted for the design of non-chemotherapeutic agents need to be identified.
Genomics
Over the last decade with the rapid expansion of genomic approaches, candidate gene and genome-wide studies have been used to discover genetic modifiers of disease severity in sickle cell patients. Based on extensive clinical research, two major SCD phenotypes including VOC and hemolysis have been described.20,21 The VOC phenotype is secondary to sickle hemoglobin polymerization, vessel occlusion, and tissue hypoxia/reperfusion injury. Acute painful episodes, ACS and avascular necrosis of the femoral head are associated with VOC pathophysiology. Genetic studies have identified mutations in the GCH1,22 MBL2,23 HMOX1,24 and MTHFR:BMP225 genes associated with VOC. By contrast, the hemolysis phenotype is caused by intravascular hemolysis, free hemoglobin release from red blood cells, and reduced nitric oxide production. The severity of anemia and complications of stroke, leg ulcers, and priapism have been associated with the hemolytic phenotype.20,21,26 Genotype–phenotype studies identified mutations in the NPRL3,27 VCAM1,28 NOS3,29 Interleukin 4 receptor,30 TGF-β/SMAD/BMP pathway, KL,31 TEK, TGFBR3, and AQP132 genes associated with hemolysis risk. de Oliveira Filho et al.33 confirmed null mutations in two glutathione S-transferase subunit-encoding genes, GSTM1 and GSTT1 associated with SCD severity. Null genotypes at these two loci increased the risk of severe complications including ACS, avascular necrosis of the femoral head, stroke, and leg ulcers 4- to 6-fold. A detailed discussion of the implications of SNPs in the various loci and SCA clinical severity is reviewed by Habara and Steinberg34 in this thematic issue of the Experimental Biology and Medicine.
The greatest impact on clinical severity in SCA is clearly illustrated by a group of disorders known as hereditary persistence of HbF (HPFH) caused by deletion and point mutations in the HBB locus.12 Persons who are heterozygous for the βS-globin mutation and a HPFH mutation display minimal clinical symptoms or complications. It is known that a subgroup of children with SCA continue to express the γ-globin gene with an average HbF level of 6% after 5 years of age attributed to SNPs in the HBB locus.13 Previous genomic analysis of the HBB locus established inherited haplotypes (SNPs inherited in unique patterns) based on different African populations. The Bantu and Benin haplotypes have been associated with low HbF levels and increased severity of clinical manifestations in sickle cell patients3,35 compared with the Senegal and Arab-Indian haplotypes with higher HbF and a milder disease phenotype.36,37 However, these haplotypes are not consistently predictive of clinical severity of SCA patients.
Candidate gene and genome-wide studies have been conducted to discover genetic modifiers of HbF expression outside the HBB locus in sickle cell patients. Twelve SNPs in four candidate genes (PDE7, PEX7, MAP3K5, and MAP7) account for 20–30% of HbF variation.38 Since genotyping performed in this early study was relatively sparse by current standards, additional studies in larger populations are required to identify true disease severity-influencing variants. Subsequent quantitative trait loci39–41 and genome-wide association studies (GWAS)42–44 discovered three major loci including -158Xmn1-HBG2 (Gγ-globin), the HBS1L-MYB region on chromosome 6q23, and BCL11A at chromosome 2p15 accounting for 13–20% of HbF variance in different hemoglobinopathy patient populations. The Xmn1-HBG2 SNP is associated with 13% of HbF variance in β-thalassemia patients; however, this locus was not replicated in African American43,45 or Tanzanian46 sickle cell patients. A group of SNPs located in the intergenic region 5′ of the HBS1L and MYB genes were associated with high HbF40,44 to support a possible mechanism of γ-globin regulation; reduced levels of the repressor transcription factor MYB was observed in primary erythroid cultures generated from individuals with HPFH.47
The most widely investigated HbF modifier is the transcription factor BCL11A; multiple SNPs in this gene has been associated with up to 15% HbF variance in several populations from different ancestry.41,45,46 Sankaran et al.48 demonstrated that high HbF levels were associated with reduced BCL11A expression. Later studies confirmed BCL11A gene inactivation in sickle cell mice inhibited developmentally regulated γ-globin gene silencing and improved several hematologic parameters including renal function and spleen size in these animals.49
Several groups have shed light on the mechanisms by which BCL11A represses γ-globin expression. This transcription factor interacts with several DNA-binding proteins such as the co-repressors LSD1/CoREST,50 DNMT1,51 GATA1/FOG1/NuRD complex,52 and Sox653 to facilitate γ-globin gene silencing through binding in the HbF silencing region located upstream of the δ-globin gene.54 Although much is known about the mechanism of γ-globin regulation by BCL11A, there are limitations to the development of this protein as a therapeutic target since it is required for B-lymphocyte maturation during normal hematopoiesis.55 Recently, an erythroid-specific enhancer was discovered in the second intron of BCL11A,56 which can be targeted to achieve lineage-specific gene silencing moving research efforts a step closer to gene therapy for SCA directed at inhibiting BCL11A in erythroid cells.
Since SNPs in the BCL11A gene accounts for about 10% of HbF variance in African Americans, efforts to discover other biomarkers of the clinical severity of SCA are needed. The ever-increasing number of potential disease-modifying SNPs identified by GWAS and the lack of replication of many of these loci underscores the concept that SCD severity and clinical phenotypes are not controlled by a single genetic event that is both necessary and sufficient to determine clinical outcome. Instead, it is likely that numerous genes each contribute a small amount toward overall clinical severity in SCA. Undoubtedly, additional genome-wide analyses will increase our understanding of SCD severity; however, a limited number of patients with accurately recorded phenotype data are available. Therefore, future strategies need to be established to identify true biomarkers of disease severity that reflect objective measures of the current state of illness and risk for complications.
Transcriptomics
Few efforts have been made to identify transcriptomic biomarkers of SCD severity, but early efforts are promising. In particular, Jison et al. found dysregulation of 112 genes in peripheral blood mononuclear cells (PBMCs) from SCA patients compared with normal subjects; many of these genes were involved in heme biosynthesis and metabolism, but this differential gene-set was also enriched with genes associated with inflammatory functions,57 angiogenesis, antioxidant and stress responses, and cell-cycle regulation. In neutrophils isolated from severe versus mild SCA patients, Kutlar and colleagues found differences in the expression of genes involved in inflammatory responses and cell-cycle progression, as well as genes involved in cellular proliferation, growth, and maintenance, and in DNA repair and replication.7 Most recently, Hounkpe et al. performed a meta-analysis of four existing blood-based transcriptomic studies of SCD, some of which included severity scores derived from various factors including age and sex, bilirubin level, history of blood transfusion, LDH level, MCV, presence of pain, priapism, reticulocyte count, sepsis, history of stroke, systolic blood pressure, and WBC count. In the subgroup of 62 patients with severity scores (44 in steady-state and 18 in acute VOC crisis), 384 differentially expressed genes were identified, a majority of which were up-regulated in the most severe cases (336 up-regulated and 48 down-regulated).58 These genes were found, in turn, to be regulated by numerous transcription factors (e.g. CREB1, FOXJ1, CEBPD, and ATF4, among others) and kinases (e.g. MAPK1, MAPK3, GSK3B, and others). Formal gene-set analysis of these data identified numerous biological pathways that were enriched among the list of differentially expressed genes, including pathways associated with heme metabolism, innate immunity, proteasome degradation, and autophagy. Numerous ontology terms were also significantly over-represented among these transcripts, including cellular response to extracellular stimulus, protein ubiquitination, type I interferon signaling pathway, porphyrin compound biosynthesis, myeloid cell development, apoptotic mitochondrial changes, and regulation of peptidase activity. Aside from the potential of transcriptome profiling to identify dynamic biomarkers of a dynamic disease state, gene-expression analyses can also implicate genomic regions to inform the identification of additional DNA variants acting to regulate those differentially expressed genes in sickle cell patients with a severe clinical phenotype. As gene expression levels are governed by both DNA sequence variations (e.g. in promoters or other regulatory elements) and environmental factors (e.g. via epigenetic modifications), the dysregulation of a transcript in association with the SCA phenotype could arise from either source of variation or a combination of both. For some such transcripts, the observed dysregulation, if not driven by some environmental factor, may therefore highlight that gene’s promoter or other regulatory elements as logical candidates for harboring symptom-associated genetic variation, which subsequently could be sought and found in a genetic study framework.
