Abstract
Sickle cell disease (SCD) is a group of inherited blood disorders that have in common a mutation in the sixth codon of the β-globin (HBB) gene on chromosome 11. However, people with the same genetic mutation display a wide range of clinical phenotypes. Fetal hemoglobin (HbF) expression is an important genetic modifier of SCD complications leading to milder symptoms and improved long-term survival. Therefore, we performed a genome-wide association study (GWAS) using a case-control experimental design in 244 African Americans with SCD to discover genetic factors associated with HbF expression. The case group consisted of subjects with HbF≥8.6% (133 samples) and control group subjects with HbF≤£3.1% (111 samples). Our GWAS results replicated SNPs previously identified in an erythroid-specific enhancer region located in the second intron of the BCL11A gene associated with HbF expression. In addition, we identified SNPs in the SPARC, GJC1, EFTUD2 and JAZF1 genes as novel candidates associated with HbF levels. To gain insights into mechanisms of globin gene regulation in the HBB locus, linkage disequilibrium (LD) and haplotype analyses were conducted. We observed strong LD in the low HbF group in contrast to a loss of LD and greater number of haplotypes in the high HbF group. A search of known HBB locus regulatory elements identified SNPs 5′ of δ-globin located in an HbF silencing region. In particular, SNP rs4910736 created a binding site for a known transcription repressor GFi1 which is a candidate protein for further investigation. Another HbF-associated SNP, rs2855122 in the cAMP response element upstream of Gγ-globin, was analyzed for functional relevance. Studies performed with siRNA-mediated CREB binding protein (CBP) knockdown in primary erythroid cells demonstrated γ-globin activation and HbF induction, supporting a repressor role for CBP. This study identifies possible molecular determinants of HbF production.
Keywords: GWAS, sickle cell disease, fetal hemoglobin, HBB locus, haplotypes, single nucleotide polymorphisms
Introduction
Sickle cell disease (SCD) is a group of inherited blood disorders that have in common a glu-val mutation in the sixth codon of the human β-globin (HBB) gene on chromosome 11. Those individuals who are homozygous for this mutation (HbSS) or compound heterozygotes for the sickle mutation and β0-thalassemia have the most severe disease phenotype and are at highest risk for developing clinical complications. SCD is characterized by acute vaso-occlusive events including pain episodes, splenic sequestration, acute chest syndrome, and stroke among others, contributing to increased morbidity and early mortality. Despite the genetic simplicity of SCD as a Mendelian single gene disorder, individuals with the same genotype display extreme clinical heterogeneity.1,2 Two major modifiers of SCD clinical phenotypes including fetal hemoglobin (HbF; α2γ2) and α-thalassemia were identified through basic research and clinical studies and subsequently confirmed by genetic studies. Many of the hematologic and clinical complications of SCD are prevented by HbF due to its ability to inhibit HbS polymerization in erythrocytes.3,4 Therefore, the discovery of genetic markers of HbF expression that can be developed as therapeutic targets for SCD has been a major ongoing research effort for decades.
The Cooperative Study of Sickle Cell Disease (CSSCD) was a multi-institutional natural history study5,6 that identified HbF as a modifier of mortality and risk factor for early death in African American patients with SCD.7 These observations have been corroborated by numerous clinical studies of European, Indian, Middle Eastern, and African SCD patients,8 highlighting the importance of understanding genetic modifiers of HbF regulation. The 81-kb HBB locus on chromosome 11 consists of five functional genes, including HBE1 (ɛ), HBG2 (Gγ), HBG1 (Aγ), HBD (δ) and HBB (β), expressed sequentially from 5′ to 3′ in a tissue- and development-specific manner during ontogeny,8 a process known as hemoglobin switching. Expression of the β-like globin genes is controlled in part by the locus control region (LCR) located 6–20 kb upstream of ɛ-globin.9,10 To achieve normal hemoglobin switching, stage-specific transcription factors bind the LCR and globin promoters during erythropoiesis to activate globin gene expression11 through a DNA looping mechanism.12 HbF comprises 80–90% of the total hemoglobin synthesized at birth, gradually decreasing to <1% by 12 months. The discovery of heritable DNA markers associated with major repressors of HBG1/HBG2 expression provides a rational approach for precision medicine in SCD.
To develop strategies of HbF induction, it is necessary to define cis-regulatory elements and DNA-protein interactions in the HBB locus that modify globin gene expression. In previous studies, up to 40% of the HbF-producing capacity in sickle cell patients was attributed to genetic determinants in Xp22.2,13 chromosome 8q14 and genes in the 6q22.3-23.4 quantitative trait locus.15,16 Thein and colleagues were the first to demonstrate single nucleotide polymorphisms (SNPs) in the HBS1L-MYB inter-genic region (HMIP) on 6q23 accounting for 18% of HbF variance in Northern Europeans,17 and later they identified the BCL11A quantitative trait locus on chromosome 218 accounting for about 15% of HbF variance in the same population. Subsequently, Uda et al.19 confirmed SNPs in the BCL11A gene associated with high HbF levels in a genome-wide association study (GWAS) of Sardinian thalassemia patients, establishing the first major repressor of HBG1/HBG2 gene expression. A third locus that contributes to HbF expression is the −158XmnI-HBG2 SNP located in the Gγ-globin promoter located at position −158 relative to the transcriptional start site.15,20
For the most part, GWAS to identify inherited HbF determinants in African American patients with SCD has been conducted using CSSCD samples.21–24 For example, Solovieff et al.23 identified SNPs in BCL11A and the ORB1B5/OR51B6 (rs4910755) genes associated with HbF levels. A subsequent meta-analysis was conducted using GWAS data generated in seven African American SCD cohorts totaling 2040 patients.25 The most significant HbF modifying SNPs were identified in BCL11A (rs766432) and the HMIP region (rs9494145), which represented 11.1% and 3.2% of the phenotypic variability in HbF expression, respectively. Recently, the first GWAS was conducted in a Tanzanian population of 1213 HbSS and HbSβ0-thalassemia patients.26 Similar to the results obtained in African Americans, SNPs in the BCL11A gene and the HMIP region were replicated in that population. Interestingly, the −158XmnI-HBG2 (rs782144) SNP was not replicated in Tanzanian patients, concordant with previous results for African Americans with SCD.23,25
The second major modifier of clinical phenotype in SCD is expression of the α-globin genes (HBA1/HBA2). Approximately one-third of sickle cell patients of African descent have coexisting α-thalassemia due to the common 3.7-kb deletion (−α3.7).1 Coexisting α-thalassemia reduces intracellular hemoglobin concentrations, thereby reducing HbS polymerization and erythrocyte hemolysis. A SNP in the NPRL3 gene on chromosome 16 was identified by GWAS and validated by targeted genotyping in an independent cohort.27 The introns of NPRL3 contain the HBA1/HBA2 gene regulatory elements consisting of hypersensitive sites (HS)-33, HS-40 and HS-48. After adjusting for HbF and α-thalassemia trait, variants of the HBA1/HBA2 gene regulatory locus in NPRL3 were associated with reduced hemolysis in SCD.
In our current study, we conducted a case (high HbF)-control (low HbF) GWAS to identify novel genetic modifiers of HBG1/HBG2 gene expression. As previously identified in several populations, SNPs in the BCL11A gene were associated with HbF levels in our population along with four additional genes suggestive of novel candidate loci. High-density SNP mapping of the 81-kb HBB locus revealed tagging-SNPs that define unique haplotypes associated with the high and low HbF phenotype. Knockdown of the transcription cofactor CREB1 binding protein (CBP) which interacts upstream of the Gγ-globin (HBG2) gene enhances HbF expression to identify possible molecular determinants of γ-globin gene regulation.
Methods and materials
DNA samples sources
A total of 254 genomic samples were analyzed including 155 samples collected during the CSSCD which were obtained after approval from the Biologic Specimen and Data Repository Information Coordinating Center at the National Heart Lung and Blood Institute. Thirty samples were obtained from the Comprehensive Sickle Cell Centers Collaborative Data (CDATA) study and the remaining samples were contributed by Dr. Samir K. Ballas, Thomas Jefferson University.
Genotype determination
The Illumina Omni1-Quad System (San Diego, CA, USA) was used to genotype the DNA samples because this chip contains 1,140,419 SNPs with 76% coverage of polymorphisms validated in people of African ancestry. Genotype data quality control and SNP calls were generated with Illumina’s Genome Studio software. For the HBB locus haplotype analysis, genotype data for SNPs rs2855121 and rs2855122 were confirmed using TaqMan® assay probes (ThermoFisher Scientific, Grand Island, NY) for real-time PCR detection.
GWAS analysis
Allele associations were calculated using the case-control association test as implemented in PLINK version 1.07.28 PLINK performs all necessary data management tasks for GWAS, including generation of summary statistics for quality control, detection of population stratification, testing for statistical association at a single SNP, multiple SNPs and haplotype levels. P values were plotted against genomic location using Haploview version 4.2.29 Allele association with case or control status was assessed by Chi-squared (χ2) test and the expected and observed distributions of SNPs were determined by statistical analysis. The expected numbers of SNPs under the null hypothesis at the nominal significance threshold were generated.
Replication Analysis
GWAS was performed at Boston University on 1645 DNA samples isolated from African Americans with SCD who participated in the CSSCD, C-DATA, PUSH, and WALK-PHASST studies. Samples were genotyped using the Illumina Human610-Quad SNP array containing 588,451 SNPs, as previously published.30 Our replication analysis was performed in 249 CSSCD patients with low HbF levels and 243 CSSCD patients with high HbF levels, using the same definitions of low and high HbF groups defined in the initial case-control GWAS study. After quality control steps, 6489 SNPs had no genotype data, 16,737 markers were excluded based on the Hardy–Weinberg equilibrium test (P£0.001), and 73,966 SNPs failed the allele frequency test (MAF<0.05). After frequency and genotyping pruning, 498,706 SNPs were tested for association with HbF levels. There was approximately a 30% overlap of samples between the discovery and replication cohorts.
