Abstract
Background. Adenomatous polyposis coli (APC) is widely known as an antagonist of the Wnt signaling pathway via the inactivation of β-catenin. An increasing number of studies have reported that APC methylation contributes to the predisposition to breast cancer (BC). However, recent studies have yielded conflicting results.
Methods. Herein, we systematically carried out a meta-analysis to assess the correlation between APC methylation and BC risk. Based on searches of the Cochrane Library, PubMed, Web of Science and Embase databases, the odds ratio (OR) with 95% confidence interval (CI) values were pooled and summarized.
Results. A total of 31 articles involving 35 observational studies with 2,483 cases and 1,218 controls met the inclusion criteria. The results demonstrated that the frequency of APC methylation was significantly higher in BC cases than controls under a random effect model (OR = 8.92, 95% CI [5.12–15.52]). Subgroup analysis further confirmed the reliable results, regardless of the sample types detected, methylation detection methods applied and different regions included. Interestingly, our results also showed that the frequency of APC methylation was significantly lower in early-stage BC patients than late-stage ones (OR = 0.62, 95% CI [0.42–0.93]).
Conclusion. APC methylation might play an indispensable role in the pathogenesis of BC and could be regarded as a potential biomarker for the diagnosis of BC.
Keywords: Breast cancer, APC, Methylation, Meta-analysis
Introduction
Breast cancer (BC) is the most common malignancy and the leading cause of cancer death among females in both well and poorly developed countries, accounting for approximately 15% of all cancer deaths in 2012 (Torre et al., 2015). It is well established that BC is a clinically and pathologically heterogeneous disease and has been categorized into five subtypes (i.e., luminal A and B, human epidermal growth receptor-2, triple negative and basal-like) based on various biological markers (Inoue & Fry, 2015). Risk factors including reproductive, hormonal and environmental factors, have been associated with an increased incidence of BC (Harrison et al., 2015). Previous studies have reported that early detection using mammography is effective and can improve the overall survival rate (Brooks et al., 2010). However, false positive mammograms always result in the over-diagnosis and over-treatment of developing BC. Therefore, no acknowledged biomarker has yet been proven to be sufficiently sensitive and specific for routine use in clinical diagnosis.
Epigenetic as well as genetic alterations are both stable and heritable and occur in tumor suppressor genes involved in tumourigenesis. The most common epigenetic alteration involving aberrant DNA methylation, a reliable and sensitive biomarker for nearly all types of cancer including breast cancer, often leads to the transcriptional silencing of tumor suppressor genes (Zmetakova et al., 2013). Several studies have demonstrated that tumor DNA derived from malignant cells can be detected in various bodily fluids and serum of BC patients and can potentially serve as a non-invasive diagnostic material (Martínez-Galán et al., 2014). A growing number of tumor suppressor genes has been shown to be directly involved in cell cycle regulation, DNA repair, cell signal transduction and angiogenesis (Dumitrescu, 2012). Notably, the promoter methylation of genes involved in the canonical Wnt signaling pathway, which regulates cell differentiation, proliferation and homeostasis, are observed more often in BC patients compared with cancer-free controls (Klarmann, Decker & Farrar, 2014).
The adenomatous polyposis coli (APC) gene is widely known as an antagonist of the Wnt signaling pathway via the inactivation of β-catenin, which is regarded as a transcriptional activator (Virmani et al., 2001). The APC gene, located at chromosome 5q21–5q22, was originally implicated in colorectal cancer (Van der Auwera et al., 2008). The inhibition or down-regulation of APC expression through APC promoter methylation contributes to the formation of colorectal cancer (Ashktorab et al., 2013). Similar to the findings in colorectal cancer, APC promoter methylation is associated with various early- or late-stage human malignancies, including BC (Matsuda et al., 2009). The promoter hypermethylation of APC is most often related to the nuclear accumulation of β-catenin, which may result in the loss of cell growth control (Sparks et al., 1998). Thus, APC promoter methylation, which acts as a non-invasive biomarker, can be used to distinguish BC patients from cancer-free controls. However, recent studies have yielded conflicting results with regard to the significant association between APC methylation and BC pathogenesis. Wojdacz et al. (2011a) reported that there was no significant difference in the frequency of APC methylation in peripheral blood leukocyte DNA between BC patients and cancer-free controls. Cho et al. (2010) also showed that the APC gene was rarely hypermethylated in blood DNA in BC patients.
Given these controversial results, we conducted this comprehensive meta-analysis of the current observational studies to evaluate the association between the aberrant methylation of the APC promoter and increased BC risk.
Materials & Methods
Search strategy
Eligible studies were identified by searching the following databases until February 2016: the Cochrane Library, PubMed, Web of Science and Embase. No language restrictions or lower data limits were imposed; only abstracts, unpublished and incomplete studies were excluded. Titles, abstracts of potential references and reference lists from relevant studies were carefully checked. We performed the search strategy using the following search terms and their various combinations: “APC,” “Adenomatous polyposis coli,” “methylation,” “breast cancer,” “breast neoplasm” and “mammary carcinoma.”
Selection criteria
The studies included in the present meta-analysis addressed the association between APC methylation and increased BC risk. Our inclusion criteria were as follows: (1) provided sufficient data on the frequency of APC methylation in BC patients and controls; (2) original observational studies in full-text form; and (3) when several studies overlapped, the most recent or large-scale article was selected. The following were exclusion criteria: (1) data based on reviews, animal models, case reports or cell line studies; (2) studies lacking key information necessary for calculations; (3) duplicated studies; and (4) studies including BC patients or controls who underwent radiotherapy and chemotherapy which may influence APC promoter methylation levels.
Data extraction
The relevant data were extracted from the eligible studies independently by two authors (D Zhou and WW Tang). Differing opinions, if any, were resolved by discussion in accordance with the original literature. The following information was extracted in a predefined table: the name of the first author, the year of publication, the country of origin, the sample type, the experimental methods used to detect APC methylation, sample size, tumor stage, tumor grade and APC methylation frequencies. Additionally, we classified stage 0, I and II as early-stage BC and stage III and IV as late-stage BC, as confirmed by the AJCC staging system. Furthermore, grades I and II were combined as low-grade BC; grade III was regarded as high-grade BC. This meta-analysis was performed following the statement of preferred reporting items set by the PRISMA Group (File S1) (Moher et al., 2009).