MicroRNAomics
The amounts and types of mRNAs produced by protein and non-protein encoding genes are regulated at the transcriptional level by inherited SNPs cis- and trans-acting regulatory elements or genes (e.g. in promoters, untranslated regions, or other regulatory domains). Recent studies have focused on posttranscriptional mechanisms of gene regulation via microRNA (miRNA)-mediated processes. miRNAs are small non-coding RNAs that regulate gene expression by interfering with mRNA translation or disrupting mRNA stability to promote degradation. The majority of miRNA studies to gain insights into clinical severity have focused on γ-globin gene regulation. Miller and colleagues59 confirmed the ability of LIN28 to silence the miRNA let-7 to mediate HbF induction in human primary erythroid cells. Likewise, miR-15a and miR-16-160 enhance γ-globin expression through inhibition of the repressor oncogene MYB. Studies by Walker et al. correlated miR-26b with steady-state HbF levels and miR-151-3p expression with the maximal tolerated dose of hydroxyurea in children with SCD.61 These studies demonstrate the potential of miRNAs as biomarkers to predict SCD severity.
The role of miRNAs in determining the severity of anemia in sickle cell patients has also been investigated. It is known that sickle red blood cells are under oxidative stress and have a deficiency of glutathione production leading to high levels of reactive oxygen species.62 Recently, Sangokoya et al.63 performed a genome-wide miRNA analysis using reticulocytes isolated from SCA patients demonstrating that miR-144 expression correlated with the level of anemia and tolerance of oxidative stress in red blood cells. It is known that miR-144 target many genes including NRF2, a transcription factor involved in regulating proteins critical for maintaining a normal cellular oxidative stress response.64 Therefore, reduced NRF2 expression is associated with excess cell stress and hemolysis supporting a mechanism for increased miR-144 levels causing anemia in sickle cell patients. Pace and colleagues recently completed genome-wide miRNA analysis of reticulocytes from SCA patients with high and low HbF levels and observed an eight-fold increase in miR-144 in the low HbF group supporting a role of NRF2 in γ-globin regulation.65 Thus, miR-144 and NRF2 not only provide a potential biomarker of severity but also support a putative mechanism for a clinical complication in SCA. Currently, miRNA mimics are being developed to treat a wide variety of diseases.66 Therefore, the discovery of miRNAs involved in the clinical severity of SCD provide potential targets for the design of mimics and antagomirs to treat sickle cell patients.
Proteomics
There are several excellent reviews on the proteome of erythrocytes and how it changes in SCD.67–70 Therefore, here we will focus specifically on the few proteomic studies that were directed at defining candidate biomarkers for clinical manifestations of sickle cell severity.
Goodman and colleagues performed the initial study to identify proteomic biomarkers for sickle cell severity.71 Using a 2D-DIGE protein profiling approach, they found 21 of ∼1000 monocyte protein spots that had significant positive or negative correlation with VOC rate. Vinculin, leukotriene A-4 hydrolase, and phosphoglycerate kinase were all highly predictive of VOC rate.71 Studies on the plasma of subjects with SCD +/− pulmonary hypertension have implicated decreased apolipoprotein A-I (apoA-I) as a potential marker for pulmonary hypertension risk.72 The same group has demonstrated that elevation of the serum amyloid A/apoA-I ratio may be a biomarker for increased VOC rate.73
Future studies should focus on whether erythrocyte proteins, which have already been shown to differ in the RBC proteome of SCA versus normal hemoglobin A subjects also differ when comparing SCA subjects with varying clinical severity.74,75 Potential target biomarkers include heat shock proteins, chaperonins, proteasomal subunits, and anti-oxidant enzymes that are discussed further in the interactomics section below.
Metabolomics
Red cell-specific or systemic metabolic derangements may underlie the pathophysiology of SCA or at least contribute to accompanying complications. A better understanding of metabolism in patients should reveal novel therapeutic avenues. In 2011, Darghouth and colleagues performed metabolomic studies on red blood cells from 28 SCA subjects versus 24 healthy controls.76 SCA metabolic signatures included alterations of redox homeostasis as well as abnormal levels of compounds from nitrogen metabolic pathways including citrulline, spermine, and spermidine.76,77 Comparison of normal old and young red blood cells revealed similarities between the metabolic phenotypes of SCA red blood cells and old erythrocytes, suggesting a role for these compounds basic amino acids and polyamines in the physiology or manifestation (biomarkers) of SCA pathology. In support of this view, correction of altered arginine metabolism has been proposed as a therapy for SCD,78 though further metabolomics studies, both discovery mode and pathway specific targeted investigations, are necessary.
In the same year, Zhang et al. from Xia’s group performed high throughput metabolomic profiling of whole blood and plasma from SCD transgenic mice and demonstrated that circulating adenosine levels and erythrocyte 2,3-biphosphoglycerate (2,3-BPG) concentrations were highly elevated in the sickle cell versus erythrocytes from control mice. They demonstrated that the same was true for erythrocytes and plasma from humans with SCA.79 Further mechanistic studies demonstrated that adenosine signaling through the adenosine A2B receptor (ADORA2BR) underlies increased 2,3-BPG production in erythrocytes from SCD transgenic mice and humans with SCA. Follow-up studies demonstrated the detrimental role of ADORA2BR activation-mediated increase in 2,3-BPG. Accumulation of 2,3-BPG in sickle erythrocytes is damaging in that it leads to increased sickling, hemolysis, and tissue damage. One of the major explanation for the pathological role of elevated adenosine signaling via ADORA2BR in sickling is represented by the role of this signaling axis in increasing 2,3-BPG. Increased 2,3-BPG mediates the stabilization of the deoxygenated T-state of sickle hemoglobin, elevates deoxygenated sickle hemoglobin, and thus promotes the formation of polymers and eventually the induction of sickling (Figure 1). Preclinical studies showed that lowering elevated adenosine by pegylated adenosine deaminase (PEG-ADA), a FDA approved drug, and ADORA2BR specific inhibitor significantly reduced sickling in SCA transgenic mice and cultured SCD patients’ erythrocytes (Figure 1).79,80 Thus, exploratory metabolomics studies led to the discovery and appreciation of the detrimental role of elevated adenosine signaling via ADORA2BR in sickling and revealed novel potential therapies for SCA.