Haplotype analysis
Genotype data were inspected and used to construct HBB locus haplotypes using Haploview 4.2, which utilizes the EM algorithm to calculate linkage disequilibrium (LD) coefficients (D′) and to infer haplotypes. Examination of genotypes was conducted to determine the conformity with Hardy–Weinberg equilibrium before Haploview analysis. The SNPs with statistically significant departures (P<0.001) and minor allele frequencies <5% were excluded from the analysis. SNPs with strong LD (D′≥0.8) were arranged into haplotype blocks following Wang’s algorithm.31 Haplotype-tagging SNPs (tagSNPs) were selected on a block-by-block basis to identify SNPs that represent non-redundant information about genomic structure.
Two phase primary erythroid cultures
Primary erythroid progenitors were generated from human peripheral blood mononuclear cells purchased from Carter BloodCare (Fort Worth, TX, USA), in accordance with guidelines of the Institutional Review Board at the University of Texas at Dallas. Cell expansion and erythroid differentiation were conducted according to a procedure previously published by our lab.32 For gene silencing experiments, SMARTpool siRNAs for CBP (M-series, GE Dharmacon, Lafayette, CO, USA) and non-targeting scrambled siRNA were transfected on day 28 of primary cell culture using a CD34+ Nucleofector kit and Nucleofector device (Amaxa, Allendale, NJ, USA) as previously published by our laboratory.32
Reverse transcription-real time polymerase chain reaction (RT-qPCR)
To quantify gene transcription levels, total RNA was extracted using RNA Stat-60™ (TEL-TEST Inc., Friendswood, TX, USA) and used for RT-qPCR of γ-globin, β-globin and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA levels as reported previously.33 Primers to quantify Actin and CBP gene expression were purchased from Qiagen (Valencia, CA, USA).
Western blot
To determine CBP gene silencing in primary erythroid progenitors, western blot analysis was performed using total protein extracts and CBP antibody (SC-583, Santa Cruz Biotechnology, Dallas, TX, USA) and Actin antibody (MAB1501, Millpore, Billerica, MA, USA), as previously published.34
Fluorescence immunocytostaining
Monolayers of erythroid cells were obtained by centrifugation onto a glass slide. Immunocytostaining with anti-γ-globin fluorescein isothiocyanate antibody (Bethyl Laboratories, Montgomery, TX, USA) was conducted as previously published.32
Enzyme-linked immunosorbent assay
After the different treatments were conducted, total protein lysates were prepared and enzyme-linked immunosorbent assay (ELISA) performed as previously published.32
Statistical analysis
Each condition was repeated independently at least three times with triplicate samples for each experiment; data are shown as the mean ± standard error of the mean (SEM). The student’s t-test was applied to compare two experimental conditions with the threshold for statistical significance at P< 0.05.
Results
GWAS confirms HbF-associated SNPs in BCL11A
The ability of HbF to ameliorate the clinical severity of SCD and β-thalassemia has been well established.35 In other diseases when a phenotype associated with morbidity and mortality has been identified, using an extremes-of-phenotype (case-control) approach gives increased power to detect SNPs associated with genetic modifiers of clinical phenotype with smaller sample sizes.36,37 Therefore, we designed a GWAS using SCD patients with extremes of HbF levels to discover novel genetic modifiers involved in HBG1/HBG2 gene expression. Based on findings from the CSSCD where improved survival was observed in SCD patients with HbF≥8.6%,7 we chose this level as the cut-off for the case group. Using hematologic reference values for infants with SCD, the average HbF level at 5 years of age is 3.3%.38 Therefore, we chose HbF≤3.1% as the upper limit for the control group, reasoning patients with HbF in this range probably have not inherited a γ-globin gene modifier.
All DNA samples analyzed in the GWAS met quality control and were genotyped on the IlluminaOmni1-Quad chip. We collected data for 254 African Americans diagnosed with HbSS and HbSβ0-thalassemia and HbF≥8.6% (case, n=133), HbF≤£3.1% (controls, n=111), and 10 intermediate HbF subjects (Table 1). We excluded 45,142 monomorphic SNPs, 546 singletons, 1346 SNPs with significant departures from Hardy–Weinberg Equilibrium (P≤ 10.001), and 181,454 SNPs with minor allele frequencies <5%, from further consideration. After frequency and genotyping pruning, 833,760 SNPs were used in the GWAS analysis.
Table 1.
Phenotype data subjects used for GWAS analysis
| Gender | Low HbF (≤3.1%) | Intermediate HbF (≥3.2 & <8.6%) | High HbF (≥8.6%) |
|---|---|---|---|
| Female* (128) | 47 | 5 | 76 |
| Male* (126) | 64 | 5 | 57 |
| Totals | 111 | 10 | 133 |
| Mean HbF (%) | 1.84 | 3.95 | 13.83 |
| HbF Range (%) | 0 to 3.1 | 3.2 to 7 | 8.6 to 60 |
GWAS, genome-wide association study; HbF, fetal hemoglobin.
All subjects are African American; total number of females and males are shown in parenthesis.
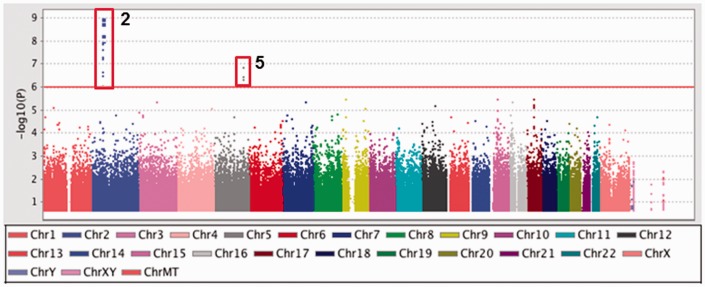
In order to determine allelic association of SNPs with HbF levels, we used the case-control association test implemented in PLINK version 1.07. Figure 1 shows the distribution of P-values (Manhattan plot) and SNPs on chromosome 2 and 5 significantly associated with HbF levels. A summary of the SNPs in the top four genes and two intergenic regions with genome-wide significance (P<1×10−6) associated with high HbF are shown in Table 2. The intronic region of the BCL11A gene contained 14 SNPs of which rs1896295, rs45606437, rs7584113, rs10172646, and rs1896294 have not been previously reported. This could reflect the differences in SNP allele frequency in African Americans compared to European or African populations. Conversely, these SNPs could have high LD with other SNPs validated in the region.
Figure 1.
Analysis of GWAS data by PLINK. The genotype data generated by the entire cohort were subjected to case-control association analysis using PLINK. Log-transformed P values (y axis) for the top 200,000 SNPs were plotted against chromosomal location (x-axis) using Haploview. The vertical red box shows significant SNPs in the BCL11A (Chr2) and SPARC (Chr5) genes
Table 2.
High-HbF associated SNPs identified by the case-control analysis
| CHR | Gene | SNP ID | Base position* | A1/A2 | Allele frequency (Case) | Allele frequency (Control) | Chi square | P value |
|---|---|---|---|---|---|---|---|---|
| 2 | BCL11A | rs1896295 | 60577590 | A/G | 0.375 | 0.1306 | 37.12 | 1.11E-09 |
| 2 | rs4671393 | 60574455 | A/G | 0.3788 | 0.1343 | 36.15 | 1.83E-09 | |
| 2 | rs6706648 | 60575544 | AG | 0.2895 | 0.5495 | 33.89 | 5.82E-09 | |
| 2 | rs766432 | 60573474 | C/A | 0.3759 | 0.1441 | 32.95 | 9.46E-09 | |
| 2 | rs1427407 | 60571547 | A/C | 0.3421 | 0.1216 | 32.09 | 1.48E-08 | |
| 2 | rs6738440 | 60575745 | G/A | 0.1955 | 0.4279 | 31.06 | 2.50E-08 | |
| 2 | rs45606437 | 60576613 | A/C | 0.2444 | 0.4865 | 31 | 2.58E-08 | |
| 2 | rs7584113 | 60574815 | A/G | 0.4135 | 0.1847 | 29.66 | 5.15E-08 | |
| 2 | rs7557939 | 60574851 | G/A | 0.4135 | 0.1847 | 29.66 | 5.15E-08 | |
| 2 | rs10195871 | 60574093 | A/G | 0.4173 | 0.1892 | 29.25 | 6.35E-08 | |
| 2 | rs10172646 | 60574261 | G/A | 0.4173 | 0.1892 | 29.25 | 6.35E-08 | |
| 2 | rs11886868 | 60573750 | G/A | 0.3947 | 0.1802 | 26.67 | 2.41E-07 | |
| 2 | rs1896294 | 60572578 | G/A | 0.3947 | 0.1818 | 26.08 | 3.28E-07 | |
| 2 | rs6709302 | 60581133 | A/G | 0.2632 | 0.4775 | 24.11 | 9.12E-07 | |
| 5 | SPARC | rs7719521 | 151029597 | A/C | 0.1241 | 0.3198 | 27.65 | 1.45E-07 |
| 5 | rs725937 | 151036904 | A/G | 0.188 | 0.3964 | 25.92 | 3.56E-07 | |
| 5 | rs6874468 | 151035888 | A/G | 0.02632 | 0.1532 | 25.3 | 4.92E-07 | |
| 17 | GJC1 | rs1122109 | 40240064 | A/C | 0.2348 | 0.07798 | 21.44 | 3.65E-06 |
| 17 | rs2108393 | 40236279 | A/G | 0.3308 | 0.1545 | 19.9 | 8.15E-06 | |
| 17 | EFTUD2 | rs16971033 | 40288000 | G/A | 0.2481 | 0.09091 | 20.43 | 6.17E-06 |
| 17 | rs2289673 | 40285496 | A/G | 0.2481 | 0.09091 | 20.43 | 6.17E-06 | |
| 17 | rs2304986 | 40283580 | A/G | 0.2727 | 0.1091 | 20.21 | 6.95E-06 | |
| 15 | Intergenic | rs776717 | 40188944 | G/A | 0.1203 | 0.2883 | 21.61 | 3.34E-06 |
| 9 | Intergenic | rs1339144 | 23208009 | A/G | 0.5337 | 0.3 | 21.55 | 3.44E-06 |
| 7 | JAZF1 | rs740127 | 28004900 | A/G | 0.2744 | 0.1171 | 18.52 | 1.68E-06 |
HbF, fetal hemoglobin; SNP, single nucleotide polymorphism; A1/A2, allele 1/allele 2.
hg18 genome coordinates.
