Abstract
Objective
Hypocellularity resulting from chondrocyte death in the aftermath of mechanical injury is thought to contribute to posttraumatic osteoarthritis. However, we observed that nonviable areas in cartilage injured by blunt impact were repopulated within 7–14 days by cells that appeared to migrate from the surrounding matrix. The aim of this study was to assess our hypothesis that the migrating cell population included chondrogenic progenitor cells that were drawn to injured cartilage by alarmins.
Methods
Osteochondral explants obtained from mature cattle were injured by blunt impact or scratching, resulting in localized chondrocyte death. Injured sites were serially imaged by confocal microscopy, and migrating cells were evaluated for chondrogenic progenitor characteristics. Chemotaxis assays were used to measure the responses to chemokines, injury-conditioned medium, dead cell debris, and high mobility group box chromosomal protein 1 (HMGB-1).
Results
Migrating cells were highly clonogenic and multipotent and expressed markers associated with chondrogenic progenitor cells. Compared with chondrocytes, these cells overexpressed genes involved in proliferation and migration and underexpressed cartilage matrix genes. They were more active than chondrocytes in chemotaxis assays and responded to cell lysates, conditioned medium, and HMGB-1. Glycyrrhizin, a chelator of HMGB-1 and a blocking antibody to receptor for advanced glycation end products (RAGE), inhibited responses to cell debris and conditioned medium and reduced the numbers of migrating cells on injured explants.
Conclusion
Injuries that caused chondrocyte death stimulated the emergence and homing of chondrogenic progenitor cells, in part via HMGB-1 release and RAGE-mediated chemotaxis. Their repopulation of the matrix could promote the repair of chondral damage that might otherwise contribute to progressive cartilage loss.
The risk of posttraumatic osteoarthritis (OA) after serious joint injuries is still as high as 70%, despite many refinements in care (1–3). This underscores the urgent need for new treatments to prevent articular cartilage loss initiated by joint damage and cartilage injury. Most macroscopic cartilage lesions do not heal and may spread locally or stimulate joint-wide cartilage degeneration (1,4). This occurs despite the presence of potentially reparative chondrogenic progenitor cells in cartilage and other intraarticular tissue (5–8) that show vigorous in vitro chondrogenic activity. It may be possible to coax these cells to be more effective in vivo, but more complete knowledge of the posttraumatic behavior and function of chondrogenic progenitor cells is needed to evaluate this potential.
Like mesenchymal stem cells (MSCs) that originate in bone marrow, progenitor cells residing in tissue are multipotent, highly clonogenic, and chemotactic (9–11). Progenitor cells migrate locally to sites of injury, where they proliferate and differentiate as needed to replace damaged tissue (12,13). Unlike MSCs, which must be capable of differentiating suitably for the regeneration of multiple tissues in different organ systems, progenitor cells do not require such pluripotency for local tissue regeneration, and the repertoire of progenitor cells is typically more limited than that of MSCs (12).
Chondrogenic progenitor cells were first identified in calf cartilage as a subpopulation of superficial zone cells required for the appositional growth of articular cartilage (5,14). This specialized cell population was isolated from other cartilage cells based on enhanced binding to fibronectin. Compared with normal chondrocytes, chondrogenic progenitor cells overexpressed the stem cell–associated factor Notch-1 and the fibronectin receptor α5β1 integrin. The cells also showed enhanced clonality in culture and multipotency when grafted to chick limb buds. Alsalameh et al (15) subsequently showed that ~4% of cells in normal human cartilage expressed the MSC markers CD105 and CD166. This frequency increased to almost 8% in OA cartilage. However, less than half of the CD105+CD166+ cells were capable of adipogenic differentiation, suggesting that CD marker status overestimated the numbers of multipotent MSC-like progenitor cells.
Using fluorescence-activated cell sorting analysis, Hattori et al demonstrated that 0.07% of cells present in the superficial zone of calf stifle cartilage were capable of Hoechst 33342 dye exclusion, whereas none of the cells from the middle or deep zones excluded the dye (7). Because expression of the multidrug transporter responsible for dye exclusion is typical of stem cells (16), the authors concluded that they had identified a stem-like progenitor cell population. A subpopulation of CD13-, CD29-, CD44-, CD73-, CD90-, and CD105-expressing cells was observed in repair tissue in OA cartilage obtained at the time of total joint replacement. The cells appeared to migrate from subchondral bone via tidemark-spanning blood vessels and expressed both the osteoblastic transcription factor runt-related transcription factor 2 (RUNX-2) and the chondrogenic transcription factor SOX9 (8,17). Concurrently, Grogan and coworkers (6) observed high numbers of chondrocytes (>45%) that were immunohistologically positive for the MSC markers Notch-1 and STRO-1 in both normal and OA cartilage. These cells were osteogenic and chondrogenic but not adipogenic, and they included a small side population (0.14%) of Hoechst dye– excluding cells.
Necrotic cell death associated with wounding releases intracellular components that serve as chemo-tactic signals for progenitor cells, stem cells, and leukocytes. Known collectively as “alarmins” or damage-associated molecular pattern molecules, they include mitochondrial DNA and formyl methionine–containing peptides and DNA-binding high mobility group proteins such as high mobility group box chromosomal protein 1 (HMGB-1) and HMGB-2 and multiple S100 proteins (18–21). Alarmin binding to receptor for advanced glycation end products (RAGE) and Toll-like receptors stimulates the migration of progenitor cells and stem cells, attracting them to injury sites, where they participate in tissue regeneration and repair (22–25). The potent antiinflammatory drug glycyrrhizic acid (glycyrrhizin) is a natural product derived from licorice that binds HMGB-1 and blocks its activities (26). Additional chemo-tactic recruitment is stimulated by CXC ligands (CXCLs) released by tissue and inflammatory cells (27,28). Several studies have shown that blunt trauma to articular cartilage induces acute chondrocyte necrosis and apoptosis (1,29–35). In an explant trauma model, we observed cells migrating over cartilage surfaces near sites of extensive chondrocyte death. These cells were isolated and evaluated for progenitor cell characteristics and chemotactic responses to injury-related alarmins.
MATERIALS AND METHODS
Explant harvest and culture
Stifle joints from young adult cattle (15–24 months old) were obtained from a local abattoir (Bud's Custom Meats). Osteochondral explants were prepared by manually sawing an ~25 × 25-mm portion of the bovine tibial plateau that included the central loaded area of the articular surface. After obtaining institutional review board approval, osteochondral explants from 3 normal tali of patients undergoing lower limb amputation for cancer were obtained and prepared in the same manner. The patients were males ages 29, 34, and 46 years who did not have a diagnosis of OA. The explants were rinsed in Hanks’ balanced salt solution (HBSS) and cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (Invitrogen Life Technologies), 50 μg/ml l-ascorbate, 100 units/ml penicillin, 100 μg/ml streptomycin, and 2.5 μg/ml Fungizone.
Injury
After 2 days in culture, the human and bovine explants were injured by blunt impact (14 J/cm2) via a 5-mm– diameter flat-ended platen, using a drop tower device as described previously (35,36). In some cases, explant cartilage was dissected free from subchondral bone immediately after impact injury. Scratch injuries were made by dragging a 26-gauge needle over the cartilage surface to create X-shaped matrix tears of ~0.5 mm in depth. Confocal imaging studies were performed essentially as previously described (35,36). Briefly, explants were stained with calcein AM (viability) and ethidium homodimer (dead cells) and submerged in culture medium. After a 30-minute incubation, the explants were imaged using a Bio-Rad 1024 confocal laser scanning microscope.