Figure 1.
Sickle cell anemia is characterized by increased plasma levels of adenosine. Adenosine signaling through the A2B receptor (AORA2B) promotes intracellular 2,3-biphosphoglycerate (2,3-BPG) accumulation and stabilization of deoxy-sickle hemoglobin, which promotes polymerization and induces red blood cell sickling. Sphingosine 1 phosphate (S1P) contributes to sickling, through unknown mechanisms. Therapeutic intervention through adenosine conversion to inosine by pegylated-adenosine deaminase or inhibition of sphingosine kinase 1 (SphK1) activity has been shown to reduce sickling and extend red blood cell lifespan
Extending from adenosine, Xia’s group used a metabolomic screening of blood and plasma and identified that sphingosine-1-P (S1P) is elevated in SCA transgenic mice and humans.81 They further demonstrated that elevated Sphingosine kinase 1 (SphK1), an enzyme enriched in erythrocytes adding phosphate to sphingosine to generate S1P, underlies increased erythrocyte S1P production contributing to sickling and disease progression.81 Inhibition or shRNA knockdown of SphK1 significantly reduced sickling by reducing S1P in the erythrocytes of SCA mice (Figure 1). More recently, they have demonstrated that elevated plasma adenosine signaling via ADORA2BR on normal and sickle human and mouse erythrocytes contributes to increased SphK1 activity in a PKA/ERK-dependent manner (Figure 1).82
Altogether, increased adenosine signaling via ADORA2BR leads to increased 2,3-BPG and S1P and in turn contributes to sickling in SCA. The molecular basis underlying elevated SphK1-mediated S1P in red blood cell sickling remains unknown. Identification of the specific molecules and signaling pathways underlying SphK1-S1P-mediated sickling in SCA will likely provide new insights for erythrocyte pathology in SCA and identify potential therapeutic strategies for preventing sickling and disease progression. Overall, metabolomics studies have revealed that adenosine, S1P, and 2,3-BPG contribute to red blood cell sickling, a central pathogenesis of the disease (Figure 1). These findings immediately implicate adenosine, S1P, and 2,3-BPG as pathogenic metabolites and suggest that lowering the elevated adenosine levels or interfering with ADORA2BR and SphK1 activation are candidate therapeutic interventions to treat SCA.
Interactomics
Goodman et al. published the first human RBC protein interactomic map and defined the Repair or Destroy (ROD) Box, which contains proteasomal proteins associated with chaperonin, heat shock proteins, and antioxidant enzymes.67 Kurdia et al. performed centrality measurements on an interactome network built from 751 RBC proteins known at that time.83 Centrality measurements indicate those nodes which are most important to the network. Of the 10 proteins altered in SCA RBCs that had the highest centrality scores, for each form of centrality measurement, nine were chaperonin, proteasomal, and anti-oxidant proteins found within the ROD Box. Since their article was published, hundreds more proteins were discovered within the RBC. We mention that the centrality software developed by Kurdia et al.83 is available and can be used on the larger data set available at this time. Ammann and Goodman utilized statistical cluster analyses to measure the similarity of nodes within the RBC interactome network using a method called Generalized Topological Overlap Measure (GTOM).84 They demonstrated that multiple SCA altered proteins in the ROD Box group (proteasomal subunits and chaperonins) fell within large clusters.84
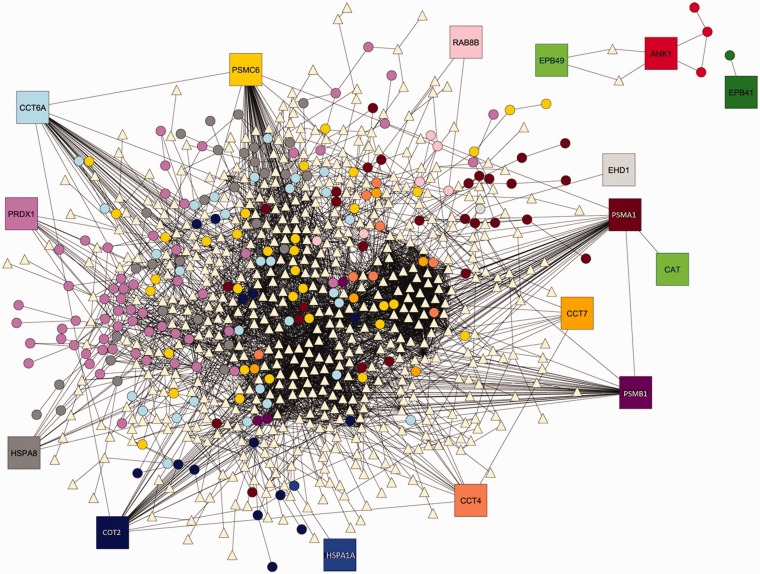
In an effort to continue RBC interactome analysis and enhance methods utilized by Kurdia et al.83 and Ammann and Goodman,84 Daescu and colleagues85 proposed the use of Voronoi diagram for graphs (VDG) as a tool for analyzing SCD dependent changes in the RBC protein interactomic map. VDG provided an efficient way to cluster nodes in the network based on their distance to the members of a predetermined subset of cluster centers called Voronoi sites. As can be seen in Figure 2, of the proteins known to be altered in SCD RBCs, chaperonins, antioxidant enzymes, and proteasomal subunits are part of major clusters while the ANK (Ankyrin) 1 and EPB 41(erythrocyte protein band 4.1) and 49 (erythrocyte protein band 4.9) clusters are smaller and disconnected from the major clusters. This in silico analysis suggests that the altered chaperonins, anti-oxidant enzymes, and proteasomal subunits are the most likely nodes (proteins) to be disruptive to the RBC network (structure-function). VDG is much faster than centrality measures and clustering algorithms. The VDG approach proved to be better at interpreting the influence that SCA affected proteins have on the global networks since those proteins were used as the Voronoi sites.85
Figure 2.