Our GWAS data also identified SNPs in the SPARC, GJC1, EFTUD2 and JAZF1 genes associated with high HbF (Table 2). SPARC (secreted protein acidic and rich in cysteine) is a widely expressed profibrotic protein linked to human obesity, insulin resistance, and diabetic retinopathy.39 GJC1 (Gap Junction Protein Gamma 1; connexin45) belongs to a group of proteins that form intercellular channels to allow exchange of small molecules between cells.40 EFTUD2 (elongation factor Tu GTP binding domain containing 2) encodes a GTPase component of the spliceosome complex.41 The fourth HbF-associated SNP was identified in the zinc finger nuclear protein JAZF1 that functions as a gene transcription repressor.42
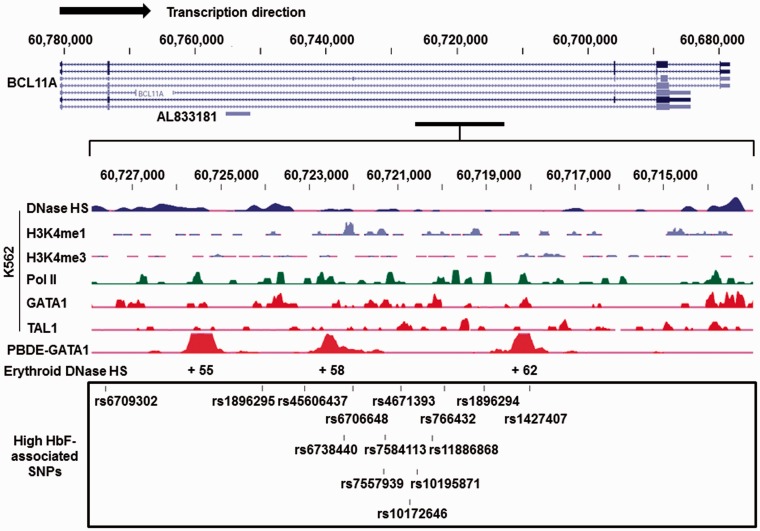
To gain insight into a possible regulatory role of the novel SNPs identified in our study, we performed an alignment across the BCL11A gene and functional genomics data generated by the ENCODE (Encyclopedia of DNA Elements) project. We focused our in silico analysis on a 15-kb region (hg19; 60,713,000 to 60,728,000) enriched with the HbF-associated SNPs (Figure 2). The ENCODE results for K562 cells showed low-level enhancer activity represented by the presence of histone H3K4me1 with minimal H3K4me3 mark detected. DNase hypersensitivity was established in this region and in vivo binding of the transcription factors GATA1 and TAL1. Moreover, three loci located at +55, +58 and +62 relative to the BCL11A gene transcription start site were detected with strong GATA1-associated enrichment in erythroblasts, corresponding to the erythroid enhancer region reported by Bauer et al.43 Two of the SNPs identified in our GWAS, including rs6738440 (+58 site) and rs1427407 (+62 site), could potentially alter recruitment of transcription factors to this region. Therefore, using a case-control GWAS of sickle cell patients with extremes of HbF levels, we confirmed previously reported SNPs and identified novel SNPs located in the erythroid enhancer of the BCL11A gene. In addition, other candidate genes that may be involved in HBG1/HBG2 gene regulation were revealed.
Figure 2.
Functional data for HbF-associated BCL11A SNPs located in an erythroid-specific enhancer region. ENCODE data for the BCL11A gene were analyzed using the (University of California, Santa Cruz) UCSC genome browser (genome.ucsc.edu). The BCL11A gene on Chromosome 2 is shown at the top with its major isoforms and AL833181 (a predicted non-coding transcript). A 15-kb region from 60,713,000 to 60,728,000 was enlarged to depict the functional ChIP-seq results in K562 cells (middle) including DNase hypersensitivity (DNase HS), H3K4me1, and H3K4me3 marks, and transcription factor RNA Polymerase II binding (Pol II), GATA1, and TAL1 binding to the region. ChIP-seq results for GATA1 in peripheral blood-derived erythroblasts (PBDE) demonstrate the +55, +58, and +62 erythroid-specific enhancer regions. The box on the bottom illustrates the top HbF-associated BCL11A SNPs identified in our GWAS indicated by the bars and respective dbSNP IDs
The case-control GWAS results presented herein was conducted with a limited sample size, and therefore we sought to replicate our findings with another dataset generated from 243 cases (HbF ≥ 8.6%) and 247 controls (HbF ≤ 3.1%) African American patients enrolled in the CSSCD.30 We replicated five SNPs in the intronic region of BCL11A (rs766432, rs10195871, rs6706648, rs6738440 and rs6709302) with genome-wide significance (Table S1). However, the BCL11A SNPs rs1896295, rs45606437, and rs1896294 were not replicated since probes for these and other gene mutations in the SPARC, JAZF1, GJC1 and EFTUD2 genes were not included on the Human610Y SNP array; this precluded our ability to replicate novel findings in our GWAS.
A permutation analysis showed that the replicated SNPs were significant only in the original partitioning of the dataset into cases and controls and not significantly associated with HbF levels in any other partitioning. This strongly suggests that the results are independent of any overlap between the original study and the replication in the CSSCD cohort.
Characterization of HBB locus haplotypes and tagSNPs
Transcriptional regulation of the HBB locus genes depends on trans-acting factors (activators and repressors), cis-acting elements (DNA motifs for binding trans-acting factors) and epigenetic changes in chromatin structure. The LCR provides enhancer activity for high-level sequential expression of the downstream β-like globin genes during developmentally regulated hemoglobin switching.12 Numerous studies support the notion that DNA sequences within 3–4 kb upstream or downstream of the HBG1/HBG2 genes influence transcription rates during erythroid maturation. We therefore performed a case-control analysis for SNPs in the HBB locus to identify divergences in genome structure between low HbF and high HbF groups. Based on the functional role of SNPs in the Gγ-globin promoter,33,44 we confirmed genotypes for rs2855121 and rs2855122 by TaqMan® assay.
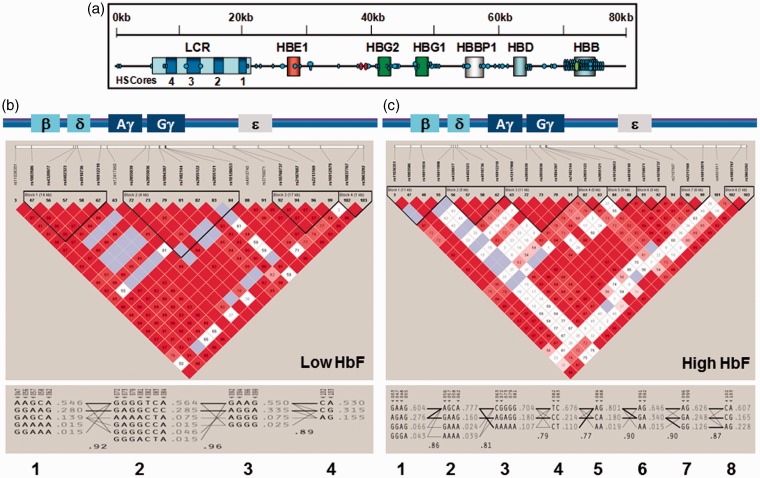
First, we performed LD and haplotype analysis of the HBB locus with genotype data generated by TaqMan and Illumina Omni-Quad 1 chip assays of the low HbF (101 subjects) and high HbF (103 subjects) groups using Haploview 4.2. There were 103 SNPs spanning the HBB locus (Figure 3(a)) producing four haplotype blocks for the low HbF cohort based upon the 4-gamete rule (Figure 3(b)). Overall there was strong LD across the HBB locus illustrated by the allelic D′values shown between the haplotype blocks (D′ > 0.8). In contrast, we observed decreased LD and more diverse haplotypes in the high HbF cohort (Figure 3(c)) with 8 haplotype blocks inferred. Interestingly, the region between rs10128653 and rs7482144 (-158XmnI-HBG2) displayed a lower allelic mean D′ < 0.8 in the high HbF group.
Figure 3.