Cell harvest
Five to seven days after injury, the explants were submerged in 0.25% trypsin–EDTA in HBSS and incubated for 10 minutes to detach migrating progenitor cells from the surface. Imaging studies performed before and after submersion in trypsin confirmed that the brief enzymatic treatment removed the surface-adherent migrating cells without disrupting the underlying superficial chondrocytes. To recover normal chondrocytes, the underlying cartilage was digested overnight with type I collagenase and Pronase E (Sigma-Aldrich) dissolved in culture medium (0.25 mg/ml each). A custom-fabricated device was used to separate the superficial one-third and lower two-thirds zones of the cartilage sample prior to collagenase/Pronase digestion. Colony-forming assays were performed as previously described (37).
Green fluorescent protein labeling
For some experiments, isolated putative chondrogenic progenitor cells were labeled with green fluorescent protein (GFP; 488 nm) by lentiviral transduction. GFP-labeled cells (1 × 105) were suspended in chilled (8°C) hydrogel consisting of 0.6% 2-kd hyaluronic acid (Easy Motion Horse) and 18% Pluronic F-127 (Sigma-Aldrich) in normal saline. Pluronic F-127 is a polyol surfactant that confers temperature-sensitive gelation to solutions (38). The cold suspension gelled on contact with warmed (37°C) explants, such that the suspended cells were held in place adjacent to a site of blunt impact. The explants were incubated for 5 days, after which they were counterstained with 0.5 μM CellTracker Red CMTPX (Invitrogen Life Technologies) and imaged using a Bio-Rad 1024 confocal microscope with a custom-built XY microscope stage driver (Condensed Matter Sciences Division). The sites were scanned to an average depth of 330 μm at 40-μm intervals. Z-axis projections of confocal images were obtained using ImageJ (rsb.info.nih. gov/ij).
Immunofluorescence staining
Immunofluorescence staining for proliferating cell nuclear antigen (PCNA) was performed on paraformaldehyde-fixed cryosections of cartilage from 3 different explants that had been cultured for 10 days after an impact injury, using an anti-PCNA monoclonal antibody (Abcam); an Alexa Fluor 488–labeled goat anti-mouse secondary antibody (Jackson ImmunoResearch) was used for detection. The sections were mounted in Vectashield mounting medium, using DAPI (Vector) to stain nuclei. Additional sections from the same explants were stained with Safranin O–fast green to reveal tissue morphology. Proteoglycin 4 (PRG4; lubricin) immunohistochemical analysis was performed on sections from the same specimens, using a mouse monoclonal antibody (MD Biosciences) and a Vectastain ABC kit (Vector). Transmitted light images and epifluorescence images were obtained using a QImage CCD camera (QImaging) mounted on an Olympus BX60 microscope.
Cell migration/chemotaxis assays
Cell migration/ chemotaxis assays were performed using a CytoSelect 24-Well Cell Invasion Assay kit (Cell Biolabs) essentially as described by the manufacturer. Putative chondrogenic progenitor cell or normal chondrocyte suspensions (3 × 105 cells in serum-free medium) were added to the upper Transwell and placed in reservoirs containing serum-free medium alone or serum-free medium with various chemokines, cell lysates, or serum-free medium conditioned by injured explants. The plates were incubated for 24 hours prior to processing. Cell lysates were transferred to fluorescence plates and read on a microplate reader (Molecular Devices). The data are presented as the percentage of migrating cells ([number of cells in the bottom chamber/number of cells seeded] × 100).
Conditioned medium was made by incubating blunt-impacted explants overnight in 10 ml of serum-free medium. The medium was concentrated 10-fold using Amicon Ultra centrifugal 10K filters (Millipore). Lysates for testing in the chemotaxis assay were obtained by repeated freeze-thawing of cells from primary cultures of bovine chondrocytes isolated from distal femurs. Glycyrrhizic acid (Sigma-Aldrich) and a rabbit polyclonal anti-RAGE antibody (Abcam) were added to culture medium at a concentration of 25 μM (39). Porcine platelet–derived growth factor (PDGF BB; R&D Systems) was diluted in medium to a concentration of 300 nM (40). The effects of glycyrrhizic acid and anti-RAGE antibody on migration in the explant model were assessed by confocal microscopy and by counting the number of migrating cells harvested from explant surfaces by trypsinization 14 days after blunt impact. The explants (n = 4/group) were treated daily starting immediately after impact.
Side population assay
Side population assays were performed essentially as previously described (7). First-passage putative chondrogenic progenitor cells and normal chondrocytes in suspension in HBSS (1 × 106/ml) were incubated at 37°C for 1.5 hours with 2.5 mg/ml of Hoechst 33342 (Sigma-Aldrich), with or without 5 mM verapamil (Sigma-Aldrich). The cells were washed in cold HBSS, filtered through a 70-μm nylon mesh, and counterstained with propidium iodide to identify dead cells. Flow cytometric analysis was performed using a BD LSR II flow cytometer with an ultraviolet laser (BD Biosciences).
Multipotent differentiation
The multipotency of putative chondrogenic progenitor cells was tested by culturing them under chondrogenic, osteogenic, and adipogenic conditions (41). For chondrogenic differentiation, 1.2 million cells were pelleted and incubated in chondrogenic medium (DMEM containing 10 ng/ml transforming growth factor β1, 0.1 μM dexamethasone, 25 μg/ml l-ascorbate, 100 μg/ml pyruvate, 50 mg/ml ITS+ Premix, and antibiotics) for 14 days. The pellets were analyzed for matrix formation, using Safranin O–fast green staining of cryosections. To induce osteogenic differentiation, 3 × 104 trypsinized migrating cells were cultured in osteogenic medium (DMEM–Ham's F-12 containing 0.1 μM dexamethasone, 100 mM β-glycerophosphate, 50 μg/ml l-ascorbate, and antibiotics) for 14 days and stained with alizarin red to detect calcium phosphate mineralization. An adipogenesis differentiation kit (Gibco) was used to induce adipogenesis, and the cells were stained with oil red O 14 days postinduction. All cultures were imaged on a Nikon CX2 inverted microscope.
Microarray
For microarray analysis, RNA was isolated from primary cultures of bovine MSCs, from freshly harvested putative chondrogenic progenitor cells, and directly from explant cartilage. Saline lavage was used to isolate MSCs from the marrow and subchondral bone of adult bovine tibiae. The lavage fluid was centrifuged, and the pelleted cells were plated in plastic dishes. Adherent cells were cultured for 5 days before harvesting. RNA was harvested from 3 independent batches of cells/explants. Cells and cartilage were homogenized in TRIzol reagent (Invitrogen Life Technologies), and total RNA was extracted using an RNeasy Mini Kit (Qiagen) according to the manufacturer's instructions. RNA (50 ng) was converted to single primer isothermal amplification–amplified complementary DNA (cDNA) using an Ovation RNA Amplification System version 2 (NuGEN). Biotinylated cDNA was placed onto Bovine Genome Arrays (Affymetrix). Arrays were scanned with an Affymetrix Model 3000, and data were collected using GeneChip operating software (MAS 5.0). Statistical analysis of the data was performed by one-way analysis of variance (ANOVA), and a heatmap and dendro-gram were generated using Partek Genomics Suite software.
Previously published work indicated that progenitor cells can be distinguished from normal chondrocytes based on up-regulation of genes expressed in MSC populations (5,6). These genes include ATP-binding cassette subfamily G member 2 (ABCG2), various CD cell markers, fetal liver kinase 1 (FLK-1), RUNX-2, and SOX9. Beta-actin was used for normalization. Real-time polymerase chain reaction (PCR) was performed to compare the expression of these markers in putative chondrogenic progenitor cells and normal chondrocytes, essentially as previously described (42).