Statistical analysis
All analyses were carried out using Review Manager 5.3 (Cochrane Collaboration) and Stata 12.0 (Stata Corporation) software. Forest plots were designed to estimate relative study-specific effects according to the 95% confidence interval (CI). The association between APC promoter methylation and BC risk was evaluated by calculating the odds ratio (OR) with corresponding 95% CI. For individual studies the OR was represented by a square and the 95% CI by a horizontal line in the centre of the forest plot. The OR and associated 95% CIs in the centre of the forest plot were plotted on a logarithmic scale. When a CI did not include 1.0, a correlation was deemed statistically significant. Heterogeneity between the included studies was quantified through Q-tests based on the chi-square test and I2 value. An I2 value >50% and a p < 0.10 denoted strong heterogeneity, an I2 value = 25–50% denoted a moderate degree of heterogeneity and an I2 value <25% or a p > 0.10 denoted mild heterogeneity (Higgins et al., 2003). A random effect model was used when statistical heterogeneity existed among studies (p < 0.1). Otherwise, the fixed effect model was employed (Li et al., 2014). Moreover, the subgroup meta-analyses were also performed according to region, experimental methods for detecting APC methylation, and sample types in order to explore the potential origin of inter-study heterogeneity. In addition, we conducted a sensitivity analysis by removing a single study to examine the stability of the results. The funnel plot, Begg’s test and Egger’s test were investigated in order to determine the degree of publication bias. The treatment effect was plotted against a measure of study size in the funnel plot. When publication bias was present, the shape of the funnel plot was asymmetric. Trim and fill analysis was used to estimate the number of potential missing studies resulting from the asymmetry of the funnel plot.
Results
Study selection and characteristics
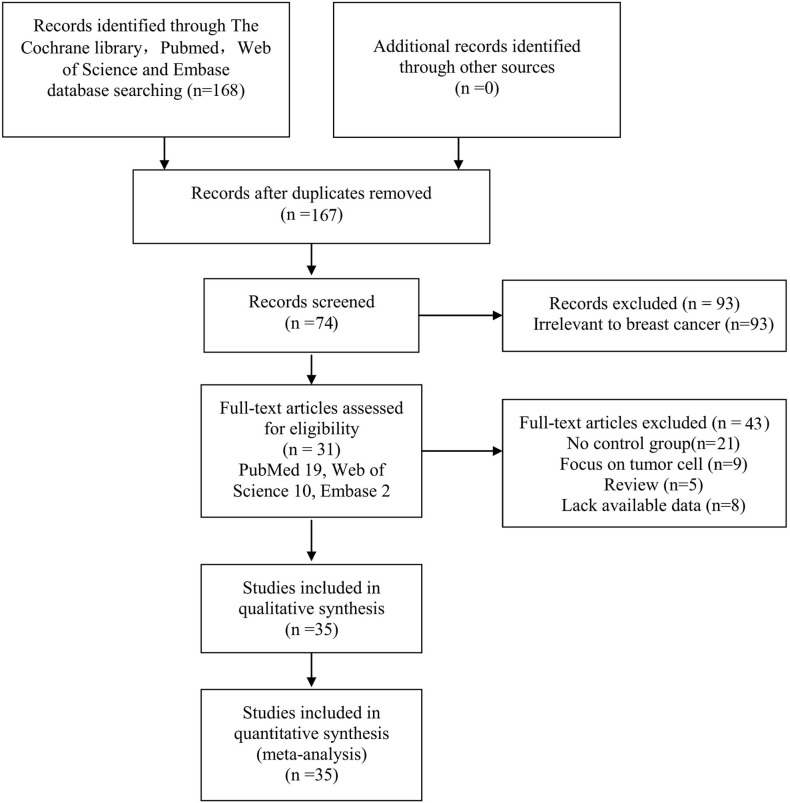
The selection process is displayed as a flow chart in Fig. 1 based on the search strategies as previously described. After a careful initial search of the abstracts, 74 potentially relevant articles were identified excluding 1 duplicate and 93 irrelevant studies. Then, we reviewed the full text articles. Among these studies, 43 were excluded (21 articles did not design a control group; 9 articles focused on BC cell lines; 8 articles lacked available data; and 5 articles were reviews). Finally, 31 studies published from 2001 to 2016 involving 35 studies were included in this systematic meta-analysis (PubMed 19, Web of Science 10, Embase 2).
Figure 1. Flow chart of the collection of studies for this meta-analysis.
The general characteristics of eligible studies were summarized and displayed in Table 1. A total of 2,483 BC patients and 1,218 controls were employed in multiple countries or regions including Asia (n = 10) (Jin et al., 2001; Jing et al., 2010; Jung et al., 2013; Lee et al., 2004; Liu et al., 2007; Park et al., 2011b; Prasad et al., 2008; Zhang et al., 2007), Europe (n = 13) (Fridrichova et al., 2015; Hoque et al., 2009; Jeronimo et al., 2008; Martins et al., 2011; Matuschek et al., 2010; Muller et al., 2003; Parrella et al., 2004; Rykova et al., 2004; Van der Auwera et al., 2009a; Van der Auwera et al., 2009b; Van der Auwera et al., 2008; Wojdacz et al., 2011b), Africa (n = 2) (Hoque et al., 2006; Swellam et al., 2015), North America (n = 9) (Brooks et al., 2010; Chen et al., 2011; Cho et al., 2010; Dulaimi et al., 2004; Lewis et al., 2005; Shinozaki et al., 2005; Taback et al., 2006; Virmani et al., 2001) and Oceania (n = 1) (Pang et al., 2014). Furthermore, the methylated APC levels in BC patients and controls were examined with 6 methods. Of these methods, methylation specific PCR (MSP) was adopted in 17 studies, quantitative real-time MSP (QMSP) was used in 9 studies, methylation specific-multiplex ligation-dependent probe amplification (MethyLight) was used in 4 studies, methylation specific-multiplex ligation-dependent probe amplification (MS-MLPA) was employed in 2 studies, methylation-sensitive high-resolution melting analysis (MS-HRM) was used in 2 studies and pyrosequencing was used in only 1 study. Furthermore, BC tissues (i.e., fresh frozen tissues, formalin fixed paraffin-embedded tissues and tissues from surgery), samples derived from blood (i.e., blood cells and serum) and needle aspirated fluid (NAF) were enrolled to assess the methylation levels of the APC promoter.