High-density SNP mapping in the HBB locus. (a) Distribution of 103 SNPs in the HBB locus interrogated by the Illumina Omni1-Quad chip and TaqMan assay. Globin genes are indicated by boxes. The schematic is not drawn to scale. Abbreviations: LCR, locus control region; HS, hypersensitive site. Symbols: Black dot, Illumina Omni1-Quad SNPs; Red dot, TaqMan SNPs -1280GATA1 (rs2588121) and -1225CRE (rs2588122); Green dot, βS-globin SNP, rs334. Haploview was used to infer haplotypes from combined Illumina and TaqMan genotype data. (b) Haplotype analysis for 101 subjects with low HbF (mean HbF = 1.84%). A schematic of the HBB locus is shown on top. The degree of LD is defined by value of D′ and LOD (logarithm of the likelihood odds ratio), which is a measure of confidence in the D′value. Red indicates strong LD (LOD > 2, D′ = 1), white indicates no LD (LOD < 2, D′ < 1), pink (LOD = 2, D′ < 1) and blue (LOD < 2, D′ = 1) indicate intermediate LD. Unless indicated in each box, the D′ = 1 and by convention D′ is multiplied by 100. SNPs with strong LD define haplotype blocks (black triangles) and the size of the region in LD is shown in parentheses. (c) Haplotype analysis for 133 subjects with high HbF (mean HbF = 13.83%). The color scheme for LD pattern is as defined in panel (b)
Haplotype analysis also identifies a subset of SNPs which carry non-redundant information (tagSNPs) which can be used to define the diversity and total number of haplotypes in a given region. We observed 14 tagSNPs in the low HbF group with 3 unique to this group (Table S2) and 18 tagSNPs in the high HbF group of which 7 were unique. These results support a diversified genomic structure in sickle cell patients with high HbF providing potential insight into molecular mechanisms of persistent HbF expression in this group.
Identification of HbF-associated SNPs located in a known HbF silencing region
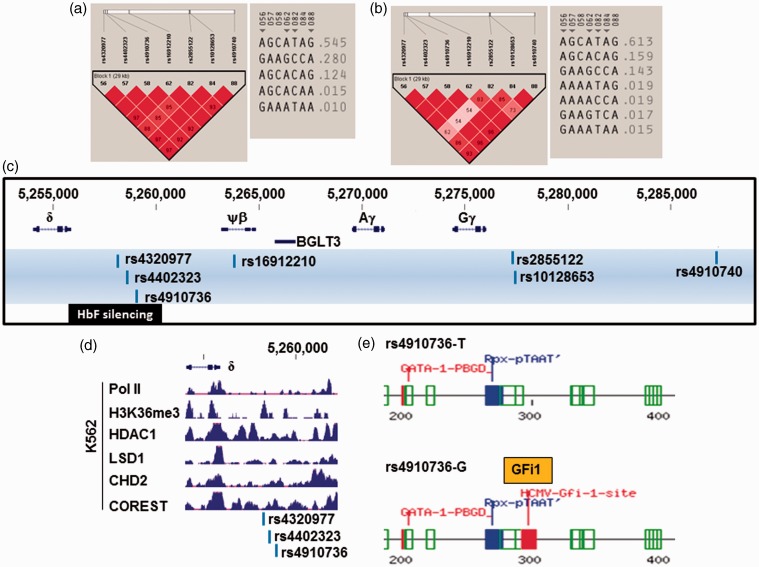
In order to identify individual SNPs associated with HbF, we performed a Chi-square (χ2) statistical analysis. We observed seven SNPs in the HBB locus with χ2 > 4.0 and P < 0.04 supporting association with high HbF (Table S3). Three SNPs were located within 30-kb of the intergenic region between ɛ-globin and Gγ-globin (rs4910740, rs10128653, and rs2855122) and four were located in the region between the ψβ-globin and δ-globin genes (rs16912210, rs4910736, rs4402323, and rs4320977). Historically, rs10128653 and rs2855122 have been implicated in forming a pre-G genetic framework that influences HbF expression in the context of HBB locus haplotypes defined in different African populations.45 These SNPs reflect changes at the DNA level, which may relate to persistent HbF expression; however, functional studies are needed to ascertain the biological relevance of these SNPs.
We next established haplotypes in the low and high HbF groups using the same 7 HbF-associated SNPs. Figure 4(a) illustrates the formation of one haplotype block with strong LD and five unique haplotypes at decreasing frequency inferred in the low HbF cohort. By contrast, analysis of the same SNPs in the high HbF group demonstrated one haplotype block with seven unique haplotypes inferred (Figure 4(b)). Interestingly, SNPs rs4910736, rs4402323, and rs4320977 located within the 3.5-kb HbF silencing region upstream of the δ-globin gene (Figure 4(c)) were identified by Sankaran et al. in δβ0-thalassemia patients.46 BCL11A was shown to bind this region to silence γ-globin expression by interacting with a transcription co-repressor complex in adult erythroid cells.
Figure 4.
HBB locus SNPs potentially involved in γ-globin gene silencing. (a) The LD profile for the low HbF group is shown on the left. The color scheme for LD pattern is as defined in Figure 3(b). The inferred haplotypes for these SNPs are depicted on the right. (b) The LD pattern and inferred haplotypes for the high HbF group. (c) The HbF-associated SNPs identified in the HBB locus between 3′ ɛ-globin and δ-globin, and the HbF silencing regions are shown. (d) K562 ChIP-seq data from the ENCODE project. Shown are the tracks for the proteins indicated between positions 5,254,000 to 5,262,140. The three HbF-associated SNPs are indicated at the bottom. (e) A 300-bp region around rs4910736 was analyzed by Tfsitescan software to identify predicted transcription factor binding sites. The results for the T allele and G allele are shown on the top and bottom, respectively. Binding sites for LIM homeodomain-containing proteins (Rpx-pTAAT) and GATA-1 were predicted regardless the presence of T or G allele indicative of the change of binding site by rs4910736. Various colors were used to represent E (expected) values for the prediction. The lower the E value, the higher the probability of transcription factor binding to the predicted site. Code: green – E values from 0.1 to 0.01; red – E values from 0.01 to 0.001; and blue – E values from 0.001 to 0.0001. Note the GFi1 predicted DNA motif has an E value in the same range as the predicted GATA-1 site
To assess the biological relevance of SNPs located in the HbF silencing region, we analyzed functional genomics data produced by the ENCODE project (Figure 4(d)). There was an absence of RNA polymerase II signal and increased binding of repressor complexes such as HDAC1, LSD1, CHD2 and COREST, supporting involvement of this region in transcription repression. Furthermore, we performed binding site prediction analysis using Tfsitescan.47 There was no difference in binding sites predicted for rs4402323 with either the C or T allele, whereas the G allele for rs4320977 created a binding site for the CCAAT/enhancer binding protein. Most interestingly, as shown in Figure 4(e), the T allele of rs4910736 was devoid of transcription factor binding sites, whereas the G allele created a Growth Factor Independence 1 (GFi1) DNA motif. GFi1 and its paralog GFi1B are critical transcriptional regulators for proliferation and maturation of hematopoietic stem cells.48–50 Although it is unclear whether GFi1/GFi1B plays a role in repressing γ-globin transcription, our analysis provides evidence for a potential GFi1 regulatory sequence in the HbF silencing region.
CBP represses γ-globin expression through the HBB locus tagSNP rs2855122
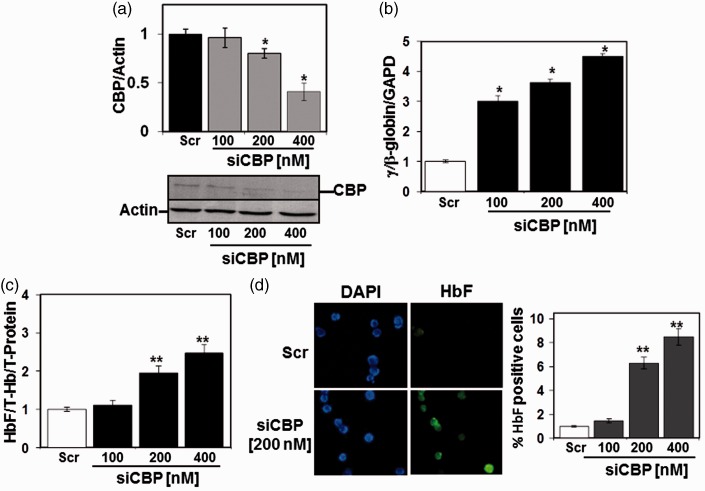
To assess the extent to which tagSNPs play a regulatory role in γ-globin transcription, we performed a functional analysis of SNP rs2855122 located in the cAMP response element (TGACGTCA) at nucleotide −1225 in the Gγ-globin promoter (G-CRE). We previously demonstrated that the trans-acting factor CREB1 binds the G-CRE to induce γ-globin expression through a p38 mitogen-activated protein kinase-dependent mechanism.51 Subsequent sequential-chromatin immunoprecipitation in K562 cells demonstrated that CREB1 and CBP bind simultaneously to the G-CRE region.51 Later studies from our lab showed a repressor role of CBP through competition with KLF4 binding in the proximal γ-globin promoter CACCC element in a luciferase reporter system.52 Therefore, we investigated the ability of CBP to repress γ-globin gene expression using siRNA gene knockdown in human erythroid progenitors at day 28 in culture when the γ-globin gene is silenced. We observed enhanced γ-globin transcription and HbF production with dose-dependent siRNA-mediated CBP gene knockdown (Figure 5(a) to (c)). In addition, the number of HbF positive cells was increased by flow cytometry supporting a repressor role of CBP in γ-globin gene expression (Figure 5(d)).
Figure 5.