Primers were purchased from Integrated DNA Technologies. The following primer sequences were used: for ABCG2, forward CCTTGGTTGTCATGGCTTCA and reverse AGTCCTGGGCAGAAGTTTTGTC; for CD105, forward CCACTGCCCCAGAGACTGCGC and reverse CCCCCACAGTGAGTGCTTAGGT; for CD90, forward CGGTGGTGTTTGGCCATGTAATGA and reverse GAGAGAGGGGAGTCCTATCCTGGT; for CD73, forward AGCTTTCCCAGCCTTCCATGCG and reverse GGGTGTCCTCTTGAGTCCTGCA; for CD39, forward CCACCCTCTCCTTCCGAGAGG and reverse TGACTGTAACCCTGGAGCTTGGCT; for CD29, forward GCGGCCTCCGGGTGGATTCC and reverse GCCGGGAAGGTCCAGGGGC; for FLK-1, forward TTCCAAGTGGCTAAGGGCAT and reverse TTTAACCACGTTCTTTTCCGACA; for RUNX-2, forward GCATGAAGCCCTATCCAGAGTCT and reverse GCTGATGGAGCTGTTGGTGTAG; for SOX9, forward CGGTGGTGTTTGGCCATGTAATGA and reverse GAGAGAGGGGAGTCCTATCCTGGT; for β-actin, forward TCGACACCGCAACCAGTTCGC and reverse CATGCCGGAGCCGTTGTCGA.
Colony-formation assay
For colony-formation assays, statistical analysis was performed using SPSS software (version 10.0.7) with one-way ANOVA and post hoc pairwise comparison. Flow cytometry data (side population) were evaluated by Student's t-test. One-way and two-way ANOVAs with the Holm-Sidak post hoc test were used to analyze migration assay data. Results are presented as the mean ± SD.
RESULTS
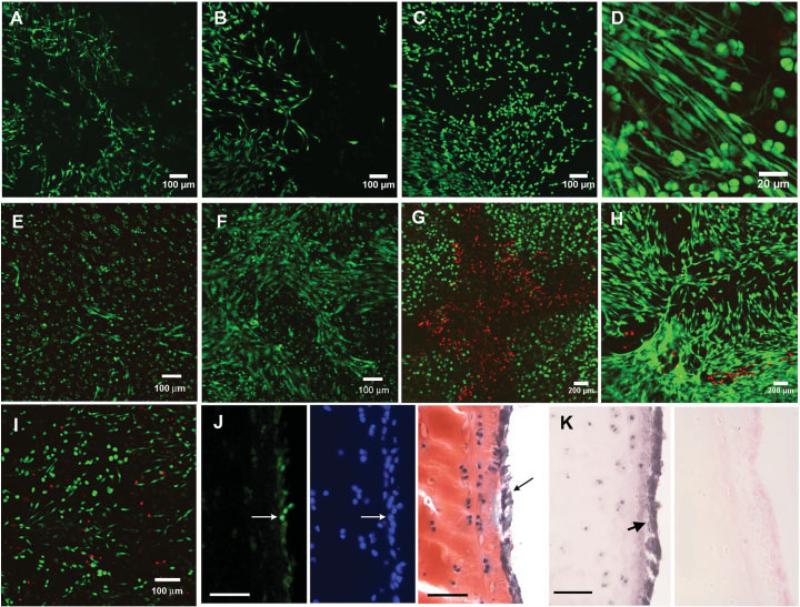
Blunt impact injury to explant surfaces caused local chondrocyte death and stimulated the emergence of migratory cells in and around impact sites. These cells began to accumulate 5 days after impact and gradually repopulated previously uninhabited areas (Figures 1A– C). Migrating cells were morphologically distinct from normal chondrocytes in that they were elongated with multiple thin cytoplasmic extensions (Figure 1D). A similar migratory reaction was observed at impact sites in explanted human tali (Figures 1E and F) and in a scratch injury (Figures 1G and H). Migrating cells were also observed on impacted explant cartilage cultured without subchondral bone (Figure 1I). Immunostaining of cryosections from impacted explants revealed that surface-migrating putative chondrogenic progenitor cells were positive for PCNA, whereas chondrocytes beneath the surface were largely negative (Figure 1J). Surface putative chondrogenic progenitor cells were also strongly positive for lubricin (Figure 1K).
Figure 1.
Migrating cells on injured cartilage. A–C, Confocal microscopy images showing live cells (green) in the same area within an impact site on the surface of an explant on day 7 (A), day 11 (B), and day 15 (C) after impact. D, Higher-magnification view showing the elongated morphology and dendritic appearance of the cells. E and F, Live cells observed on a human talus specimen (obtained from a 36-year-old man) on day 6 (E) and day 10 (F) after impact. G and H, Dead cells (red) and live cells in a bovine explant with a cross-shaped needle scratch. Images were obtained immediately after the injury (G) and 14 days later (H). I, Appearance of migrating cells on the surface of cartilage dissected free from subchondral bone immediately after impact. J, Left, Green immunofluorescence staining for proliferating cell nuclear antigen, showing positive cells (arrow) on the surface of a cartilage explant. Middle, Blue staining in the same section, showing all nuclei. Right, Surface-migrating putative chondrogenic progenitor cells (arrow) in a consecutive section stained with Safranin O–fast green. K, Immunohistochemical staining for lubricin. Arrow (left) indicates strongly positive migrating cells in an impact site. Right, Negative control. Bars in J and K = 100 μm.
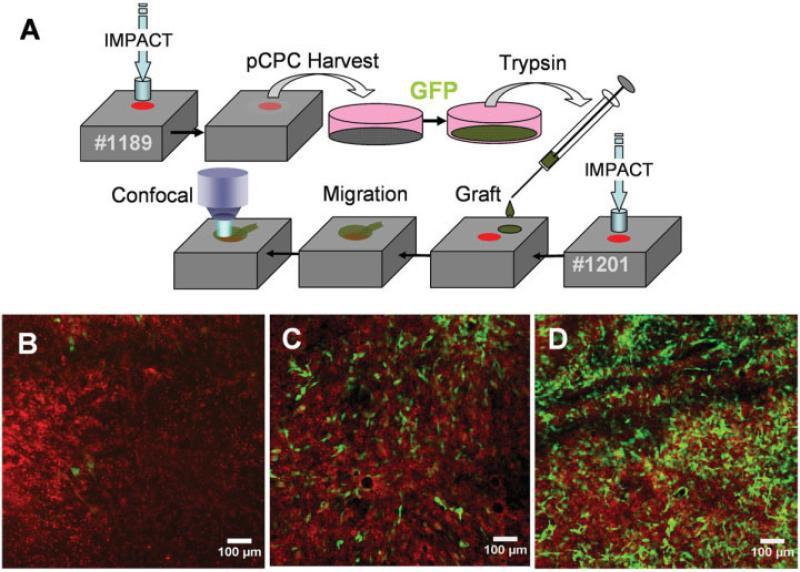
Five to seven days after impact injury, surface-adherent migrating cells were detached from the injured explants by trypsin treatment. Some cells were transduced with GFP and grafted ~4 mm away from a freshly made impact site on another explant (Figure 2A). The number of labeled cells in the impact site increased dramatically from day 2 to day 12 (Figures 2B–D). The clonogenic activity of trypsinized cells was compared with that of chondrocytes from the upper one-third of the cartilage, which included the superficial and transitional zones, and from the bottom two-thirds, which included the transitional and deep zones (Figure 3). Primary cultures were established, and the cells were harvested after 5–7 days in culture for colony-formation assays. Trypsinized cells in monolayer culture grew more rapidly than did chondrocytes (Figures 3A–D). These cultures were passaged, seeded in cloning dishes, and incubated for 10 days. Trypsinized cells showed the most vigorous colony formation in terms of both the total number of colonies and the average colony size (Figures 3E–G). Trypsinized cells and chondrocytes from the upper one-third of the cartilage matrix showed significantly higher numbers of colonies (150 and 120, respectively) compared with chondrocytes from the bottom two-thirds of the matrix (20 colonies) (P = 0.001). The average colony size of trypsinized cells (20 mm2) was significantly greater than that of chondrocytes from the upper one-third or lower two-thirds of the matrix, both of which showed average colony sizes of <5 mm2 (P = 0.001). However, ~1% of the colonies formed by chondrocytes from the upper one-third of the matrix showed areas of ≥20 mm2.