Table 1. General characteristics of eligible studies.
| Author | Year | County/region | Method | Sample type | M/N | Stage (M/N) | Grade (N/M) | |||
|---|---|---|---|---|---|---|---|---|---|---|
| BC | Control | Early | Late | Low | High | |||||
| Brooks JD | 2010 | USA | QMSP | Serum | 1/50 | 6/148 | – | – | – | – |
| Chen KM | 2011 | USA | MS-MLPA | FFT | 12/17 | 1/10 | – | – | – | – |
| Cho YH | 2010 | USA | MethyLight | FFT | 21/40 | 12/27 | – | – | – | – |
| Dulaimi E 1 | 2004 | USA | MSP | Surgery | 15/34 | 0/6 | 14/29 | 1/5 | 6/13 | 9/18 |
| Dulaimi E 2 | Serum | 10/34 | 0/20 | 9/29 | 1/5 | 4/13 | 6/18 | |||
| Fridrichova I | 2015 | Slovak Republic | Pyro | FFPET | 144/206 | 0/9 | – | – | – | – |
| Hoque MO | 2006 | West Africa | QMSP | Blood | 8/47 | 0/38 | – | – | – | – |
| Hoque MO | 2009 | Italy | QMSP | FFPET | 56/112 | 3/32 | – | – | – | – |
| Jeronimo C | 2008 | Portugal | QMSP | FFPET | 55/66 | 10/12 | – | – | – | – |
| Jin Z | 2001 | Japan | MSP | Surgery | 18/50 | 0/21 | 13/36 | 4/10 | – | – |
| Jing F | 2010 | China | MSP | Serum | 14/50 | 0/50 | – | – | 7/25 | 12/25 |
| Jung EJ | 2013 | Korea | MS-MLPA | Surgery | 19/60 | 0/60 | 17/53 | 2/7 | 13/40 | 6/20 |
| Lee A | 2004 | Korea | MSP | NAF | 14/33 | 0/19 | 13/31 | 1/2 | – | – |
| Lewis CM | 2005 | USA | MSP | NAF | 15/27 | 14/55 | – | – | – | – |
| Liu Z | 2007 | China | MSP | Surgery | 28/76 | 0/76 | 15/54 | 13/22 | 15/48 | 13/28 |
| Martins AT | 2011 | Portugal | QMSP | NAF | 144/178 | 18/33 | – | – | – | – |
| Matuschek C | 2010 | Germany | MethyLight | Blood | 25/85 | 2/22 | 5/42 | 16/35 | – | – |
| Müller HM | 2003 | Austria | MethyLight | Serum | 6/26 | 0/10 | – | – | – | – |
| Pang JM | 2014 | Australia | MS-HRM | FFPET | 39/80 | 0/15 | – | – | – | – |
| Park SY | 2011 | South Korea | MethyLight | FFPET | 31/85 | 2/30 | – | – | 13/30 | 6/20 |
| Parrella P | 2004 | Italy | MSP | Tissue | 15/54 | 1/10 | – | – | – | – |
| Prasad CP 1 | 2008 | India | MSP | Surgery | 6/32 | 0/5 | 2/19 | 4/13 | – | – |
| Prasad CP 2 | Serum | 11/50 | 0/50 | – | – | 4/28 | 7/22 | |||
| Rykova EY | 2004 | Russia | MSP | Blood | 4/10 | 0/6 | – | – | – | – |
| Shinozaki M | 2005 | USA | MSP | FFPET | 74/151 | 0/10 | – | – | – | – |
| Swellam M | 2015 | Egypt | MSP | Serum | 113/121 | 0/66 | 81/86 | 32/35 | 84/89 | 29/32 |
| Taback B | 2006 | USA | QMSP | Blood | 1/33 | 0/10 | – | – | – | – |
| Van der A I | 2009 | Belgium | QMSP | FFT | 60/100 | 0/9 | – | – | – | – |
| Van der A I 1 | 2008 | Belgium | MSP | FFPET | 28/51 | 3/27 | – | – | – | – |
| Van der A I 2 | QMSP | FFT | 53/54 | 7/9 | – | – | – | – | ||
| Van der A I | 2009 | Belgium | QMSP | Blood | 15/78 | 1/19 | – | – | – | – |
| Virmani AK | 2001 | USA | MSP | Surgery | 19/45 | 3/28 | – | – | – | – |
| Wojdacz TK | 2011 | Denmark | MS-HRM | Blood | 24/180 | 13/108 | – | – | – | – |
| Zhang JJ 1 | 2007 | China | MSP | Surgery | 38/84 | 0/84 | 30/66 | 8/18 | – | – |
| Zhang JJ 2 | Serum | 26/84 | 0/10 | 20/66 | 6/18 | – | – | |||
| Total | 1162/2483 | 96/1218 | 219/511 | 88/170 | 146/286 | 88/183 | ||||
Notes.