CBP gene silencing induces γ-globin expression in primary erythroid cells. Using a two-phase liquid cell culture system, erythroid progenitors were generated and transfected at day 28 with siRNA against CBP and changes in γ-globin and HbF expression were quantified (see Materials and methods section). (a) RT-qPCR data for CBP mRNA normalized to Actin. Representative western blots for CBP and Actin expression after siRNA treatment are shown on the bottom; *P < 0.05. (b) RT-qPCR analysis of γ-globin, β-globin and GAPDH mRNA showing the fold change of γ-globin expression under the different conditions. (c) ELISA was performed with HbF and total hemoglobin (Hb) antibodies. Raw data were normalized by total hemoglobin (T-Hb) and total protein (T-Protein) for each sample. (d) Immunocytostaining with anti-γ-globin fluorescein isothiocyanate antibody demonstrated an increase in HbF-producing cells (photomicrograph on left). Quantification of the % HbF-positive cells under the different conditions is shown in the graph; **P < 0.001
Discussion
Mutations in the HBB locus, including those located in the HBG1/HBG2 gene promoters,53,54 and quantitative trait loci at distant sites on chromosomes 2, 6, 8, and the X chromosome have been associated with high HbF expression in humans.13–19,23,25 GWAS in several populations have identified SNPs in the BCL11A gene and the HMIP and −158XmnI-HBG2 regions associated with HbF expression. In African American patients with SCD, the BCL11A SNPs show the strongest effect on HBG1/HBG2 expression, accounting for approximately 11% of HbF variance.23,25 This suggests, however, that other genetic modifiers of HbF remain to be discovered. In our study, we performed GWAS using African American sickle cell patients and a case (high HbF)-control (low HbF) experimental design. We identified multiple SNPs in BCL11A, replicating previous studies.18,19,23,25 BCL11A is a stage-specific repressor of γ-globin expression in human primary erythroid progenitors55 and mediates HBG1/HBG2 gene silencing in adult sickle cell transgenic mice.56 The gene silencing effect involves reconfiguration of the HBB locus through interactions of BCL11A with GATA1, FOG1, and SOX6, as well as the NuRD, histone deacetylase remodeling complex.55,57 Genetic studies in a Maltese family demonstrated that KLF1 p.K288X carriers had reduced BCL11A gene expression58 and elevated HbF levels. Subsequent studies established the ability of KLF1 to activate BCL11A in human erythroid progenitors and transgenic mice.58,59 Recently, an erythroid-specific enhancer was discovered in the second intron of BCL11A43; four SNPs (rs1427407, rs6706648, rs6738440, and rs7606173) were associated with HbF levels in SCD patients of African and Arab–Indian descent.60,61 Therefore, the erythroid enhancer is an attractive DNA element for lineage-specific BCL11A gene silencing as a strategy for gene therapy in SCD and β-thalassemia.
Genome-wide studies by Thein and colleagues identified the HMIP region on chromosome 6q23 as a modifier of HbF production in a European population.17,62 A meta-analysis of GWAS data from over 2000 African American sickle cell patients replicated the association of HbF levels with the HMIP region.25 More recently, SNPs in the HMIP region were shown to be associated with HbF levels in SCD patients from Tanzania,26 Northern Brazil, and Cameroon.63,64 The ability of MYB to regulate HbF production in erythroid cells65 has been demonstrated. Subsequently, Stadhouders et al. described an erythroid-specific enhancer located in the HMIP region that regulates MYB expression.66 Furthermore, our group demonstrated differences in haplotype structures in the core haplotype block 2 in the HMIP locus among European, African, and African American SCD subjects.67 In our current study, we did not observe SNPs in the HMIP region associated with HbF level, most likely due to small sample size.
Studies in healthy individuals with elevated HbF identified SNPs in the promoters of the HBG1 and HBG2 genes that produce non-deletional hereditary persistence of HbF; however, molecular mechanisms or trans-activating factors that bind these mutations have not been identified. The most extensively studied is the XmnI mutation (C to T) at position −158 Gγ-globin.17,25,26 Individuals carrying this mutation (rs7482144) have a delay in the Gγ- to Aγ-globin switch.68 Similar to other GWAS findings in sickle cell patients of African descent,23,25,26 we did not observe an association of the −158XmnI-HBG2 mutation with HbF levels in our cohort.
In addition to known loci, we identified SNPs in the SPARC, GJC1, EFTUD2, and JAZF1 genes associated with HbF expression. Multiple SNPs were found in the SPARC gene, which is expressed in bone marrow stromal and hematopoietic cells. Gene expression analysis of stem cells isolated from patients with −5q syndrome showed decreased SPARC expression among other proteins associated with lower platelet counts and impaired ability to form erythroid burst-forming units.69 Recent studies in SPARC-null mice demonstrated that this protein plays a crucial role in the regulation of early B lymphopoiesis70; however, SPARC has not been demonstrated to regulate globin gene expression. We also identified the transcription repressor JAZF1 associated with high HbF levels, although at a lower genome-wide significance. SNPs in JAZF1 have been associated with type 2 diabetes through a non-coding signal in the first intron, which regulates islet cell function.71
The case-control GWAS design presented herein was conducted with a limited sample size. We therefore sought to replicate our findings with another dataset of 243 cases (HbF ≥ 8.6%) and 247 controls (HbF ≤ 3.1%) drawn from African Americans enrolled in the CSSCD.30 Five SNPs in the intronic region of BCL11A including rs766432 and rs6706648 were identified with genome-wide significance. However, BCL11A SNPs identified in other GWAS studies18,19 associated with HbF were not replicated most likely due to a lower SNP density on this array. Furthermore, early GWAS arrays were biased towards European and Asian populations prior to the publication of the HapMap Phase 2.72 The Illumina OmniQuad1.0 array used in our GWAS study has a higher density of SNP coverage of the human genome and contains SNPs unique to individuals of African descent. These differences might provide a basis for our ability to discover unique HbF-associated SNPs; however, additional replication studies with a larger sample size are needed to corroborate a role of the SPARC, GJC1, EFTUD2, and JAZF1 genes in γ-globin regulation.
In contrast to other published GWAS studies, we performed a detailed analysis of the HBB locus to define haplotypes associated with high and low HbF expression. Historically, five major β-haplotypes, including Senegal, Benin, Central African Republic (Bantu), Cameroon, and Asian (Indian/Saudi-Arabian),35,54,73–76 have been defined in African SCD populations; however, these haplotypes are not predictive of HbF levels. We recently published data demonstrating that β-haplotypes generated using traditional methods such as restriction fragment length polymorphisms are insufficient to define the genomic structure in the HBB locus.77 These findings were germane to our experimental design using a case-control approach to identify SNPs in the HBB locus which did not reach significance at the genome-wide level.
When comparing haplotypes in the case-control groups, we observed more inferred haplotypes in the high-HbF group, supporting lower LD in the HBB locus, particularly in the region between the Gγ-globin and δ-globin genes recently demonstrated to be involved in γ-globin regulation through long-range chromatin looping mediated by transcription factors and non-coding RNA transcripts.57,78,79 Kiefer et al. discovered the BGLT3, non-coding RNA transcript encoded by DNA sequences between the Aγ- and ψβ-globin genes primarily expressed in γ-globin producing cells.78 By contrast, BGLT3 is not transcribed in late erythropoiesis when β-globin expression is high due to the binding of a repressor complex consisting of ETO2 and BCL11A. Subsequently, Xu et al. demonstrate that a repressor complex consisting of BCL11A, HDAC1, LSD1, and CoREST binds the inter-genetic region between the Aγ- and δ-globin genes.79 Sankaran and colleagues identified a putative HbF silencing region 3.5 kb upstream of δ-globin where the BCL11A repressor complex binds.46 Additional studies localized the HbF silencing region to a 2.4-kb sequence outside the BCL11A binding region supporting other mechanisms of γ-globin regulation.80
Our HBB locus analysis identified the novel SNP rs4910736 that creates a GFi1 binding site. The nuclear proteins GFi1 and GFi1B can recruit LSD1 and CoREST to the targeted chromatin region and interact with HDAC1 and G9a (H3K9 methyltransferase). The formation of protein complexes between GFi1/GFi1B mediates histone modifications associated with gene transcription silencing and regulation of hematopoiesis.48–50,81 Studies by Vassen and colleagues demonstrated increased transcription of the embryonic and fetal globin genes and delayed erythroid maturation in Gfi1B-null mice82 providing evidence that GFi1/GFi1B are involved in murine globin gene regulation.
Lastly, we performed functional analysis of the HBB locus SNP rs2855122 located in G-CRE; we recently characterized a multi-protein complex composed of trans-activators including CREB1 and ATF2 and the repressor HDAC1 that bind the G-CRE.34 Studies conducted herein with the CREB1 binding partner CBP, support a repressor role of CBP in γ-globin expression. However, additional in vivo functional genomics studies are warranted to fully define molecular mechanisms of γ-globin gene regulation mediated by novel HbF-associated SNPs identified in our study.
Acknowledgements
This work was supported by grant R56DK083921 from the National Institute of Diabetes Digestive and Kidney Diseases to Dr. Betty Pace and grants HL87681 and HL101212 from the National Heart Lung and Blood Institute to Dr. Martin Steinberg. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Authors’ contributions
All authors participated in the design of the GWAS, writing, and review of the manuscript; LL conducted the majority of the experiments including the haplotype analysis and functional studies in primary erythroid cultures. AP conducted genotype data pruning and genotype-phenotype analysis. MS and LD processed the DNA samples to generate the genotyping raw data pruning. MS and PS contributed genotype data and assisted for the replication analysis; CH and SB contributed to study design and DNA samples screening; BSP designed and supervised all experimental studies and contributed to editing and writing the manuscript.
Declaration of conflicting interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
References
- 1.Steinberg MH, Forget BG, Higgs DR, Weatherall DJ (eds) Disorders of hemoglobin: genetics, pathophysiology, and clinical management. 2nd ed. Cambridge, UK: Cambridge University Press, 2009.