Figure 2.
Migration of grafted putative chondrogenic progenitor cells (pCPC). A, Procedure for harvesting and grafting putative chondrogenic progenitor cells. The boxes represent 2 different explants (specimen no. 1189 and specimen no. 1201). Explant no. 1189 was impacted and incubated for 5 days to allow putative chondrogenic progenitor cells to emerge. These cells were harvested and placed in monolayer culture for green fluorescent protein (GFP) transduction. Labeled cells were trypsinized, suspended in a temperature-sensitive hydrogel, and grafted onto explant no. 1201, which had been impacted a few hours earlier. B–D, The impact site was imaged by confocal microscopy at various times after grafting. Grafted GFP-labeled cells (green) can be seen against the background of host cells labeled with a red tracking stain. Exactly the same field within the impact site was imaged 2 days (B), 5 days (C), and 12 days (D) after grafting.
Figure 3.
Colony formation by migrating progenitor cells and chondrocytes. A–D, Light microscopy images of a single colony of progenitor cells 2 days (A), 3 days (B), and 6 days (C) after seeding, and a chondrocyte colony cultured for 13 days (D). E, Macroscopic image of cloning plates seeded with chondrocytes from the deep and superficial (Sup) zones or progenitor cells after 10 days of growth. F and G, Total number of colonies (F) and average colony area (G), as measured using ImageJ. Progenitor cells and superficial chondrocytes showed higher numbers of colonies compared with deep chondrocytes. However, the colony area was much larger for progenitor cells than for chondrocytes from either zone. Values are the mean ± SD of 4–5 different batches of cells. Horizontal bars indicate significant differences (P = 0.001).
Putative chondrogenic progenitor cells were cultured in chondrogenic, osteogenic, or adipogenic medium for 14 days in order to evaluate their differentiation potential. After the induction of chondrogenic differentiation, cultured pellets were fixed and stained with Safranin O–fast green, revealing a proteoglycan-rich matrix throughout the pellets (Figure 4A). Similarly, most cells in osteogenic medium deposited a calcium phosphate–rich mineralized matrix, as detected by alizarin red staining (Figure 4B). However, few cells (<1%) stained with oil red O after 2 weeks of culture in adipogenic medium (Figure 4C).
Figure 4.
Stem cell–like characteristics of putative chondrogenic progenitor cells (pCPC). A–C, Putative chondrogenic progenitor cells cultured under chondrogenic (A), osteogenic (B), and adipogenic (C) conditions. The pellet culture showed intense red Safranin O–fast green staining, indicating the presence of cartilage proteoglycans. Deposition of calcium phosphate was detected by staining with alizarin red (dark red spots) (B). Few cells stained with oil red O after 2 weeks of culture in adipogenic medium (C). D, Side populations in progenitor cells and chondrocytes, as determined by fluorescence-activated cell sorting analysis. As expected, verapamil (V), an ATP-binding cassette subfamily G member transport inhibitor, ablated the side population. E, Marker gene expression in putative chondrogenic progenitor cells relative to normal chondrocytes (NCs), as determined by real-time polymerase chain reaction analysis (dark columns) and microarray analysis (light columns). F, Heatmap and dendrogram summarizing microarray data for the indicated cell populations (triplicate analyses). Colored bars in the heatmap show genes that were expressed at higher or lower levels than the median value (green and red, respectively). The dendrogram shows that chondrogenic progenitor cells and mesenchymal stem cells (MSCs) were more closely related to each other than to NCs. Bars in D and E show the mean ± SD.
Flow cytometric analysis of Hoechst 33342– stained normal chondrocytes revealed few side population cells (mean ± SD 0.032 ± 0.012%) (Figure 4D). The side population was significantly larger in trypsinized cells (0.22 ± 0.07%; P = 0.001). Verapamil treatment reduced side populations to <0.005%, indicating that stain efflux depended on the stem cell–associated ABCG2 transporter. Real-time PCR analysis revealed substantially higher expression of stem cell marker genes in putative chondrogenic progenitor cells compared with normal chondrocytes (Figure 4E). ABCG2 expression was increased by >4-fold, SOX9 expression was increased by 3.8-fold, and FLK-1 expression was increased by 2.8-fold. CD105, CD73, CD39, and CD29 expression was increased by <2-fold in putative chondrogenic progenitor cells, and CD90 expression was 2-fold higher in normal chondrocytes than in putative chondrogenic progenitor cells. Microarray analysis showed elevated expression of the progenitor cell markers Notch-1 (7.4-fold) and CD44 (12-fold) in putative chondrogenic progenitor cells compared with normal chondrocytes.
Statistical analysis of microarrays indicated that overall gene expression in putative chondrogenic progenitor cells was more similar to that of MSCs than to that of normal chondrocytes (Figure 4F). Individual genes that showed statistically significant differences (P < 0.05) in expression of >2-fold in putative chondrogenic progenitor cells versus normal chondrocytes or MSCs are listed in Table 1. The listed genes were selected for relevance to inflammation, proliferation, migration, and chondrogenic differentiation. The pro-inflammatory cytokine interleukin-6 (IL-6) was among the most highly up-regulated genes in chondrogenic progenitor cells compared with normal chondrocytes (130-fold increase). Chemokines involved in stem cell and leukocyte chemotaxis (28,43) were also strikingly up-regulated in putative chondrogenic progenitor cells relative to normal chondrocytes: CXCL12 expression was >25-fold greater, and CXCL8 (IL-8) expression was increased by >35-fold. Also relative to normal chondrocytes, matrix metalloproteinase 1 (MMP-1) and MMP-13 genes were overexpressed in putative chondrogenic progenitor cells by 18-fold and 4.3-fold, respectively, and the proliferation-related cyclin B1 and cyclin D1 genes were overexpressed by 35-fold and 9-fold, respectively. Genes encoding the cartilage extracellular matrix components type X collagen (COL10A1), type IX collagen (COL9A2), type II collagen (COL2A1), aggrecan (ACAN), and cartilage oligomeric matrix protein (COMP) were among the most down-regulated genes in putative chondrogenic progenitor cells.
Table 1.