Voronoi regions induced by the nodes corresponding to the proteins altered by sickle cell disease. Sixteen proteins (Voronoi sites) altered by sickle cell disease74 yielded 16 Voronoi clusters. Each Voronoi site, shown as a square labeled with its gene symbol, and the nodes of its induced Voronoi region, are distinctly marked. Chaperonins, antioxidant enzymes, and proteasomal subunits were part of major clusters while the ANK (Ankyrin) 1 and EPB 41(erythrocyte protein band 4.1) and 49 (erythrocyte protein band 4.9) clusters were smaller and disconnected from the major clusters. Triangle shaped nodes belong to more than one cluster. Modified from Zivanik et al.85 with permission of the copyright holder
Following the recent strides in the field of proteomics analyses of red blood cells, D’Alessandro et al.86 and, more recently, Daescu and colleagues85 compiled a list of 1789 and 1834 proteins, respectively, by assembling data from Roux-Dalvai et al.,87 Pasini et al,88 and Goodman et al.67 Despite technical differences in these analyses that may affect the clinical validity of these datasets for diagnostic purposes,89 a few recurring patterns have been highlighted by all of the in silico elaborations of red blood cell proteomics data. Again the chaperonin, proteasomal, and anti-oxidant proteins altered in SCA were all part of major clusters. Our recent review of the erythrocyte proteome indicates that it contains about 2300 unique proteins70 and this number could grow further leading to enhanced interactome maps of the RBC. The VDG software developed by Daescu and colleagues85 can be used to produce updated results whenever significant changes are made to the network, either by addition of newly discovered proteins or by changing the status of some proteins to SCA affected proteins. Such a workflow may be useful to elaborate and integrate90 data from various omics platforms (e.g. proteomics, transcriptomics, metabolomics91) with the goal of evaluating how the red blood cell interactome can be affected by normal and SCD pathological erythropoiesis. Defining key biological nodes is a means of identifying which cellular components may require verification and validation as biomarkers.
Integrated omics and validated biomarker panels
So far there are no published validated individual or panels of biomarkers for clinical manifestations of SCD. Therefore, longitudinal clinical trials to determine the efficacy of biomarkers in predicting sickle cell severity early in life have not been performed. The reason for needing validated individual or panels of biomarkers, which can predict sickle cell severity over the first year of life, is that clinical symptoms typically presents between 6 and 12 months after birth when fetal hemoglobin reaches low levels. An important step towards reaching the goal of early severity prediction is the development of a panel of diverse validated biomarkers derived from metabolomic, transcriptomic, and proteomic studies. These omic layers are not redundant, and a “multiomic” profile (multi-biomarker test) may be required to explain maximal variance in clinical phenotypes.
The utilization of platforms that can integrate validated biomarkers from proteomics, transcriptomics, metabolomics, and other omic platforms should allow the identification of functionally linked panels of validated biomarkers. A recently developed systems biology visualization tool called 3Omics can be used to integrate the transcriptomic, proteomic, and metabolomic data into inter-omic networks, as described by Kuo et al.92 This powerful web-based platform may allow a look at how the sickle cell severity-related transcriptomic, proteomic, and metabolomic expression profiles can be integrated. Such an approach has already increased the coherence and robustness of newly discovered multiomic panels of validated biomarkers for breast cancer.93 Simply stated, integrated interactomics has the potential to allow the assembly of panels of validated functionally linked biomarkers for validation in future longitudinal clinical trials to ascertain whether they can be used as prognostic predictors of sickle cell severity early in life; or for therapeutic intervention to prevent disease progression later in life.
Precision medicine for SCD
In this review, we have provided information on candidate biomarkers for various clinical aspects of sickle cell severity. All must undergo verification and then validation before they can become clinically useful, but they do represent progress toward the goal of precision medicine for SCD.
Rather than becoming involved in parsing the differences and similarities of terms between precision and personalized medicine; we use the term precision medicine in a simple manner. To conduct medicine with precision one must understand the genetics and epigenetics of individuals and as much as possible about the environment in which they live. These factors will influence amounts and modifications of components of the genome, transcriptome, microRNAome, proteome, and metabolome. These changes in amounts and modifications to DNA, RNA, protein, and various metabolites can potentially explain the etiology or a clinical manifestation of that disease; and may supply a way to predict the occurrence or severity of a disease. So with this simplified definition, we can discuss the components of Precision Medicine. You need the use of multiomic technologies, sometimes called omics stacks, to measure disease based changes in the amount and modification in the various cellular molecules and macromolecules. You also need a way to computationally assess how these biologic modifications intersect with changes in the patient’s environmental exposure (exposome) and medical history. This requires analysis of big data with appropriate algorithms, which in theory should lead to treatment of individuals or groups with common characteristics, with greater precision.
In the case of SCD, significant effort has gone into attempting to define a composite measure of sickle cell severity, which can be used to compare changes in either individual or panels of biomarkers. There has been no consensus in the field on a composite definition of sickle severity and this is reviewed in an accompanying article in this thematic issue.34 Furthermore, we believe that each clinical manifestation of sickle cell severity will have its own set of non-overlapping biomarkers, in addition to those that are in common, and that the former will be of greater interest for prediction of severity and the application of appropriate therapeutic approaches or precision medicine.
Conclusion
In conclusion to reach the point where we can offer precision medicine for infants, children and adults based on their clinical manifestations of sickle cell severity will require a consortium of researchers with expertise in all of the multiomic platforms, including integrated interactomics, and in the utilization of algorithms, statistics, and computational science for handling big data. It will also require many clinical sites to obtain the necessary number of subjects. Finally, it will need the financial support of national and international funding agencies that make reaching this goal a high priority.
Acknowledgements
Daescu′s work is partially supported by NSF award IIP1439718 and CPRIT award RP150164. The work on metabolomic profiling in SCD is funded by NIH grants HL119549 (to YX) and P01HL114457 (Project 3 to YX).
Authors contributions
SRG drafted the Abstract, Introduction, Proteomics, Precision Medicine for Sickle Cell Disease, and Conclusion sections. BSP drafted the Fetal Hemoglobin and Sickle Cell Severity, Genomics, and MicroRNAomics sections. KCH, AD, and YX drafted the section on Metabolomics. OD drafted the Interactomics; and Integrated Omics and Validated Biomarker Panels sections. SJG drafted the Transcriptomics section. All authors read, revised, and approved the manuscript prior to submission.