- 2.Galanello R, Origa R. Beta-thalassemia. Orphanet J Rare Dis 2010; 5: 11–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akinsheye I, Solovieff N, Ngo D, Malek A, Sebastiani P, Steinberg MH, Chui DH. Fetal hemoglobin in sickle cell anemia: molecular characterization of the unusually high fetal hemoglobin phenotype in African Americans. Am J Hematol 2012; 87: 217–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Steinberg MH, Sebastiani P. Genetic modifiers of sickle cell disease. Am J Hematol 2012; 87: 795–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gaston M, Smith J, Gallagher D, Flournoy-Gill Z, West S, Bellevue R, Farber M, Grover R, Koshy M, Ritchey AK, et al. Recruitment in the Cooperative Study of Sickle Cell Disease (CSSCD). Control Clin Trial 1987; 8: 131S–40S. [DOI] [PubMed] [Google Scholar]
- 6.Platt OS, Thorington BD, Brambilla DJ, Milner PF, Rosse WF, Vichinsky E, Kinney TR. Pain in sickle cell disease. Rates and risk factors. N Engl J Med 1991; 325: 11–6. [DOI] [PubMed] [Google Scholar]
- 7.Platt OS, Brambilla DJ, Rosse WF, Milner PF, Castro O, Steinberg MH, Klug PP. Mortality in sickle cell disease. Life expectancy and risk factors for early death. N Engl J Med 1994; 330: 1639–44. [DOI] [PubMed] [Google Scholar]
- 8.Stamatoyannopoulos G, Grosveld F. Hemoglobin switching. In: Stamatoyannopoulos G, Majerus PW, Perlmutter RM, Varmus H (eds) The molecular basis of blood diseases, 3rd ed Philadelphia: Saunders, 2001. [Google Scholar]
- 9.Forrester WC, Thompson C, Elder JT, Groudine M. A developmentally stable chromatin structure in the human beta-globin gene cluster. Proc Natl Acad Sci U S A 1986; 83: 1359–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tuan D, Abeliovich A, Lee-Oldham M, Lee D. Identification of regulatory elements of human beta-like globin genes. Prog Clin Biol Res 1987; 251: 211–20. [PubMed] [Google Scholar]
- 11.Engel JD, Tanimoto K. Looping, linking, and chromatin activity: new insights into beta-globin locus regulation. Cell 2000; 100: 499–502. [DOI] [PubMed] [Google Scholar]
- 12.Stamatoyannopoulos G. Control of globin gene expression during development and erythroid differentiation. Exp Hematol 2005; 33: 259–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang YP, Maier-Redelsperger M, Smith KD, Contu L, Ducroco R, de Montalembert M, Belloy M, Elion J, Dover GJ, Girot R. The relative importance of the X-linked FCP locus and beta-globin haplotypes in determining haemoglobin F levels: a study of SS patients homozygous for beta S haplotypes. Br J Haematol 1997; 96: 806–14. [DOI] [PubMed] [Google Scholar]
- 14.Garner C, Silver N, Best S, Menzel S, Martin C, Spector TD, Thein SL. Quantitative trait locus on chromosome 8q influences the switch from fetal to adult hemoglobin. Blood 2004; 104: 2184–6. [DOI] [PubMed] [Google Scholar]
- 15.Craig JE, Rochette J, Fisher CA, Weatherall DJ, Marc S, Lathrop GM, Demenais F, Thein S. Dissecting the loci controlling fetal haemoglobin production on chromosomes 11p and 6q by the regressive approach. Nat Genet 1996; 12: 58–64. [DOI] [PubMed] [Google Scholar]
- 16.Wyszynski DF, Baldwin CT, Cleves MA, Amirault Y, Nolan VG, Farrell JJ, Bisbee A, Kutlar A, Farrer LA, Steinberg MH. Polymorphisms near a chromosome 6q QTL area are associated with modulation of fetal hemoglobin levels in sickle cell anemia. Cell Mol Biol (Noisy-le-grand) 2004; 50: 23–33. [PubMed] [Google Scholar]
- 17.Thein SL, Menzel S, Peng X, Best S, Jiang J, Close J, Silver N, Gerovasilli A, Ping C, Yamaguchi M, Wahlberg K, Ulug P, Spector TD, Garner C, Matsuda F, Farrall M, Lathrop M. Intergenic variants of HBS1L-MYB are responsible for a major quantitative trait locus on chromosome 6q23 influencing fetal hemoglobin levels in adults. Proc Natl Acad Sci U S A 2007; 104: 11346–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Menzel S, Garner C, Gut I, Matsuda F, Yamaguchi M, Heath S, Foglio M, Zelenika D, Boland A, Rooks H, Best S, Spector TD, Farrall M, Lathrop M, Thein SL. A QTL influencing F cell production maps to a gene encoding a zinc-finger protein on chromosome 2p15. Nat Genet 2007; 39: 1197–9. [DOI] [PubMed] [Google Scholar]
- 19.Uda M, Galanello R, Sanna S, Lettre G, Sankaran VG, Chen W, Usala G, Busonero F, Maschio A, Albai G, Piras MG, Sestu N, Lai S, Dei M, Mulas A, Crisponi L, Naitza S, Asunis I, Deiana M, Nagaraja R, Perseu L, Satta S, Cipollina MD, Sollaino C, Moi P, Hirschhorn JN, Orkin SH, Abecasis GR, Schlessinger D, Cao A. Genome-wide association study shows BCL11A associated with persistent fetal hemoglobin and amelioration of the phenotype of beta-thalassemia. Proc Natl Acad Sci U S A 2008; 105: 1620–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ballas SK, Talacki CA, Adachi K, Schwartz E, Surrey S, Rappaport E. The Xmn I site (-158, C—-T) 5’ to the G gamma gene: correlation with the Senegalese haplotype and G gamma globin expression. Hemoglobin 1991; 15: 393–405. [DOI] [PubMed] [Google Scholar]
- 21.Sebastiani P, Wang L, Nolan VG, Melista E, Ma Q, Baldwin CT, Steinberg MH. Fetal hemoglobin in sickle cell anemia: Bayesian modeling of genetic associations. Am J Hematol 2008; 83: 189–95. [DOI] [PubMed] [Google Scholar]
- 22.Sedgewick AE, Timofeev N, Sebastiani P, So JC, Ma ES, Chan LC, Fucharoen G, Fucharoen S, Barbosa CG, Vardarajan BN, Farrer LA, Baldwin CT, Steinberg MH, Chui DH. BCL11A is a major HbF quantitative trait locus in three different populations with beta-hemoglobinopathies. Blood Cells Mol Dis 2008; 41: 255–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Solovieff N, Milton JN, Hartley SW, Sherva R, Sebastiani P, Dworkis DA, Klings ES, Farrer LA, Garrett ME, Ashley-Koch A, Telen MJ, Fucharoen S, Ha SY, Li CK, Chui DH, Baldwin CT, Steinberg MH. Fetal hemoglobin in sickle cell anemia: genome-wide association studies suggest a regulatory region in the 5’ olfactory receptor gene cluster. Blood 2010; 115: 1815–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lettre G, Sankaran VG, Bezerra MA, Araujo AS, Uda M, Sanna S, Cao A, Schlessinger D, Costa FF, Hirschhorn JN, Orkin SH. DNA polymorphisms at the BCL11A, HBS1L-MYB, and beta-globin loci associate with fetal hemoglobin levels and pain crises in sickle cell disease. Proc Natl Acad Sci U S A 2008; 105: 11869–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bae HT, Baldwin CT, Sebastiani P, Telen MJ, Ashley-Koch A, Garrett M, Hooper WC, Bean CJ, Debaun MR, Arking DE, Bhatnagar P, Casella JF, Keefer JR, Barron-Casella E, Gordeuk V, Kato GJ, Minniti C, Taylor J, Campbell A, Luchtman-Jones L, Hoppe C, Gladwin MT, Zhang Y, Steinberg MH. Meta-analysis of 2040 sickle cell anemia patients: BCL11A and HBS1L-MYB are the major modifiers of HbF in African Americans. Blood 2012; 120: 1961–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mtatiro SN, Singh T, Rooks H, Mgaya J, Mariki H, Soka D, Mmbando B, Msaki E, Kolder I, Thein SL, Menzel S, Cox SE, Makani J, Barrett JC. Genome wide association study of fetal hemoglobin in sickle cell anemia in Tanzania. PLoS One 2014; 9: e111464–e111464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Milton JN, Rooks H, Drasar E, McCabe EL, Baldwin CT, Melista E, Gordeuk VR, Nouraie M, Kato GR, Minniti C, Taylor J, Campbell A, Luchtman-Jones L, Rana S, Castro O, Zhang Y, Thein SL, Sebastiani P, Gladwin MT, Steinberg MH. Genetic determinants of haemolysis in sickle cell anaemia. Br J Haematol 2013; 161: 270–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81: 559–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 2005; 21: 263–5. [DOI] [PubMed] [Google Scholar]