Relative gene expression of putative chondrogenic progenitor cells (CPCs) versus normal chondrocytes (NCs) and mesenchymal stem cells (MSCs)*
| Gene symbol | Δ | P | Gene symbol | Δ | P |
|---|---|---|---|---|---|
| CPCs vs. NCs | CPCs vs. MSCs | ||||
| IL6 | 130 | 3.6 × 10–5 | PRG4 | 78 | 7.6 × 10–5 |
| DOCK10 | 62 | 4.4 × 10–7 | MMP3 | 23 | 1.0 × 10–3 |
| CXCL8 | 36 | 3.0 × 10–3 | ACAN | 17 | 5.4 × 10–4 |
| CCNB1 | 35 | 5.8 × 10–5 | S100A1 | 16 | 2.7 × 10–4 |
| CXCL12 | 28 | 2.4 × 10–5 | MMP1 | 15 | 5.2 × 10–3 |
| CSF1 | 21 | 9.3 × 10–6 | HAPLN1 | 13 | 1.0 × 10–5 |
| MMP1 | 18 | 3.9 × 10–3 | CXCL2 | 12 | 2.0 × 10–2 |
| CD44 | 12 | 6.1 × 10–3 | IL8 | 10 | 2.5 × 10–2 |
| ADAMTS4 | 10 | 2.9 × 10–5 | COL2A1 | 6.9 | 8.0 × 10–5 |
| IL1RN | 9.1 | 1.3 × 10–4 | SOD3 | 6.7 | 1.3 × 10–4 |
| CCND1 | 9.0 | 2.3 × 10–5 | COL5A3 | 6.2 | 1.5 × 10–3 |
| NOTCH1 | 7.4 | 9.4 × 10–7 | FGF2 | 5.1 | 8.6 × 10–5 |
| ADAM9 | 6.4 | 1.6 × 10–3 | IL16 | 3.3 | 8.5 × 10–3 |
| IGFBP3 | 5.6 | 2.7 × 10–2 | CD83 | 3.2 | 1.5 × 10–2 |
| MMP13 | 4.3 | 1.2 × 10–2 | PLAT | 3.0 | 4.2 × 10–2 |
| HMMR | 4.2 | 1.4 × 10–4 | SOCS1 | 2.7 | 2.2 × 10–2 |
| TLR3 | 3.7 | 3.7 × 10–2 | IL1A | 2.7 | 2.2 × 10–2 |
| ITGB5 | 3.6 | 4.0 × 10–2 | S100B | 2.6 | 1.6 × 10–3 |
| COL6A1 | 3.1 | 1.1 × 10–2 | PDGFRA | 2.5 | 8.4 × 10–3 |
| ADAM8 | 3.0 | 2.0 × 10–2 | SOD2 | 2.3 | 3.4 × 10–2 |
| COL10A1 | –192 | 7.0 × 10–6 | NID1 | –11 | 4.4 × 10–2 |
| CHAD | –136 | 1.5 × 10–8 | IGFBP7 | –10 | 6.7 × 10–3 |
| COL9A2 | –38 | 4.9 × 10–6 | PPARG | –9.8 | 5.3 × 10–3 |
| COL2A1 | –12 | 9.8 × 10–6 | THBS1 | –6.5 | 4.2 × 10–2 |
| TIMP4 | –8.3 | 9.4 × 10–5 | GPC4 | –6.2 | 1.8 × 10–2 |
| ACAN | –8.0 | 5.2 × 10–3 | IGFBP3 | –5.4 | 4.6 × 10–2 |
| INSR | –6.0 | 1.7 × 10–3 | IGFBP2 | –3.6 | 1.0 × 10–2 |
| COMP | –5.4 | 4.9 × 10–4 | IL1RN | –3.6 | 4.3 × 10–3 |
| TIMP4 | –4.8 | 9.7 × 10–4 | PLAU | –3.0 | 2.0 × 10–3 |
| FGF2 | –3.9 | 3.2 × 10–4 | DNMT3A | –2.7 | 1.1 × 10–2 |
| ACAN | –3.7 | 3.2 × 10–2 | IGFBP4 | –2.6 | 2.6 × 10–2 |
| PTH1R | –3.1 | 1.8 × 10–2 | NOTCH1 | –2.3 | 6.2 × 10–4 |
| DNMT3A | – 2.9 | 2.3 × 10–2 |
The genes shown are those that were expressed at levels ≥2-fold higher or lower compared with CPCs.
However, compared with MSCs, putative chondrogenic progenitor cells overexpressed the superficial chondrocyte marker PRG4 (lubricin) and the chondrocyte-associated genes S100A1 (16-fold) and S100B (2.6-fold). Moreover, ACAN and COL2A1 expression was much higher in putative chondrogenic progenitor cells than in MSCs (17-fold and 6.9-fold increases, respectively). In contrast, expression of the IL-1 receptor antagonist gene (IL1RN) was substantially down-regulated in putative chondrogenic progenitor cells compared with MSCs (−3.6-fold), as was the expression of several insulin-like growth factor binding proteins, including IGFBP7 (−10-fold), IGFBP3 (−5.4-fold), and IGFBP2 (−3.6-fold).
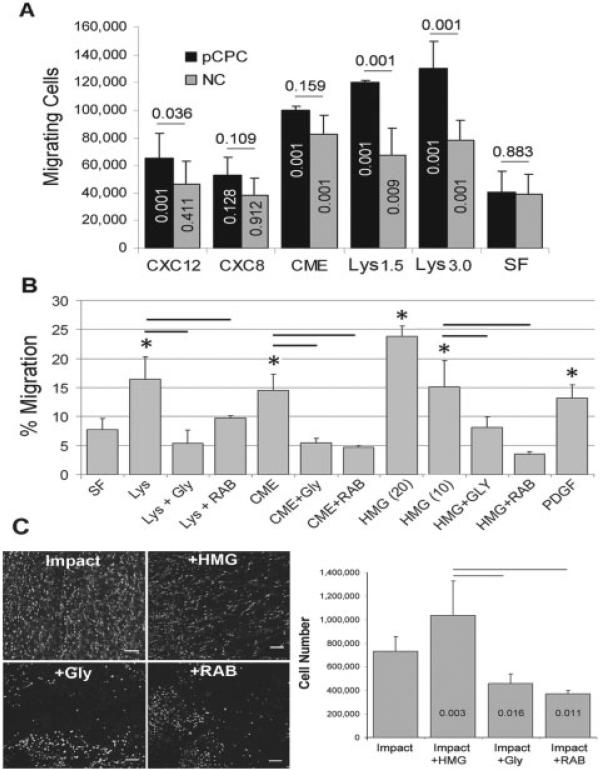
In general, putative chondrogenic progenitor cells were significantly more active in Transwell chemo-taxis assays than were normal chondrocytes (P = 0.001); however, this was chemotactic factor dependent (Figure 5A). Compared with untreated control medium, CXCL12 significantly increased putative chondrogenic progenitor cell chemotaxis (P = 0.001) but had no effect on normal chondrocytes (P = 0.411). Neither putative chondrogenic progenitor cells nor normal chondrocytes responded to CXCL8 (P = 0.128 and P = 0.912, respectively), but conditioned medium from impact-injured explants induced chemotaxis to a similar degree in both putative chondrogenic progenitor cells and normal chondrocytes (P = 0.001). The response to cell lysates was significant for putative chondrogenic progenitor cells and normal chondrocytes (P = 0.001), but the putative chondrogenic progenitor cell response was significantly greater than the normal chondrocyte response (P = 0.001).
Figure 5.