Declaration of conflicting interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
References
- 1.Platt OS, Thorington BD, Brambilla DJ, Milner PF, Rosse WF, Vichinsky E, Kinney TR. Pain in sickle cell disease: rates and risk factors. New Engl J Med 1991; 325: 11–6. [DOI] [PubMed] [Google Scholar]
- 2.Platt OS, Brambilla DJ, Rosse WF, Milner PF, Castro O, Steinberg MH, Klug PP. Mortality in sickle cell disease. Life expectancy and risk factors for early death. N Engl J Med 1994; 330: 1639–44. [DOI] [PubMed] [Google Scholar]
- 3.Powars DR, Meiselman HJ, Fisher TC, Hiti A, Johnson C. Beta-S gene cluster haplotypes modulate hematologic and hemorheologic expression in sickle cell anemia Use in predicting clinical severity. Am J Pediatr Hematol Oncol 1994; 16: 55–61. [PubMed] [Google Scholar]
- 4.Quinn CT, Miller ST. Risk factors and prediction of outcomes in children and adolescents who have sickle cell anemia. Hematol Oncol Clin N Am 2004; 18: 1339–54. [DOI] [PubMed] [Google Scholar]
- 5.Quinn CT, Rogers ZR, Buchanan GR. Survival of children with sickle cell disease. Blood 2004; 103: 4023–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Quinn CT, Shull EP, Ahmad N, Lee NJ, Rogers ZR, Buchanan GR. Prognostic significance of early vaso-occlusive complications in children with sickle cell anemia. Blood 2007; 109: 40–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kutlar A. Sickle cell disease a multigenic perspective of a single gene disorder. Hemoglobin 2007; 31: 209–24. [DOI] [PubMed] [Google Scholar]
- 8.Fertrin KY, Costa FF. Genomic polymorphisms in sickle cell disease: implications for clinical diversity and treatment. Expert Rev Hematol 2010; 3: 443–58. [DOI] [PubMed] [Google Scholar]
- 9.Steinberg MH, Sebastiani P. Genetic modifiers of sickle cell disease. Am J Hematol 2012; 87: 795–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bauer DE, Kamran SC, Orkin SH. Reawakening fetal hemoglobin: prospects for new therapies for the β-globin disorders. Blood 2013; 120: 2945–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Castro O, Brambilla DJ, Thorington B, Reindorf CA, Scott RB, Gillette P, Vera JC, Levy PS. The acute chest syndrome in Sickle Cell disease: incidence and risk factors. Blood 1994; 84: 643–49. [PubMed] [Google Scholar]
- 12.Stamatoyannopoulos G, Grosveld F. Hemoglobin switching, in the molecular basis of blood disease, 3rd ed Philadelphia: Saunders, 2001. [Google Scholar]
- 13.Brown AK, Sleeper LA, Miller ST, Pegelow CH, Gill FM, Waclawiw MA. Reference values and hematologic changes from birth to 5 years in patients with sickle cell disease. Cooperative Study of Sickle Cell Disease. Arch Pediatr Adolesc Med 1994; 148: 796–804. [DOI] [PubMed] [Google Scholar]
- 14.Poillon WN, Kim BC, Rodgers GP, Noguchi CT, Schechter AN. Sparing effect of hemoglobin F and hemoglobin A2 on the polymerization of hemoglobin S at physiologic ligand saturations. Proc Natl Acad Sci USA 1993; 90: 5039–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Charache S, Terrin ML, Moore RD, Dover GJ, Barton FB, Eckert SV, McMahon RP, Bonds DR. Effect of hydroxyurea on the frequency of painful crises in sickle cell anemia. Investigators of the Multicenter Study of Hydroxyurea in Sickle Cell Anemia. N Engl J Med 1995; 332: 1317–22. [DOI] [PubMed] [Google Scholar]
- 16.Steinberg MH, Lu ZH, Barton FB, Terrin ML, Charache S, Dover GJ. Fetal hemoglobin in sickle cell anemia: determinants of response to hydroxyurea. Multicenter Study of Hydroxyurea. Blood 1997; 89: 1078–88. [PubMed] [Google Scholar]
- 17.Wang WC, Ware RE, Miller ST, Iyer RV, Casella JF, Minniti CP, Rana S, Thornburg CD, Rogers ZR, Kalpatthi RV, Barredo JC, Brown RC, Sarnaik SA, Howard TH, Wynn LW, Kutlar A, Armstrong FD, Files BA, Goldsmith JC, Waclawiw MA, Huang X, Thompson BW. BABY HUG investigators. Hydroxycarbamide in very young children with sickle-cell anaemia: a multicentre, randomised, controlled trial (BABY HUG). Lancet 2011; 377: 1663–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McGann PT, Ware RE. Hydroxyurea therapy for sickle cell anemia. Expert Opin Drug Saf 2015; 14: 1749–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.http://www.nhlbi.nih.gov/health-pro/guidelines/sickle-cell-disease-guidelines (accessed 7 March 2016).
- 20.Kato GJ, Gladwin MT, Steinberg MH. Deconstructing sickle cell disease: reappraisal of the role of hemolysis in the development of clinical subphenotypes. Blood Rev 2007; 21: 37–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taylor JG, 6th, Nolan VG, Mendelsohn L, Kato GJ, Gladwin MT, Steinberg MH. Chronic hyper-hemolysis in sickle cell anemia: association of vascular complications and mortality with less frequent vasoocclusive pain. PLoS One 2008; 3: e2095–e2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Belfer I, Youngblood V, Darbari DS, Wang Z, Diaw L, Freeman L, Desai K, Dizon M, Allen D, Cunnington C, Channon KM, Milton J, Hartley SW, Nolan V, Kato GJ, Steinberg MH, Goldman D, Taylor JG., 6th A GCH1 haplotype confers sex-specific susceptibility to pain crises and altered endothelial function in adults with sickle cell anemia. Am J Hematol 2014; 89: 187–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mendonça TF, Oliveira MC, Vasconcelos LR, Pereira LM, Moura P, Bezerra MA, Santos MN, Araújo AS, Cavalcanti M. Association of variant alleles of MBL2 gene with vasoocclusive crisis in children with sickle cell anemia. Blood Cells Mol Dis 2010; 44: 224–8. [DOI] [PubMed] [Google Scholar]
- 24.Bean CJ, Boulet SL, Ellingsen D, Pyle ME, Barron-Casella EA, Casella JF, Payne AB, Driggers J, Trau HA, Yang G, Jones K, Ofori-Acquah SF, Hooper WC, DeBaun MR. Heme oxygenase-1 gene promoter polymorphism is associated with reduced incidence of acute chest syndrome among children with sickle cell disease. Blood 2012; 120: 3822–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adams GT, Snieder H, McKie VC, Clair B, Brambilla D, Adams RJ, Kutlar F, Kutlar A. Genetic risk factors for cerebrovascular disease in children with sickle cell disease: design of a case-control association study and genomewide screen. BMC Med Genet 2003; 4: 6–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elliott L, Ashley-Koch AE, De Castro L, Jonassaint J, Price J, Ataga KI, Levesque MC, Brice Weinberg J, Eckman JR, Orringer EP, Vance JM, Telen MJ. Genetic polymorphisms associated with priapism in sickle cell disease. Br J Haematol 2007; 137: 262–7. [DOI] [PubMed] [Google Scholar]