- 30.Milton JN, Gordeuk VR, Taylor JGt, Gladwin MT, Steinberg MH, Sebastiani P. Prediction of fetal hemoglobin in sickle cell anemia using an ensemble of genetic risk prediction models. Circ Cardiovasc Genet 2014; 7: 110–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang N, Akey JM, Zhang K, Chakraborty R, Jin L. Distribution of recombination crossovers and the origin of haplotype blocks: the interplay of population history, recombination, and mutation. Am J Hum Genet 2002; 71: 1227–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muralidhar SA, Ramakrishnan V, Kalra IS, Li W, Pace BS. Histone deacetylase 9 activates gamma-globin gene expression in primary erythroid cells. J Biol Chem 2011; 286: 2343–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sangerman J, Lee MS, Yao X, Oteng E, Hsiao CH, Li W, Zein S, Ofori-Acquah SF, Pace BS. Mechanism for fetal hemoglobin induction by histone deacetylase inhibitors involves gamma-globin activation by CREB1 and ATF-2. Blood 2006; 108: 3590–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu L, Karmakar S, Dhar R, Mahajan M, Choudhury A, Weissman S, Pace BS. Regulation of Ggamma-globin gene by ATF2 and its associated proteins through the cAMP-response element. PLoS One 2013; 8: e78253–e78253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stamatoyannopoulos G. The molecular basis of blood diseases, 3rd ed Philadelphia, PA: W.B. Saunders, 2001. [Google Scholar]
- 36.Daly AK. Candidate gene case-control studies. Pharmacogenomics 2003; 4: 127–39. [DOI] [PubMed] [Google Scholar]
- 37.Guey LT, Kravic J, Melander O, Burtt NP, Laramie JM, Lyssenko V, Jonsson A, Lindholm E, Tuomi T, Isomaa B, Nilsson P, Almgren P, Kathiresan S, Groop L, Seymour AB, Altshuler D, Voight BF. Power in the phenotypic extremes: a simulation study of power in discovery and replication of rare variants. Genetic Epidemiol 2011; 35: 236–46. [DOI] [PubMed] [Google Scholar]
- 38.Brown AK, Sleeper LA, Miller ST, Pegelow CH, Gill FM, Waclawiw MA. Reference values and hematologic changes from birth to 5 years in patients with sickle cell disease. Cooperative Study of Sickle Cell Disease. Arch Pediatr Adolesc Med 1994; 148: 796–804. [DOI] [PubMed] [Google Scholar]
- 39.Kos K, Wilding JP. SPARC: a key player in the pathologies associated with obesity and diabetes. Nat Rev Endocrinol 2010; 6: 225–35. [DOI] [PubMed] [Google Scholar]
- 40.Kanter HL, Saffitz JE, Beyer EC. Molecular cloning of two human cardiac gap junction proteins, connexin40 and connexin45. J Mol Cell Cardiol 1994; 26: 861–8. [DOI] [PubMed] [Google Scholar]
- 41.Lines MA, Huang L, Schwartzentruber J, Douglas SL, Lynch DC, Beaulieu C, Guion-Almeida ML, Zechi-Ceide RM, Gener B, Gillessen-Kaesbach G, Nava C, Baujat G, Horn D, Kini U, Caliebe A, Alanay Y, Utine GE, Lev D, Kohlhase J, Grix AW, Lohmann DR, Hehr U, Bohm D, Majewski J, Bulman DE, Wieczorek D, Boycott KM. Haploinsufficiency of a spliceosomal GTPase encoded by EFTUD2 causes mandibulofacial dysostosis with microcephaly. Am J Hum Genet 2012; 90: 369–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fogarty MP, Panhuis TM, Vadlamudi S, Buchkovich ML, Mohlke KL. Allele-specific transcriptional activity at type 2 diabetes-associated single nucleotide polymorphisms in regions of pancreatic islet open chromatin at the JAZF1 locus. Diabetes 2013; 62: 1756–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bauer DE, Kamran SC, Lessard S, Xu J, Fujiwara Y, Lin C, Shao Z, Canver MC, Smith EC, Pinello L, Sabo PJ, Vierstra J, Voit RA, Yuan GC, Porteus MH, Stamatoyannopoulos JA, Lettre G, Orkin SH. An erythroid enhancer of BCL11A subject to genetic variation determines fetal hemoglobin level. Science 2013; 342: 253–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kodeboyina S, Balamurugan P, Liu L, Pace BS. cJun modulates Ggamma-globin gene expression via an upstream cAMP response element. Blood Cells Mol Dis 2010; 44: 7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pissard S, Beuzard Y. A potential regulatory region for the expression of fetal hemoglobin in sickle cell disease. Blood 1994; 84: 331–8. [PubMed] [Google Scholar]
- 46.Sankaran VG, Xu J, Byron R, Greisman HA, Fisher C, Weatherall DJ, Sabath DE, Groudine M, Orkin SH, Premawardhena A, Bender MA. A functional element necessary for fetal hemoglobin silencing. N Engl J Med 2011; 365: 807–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ghosh D. Object-oriented transcription factors database (ooTFD). Nucleic Acids Res 2000; 28: 308–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moroy T, Vassen L, Wilkes B, Khandanpour C. From cytopenia to leukemia: the role of Gfi1 and Gfi1b in blood formation. Blood 2015; 126: 2561–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van der Meer LT, Jansen JH, van der Reijden BA. Gfi1 and Gfi1b: key regulators of hematopoiesis. Leukemia 2010; 24: 1834–43. [DOI] [PubMed] [Google Scholar]
- 50.Zeng H, Yucel R, Kosan C, Klein-Hitpass L, Moroy T. Transcription factor Gfi1 regulates self-renewal and engraftment of hematopoietic stem cells. EMBO J 2004; 23: 4116–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ramakrishnan VPB. Regulation of γ-globin gene expression involves signaling through the p38 MAPK/CREB1 pathway. Blood Cells Mol Dis 2011; 47: 12–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kalra IS, Alam MM, Choudhary PK, Pace BS. Kruppel-like Factor 4 activates HBG gene expression in primary erythroid cells. Br J Haematol 2011; 154: 248–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miller BA, Salameh M, Ahmed M, Olivieri N, Antognetti G, Orkin SH, Huisman TH, Nathan DG. Analysis of hemoglobin F production in Saudi Arabian families with sickle cell anemia. Blood 1987; 70: 716–20. [PubMed] [Google Scholar]
- 54.Miller BA, Olivieri N, Salameh M, Ahmed M, Antognetti G, Huisman TH, Nathan DG, Orkin SH. Molecular analysis of the high-hemoglobin-F phenotype in Saudi Arabian sickle cell anemia. N Engl J Med 1987; 316: 244–50. [DOI] [PubMed] [Google Scholar]
- 55.Sankaran VG, Menne TF, Xu J, Akie TE, Lettre G, Van Handel B, Mikkola HK, Hirschhorn JN, Cantor AB, Orkin SH. Human fetal hemoglobin expression is regulated by the developmental stage-specific repressor BCL11A. Science 2008; 322: 1839–42. [DOI] [PubMed] [Google Scholar]
- 56.Xu J, Peng C, Sankaran VG, Shao Z, Esrick EB, Chong BG, Ippolito GC, Fujiwara Y, Ebert BL, Tucker PW, Orkin SH. Correction of sickle cell disease in adult mice by interference with fetal hemoglobin silencing. Science 2011; 334: 993–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu J, Sankaran VG, Ni M, Menne TF, Puram RV, Kim W, Orkin SH. Transcriptional silencing of {gamma}-globin by BCL11A involves long-range interactions and cooperation with SOX6. Genes Dev 2010; 24: 783–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Borg J, Papadopoulos P, Georgitsi M, Gutierrez L, Grech G, Fanis P, Phylactides M, Verkerk AJ, van der Spek PJ, Scerri CA, Cassar W, Galdies R, van Ijcken W, Ozgur Z, Gillemans N, Hou J, Bugeja M, Grosveld FG, von Lindern M, Felice AE, Patrinos GP, Philipsen S. Haploinsufficiency for the erythroid transcription factor KLF1 causes hereditary persistence of fetal hemoglobin. Nat Genet 2010; 42: 801–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou D, Liu K, Sun CW, Pawlik KM, Townes TM. KLF1 regulates BCL11A expression and gamma- to beta-globin gene switching. Nat Genet 2010; 42: 742–4. [DOI] [PubMed] [Google Scholar]
- 60.Pule GD, Ngo Bitoungui VJ, Chetcha Chemegni B, Kengne AP, Antonarakis S, Wonkam A. Association between variants at BCL11A erythroid-specific enhancer and fetal hemoglobin levels among sickle cell disease patients in Cameroon: implications for future therapeutic interventions. OMICS 2015; 19: 627–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sebastiani P, Farrell JJ, Alsultan A, Wang S, Edward HL, Shappell H, Bae H, Milton JN, Baldwin CT, Al-Rubaish AM, Naserullah Z, Al-Muhanna F, Alsuliman A, Patra PK, Farrer LA, Ngo D, Vathipadiekal V, Chui DH, Al-Ali AK, Steinberg MH. BCL11A enhancer haplotypes and fetal hemoglobin in sickle cell anemia. Blood Cells Mol Dis 2015; 54: 224–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Creary LE, Ulug P, Menzel S, McKenzie CA, Hanchard NA, Taylor V, Farrall M, Forrester TE, Thein SL. Genetic variation on chromosome 6 influences F cell levels in healthy individuals of African descent and HbF levels in sickle cell patients. PLoS One 2009; 4: e4218–e4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wonkam A, Ngo Bitoungui VJ, Vorster AA, Ramesar R, Cooper RS, Tayo B, Lettre G, Ngogang J. Association of variants at BCL11A and HBS1L-MYB with hemoglobin F and hospitalization rates among sickle cell patients in Cameroon. PLoS One 2014; 9: e92506–e92506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cardoso GL, Diniz IG, Silva AN, Cunha DA, Silva Junior JS, Uchoa CT, Santos SE, Trindade SM, Mdo Cardoso S, Guerreiro JF. DNA polymorphisms at BCL11A, HBS1L-MYB and Xmn1-HBG2 site loci associated with fetal hemoglobin levels in sickle cell anemia patients from Northern Brazil. Blood Cells Mol Dis 2014; 53: 176–9. [DOI] [PubMed] [Google Scholar]