Chemotactic activity. A, Migrating cell responses to CXCL12, CXCL8, conditioned medium (CME), cell lysates (Lys) made from 1.5 × 106 cells and 3.0 × 106 cells, or serum-free medium (SF). Numbers above the bars indicate P values for differences between putative chondrogenic progenitor cells (pCPC) and normal chondrocytes (NC). Numbers within the columns are P values for differences between treatments and serum-free medium. Bars show the mean ± SD. B, Effects of glycyrrhizin (Gly) and anti–receptor for advanced glycation end products (anti-RAGE) antibody (RAB) on responses to CME, Lys (3.0 × 106 cells), and high mobility group box chromosomal protein 1 (HMGB-1) 10 nM and 20 nM on putative chondrogenic progenitor cell chemotaxis. Bars show the mean ± SD (n = 3–9). * = P < 0.005 for treated versus serum-free medium, by one-way analysis of variance. C, Left, Confocal microscopy images showing an untreated impacted control explant and explants treated with HMGB-1 (HMG), glycyrrhizin, and anti-RAGE antibody. Bars = 100 μm. Right, Numbers of migrating cells in untreated control implant and implants treated with HMGB-1, glycyrrhizin, and anti-RAGE antibody. Numbers within the columns are P values versus impact only. Values are the mean ± SD (n = 4/group). Horizontal bars indicate significant differences (P = 0.001).
In addition to cell lysates and conditioned medium, putative chondrogenic progenitor cells responded strongly to purified HMGB-1 (Figure 5B). HMGB-1 at 10 nM or 20 nM significantly enhanced chemotaxis compared with controls (P = 0.001). The stimulatory effects of 10 nM HMGB-1 and the effects of conditioned medium and cell lysates were significantly suppressed by glycyrrhizin and by an anti-RAGE antibody (P = 0.001). The migratory activity stimulated by lysates, conditioned medium, and 10 nM HMGB-1 was similar to the activity stimulated by PDGF (250 nM), a well-known stem cell chemotactic factor. Glycyrrhizin and anti-RAGE antibody also significantly inhibited putative chondrogenic progenitor cell migration in impacted explants (P = 0.016 and P = 0.011, respectively) (Figure 5C).
DISCUSSION
The results of these experiments demonstrated that the migrating cells we observed on injured bovine osteochondral explants closely resembled chondrogenic progenitor cells previously identified in normal and human OA cartilage. The chemotactic activity, clonogenicity, limited multipotency, and side population of the cells were all notably consistent with published descriptions of progenitor cells from cartilage and other tissue (6,8,14,15,17).
In vitro chemotaxis assays confirmed that media conditioned by impacted cartilage or cell lysates were relatively strong chemoattractants for putative chondrogenic progenitor cells. Furthermore, a scratch injury that caused localized chondrocyte death much like that observed in impact sites provoked the same response as impact injury, indicating that chondrocyte death was the main cause of putative chondrogenic progenitor cell activation in this system. The strong blocking effects of glycyrrhizin in the migration assays implicated the nuclear protein HMGB-1 as a primary chemoattractant in these complex mixtures. A blocking antibody to the RAGE receptor also significantly diminished migration, indicating that the effects of HMGB-1 were mediated in part by RAGE. The inhibitory effects of glycyrrhizin and anti-RAGE on putative chondrogenic progenitor cell migration/proliferation in the explant system were also significant; however, the treatments were less potent than those used in Transwell migration assays, suggesting that other factors contributed to the chemotactic activity. Indeed, our data suggested that PDGF might play such a role. A similar migratory response was observed in explanted human tali specimens with the same impact injuries as those in bovine explants, indicating that putative chondrogenic progenitor cell activation was not unique to the bovine system.
Putative chondrogenic progenitor cells appeared after injury when cartilage was cultured separately from subchondral bone. Cell populations derived from the top one-third of the cartilage matrix consistently yielded a few colonies that were as large as those formed by putative chondrogenic progenitor cells, whereas cells from the bottom two-thirds of the matrix never formed such colonies. Moreover, PRG4, which is expressed at high levels by superficial chondrocytes, was one of the most highly up-regulated genes in putative chondrogenic progenitor cells relative to MSCs. These findings strongly suggest that putative chondrogenic progenitor cells resided in the superficial zone before cartilage injury.
Compared with normal chondrocytes, putative chondrogenic progenitor cells overexpressed a few (but not all) stem cell–associated markers. Although ABCG2, RUNX2, and NOTCH1 expression was increased by ≥4-fold in putative chondrogenic progenitor cells, increases in CD markers were minor (<2-fold) and were not likely to be biologically significant. These results are mainly consistent with those of previous studies (5,7,15,17), but the lack of substantive increases in CD marker expression suggests that the putative chondrogenic progenitor cells studied here might differ from progenitor cells of subchondral origin that more clearly overexpress these markers (17). Relative to normal chondrocytes, chondrogenic progenitor cells underex-pressed chondrocyte-associated genes such as COL2A1 and ACAN. However, compared with MSCs, putative chondrogenic progenitor cells overexpressed COL2A1, ACAN, PRG4, S100A1, and S100B, all of which are considered to be chondrocyte markers (44). Thus, although they closely resembled MSCs in their overall pattern of gene expression, putative chondrogenic progenitor cells retained some chondrocyte-like features and were distinguishable from MSCs on that basis.
The expression of several metalloproteinases (MMP1, MMP13, ADAMTS4, ADAM8, and ADAM9) was significantly higher in putative chondrogenic progenitor cells than in normal chondrocytes. This was consistent with the enhanced chemotactic activity of putative chondrogenic progenitor cells compared with normal chondrocytes observed in our Transwell-based assays, which measured movement through a collagen matrix. With the notable exceptions of MMP1 and MMP13, the expression of which was up-regulated by >4-fold in putative chondrogenic progenitor cells, metalloproteinases were expressed at similar levels by MSCs, whose physiologic functions include migration through extracellular matrices.
The accumulation of hundreds of putative chon drogenic progenitor cells at injury sites was unlikely to be attributable solely to migration from the surrounding matrix. Putative chondrogenic progenitor cells grew rapidly in culture, and proliferation on cartilage surfaces is likely to explain the rapid repopulation of impact sites in the explant model. This was corroborated by immunofluorescence staining, which identified relatively high numbers of PCNA-positive cells among surface-migrating cells. Consistent with their highly clonogenic, proliferative phenotype, both MSCs and putative chondrogenic progenitor cells significantly overexpressed colony-stimulating factors, cyclins, and other growth-related genes.
The circumstances of the explant experiment dictated that we use different techniques to harvest RNA from chondrogenic progenitor cells and from normal chondrocytes. Thus, chondrogenic progenitor cell RNA was obtained from cells freshly removed from explant surfaces by trypsinization, normal chondrocyte RNA was extracted directly from cartilage, and MSC RNA was obtained from primary monolayer cultures. Although the trypsin treatment of chondrogenic progenitor cells was brief (~10 minutes), it might have led to changes in the expression of some early-response genes. Moreover, procedural losses associated with RNA extraction from cartilage might have contributed to variability among the normal chondrocyte samples, which was higher than that in putative chondrogenic progenitor cell or MSC samples. Last, the effects of isolation and short-term culture on MSC gene expression are unknown and might have affected many of the genes that appeared to be differentially regulated compared with putative chondrogenic progenitor cells and normal chondrocytes.
The physiologic functions of chondrogenic progenitor cells and their effects on healing in injured joints remain unknown. Although chondrogenic progenitor cells efficiently repopulated damaged cartilage in our model, their relatively high levels of chemokine and cytokine expression and excessive metalloproteinase production could contribute to synovitis and cartilage degeneration in injured joints. In contrast, chondrogenic progenitor cells may be involved in the early stages of cartilage repair: confocal studies and histology showed that putative chondrogenic progenitor cells formed a continuous sheet over injured cartilage surfaces after ≥1 week in culture. The relatively high level of PRG4 expression by chondrogenic progenitor cells observed at the RNA and protein levels suggests that they help to restore the surface-protective lubricant coating on damaged cartilage, a process that may be involved in the repair of superficial defects (1,45–49). The identification of HMGB-1 as an activator suggests a method to control chondrogenic progenitor cell responses to promote healing. Whether the goal should be to thwart or augment these responses will likely be decided by findings from in vivo models, in which treatments can be evaluated in a more physiologic setting.