- 27.Milton JN, Rooks H, Drasar E, McCabe EL, Baldwin CT, Melista E, Gordeuk VR, Nouraie M, Kato GR, Minniti C, Taylor J, Campbell A, Luchtman-Jones L, Rana S, Castro O, Zhang Y, Thein SL, Sebastiani P, Gladwin MT, Walk-PHAAST Investigators, Steinberg MH. Genetic determinants of haemolysis in sickle cell anaemia. Br J Haematol 2013; 161: 270–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dworkis DA, Klings ES, Solovieff N, Li G, Milton JN, Hartley SW, Melista E, Parente J, Sebastiani P, Steinberg MH, Baldwin CT. Severe sickle cell anemia is associated with increased plasma levels of TNF-R1 and VCAM-1. Am J Hematol 2011; 86: 220–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coelho A, Dias A, Morais A, Nunes B, Ferreira E, Picanço I, Faustino P, Lavinha J. Genetic variation in CD36, HBA, NOS3 and VCAM1 is associated with chronic haemolysis level in sickle cell anaemia: a longitudinal study. Eur J Haematol 2014; 92: 237–43. [DOI] [PubMed] [Google Scholar]
- 30.Hoppe C, Klitz W, D'Harlingue K, Cheng S, Grow M, Steiner L, Noble J, Adams R, Styles L. Stroke Prevention Trial in Sickle Cell Anemia (STOP) Investigators. Confirmation of an association between the TNF(-308) promoter polymorphism and stroke risk in children with sickle cell anemia. Stroke 2007; 38: 2241–6. [DOI] [PubMed] [Google Scholar]
- 31.Adewoye AH, Nolan VG, Ma Q, Baldwin C, Wyszynski DF, Farrell JJ, Farrer LA, Steinberg MH. Association of polymorphisms of IGF1R and genes in the transforming growth factor- beta/bone morphogenetic protein pathway with bacteremia in sickle cell anemia. Clin Infect Dis 2006; 43: 593–8. [DOI] [PubMed] [Google Scholar]
- 32.Nolan VG, Adewoye A, Baldwin C, Wang L, Ma Q, Wyszynski DF, Farrell JJ, Sebastiani P, Farrer LA, Steinberg MH. Sickle cell leg ulcers: associations with haemolysis and SNPs in Klotho, TEK and genes of the TGF-beta/BMP pathway. Br J Haematol 2006; 133: 570–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Oliveira Filho RA, Silva GJ, de Farias Domingos I, Hatzlhofer BL, da Silva Araújo A, de Lima Filho JL, Bezerra MA, Martins DB, de Araújo RF. Association between the genetic polymorphisms of glutathione S-transferase (GSTM1 and GSTT1) and the clinical manifestations in sickle cell anemia. Blood Cells Mol Dis 2013; 51: 76–9. [DOI] [PubMed] [Google Scholar]
- 34.Habara A, Steinberg MH. Genetic basis of heterogeneity and severity in sickle cell disease. Exp Biol Med (in press). [DOI] [PMC free article] [PubMed]
- 35.Nagel RL, Rao SK, Dunda-Belkhodja O, Connolly MM, Fabry ME, Georges A, Krishnamoorthy R, Labie D. The hematologic characteristics of sickle cell anemia bearing the Bantu haplotype: the relationship between G gamma and HbF level. Blood 1987; 69: 1026–30. [PubMed] [Google Scholar]
- 36.Nagel RL, Erlingsson S, Fabry ME, Croizat H, Susuka SM, Lachman H, Sutton M, Driscoll C, Bouhassira E, Billett HH. The Senegal DNA haplotype is associated with the amelioration of anemia in African-American sickle cell anemia patients. Blood 1991; 77: 1371–5. [PubMed] [Google Scholar]
- 37.Kutlar A, Hattori Y, Bakioglu I, Kutlar F, Kamel K, Huisman TH. Hematological observations on Arabian SS patients with a homozygosity or heterozygosity for a beta S chromosome with haplotype #31. Hemoglobin 1985; 9: 545–57. [DOI] [PubMed] [Google Scholar]
- 38.Wyszynski DF, Baldwin CT, Cleves MA, Amirault Y, Nolan VG, Farrell JJ, Bisbee A, Kutlar A, Farrer LA, Steinberg MH. Polymorphisms near a chromosome 6q QTL area are associated with modulation of fetal hemoglobin levels in sickle cell anemia. Cell Mol Biol (Noisy-le-grand) 2004; 50: 23–33. [PubMed] [Google Scholar]
- 39.Garner C, Menzel S, Martin C, Silver N, Best S, Spector TD, Thein SL. Interaction between two quantitative trait loci affects fetal haemoglobin expression. Ann Hum Genet 2005; 69(Pt 6): 707–14. [DOI] [PubMed] [Google Scholar]
- 40.Thein SL, Menzel S, Peng X, Best S, Jiang J, Close J, Silver N, Gerovasilli A, Ping C, Yamaguchi M, Wahlberg K, Ulug P, Spector TD, Garner C, Matsuda F, Farrall M, Lathrop M. Intergenic variants of HBS1L-MYB are responsible for a major quantitative trait locus on chromosome 6q23 influencing fetal hemoglobin levels in adults. Proc Natl Acad Sci USA 2007; 104: 11346–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Menzel S, Garner C, Gut I, Matsuda F, Yamaguchi M, Heath S, Foglio M, Zelenika D, Boland A, Rooks H, Best S, Spector TD, Farrall M, Lathrop M, Thein SL. A QTL influencing F cell production maps to a gene encoding a zinc-finger protein on chromosome 2p15. Nat Genet 2007; 39: 1197–9. [DOI] [PubMed] [Google Scholar]
- 42.Uda M1, Galanello R, Sanna S, Lettre G, Sankaran VG, Chen W, Usala G, Busonero F, Maschio A, Albai G, Piras MG, Sestu N, Lai S, Dei M, Mulas A, Crisponi L, Naitza S, Asunis I, Deiana M, Nagaraja R, Perseu L, Satta S, Cipollina MD, Sollaino C, Moi P, Hirschhorn JN, Orkin SH, Abecasis GR, Schlessinger D, Cao A. Genome-wide association study shows BCL11A associated with persistent fetal hemoglobin and amelioration of the phenotype of beta-thalassemia. Proc Natl Acad Sci USA 2008; 105: 1620–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Solovieff N, Milton JN, Hartley SW, Sherva R, Sebastiani P, Dworkis DA, Klings ES, Farrer LA, Garrett ME, Ashley-Koch A, Telen MJ, Fucharoen S, Ha SY, Li CK, Chui DH, Baldwin CT, Steinberg MH. Fetal hemoglobin in sickle cell anemia: genome-wide association studies suggest a regulatory region in the 5' olfactory receptor gene cluster. Blood 2010; 115: 1815–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nuinoon M, Makarasara W, Mushiroda T, Setianingsih I, Wahidiyat PA, Sripichai O, Kumasaka N, Takahashi A, Svasti S, Munkongdee T, Mahasirimongkol S, Peerapittayamongkol C, Viprakasit V, Kamatani N, Winichagoon P, Kubo M, Nakamura Y, Fucharoen S. A genome-wide association identified the common genetic variants influence disease severity in beta0-thalassemia/hemoglobin E. Hum Genet 2010; 127: 303–14. [DOI] [PubMed] [Google Scholar]