- MSP
- Methylation specific PCR
- QMSP
- Quantitative real-time MSP
- Pyro
- Pyrosequencing
- MS-HRM
- Methylation-sensitive high-resolution melting analysis
- FFPET
- Formalin fixed paraffin-embedded tissue
- FFT
- Fresh frozen tissue
- NAF
- Needle aspirate fluid
- MS-MLPA
- Methylation specific-multiplex ligation-dependent probe amplification
- M
- Number of APC promoter methylated patients
- N
- Number of control
Meta-analysis
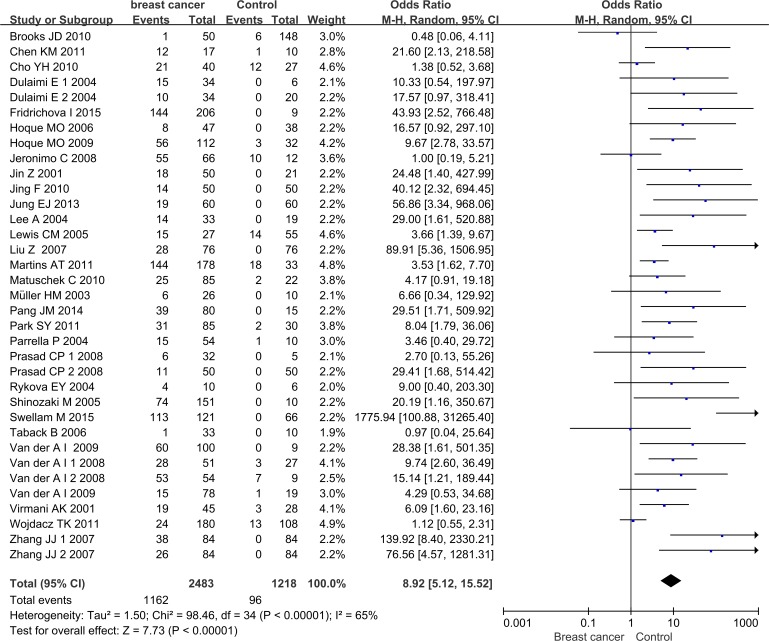
The pooled results of this meta-analysis reflected the association between APC promoter methylation and BC pathogenesis (Fig. 2). Due to the existence of significant heterogeneity among the included studies (p < 0.00001, I2 = 65%), the random effect model was adopted to evaluate the combined effects of APC promoter methylation. The overall analysis indicated that the frequency of APC promoter methylation was remarkably higher in BC patients than in cancer-free controls. The combined OR for 35 included relevant studies showed that APC methylation was significantly correlated with increased BC risk and the absence of APC expression played an important role in BC pathogenesis (OR = 8.92, 95% CI [5.12–15.52]).
Figure 2. Forest plot of APC promoter methylation and breast cancer risk based on the random effects model.
The small squares and horizontal lines represent the OR and 95% CI of individual studies. If the 95% CI included 1, the difference in APC methylation between patients with breast cancer and controls was not significant. The centre of the diamond represents the combined treatment effect (calculated as a weighted average of individual ORs) and the horizontal tips represent the 95% CI. OR represents the odds ratio. 95% CI represents the 95% confidence interval.
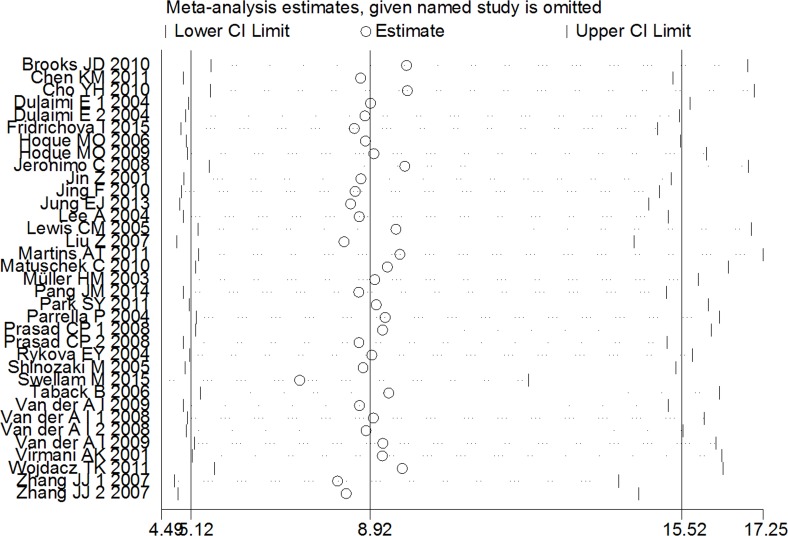
Sensitivity analysis
A sensitivity analysis was conducted by omitting one individual study every time to evaluate the stability of the pooled OR and to choose the heterogeneous study. As shown in Fig. 3, the combined OR between APC methylation and increased BC risk was indeed reliable without heterogeneous studies.
Figure 3. Sensitive analysis of pooled OR based on the random effects model.
The results were calculated by omitting each study in turn. The circles represent the individual studies in this meta-analysis. The two ends of the dotted lines represent the 95% CI. OR represents the odds ratio. 95% CI represents the 95% confidence interval.
Subgroup analysis
Due to the significant existence of inter-study heterogeneity (p < 0.00001, I2 = 65%), subgroup analysis based on region, experimental methods for the detection of APC methylation and sample types were carried out to appraise the sources of the heterogeneity (Table 2). With regard to subgroup analysis based on region, heterogeneity in Asian subgroups disappeared completely (I2 = 0%) and the pooled OR value was 24.48 [10.94, 54.74]. The I2 value representing heterogeneity in the European and North American subgroups decreased by 50% and 42%, compared with the overall I2 value. Furthermore, their OR values also decreased to 4.63 [2.44, 8.78] and 3.79 [1.70, 8.44]. In the African subgroup, the OR was 172.05 [1.76, 16792.96] with higher heterogeneity (I2 = 80%) due to the small subset containing only 2 studies. These results indicated that the heterogeneity might result from different regions and APC methylation was remarkably related to increased BC risk without geographical restrictions. For the subgroup analyses based on sample types, the blood or serum group (OR = 9.44, 95% CI [2.56–34.83]) made the largest contribution to the heterogeneity (I2 = 78%). In the tissue subgroup, the OR was 9.93 [5.10, 19.34] with lower heterogeneity (I2 = 50%). Moreover, heterogeneity in the NAF subgroups could be ignored (I2 = 6%). These results confirmed the stable association between APC methylation and BC risk in different sample types. For studies based on the methods used to detect the methylation of the APC promoter, the combined OR value was 18.18 for MSP (95% CI [7.96–41.52]), 3.93 for QMSP (95% CI [1.78–8.69]), 3.29 for MethyLight (95% CI [1.27–8.52]) and 31.81 for MS-MLPA (95% CI [5.30–191.06]). Heterogeneity in the QMSP (I2 = 36%) and MS-MLPA (I2 = 0%) subgroups was far lower than that of the MethyLight and MS-HRM subgroups (I2 = 83%).