- 65.Jiang J, Best S, Menzel S, Silver N, Lai MI, Surdulescu GL, Spector TD, Thein SL. cMYB is involved in the regulation of fetal hemoglobin production in adults. Blood 2006; 108: 1077–83. [DOI] [PubMed] [Google Scholar]
- 66.Stadhouders R, Aktuna S, Thongjuea S, Aghajanirefah A, Pourfarzad F, van Ijcken W, Lenhard B, Rooks H, Best S, Menzel S, Grosveld F, Thein SL, Soler E. HBS1L-MYB intergenic variants modulate fetal hemoglobin via long-range MYB enhancers. J Clin Invest 2014; 124: 1699–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Menzel S, Rooks H, Zelenika D, Mtatiro SN, Gnanakulasekaran A, Drasar E, Cox S, Liu L, Masood M, Silver N, Garner C, Vasavda N, Howard J, Makani J, Adekile A, Pace B, Spector T, Farrall M, Lathrop M, Thein SL. Global genetic architecture of an erythroid quantitative trait locus, HMIP-2. Ann Hum Genet 2014; 78: 434–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Peri KG, Gagnon J, Gagnon C, Bard H. Association of -158 (C–>T) (XmnI) DNA polymorphism in G gamma-globin promoter with delayed switchover from fetal to adult hemoglobin synthesis. Pediatr Res 1997; 41: 214–7. [DOI] [PubMed] [Google Scholar]
- 69.Lehmann S, O’Kelly J, Raynaud S, Funk SE, Sage EH, Koeffler HP. Common deleted genes in the 5q- syndrome: thrombocytopenia and reduced erythroid colony formation in SPARC null mice. Leukemia 2007; 21: 1931–6. [DOI] [PubMed] [Google Scholar]
- 70.Luo Z, Zhou Y, Luo P, Zhao Q, Xiao N, Yu Y, Yan Q, Lu G, Cheng L. SPARC deficiency affects bone marrow stromal function, resulting in impaired B lymphopoiesis. J Leukoc Biol 2014; 96: 73–82. [DOI] [PubMed] [Google Scholar]
- 71.Zeggini E, Scott LJ, Saxena R, Voight BF, Marchini JL, Hu T, de Bakker PI, Abecasis GR, Almgren P, Andersen G, Ardlie K, Bostrom KB, Bergman RN, Bonnycastle LL, Borch-Johnsen K, Burtt NP, Chen H, Chines PS, Daly MJ, Deodhar P, Ding CJ, Doney AS, Duren WL, Elliott KS, Erdos MR, Frayling TM, Freathy RM, Gianniny L, Grallert H, Grarup N, Groves CJ, Guiducci C, Hansen T, Herder C, Hitman GA, Hughes TE, Isomaa B, Jackson AU, Jorgensen T, Kong A, Kubalanza K, Kuruvilla FG, Kuusisto J, Langenberg C, Lango H, Lauritzen T, Li Y, Lindgren CM, Lyssenko V, Marvelle AF, Meisinger C, Midthjell K, Mohlke KL, Morken MA, Morris AD, Narisu N, Nilsson P, Owen KR, Palmer CN, Payne F, Perry JR, Pettersen E, Platou C, Prokopenko I, Qi L, Qin L, Rayner NW, Rees M, Roix JJ, Sandbaek A, Shields B, Sjogren M, Steinthorsdottir V, Stringham HM, Swift AJ, Thorleifsson G, Thorsteinsdottir U, Timpson NJ, Tuomi T, Tuomilehto J, Walker M, Watanabe RM, Weedon MN, Willer CJ, Illig T, Hveem K, Hu FB, Laakso M, Stefansson K, Pedersen O, Wareham NJ, Barroso I, Hattersley AT, Collins FS, Groop L, McCarthy MI, Boehnke M, Altshuler D. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet 2008; 40: 638–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, Belmont JW, Boudreau A, Hardenbol P, Leal SM, Pasternak S, Wheeler DA, Willis TD, Yu F, Yang H, Zeng C, Gao Y, Hu H, Hu W, Li C, Lin W, Liu S, Pan H, Tang X, Wang J, Wang W, Yu J, Zhang B, Zhang Q, Zhao H, Zhou J, Gabriel SB, Barry R, Blumenstiel B, Camargo A, Defelice M, Faggart M, Goyette M, Gupta S, Moore J, Nguyen H, Onofrio RC, Parkin M, Roy J, Stahl E, Winchester E, Ziaugra L, Altshuler D, Shen Y, Yao Z, Huang W, Chu X, He Y, Jin L, Liu Y, Sun W, Wang H, Wang Y, Xiong X, Xu L, Waye MM, Tsui SK, Xue H, Wong JT, Galver LM, Fan JB, Gunderson K, Murray SS, Oliphant AR, Chee MS, Montpetit A, Chagnon F, Ferretti V, Leboeuf M, Olivier JF, Phillips MS, Roumy S, Sallee C, Verner A, Hudson TJ, Kwok PY, Cai D, Koboldt DC, Miller RD, Pawlikowska L, Taillon-Miller P, Xiao M, Tsui LC, Mak W, Song YQ, Tam PK, Nakamura Y, Kawaguchi T, Kitamoto T, Morizono T, Nagashima A, Ohnishi Y, Sekine A, Tanaka T, Tsunoda T, Deloukas P, Bird CP, Delgado M, Dermitzakis ET, Gwilliam R, Hunt S, Morrison J, Powell D, Stranger BE, Whittaker P, Bentley DR, Daly MJ, de Bakker PI, Barrett J, Chretien YR, Maller J, McCarroll S, Patterson N, Pe’er I, Price A, Purcell S, Richter DJ, Sabeti P, Saxena R, Schaffner SF, Sham PC, Varilly P, Stein LD, Krishnan L, Smith AV, Tello-Ruiz MK, Thorisson GA, Chakravarti A, Chen PE, Cutler DJ, Kashuk CS, Lin S, Abecasis GR, Guan W, Li Y, Munro HM, Qin ZS, Thomas DJ, McVean G, Auton A, Bottolo L, Cardin N, Eyheramendy S, Freeman C, Marchini J, Myers S, Spencer C, Stephens M, Donnelly P, Cardon LR, Clarke G, Evans DM, Morris AP, Weir BS, Mullikin JC, Sherry ST, Feolo M, Skol A, Zhang H, Matsuda I, Fukushima Y, Macer DR, Suda E, Rotimi CN, Adebamowo CA, Ajayi I, Aniagwu T, Marshall PA, Nkwodimmah C, Royal CD, Leppert MF, Dixon M, Peiffer A, Qiu R, Kent A, Kato K, Niikawa N, Adewole IF, Knoppers BM, Foster MW, Clayton EW, Watkin J, Muzny D, Nazareth L, Sodergren E, Weinstock GM, Yakub I, Birren BW, Wilson RK, Fulton LL, Rogers J, Burton J, Carter NP, Clee CM, Griffiths M, Jones MC, McLay K, Plumb RW, Ross MT, Sims SK, Willey DL, Chen Z, Han H, Kang L, Godbout M, Wallenburg JC, L’Archeveque P, Bellemare G, Saeki K, An D, Fu H, Li Q, Wang Z, Wang R, Holden AL, Brooks LD, McEwen JE, Guyer MS, Wang VO, Peterson JL, Shi M, Spiegel J, Sung LM, Zacharia LF, Collins FS, Kennedy K, Jamieson R, Stewart J. A second generation human haplotype map of over 3.1 million SNPs. Nature 2007; 449: 851–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kutlar A, Hattori Y, Bakioglu I, Kutlar F, Kamel K, Huisman TH. Hematological observations on Arabian SS patients with a homozygosity or heterozygosity for a beta S chromosome with haplotype #31. Hemoglobin 1985; 9: 545–57. [DOI] [PubMed] [Google Scholar]
- 74.Nagel RL, Erlingsson S, Fabry ME, Croizat H, Susuka SM, Lachman H, Sutton M, Driscoll C, Bouhassira E, Billett HH. The Senegal DNA haplotype is associated with the amelioration of anemia in African-American sickle cell anemia patients. Blood 1991; 77: 1371–5. [PubMed] [Google Scholar]
- 75.Nagel RL, Rao SK, Dunda-Belkhodja O, Connolly MM, Fabry ME, Georges A, Krishnamoorthy R, Labie D. The hematologic characteristics of sickle cell anemia bearing the Bantu haplotype: the relationship between G gamma and HbF level. Blood 1987; 69: 1026–30. [PubMed] [Google Scholar]
- 76.Powars DR, Meiselman HJ, Fisher TC, Hiti A, Johnson C. Beta-S gene cluster haplotypes modulate hematologic and hemorheologic expression in sickle cell anemia. Use in predicting clinical severity. Am J Pediatr Hematol Oncol 1994; 16: 55–61. [PubMed] [Google Scholar]
- 77.Liu L, Muralidhar S, Singh M, Sylvan C, Kalra IS, Quinn CT, Onyekwere OC, Pace BS. High-density SNP genotyping to define beta-globin locus haplotypes. Blood Cells Mol Dis 2009; 42: 16–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kiefer CM, Lee J, Hou C, Dale RK, Lee YT, Meier ER, Miller JL, Dean A. Distinct Ldb1/NLI complexes orchestrate gamma-globin repression and reactivation through ETO2 in human adult erythroid cells. Blood 2011; 118: 6200–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xu J, Bauer DE, Kerenyi MA, Vo TD, Hou S, Hsu YJ, Yao H, Trowbridge JJ, Mandel G, Orkin SH. Corepressor-dependent silencing of fetal hemoglobin expression by BCL11A. Proc Natl Acad Sci U S A 2013; 110: 6518–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hein MS, Swanson KC, Lundquist PA, Yungerberg JA, Coon LM, Dawson BD, Jevremovic D, Oliveira AM, Hoyer JD, Oliveira JL. Deletional HPFH Vs. delta beta thalassemia: closing in on a possible Hb F silencer location. Blood 2015; 126: 3372–3372. [Google Scholar]
- 81.Thambyrajah R, Mazan M, Patel R, Moignard V, Stefanska M, Marinopoulou E, Li Y, Lancrin C, Clapes T, Moroy T, Robin C, Miller C, Cowley S, Gottgens B, Kouskoff V, Lacaud G. GFI1 proteins orchestrate the emergence of haematopoietic stem cells through recruitment of LSD1. Nat Cell Biol 2016; 18: 21–32. [DOI] [PubMed] [Google Scholar]
- 82.Vassen L, Beauchemin H, Lemsaddek W, Krongold J, Trudel M, Moroy T. Growth factor independence 1b (gfi1b) is important for the maturation of erythroid cells and the regulation of embryonic globin expression. PLoS One 2014; 9: e96636–e96636. [DOI] [PMC free article] [PubMed] [Google Scholar]