ACKNOWLEDGMENTS
We thank Dr. Tae-Hong Lim for providing his hydro-gel formulation, Dr. Prem S. Ramakrishnan for developing a cartilage dissection jig, and Sean M. Martin for his indispensible help with migration assays.
Supported by the US Department of Defense (grant W81XWH-10-1-0702a) and the World Arthrosis Organization.
Footnotes
AUTHOR CONTRIBUTIONS
All authors were involved in drafting the article or revising it critically for important intellectual content, and all authors approved the final version to be published. Dr. Martin had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study conception and design. Seol, McCabe, Buckwalter, Martin.
Acquisition of data. Seol, McCabe, Choe, Zheng, Yu, Jang, Walter, Lehman, Ding.
Analysis and interpretation of data. Seol, McCabe, Martin.
REFERENCES
- 1.Buckwalter JA, Brown TD. Joint injury, repair, and remodeling: roles in post-traumatic osteoarthritis. Clin Orthop Relat Res. 2004;423:7–16. [PubMed] [Google Scholar]
- 2.Marsh JL, Borrelli J, Jr, Dirschl DR, Sirkin MS. Fractures of the tibial plafond. Instr Course Lect. 2007;56:331–52. [PubMed] [Google Scholar]
- 3.Marsh JL, Weigel DP, Dirschl DR. Tibial plafond fractures: how do these ankles function over time? J Bone Joint Surg Am. 2003;85-A:287–95. [PubMed] [Google Scholar]
- 4.Hunziker EB. The elusive path to cartilage regeneration. Adv Mater. 2009;21:3419–24. doi: 10.1002/adma.200801957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dowthwaite GP, Bishop JC, Redman SN, Khan IM, Rooney P, Evans DJ, et al. The surface of articular cartilage contains a progenitor cell population. J Cell Sci. 2004;117:889–97. doi: 10.1242/jcs.00912. [DOI] [PubMed] [Google Scholar]
- 6.Grogan SP, Miyaki S, Asahara H, D'Lima DD, Lotz MK. Mesenchymal progenitor cell markers in human articular cartilage: normal distribution and changes in osteoarthritis. Arthritis Res Ther. 2009;11:R85. doi: 10.1186/ar2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hattori S, Oxford C, Reddi AH. Identification of superficial zone articular chondrocyte stem/progenitor cells. Biochem Biophys Res Commun. 2007;358:99–103. doi: 10.1016/j.bbrc.2007.04.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khan IM, Williams R, Archer CW. One flew over the progenitor's nest: migratory cells find a home in osteoarthritic cartilage. Cell Stem Cell. 2009;4:282–4. doi: 10.1016/j.stem.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 9.Hilfiker A, Kasper C, Hass R, Haverich A. Mesenchymal stem cells and progenitor cells in connective tissue engineering and regenerative medicine: is there a future for transplantation? Langenbecks Arch Surg. 2011;396:489–97. doi: 10.1007/s00423-011-0762-2. [DOI] [PubMed] [Google Scholar]
- 10.Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, et al. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143–7. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 11.Prockop DJ. Repair of tissues by adult stem/progenitor cells (MSCs): controversies, myths, and changing paradigms. Mol Ther. 2009;17:939–46. doi: 10.1038/mt.2009.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Quesenberry PJ, Colvin G, Dooner G, Dooner M, Aliotta JM, Johnson K. The stem cell continuum: cell cycle, injury, and phenotype lability. Ann N Y Acad Sci. 2007;1106:20–9. doi: 10.1196/annals.1392.016. [DOI] [PubMed] [Google Scholar]
- 13.Spees JL, Whitney MJ, Sullivan DE, Lasky JA, Laboy M, Ylostalo J, et al. Bone marrow progenitor cells contribute to repair and remodeling of the lung and heart in a rat model of progressive pulmonary hypertension. FASEB J. 2008;22:1226–36. doi: 10.1096/fj.07-8076com. [DOI] [PubMed] [Google Scholar]
- 14.Williams RJ, III, Harnly HW. Microfracture: indications, technique, and results. Instr Course Lect. 2007;56:419–28. [PubMed] [Google Scholar]
- 15.Alsalameh S, Amin R, Gemba T, Lotz M. Identification of mesenchymal progenitor cells in normal and osteoarthritic human articular cartilage. Arthritis Rheum. 2004;50:1522–32. doi: 10.1002/art.20269. [DOI] [PubMed] [Google Scholar]
- 16.Golebiewska A, Brons NH, Bjerkvig R, Niclou SP. Critical appraisal of the side population assay in stem cell and cancer stem cell research. Cell Stem Cell. 2011;8:136–47. doi: 10.1016/j.stem.2011.01.007. [DOI] [PubMed] [Google Scholar]
- 17.Koelling S, Kruegel J, Irmer M, Path JR, Sadowski B, Miro X, et al. Migratory chondrogenic progenitor cells from repair tissue during the later stages of human osteoarthritis. Cell Stem Cell. 2009;4:324–35. doi: 10.1016/j.stem.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 18.Klune JR, Dhupar R, Cardinal J, Billiar TR, Tsung A. HMGB1: endogenous danger signaling. Mol Med. 2008;14:476–84. doi: 10.2119/2008-00034.Klune. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Q, O'Hearn S, Kavalukas SL, Barbul A. Role of high mobility group box 1 (HMGB1) in wound healing. J Surg Res. 2012;176:343–7. doi: 10.1016/j.jss.2011.06.069. [DOI] [PubMed] [Google Scholar]
- 20.Jeannin P, Jaillon S, Delneste Y. Pattern recognition receptors in the immune response against dying cells. Curr Opin Immunol. 2008;20:530–7. doi: 10.1016/j.coi.2008.04.013. [DOI] [PubMed] [Google Scholar]
- 21.Foell D, Wittkowski H, Roth J. Mechanisms of disease: a ‘DAMP’ view of inflammatory arthritis. Nat Clin Pract Rheumatol. 2007;3:382–90. doi: 10.1038/ncprheum0531. [DOI] [PubMed] [Google Scholar]
- 22.Meng E, Guo Z, Wang H, Jin J, Wang J, Wang H, et al. High mobility group box 1 protein inhibits the proliferation of human mesenchymal stem cells and promotes their migration and differentiation along osteoblastic pathway. Stem Cells Dev. 2008;17:805–13. doi: 10.1089/scd.2007.0276. [DOI] [PubMed] [Google Scholar]
- 23.Stich S, Loch A, Leinhase I, Neumann K, Kaps C, Sittinger M, et al. Human periosteum-derived progenitor cells express distinct chemokine receptors and migrate upon stimulation with CCL2, CCL25, CXCL8, CXCL12, and CXCL13. Eur J Cell Biol. 2008;87:365–76. doi: 10.1016/j.ejcb.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 24.Chavakis E, Urbich C, Dimmeler S. Homing and engraftment of progenitor cells: a prerequisite for cell therapy. J Mol Cell Cardiol. 2008;45:514–22. doi: 10.1016/j.yjmcc.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 25.Palumbo R, Galvez BG, Pusterla T, De Marchis F, Cossu G, Marcu KB, et al. Cells migrating to sites of tissue damage in response to the danger signal HMGB1 require NF-[H9260]B activation. J Cell Biol. 2007;179:33–40. doi: 10.1083/jcb.200704015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Girard JP. A direct inhibitor of HMGB1 cytokine. Chem Biol. 2007;14:345–7. doi: 10.1016/j.chembiol.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 27.Kitaori T, Ito H, Schwarz EM, Tsutsumi R, Yoshitomi H, Oishi S, et al. Stromal cell–derived factor 1/CXCR4 signaling is critical for the recruitment of mesenchymal stem cells to the fracture site during skeletal repair in a mouse model. Arthritis Rheum. 2009;60:813–23. doi: 10.1002/art.24330. [DOI] [PubMed] [Google Scholar]