Table 2. Subgroup analysis for the relationship between APC promoter methylation and breast cancer.
| Subgroup | No | BC M/N | Control M/N | OR (95% CI) | Heterogeneity test | ||
|---|---|---|---|---|---|---|---|
| I2 | p | Chi2 | |||||
| Sample types | |||||||
| Tissue | 19 | 731/1397 | 42/480 | 9.93 [5.10, 19.34] | 50% | 0.006 | 36.34 |
| Blood or Serum | 13 | 258/848 | 22/631 | 9.44 [2.56, 34.83] | 78% | <0.00001 | 55.34 |
| NAF | 3 | 173/238 | 32/107 | 3.95 [2.10, 7.42] | 6% | 0.34 | 2.13 |
| Region | |||||||
| Asia | 10 | 205/604 | 2/479 | 24.48 [10.94, 54.74] | 0% | 0.53 | 8.07 |
| Europe | 13 | 629/1200 | 58/306 | 4.63 [2.44, 8.78] | 50% | 0.02 | 24.18 |
| North America | 9 | 168/430 | 36/314 | 3.79 [1.70, 8.44] | 42% | 0.09 | 13.76 |
| Africa | 2 | 121/168 | 0/104 | 172.05 [1.76, 16792.96] | 80% | 0.02 | 5.07 |
| Oceania | 1 | 39/80 | 0/15 | 29.51 [1.71, 509.92] | NA | NA | NA |
| Methods | |||||||
| MSP | 17 | 448/986 | 21/617 | 18.18 [7.96, 41.52] | 54% | 0.004 | 35.03 |
| QMSP | 9 | 393/718 | 45/310 | 3.93 [1.78, 8.69] | 39% | 0.11 | 13.20 |
| MethyLight | 4 | 83/236 | 16/89 | 3.29 [1.27, 8.52] | 36% | 0.20 | 4.66 |
| MS-MLPA | 2 | 31/77 | 1/70 | 31.81 [5.30, 191.06] | 0% | 0.58 | 0.31 |
| MS-HRM | 2 | 63/260 | 13/123 | 4.49 [0.14, 146.62] | 83% | 0.02 | 5.76 |
| Pyro | 1 | 144/206 | 0/9 | 43.93 [2.52, 766.48] | NA | NA | NA |
Notes.
- NAF
- Needle aspirate fluid
- MSP
- Methylation specific PCR
- QMSP
- Quantitative real-time MSP
- Pyro
- Pyrosequencing
- MS-MLPA
- Methylation specific-multiplex ligation-dependent probe amplification
- MS-HRM
- Methylation-sensitive high-resolution melting analysis
- NA
- Not available
- M
- Number of APC promoter methylated patients
- N
- Number of control
To assess the association between APC methylation and tumor stage, 11 studies comprising 681 BC patients were pooled to calculate the OR. The results showed that the frequency of APC promoter methylation was significantly lower in early-stage patients than in late-stage patients (OR = 0.62, 95% CI [0.42–0.93], I2 = 34%). Meanwhile, the OR of 8 studies revealed that the association between APC methylation and tumor grade was not statistically significant (OR = 0.78, 95% CI [0.51–1.21], I2 = 0%).
Publication bias
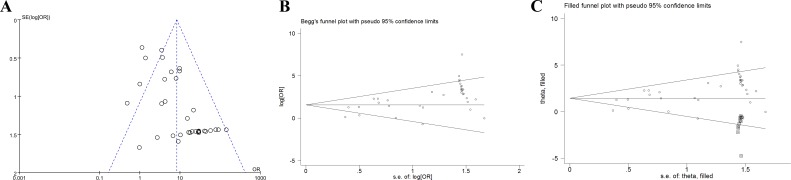
We used the funnel plot, Begg’s test and Egger’s test to evaluate the degree of publication bias. The shape of the funnel plot had no obvious asymmetry (Fig. 4A). Moreover, Begg’s test (Pr > |z| = 0.239 > 0.05) suggested no significant publication bias (Fig. 4B). Interestingly, Egger’s test revealed evident statistical proof for the existence of publication bias (p > |t| = 0.000 < 0.05). Therefore, we carried out trim and fill analysis to identify and revise the bias. As shown in Fig. 4C, 12 adjusted studies were added to the initial meta-analysis. The corrected OR was still highly significant for the association between APC methylation and BC risk (OR = 1.444, 95% CI [1.081–1.965]), further proving the stability of our meta-analysis.
Figure 4. Publication bias analysis.
(A) The funnel plot of APC methylation and breast cancer risk. The log of OR against the standard error of the log of the OR was plotted in this graph.(B) The Begg’s plot of APC methylation and breast cancer risk. The circles represent the individual studies in this meta-analysis. The line in the centre represents the pooled OR. (C) The Begg’s plot of publication bias after trim-and-fill analysis. The circles represent the included studies. The diamonds represent the presumed missing studies. OR represents the odds ratio.
Discussion
To the best of our knowledge, this is the first meta-analysis to systematically evaluate the association between APC promoter methylation and BC pathogenesis. BC is a significant clinical and public health problem and is mainly attributed to epigenetic and genetic changes. Epigenetic alternation involving DNA methylation is a relatively early event that serves as a tumor molecular biomarker candidate in BC and can be detected in all pathological tumor stages. The APC gene is considered to be a tumor suppressor gene, and the silencing of its expression may result in cell-to-cell adhesion disorders and the disruption of the Wnt signaling pathway. APC methylation, a contributing factor to the absence of APC expression, is often linked to β-catenin accumulation and TCF/LEF-induced transcription (Klarmann, Decker & Farrar, 2008). Numerous studies have reported that APC methylation is highly specific for BC and can be used as a biomarker in the diagnosis of BC (Dumitrescu, 2012; Van der Auwera et al., 2008). Zhang, Li & Lang (2015) found that β-catenin overexpression was significantly associated with an unfavourable prognosis in patients with breast cancer. However, the role of APC methylation in BC pathogenesis remains controversial.