- 45.Bae HT, Baldwin CT, Sebastiani P, Telen MJ, Ashley-Koch A, Garrett M, Hooper WC, Bean CJ, Debaun MR, Arking DE, Bhatnagar P, Casella JF, Keefer JR, Barron-Casella E, Gordeuk V, Kato GJ, Minniti C, Taylor J, Campbell A, Luchtman-Jones L, Hoppe C, Gladwin MT, Zhang Y, Steinberg MH. Meta-analysis of 2040 sickle cell anemia patients: BCL11A and HBS1L-MYB are the major modifiers of HbF in African Americans. Blood 2012; 120: 1961–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mtatiro SN, Singh T, Rooks H, Mgaya J, Mariki H, Soka D, Mmbando B, Msaki E, Kolder I, Thein SL, Menzel S, Cox SE, Makani J, Barrett JC. Genome wide association study of fetal hemoglobin in sickle cell anemia in Tanzania. PLoS One 2014; 9: e111464–e111464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stadhouders R, Aktuna S, Thongjuea S, Aghajanirefah A, Pourfarzad F, van Ijcken W, Lenhard B, Rooks H, Best S, Menzel S, Grosveld F, Thein SL, Soler E. HBS1L-MYB intergenic variants modulate fetal hemoglobin via long-range MYB enhancers. J Clin Invest 2014; 124: 1699–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sankaran VG, Menne TF, Xu J, Akie TE, Lettre G, Van Handel B, Mikkola HK, Hirschhorn JN, Cantor AB, Orkin SH. Human fetal hemoglobin expression is regulated by the developmental stage-specific repressor BCL11A. Science 2008; 322: 1839–42. [DOI] [PubMed] [Google Scholar]
- 49.Xu J, Peng C, Sankaran VG, Shao Z, Esrick EB, Chong BG, Ippolito GC, Fujiwara Y, Ebert BL, Tucker PW, Orkin SH. Correction of sickle cell disease in adult mice by interference with fetal hemoglobin silencing. Science 2011;18;334:993–6. [DOI] [PMC free article] [PubMed]
- 50.Xu J, Bauer DE, Kerenyi MA, Vo TD, Hou S, Hsu YJ, Yao H, Trowbridge JJ, Mandel G, Orkin SH. Corepressor-dependent silencing of fetal hemoglobin expression by BCL11A. Proc Natl Acad Sci USA 2013; 110: 6518–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roosjen M, McColl B, Kao B, Gearing LJ, Blewitt ME, Vadolas J. Transcriptional regulators Myb and BCL11A interplay with DNA methyltransferase 1 in developmental silencing of embryonic and fetal β-like globin genes. FASEB J 2014; 28: 1610–20. [DOI] [PubMed] [Google Scholar]
- 52.Amaya M, Desai M, Gnanapragasam MN, Wang SZ, Zu Zhu S, Williams DC, Jr, Ginder GD. Mi2β-mediated silencing of the fetal γ-globin gene in adult erythroid cells. Blood 2013; 121: 3493–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu J, Sankaran VG, Ni M, Menne TF, Puram RV, Kim W, Orkin SH. Transcriptional silencing of {gamma}-globin by BCL11A involves long-range interactions and cooperation with SOX6. Genes Dev 2010; 24: 783–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sankaran VG, Xu J, Byron R, Greisman HA, Fisher C, Weatherall DJ, Sabath DE, Groudine M, Orkin SH, Premawardhena A, Bender MA. A functional element necessary for fetal hemoglobin silencing. N Engl J Med 2011; 365: 807–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu Y, Wang J, Khaled W, Burke S, Li P, Chen X, Yang W, Jenkins NA, Copeland NG, Zhang S, Liu P. Bcl11a is essential for lymphoid development and negatively regulates p53. J Exp Med 2012; 209: 2467–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bauer DE, Kamran SC, Lessard S, Xu J, Fujiwara Y, Lin C, Shao Z, Canver MC, Smith EC, Pinello L, Sabo PJ, Vierstra J, Voit RA, Yuan GC, Porteus MH, Stamatoyannopoulos JA, Lettre G, Orkin SH. An erythroid enhancer of BCL11A subject to genetic variation determines fetal hemoglobin level. Science 2013; 342: 253–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jison ML, Munson PJ, Barb JJ, Suffredini AF, Talwar S, Logun C, Raghavachari N, Beigel JH, Shelhamer JH, Danner RL, Gladwin MT. Blood mononuclear cell gene expression profiles characterize the oxidant, hemolytic, and inflammatory stress of sickle cell disease. Blood 2004; 104: 270–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hounkpe BW, Fiusa MM, Colella MP, Nilkenes Gomes da Costa L, Benatti Rde O, Olalla Saad ST, Costa FF, Dos Santos MN, De Paula EV. Role of innate immunity-triggered pathways in the pathogenesis of Sickle Cell Disease: a meta-analysis of gene expression studies. Sci Rep 2015; 5: 17822–17822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee YT, de Vasconcellos JF, Yuan J, Byrnes C, Noh SJ, Meier ER, Kim KS, Rabel A, Kaushal M, Muljo SA, Miller JL. LIN28B-mediated expression of fetal hemoglobin and production of fetal-like erythrocytes from adult human erythroblasts ex vivo. Blood 2013; 122: 1034–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sankaran VG, Menne TF, Scepanovic D, Vergilio JA, Ji P, Kim J, Thiru P, Orkin SH, Lander ES, Lodish HF. MicroRNA-15a and -16-1 act via MYB to elevate fetal hemoglobin expression in human trisomy 13. Proc Natl Acad Sci USA 2011; 108: 1519–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Walker AL, Steward S, Howard TA, Mortier N, Smeltzer M, Wang YD, Ware RE. Epigenetic and molecular profiles of erythroid cells after hydroxyurea treatment in sickle cell anemia. Blood 2011; 118: 5664–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nur E, Verwijs M, de Waart DR, Schnog JJ, Otten HM, Brandjes DP, Biemond BJ, Elferink RP. CURAMA Study Group. Increased efflux of oxidized glutathione (GSSG) causes glutathione depletion and potentially diminishes antioxidant defense in sickle erythrocytes. Biochim Biophys Acta 2011; 1812: 1412–7. [DOI] [PubMed] [Google Scholar]
- 63.Sangokoya C, Telen MJ, Chi JT. microRNA miR-144 modulates oxidative stress tolerance and associates with anemia severity in sickle cell disease. Blood 2010; 116: 4338–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.O'Connell MA, Hayes JD. The Keap1/Nrf2 pathway in health and disease: from the bench to the clinic. Biochem Soc Trans 2015; 43: 687–9. [DOI] [PubMed] [Google Scholar]
- 65.Li B, Torres CM, Takezaki M, Neunert C, Kutlar A, Pace BS. Micro RNA profiling to identify regulator of fetal hemoglobin expression in sickle cell disease. Blood 2014; 124: 49–49.24797299 [Google Scholar]
- 66.Bader AG. miR-34—a microRNA replacement therapy is headed to the clinic. Front Genet 2012; 3: Article 120–Article 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Goodman SR, Kurdia A, Ammann L, Kakhniashvili D, Daescu O. The human red blood cell proteome and interactome. Exp Biol Med 2007; 232: 1391–408. [DOI] [PubMed] [Google Scholar]
- 68.Liumbruno G, D'Alessandro A, Grazzini G, Zolla L. Blood-related proteomics. J Proteomics 2010; 73: 483–507. [DOI] [PubMed] [Google Scholar]