- 28.Kucia M, Jankowski K, Reca R, Wysoczynski M, Bandura L, Allendorf DJ, et al. CXCR4-SDF-1 signalling, locomotion, chemo-taxis and adhesion. J Mol Histol. 2004;35:233–45. doi: 10.1023/b:hijo.0000032355.66152.b8. [DOI] [PubMed] [Google Scholar]
- 29.Carter DR, Beaupre GS, Wong M, Smith RL, Andriacchi TP, Schurman DJ. The mechanobiology of articular cartilage development and degeneration. Clin Orthop Relat Res. 2004;427(Suppl):S69–77. doi: 10.1097/01.blo.0000144970.05107.7e. [DOI] [PubMed] [Google Scholar]
- 30.Duda GN, Eilers M, Loh L, Hoffman JE, Kaab M, Schaser K. Chondrocyte death precedes structural damage in blunt impact trauma. Clin Orthop Relat Res. 2001;393:302–9. doi: 10.1097/00003086-200112000-00035. [DOI] [PubMed] [Google Scholar]
- 31.Isaac DI, Meyer EG, Haut RC. Chondrocyte damage and contact pressures following impact on the rabbit tibiofemoral joint. J Biomech Eng. 2008;130:041018. doi: 10.1115/1.2948403. [DOI] [PubMed] [Google Scholar]
- 32.Phillips DM, Haut RC. The use of a non-ionic surfactant (P188) to save chondrocytes from necrosis following impact loading of chondral explants. J Orthop Res. 2004;22:1135–42. doi: 10.1016/j.orthres.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 33.Rundell SA, Baars DC, Phillips DM, Haut RC. The limitation of acute necrosis in retro-patellar cartilage after a severe blunt impact to the in vivo rabbit patello-femoral joint. J Orthop Res. 2005;23:1363–9. doi: 10.1016/j.orthres.2005.06.001.1100230618. [DOI] [PubMed] [Google Scholar]
- 34.Beecher BR, Martin JA, Pedersen DR, Heiner AD, Buckwalter JA. Antioxidants block cyclic loading induced chondrocyte death. Iowa Orthop J. 2007;27:1–8. [PMC free article] [PubMed] [Google Scholar]
- 35.Martin JA, McCabe D, Walter M, Buckwalter JA, McKinley TO. N-acetylcysteine inhibits post-impact chondrocyte death in osteochondral explants. J Bone Joint Surg Am. 2009;91:1890–7. doi: 10.2106/JBJS.H.00545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goodwin W, McCabe D, Sauter E, Reese E, Walter M, Buckwalter JA, et al. Rotenone prevents impact-induced chondrocyte death. J Orthop Res. 2010;28:1057–63. doi: 10.1002/jor.21091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moussavi-Harami F, Mollano A, Martin JA, Ayoob A, Domann FE, Gitelis S, et al. Intrinsic radiation resistance in human chondrosarcoma cells. Biochem Biophys Res Commun. 2006;346:379–85. doi: 10.1016/j.bbrc.2006.05.158. [DOI] [PubMed] [Google Scholar]
- 38.Lee JW, Lim TH, Park JB. Intradiscal drug delivery system for the treatment of low back pain. J Biomed Mater Res A. 2010;92:378–85. doi: 10.1002/jbm.a.32377. [DOI] [PubMed] [Google Scholar]
- 39.Mollica L, De Marchis F, Spitaleri A, Dallacosta C, Pennacchini D, Zamai M, et al. Glycyrrhizin binds to high-mobility group box 1 protein and inhibits its cytokine activities. Chem Biol. 2007;14:431–41. doi: 10.1016/j.chembiol.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 40.Fiedler J, Etzel N, Brenner RE. To go or not to go: migration of human mesenchymal progenitor cells stimulated by isoforms of PDGF. J Cell Biochem. 2004;93:990–8. doi: 10.1002/jcb.20219. [DOI] [PubMed] [Google Scholar]
- 41.Jiang Y, Jahagirdar BN, Reinhardt RL, Schwartz RE, Keene CD, Ortiz-Gonzalez XR, et al. Pluripotency of mesenchymal stem cells derived from adult marrow. Nature. 2002;418:41–9. doi: 10.1038/nature00870. [DOI] [PubMed] [Google Scholar]
- 42.Seol D, Choe H, Zheng H, Jang K, Ramakrishnan PS, Lim TH, et al. Selection of reference genes for normalization of quantitative real-time PCR in organ culture of the rat and rabbit inter-vertebral disc. BMC Res Notes. 2011;4:162. doi: 10.1186/1756-0500-4-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kucia M, Reca R, Miekus K, Wanzeck J, Wojakowski W, Janowska-Wieczorek A, et al. Trafficking of normal stem cells and metastasis of cancer stem cells involve similar mechanisms: pivotal role of the SDF-1-CXCR4 axis. Stem Cells. 2005;23:879–94. doi: 10.1634/stemcells.2004-0342. [DOI] [PubMed] [Google Scholar]
- 44.Wolff DA, Stevenson S, Goldberg VM. S-100 protein immuno-staining identifies cells expressing a chondrocytic phenotype during articular cartilage repair. J Orthop Res. 1992;10:49–57. doi: 10.1002/jor.1100100106. [DOI] [PubMed] [Google Scholar]
- 45.Bao JP, Chen WP, Wu LD. Lubricin: a novel potential biotherapeutic approaches for the treatment of osteoarthritis. Mol Biol Rep. 2011;38:2879–85. doi: 10.1007/s11033-010-9949-9. [DOI] [PubMed] [Google Scholar]
- 46.Jay GD, Fleming BC, Watkins BA, McHugh KA, Anderson SC, Zhang LX, et al. Prevention of cartilage degeneration and restoration of chondroprotection by lubricin tribosupplementation in the rat following anterior cruciate ligament transection. Arthritis Rheum. 2010;62:2382–91. doi: 10.1002/art.27550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Teeple E, Elsaid KA, Jay GD, Zhang L, Badger GJ, Akelman M, et al. Effects of supplemental intra-articular lubricin and hyaluronic acid on the progression of posttraumatic arthritis in the anterior cruciate ligament-deficient rat knee. Am J Sports Med. 2011;39:164–72. doi: 10.1177/0363546510378088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hunziker EB, Rosenberg LC. Repair of partial-thickness defects in articular cartilage: cell recruitment from the synovial membrane. J Bone Joint Surg Am. 1996;78:721–33. doi: 10.2106/00004623-199605000-00012. [DOI] [PubMed] [Google Scholar]
- 49.Wang Q, Breinan HA, Hsu HP, Spector M. Healing of defects in canine articular cartilage: distribution of nonvascular [H9251]-smooth muscle actin-containing cells. Wound Repair Regen. 2000;8:145–58. doi: 10.1046/j.1524-475x.2000.00145.x. [DOI] [PubMed] [Google Scholar]