To resolve these contradictory results, we gathered relevant studies and carried out this meta-analysis using systematic statistical methods. Herein, we included a total of 35 studies with 2,483 cases and 1,218 controls published from 2001 to 2016. Our results based on the pooled OR revealed that the level of APC methylation was observably higher in BC patients compared to cancer-free controls, which indicated that APC methylation could serve as a potential biomarker for BC diagnosis, regardless of the various sample types detected, APC methylation detection methods applied and cases employed in different regions.
Then, we conducted subgroup analysis to identify the sources of the heterogeneity and found that various sample types, methylation detection methods and cases employed in different regions all contributed to the heterogeneity. In subgroup analysis based on sample types, the results showed that APC methylation was significantly related to BC pathogenesis, whether in tissue, blood or serum and NAF. Cell-free DNA in serum and plasma, which mostly originates from tumor cell degradation, can be collected and examined for epigenetic alterations with various malignancies (Anker et al., 1999). The sample materials including blood or serum, used for extracting DNA are often stored for different time periods which will produce false positives and false negatives. Thus, blood samples should be examined as rapidly as possible after being collected. Therefore, the accuracy of cell-free DNA largely depends on the standardized storage conditions. NAF is a rapid, minimally invasive and cheap diagnostic means with high sensitivity. The accuracy of NAF mainly relies on the experience of the cytopathologist which may result in an increasing trend for false negatives (Jeronimo et al., 2003). In subgroup analysis based on methylation detection methods, significant associations were observed when examined using MSP, QMSP, MethyLight and MS-MLPA, except for MS-HRM. Among these, the pooled OR derived from studies using MS-MLPA was the maximum with no heterogeneity. The diagnostic accuracy of MS-MLPA was not affected by sample types (Park et al., 2011a). Cut-off values and primers based on different CPG islands which were used in different studies, contributed to the heterogeneity of other methods. In subgroup analysis based on different regions, APC methylation was significantly correlated with BC patients in all included regions. The results indicated that although the genetic factors, environments and life styles were totally different, the correlation was still strong and stable. Therefore, an appropriate APC methylation detection method considering the regions and sample types employed is essential for routine clinical diagnosis. Additionally, we found that the status of APC methylation increased notably in late-stage patients compared with early-stage ones, which indicated that APC methylation might be closely related to the malignant evolution of BC.
As mentioned above, Wojdacz et al. (2011b) examined the use of methylation biomarkers as screening tools for BC diagnosis. They found no significant difference in the frequency between 180 BC patients and 108 healthy controls and a weak association between APC methylation and BC pathogenesis. This discrepancy mainly resulted from the methylation detection method. They used MS-HRM which may yield heterogeneous methylation values derived from the primer and cut-off values, and it tended to produce a lower evaluation of methylation when applying less methylated samples (Migheli et al., 2013).
Surprisingly, only Egger’s linear regression showed an obvious publication bias other than Begg’s test and funnel plots. Egger et al. (1997) suggested that Egger’s test was more sensitive than Begg’s test. The publication bias mainly resulted from the inclusion criteria. Only full-text published studies were collected in this meta-analysis. Therefore, unpublished studies and conference abstracts were not included. Additionally, other study characteristics including the source of funding and prevailing theories at the time of publication, can contribute to publication bias. However, we included a large number of BC patients (n = 2,483) to ensure the reliability of the meta-analysis and minimize the potential publication bias.
Although the meta-analysis indeed confirmed the significance of a correlation between APC methylation and BC pathogenesis, several limitations should be considered. First, the sample sizes used in several studies were small, which may have increased the risk of publication bias and limited the results of the meta-analysis. Second, the quality of the selected studies varied, as we included high-quality and low-quality studies. Therefore, heterogeneity likely existed. Third, the cut-off points of APC methylation and the primers based on CPG islands were difficult to unify. Thus, we were unable to calculate the pooled sensitivity and specificity of APC methylation.
In conclusion, the results of our meta-analysis highlight the clinical significance and scientific value of APC promoter methylation in the diagnosis of BC. Consequently, APC methylation is a potential biomarker for monitoring BC development. However, given the limitations listed above, high-quality studies with large-scale and consistent standards should be carried out. The guidelines for the reporting of tumor marker studies recommended by the National Cancer Institute are necessary for adaptation to high-quality studies (McShane et al., 2005).
Supplemental Information
Funding Statement
This study was supported by grants from the National Natural Science Foundation of China (No. 31400699) and the Natural Science Foundation of Fujian Province of China (No. 2014J01142). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Contributor Information
Han-Xiang An, Email: anhanxiang@yahoo.com.
Yun Zhang, Email: zhangy@fjirsm.ac.cn.
Additional Information and Declarations
Competing Interests
The authors declare there are no competing interests.
Author Contributions
Dan Zhou conceived and designed the experiments, performed the experiments, wrote the paper, prepared figures and/or tables.
Weiwei Tang performed the experiments, prepared figures and/or tables.
Wenyi Wang analyzed the data.
Xiaoyan Pan contributed reagents/materials/analysis tools.
Han-Xiang An and Yun Zhang conceived and designed the experiments, reviewed drafts of the paper.