- 69.Yuditskaya S, Suffredini AF, Kato GJ. The proteome of sickle cell disease: Insights from exploratory proteomic profiling. Exp Rev Proteom 2010; 7: 833–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Goodman SR, Daescu O, Kakhniashvili DG, Zivanic M. The proteomics and interactomics of human erythrocytes. Exp Biol Med 2013; 238: 509–18. [DOI] [PubMed] [Google Scholar]
- 71.Hryniewicz-Jankowska A, Choudhary PK, Ammann L, Quinn CT, Goodman SR. Monocyte protein signatures of disease severity in sickle cell anemia. Exp Biol Med 2009; 234: 210–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yuditskaya S, Tumblin A, Hoehn GT, Wang G, Drake SK, Xu X, Ying Chi AH, Remaley AT, Shen RF, Munson PJ, Suffredini AF, Kato GJ. Proteomic identification of altered apolipoprotein patterns in pulmonary hypertension and vasculopathy of sickle cell disease. Blood 2009; 113: 1122–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tumblin A, Tailor A, Hoehn GT, Kyle Mack A, Mendelsohn L, Freeman L, Xu X, Remaley AT, Munson PJ, Suffredini AF, Kato GJ. Apolipoprotein A-I and serum amyloid a plasma levels are biomarkers of acute painful episodes in patients with sickle cell disease. Haematology 2010; 95: 1467–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kakhniashvili DG, Griko NB, Bulla LA, Jr, Goodman SR. The proteomics of sickle cell disease: Profiling of erythrocyte membrane proteins by 2D-DIGE and tandem mass spectrometry. Exp Biol Med 2005; 230: 787–92. [DOI] [PubMed] [Google Scholar]
- 75.Basu A, Saha S, Karmakar S, Chakravarty S, Banerjee D, Dash BP, Chakrabarti A. 2D DIGE based proteomics study of erythrocyte cytosol in sickle cell disease: Altered proteostasis and oxidative stress. Proteomics 2013; 13: 3233–42. [DOI] [PubMed] [Google Scholar]
- 76.Darghouth D, Koehl B, Madalinski G, Heilier J-F, Bovee P, Xu Y, Olivier M-F, Bartolucci P, Benkerrou M, Pissard S, Colin Y, Galacteros F, Bosman G, Junot C, Roméo P-H. Pathophysiology of sickle cell disease is mirrored by the red blood cell metabolome. Blood 2011; 117: e57–66. [DOI] [PubMed] [Google Scholar]
- 77.Darghouth D, Koehl B, Junot C, Roméo P-H. Metabolomic analysis of normal and sickle cell erythrocytes. Transf Clin Biol 2010; 17: 148–50. [DOI] [PubMed] [Google Scholar]
- 78.Morris CR. Alterations of the arginine metabolome in sickle cell disease a growing rationale for arginine therapy. Hematol/Oncol Clin North Am 2014; 28: 301–21. [DOI] [PubMed] [Google Scholar]
- 79.Zhang Y, Dai Y, Wen J, Zhang W, Grenz A, Sun H, Tao L, Lu G, Alexander DC, Milburn MV, Carter Dawson L, Lewis DE, Zhang W, Eltzschig HK, Kellems RE, Blackburn MR, Juneja HS, Xia Y. Detrimental effects of adenosine signaling in sickle cell disease. Nat Med 2011; 17: 79–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang Y, Xia Y. Adenosine signaling in normal and sickle erythrocytes and beyond. Microb Infect 2012; 14: 863–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang Y, Berka V, Song A, Sun K, Wang W, Zhang W, Ning C, Xia Y. Elevated sphingosine-1-phosphate promotes sickling and sickle cell disease progression. J Clin Invest 2014; 124: 2750–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sun K, Zhang Y, Bogdanov MV, Wu H, Song A, Li J, Dowhan W, Idowu M, Juneja HS, Molina JG, Blackburn MR, Kellems RE, Xia Y. Elevated adenosine signaling via adenosine A2B receptor induces normal and sickle erythrocyte sphingosine kinase 1 activity. Blood 2015; 125: 1643–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kurdia A, Daescu O, Ammann L, Kakhniashvili D, Goodman SR. Centrality measures for the human red blood cell interactome. IEEE Proc 2007, pp. 8–101. [DOI] [PubMed] [Google Scholar]
- 84.Ammann L, Goodman SR. Cluster analysis for the impact of sickle cell disease on the human erythrocyte protein interactome. Exp Biol Med 2009; 234: 703–11. [DOI] [PubMed] [Google Scholar]
- 85.Zivanic M, Daescu O, Kurdia A, Goodman SR. The Voronoi diagram for graphs and its application in the sickle cell disease research. J Comput Sci 2012; 3: 335–43. [Google Scholar]
- 86.D'Alessandro A, Righetti PG, Zolla L. The red blood cell proteome and interactome: an update. J Proteome Res 2010; 9: 144–63. [DOI] [PubMed] [Google Scholar]
- 87.Roux-Dalvai F, de Peredo AG, Simó C, Guerrier L, Bouyssié D, Zanella A, Citterio A, Burlet-Schlitz O, Boscetti E, Righetti PG, Monsarrat B. Extensive analysis of the cytoplasmic proteome of human erythrocytes using the peptide ligand library technology and advanced mass spectrometry. Mol Cell Proteom 2008; 7: 2254–69. [DOI] [PubMed] [Google Scholar]
- 88.Pasini EM, Kirkegaard M, Mortensen P, Lutz HU, Thomas AW, Mann M. In-depth analysis of the membrane and cytosolic proteome of red blood cells. Blood 2006; 108: 791–801. [DOI] [PubMed] [Google Scholar]
- 89.Hegedűs T, Chaubey PM, Várady G, Szabó E, Sarankó H, Hofstetter L, Roschitzki B, Stieger B, Sarkadi B. Inconsistencies in the red blood cell membrane proteome analysis: generation of a database for research and diagnostic applications. Database 2015; 2015: bav056–bav056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cabezas-Wallscheid N, Klimmeck D, Hansson J, Lipka DB, Reyes A, Wang Q, Weichenhan D, Lier A, von Paleske L, Renders S, Wünsche P, Zeisberger P, Brocks D, Gu L, Herrmann C, Haas S, Essers MA, Brors B, Eils R, Huber W, Milsom MD, Plass C, Krijgsveld J, Trumpp A. Identification of regulatory networks in HSCs and their immediate progeny via integrated proteome, transcriptome, and DNA methylome analysis. Cell Stem Cell 2014; 15: 507–22. [DOI] [PubMed] [Google Scholar]
- 91.Oburoglu L, Tardito S, Fritz V, de Barros SC, Merida P, Craveiro M, Mamede J, Cretenet G, Mongellaz C, An X, Klysz D, Touhami J, Boyer-Clavel M, Battini JL, Dardalhon V, Zimmermann VS, Mohandas N, Gottlieb E, Sitbon M, Kinet S, Taylor N. Glucose and glutamine metabolism regulate human hematopoietic stem cell lineage specification. Cell Stem Cell 2014; 15: 169–84. [DOI] [PubMed] [Google Scholar]
- 92.Kuo T-C, Tian T-F, Tseng YJ. 3Omics: A web-based systems biology tool for analysis, integration and visualization of human transcriptomic, proteomic and metabolomic data. BMC Syst Biol 2013; 7: 64–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Su G, Burant CF, Beecher CW, Athey BD, Meng F. Integrated metabolome and transcriptome analysis of the NCI60 dataset. BMC Bioinform 2011; 12: 36–36. [DOI] [PMC free article] [PubMed] [Google Scholar]