Data Availability
References
- Anker et al. (1999).Anker P, Mulcahy H, Chen XQ, Stroun M. Detection of circulating tumour DNA in the blood (plasma/serum) of cancer patients. Cancer and Metastasis Reviews. 1999;18:65–73. doi: 10.1023/A:1006260319913. [DOI] [PubMed] [Google Scholar]
- Ashktorab et al. (2013).Ashktorab H, Rahi H, Wansley D, Varma S, Shokrani B, Lee E, Daremipouran M, Laiyemo A, Goel A, Carethers JM, Brim H. Toward a comprehensive and systematic methylome signature in colorectal cancers. Epigenetics. 2013;8:807–815. doi: 10.4161/epi.25497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks et al. (2010).Brooks JD, Cairns P, Shore RE, Klein CB, Wirgin I, Afanasyeva Y, Zeleniuch-Jacquotte A. DNA methylation in pre-diagnostic serum samples of breast cancer cases: results of a nested case-control study. Cancer Epidemiology. 2010;34:717–723. doi: 10.1016/j.canep.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen et al. (2011).Chen KM, Stephen JK, Raju U, Worsham MJ. Delineating an epigenetic continuum for initiation, transformation and progression to breast cancer. Cancers (Basel) 2011;3:1580–1592. doi: 10.3390/cancers3021580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho et al. (2010).Cho YH, Yazici H, Wu HC, Terry MB, Gonzalez K, Qu M, Dalay N, Santella RM. Aberrant promoter hypermethylation and genomic hypomethylation in tumor, adjacent normal tissues and blood from breast cancer patients. Anticancer Research. 2010;30:2489–2496. [PMC free article] [PubMed] [Google Scholar]
- Dulaimi et al. (2004).Dulaimi E, Hillinck J, Ibanez de Caceres I, Al-Saleem T, Cairns P. Tumor suppressor gene promoter hypermethylation in serum of breast cancer patients. Clinical Cancer Research. 2004;10:6189–6193. doi: 10.1158/1078-0432.CCR-04-0597. [DOI] [PubMed] [Google Scholar]
- Dumitrescu (2012).Dumitrescu RG. Epigenetic markers of early tumor development. Methods in Molecular Biology. 2012;863:3–14. doi: 10.1007/978-1-61779-612-8_1. [DOI] [PubMed] [Google Scholar]
- Egger et al. (1997).Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridrichova et al. (2015).Fridrichova I, Smolkova B, Kajabova V, Zmetakova I, Krivulcik T, Mego M, Cierna Z, Karaba M, Benca J, Pindak D, Bohac M, Repiska V, Danihel L. CXCL12 and ADAM23 hypermethylation are associated with advanced breast cancers. Translational Research. 2015;165:717–730. doi: 10.1016/j.trsl.2014.12.006. [DOI] [PubMed] [Google Scholar]
- Harrison et al. (2015).Harrison K, Hoad G, Scott P, Simpson L, Horgan GW, Smyth E, Heys SD, Haggarty P. Breast cancer risk and imprinting methylation in blood. Clin Epigenetics. 2015;7 doi: 10.1186/s13148-015-0125-x. Article 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins et al. (2003).Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoque et al. (2006).Hoque MO, Feng Q, Toure P, Dem A, Critchlow CW, Hawes SE, Wood T, Jeronimo C, Rosenbaum E, Stern J, Yu M, Trink B, Kiviat NB, Sidransky D. Detection of aberrant methylation of four genes in plasma DNA for the detection of breast cancer. Journal of Clinical Oncology. 2006;24:4262–4269. doi: 10.1200/JCO.2005.01.3516. [DOI] [PubMed] [Google Scholar]
- Hoque et al. (2009).Hoque MO, Prencipe M, Poeta ML, Barbano R, Valori VM, Copetti M, Gallo AP, Brait M, Maiello E, Apicella A, Rossiello R, Zito F, Stefania T, Paradiso A, Carella M, Dallapiccola B, Murgo R, Carosi I, Bisceglia M, Fazio VM, Sidransky D, Parrella P. Changes in CpG islands promoter methylation patterns during ductal breast carcinoma progression. Cancer Epidemiology, Biomarkers & Prevention. 2009;18:2694–2700. doi: 10.1158/1055-9965.EPI-08-0821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue & Fry (2015).Inoue K, Fry EA. Aberrant splicing of estrogen receptor, HER2, and CD44 genes in breast cancer. Genetics and Epigenetics. 2015;7:19–32. doi: 10.4137/GEG.S35500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeronimo et al. (2003).Jeronimo C, Costa I, Martins MC, Monteiro P, Lisboa S, Palmeira C, Henrique R, Teixeira MR, Lopes C. Detection of gene promoter hypermethylation in fine needle washings from breast lesions. Clinical Cancer Research. 2003;9:3413–3417. [PubMed] [Google Scholar]
- Jeronimo et al. (2008).Jeronimo C, Monteiro P, Henrique R, Dinis-Ribeiro M, Costa I, Costa VL, Filipe L, Carvalho AL, Hoque MO, Pais I, Leal C, Teixeira MR, Sidransky D. Quantitative hypermethylation of a small panel of genes augments the diagnostic accuracy in fine-needle aspirate washings of breast lesions. Breast Cancer Research and Treatment. 2008;109:27–34. doi: 10.1007/s10549-007-9620-x. [DOI] [PubMed] [Google Scholar]
- Jin et al. (2001).Jin Z, Tamura G, Tsuchiya T, Sakata K, Kashiwaba M, Osakabe M, Motoyama T. Adenomatous polyposis coli (APC) gene promoter hypermethylation in primary breast cancers. British Journal of Cancer. 2001;85:69–73. doi: 10.1054/bjoc.2001.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing et al. (2010).Jing F, Yuping W, Yong C, Jie L, Jun L, Xuanbing T, Lihua H. CpG island methylator phenotype of multigene in serum of sporadic breast carcinoma. Tumour Biology. 2010;31:321–331. doi: 10.1007/s13277-010-0040-x. [DOI] [PubMed] [Google Scholar]
- Jung et al. (2013).Jung EJ, Kim IS, Lee EY, Kang JE, Lee SM, Kim DC, Kim JY, Park ST. Comparison of methylation profiling in cancerous and their corresponding normal tissues from korean patients with breast cancer. Annals of Laboratory Medicine. 2013;33:431–440. doi: 10.3343/alm.2013.33.6.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klarmann, Decker & Farrar (2008).Klarmann GJ, Decker A, Farrar WL. Epigenetic gene silencing in the Wnt pathway in breast cancer. Epigenetics. 2008;3:59–63. doi: 10.4161/epi.3.2.5899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klarmann, Decker & Farrar (2014).Klarmann GJ, Decker A, Farrar WL. Epigenetic gene silencing in the Wnt pathway in breast cancer. Epigenetics. 2014;3:59–63. doi: 10.4161/epi.3.2.5899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee et al. (2004).Lee A, Kim Y, Han K, Kang CS, Jeon HM, Shim SI. Detection of tumor markers including carcinoembryonic antigen, APC, and cyclin D2 in fine-needle aspiration fluid of breast. Archives of Pathology and Laboratory Medicine. 2004;128:1251–1256. doi: 10.1043/1543-2165(2004)128<1251:DOTMIC>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Lewis et al. (2005).Lewis CM, Cler LR, Bu DW, Zochbauer-Muller S, Milchgrub S, Naftalis EZ, Leitch AM, Minna JD, Euhus DM. Promoter hypermethylation in benign breast epithelium in relation to predicted breast cancer risk. Clinical Cancer Research. 2005;11:166–172. [PubMed] [Google Scholar]
- Li et al. (2014).Li S, Zeng XT, Ruan XL, Weng H, Liu TZ, Wang X, Zhang C, Meng Z, Wang XH. Holmium laser enucleation versus transurethral resection in patients with benign prostate hyperplasia: an updated systematic review with meta-analysis and trial sequential analysis. PLoS ONE. 2014;9:e2203. doi: 10.1371/journal.pone.0101615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu et al. (2007).Liu Z, Yang L, Cui DX, Liu BL, Zhang XB, Ma WF, Zhang Q. Methylation status and protein expression of adenomatous polyposis coli (APC) gene in breast cancer. Ai Zheng. 2007;26:586–590. [PubMed] [Google Scholar]
- Martínez-Galán et al. (2014).Martínez-Galán J, Torres B, Del Moral R, Muñnoz-Gámez JA, Martín-Oliva D, Villalobos M, Núñez MI, Luna Jde D, Oliver FJ, Ruiz de Almodóvar JM. Quantitative detection of methylated ESR1 and 14-3-3-σ gene promoters in serum as candidate biomarkers for diagnosis of breast cancer and evaluation of treatment efficacy. Cancer Biology and Therapy. 2014;7:958–965. doi: 10.4161/cbt.7.6.5966. [DOI] [PubMed] [Google Scholar]
- Martins et al. (2011).Martins AT, Monteiro P, Ramalho-Carvalho J, Costa VL, Dinis-Ribeiro M, Leal C, Henrique R, Jeronimo C. High RASSF1A promoter methylation levels are predictive of poor prognosis in fine-needle aspirate washings of breast cancer lesions. Breast Cancer Research and Treatment. 2011;129:1–9. doi: 10.1007/s10549-010-1160-0. [DOI] [PubMed] [Google Scholar]
- Matsuda et al. (2009).Matsuda Y, Schlange T, Oakeley EJ, Boulay A, Hynes NE. WNT signaling enhances breast cancer cell motility and blockade of the WNT pathway by sFRP1 suppresses MDA-MB-231 xenograft growth. Breast Cancer Research. 2009;11 doi: 10.1186/bcr2317. Article R32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matuschek et al. (2010).Matuschek C, Bolke E, Lammering G, Gerber PA, Peiper M, Budach W, Taskin H, Prisack HB, Schieren G, Orth K, Bojar H. Methylated APC and GSTP1 genes in serum DNA correlate with the presence of circulating blood tumor cells and are associated with a more aggressive and advanced breast cancer disease. European Journal of Medical Research. 2010;15:277–286. doi: 10.1186/2047-783X-15-7-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McShane et al. (2005).McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM, Statistics Subcommittee of the NCIEWGoCD Reporting recommendations for tumor marker prognostic studies (REMARK) Journal of the National Cancer Institute. 2005;97:1180–1184. doi: 10.1093/jnci/dji237. [DOI] [PubMed] [Google Scholar]
- Migheli et al. (2013).Migheli F, Stoccoro A, Coppede F, Wan Omar WA, Failli A, Consolini R, Seccia M, Spisni R, Miccoli P, Mathers JC, Migliore L. Comparison study of MS-HRM and pyrosequencing techniques for quantification of APC and CDKN2A gene methylation. PLoS ONE. 2013;8:e2203. doi: 10.1371/journal.pone.0052501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moher et al. (2009).Moher D, Liberati A, Tetzlaff J, Altman DG, Group P. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Journal of Clinical Epidemiology. 2009;62:1006–1012. doi: 10.1016/j.jclinepi.2009.06.005. [DOI] [PubMed] [Google Scholar]
- Muller et al. (2003).Muller HM, Widschwendter A, Fiegl H, Ivarsson L, Goebel G, Perkmann E, Marth C, Widschwendter M. DNA methylation in serum of breast cancer patients: an independent prognostic marker. Cancer Research. 2003;63:7641–7645. [PubMed] [Google Scholar]
- Pang et al. (2014).Pang JM, Deb S, Takano EA, Byrne DJ, Jene N, Boulghourjian A, Holliday A, Millar E, Lee CS, O’Toole SA, Dobrovic A, Fox SB. Methylation profiling of ductal carcinoma in situ and its relationship to histopathological features. Breast Cancer Research. 2014;16 doi: 10.1186/s13058-014-0423-9. Article 423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park et al. (2011a).Park CK, Kim J, Yim SY, Lee AR, Han JH, Kim CY, Park SH, Kim TM, Lee SH, Choi SH, Kim SK, Kim DG, Jung HW. Usefulness of MS-MLPAfor detection of MGMT promoter methylation in the evaluation of pseudoprogression in glioblastoma patients. Neuro-Oncology. 2011a;13:195–202. doi: 10.1093/neuonc/noq162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park et al. (2011b).Park SY, Kwon HJ, Lee HE, Ryu HS, Kim SW, Kim JH, Kim IA, Jung N, Cho NY, Kang GH. Promoter CpG island hypermethylation during breast cancer progression. Virchows Archiv. 2011b;458:73–84. doi: 10.1007/s00428-010-1013-6. [DOI] [PubMed] [Google Scholar]
- Parrella et al. (2004).Parrella P, Poeta ML, Gallo AP, Prencipe M, Scintu M, Apicella A, Rossiello R, Liguoro G, Seripa D, Gravina C, Rabitti C, Rinaldi M, Nicol T, Tommasi S, Paradiso A, Schittulli F, Altomare V, Fazio VM. Nonrandom distribution of aberrant promoter methylation of cancer-related genes in sporadic breast tumors. Clinical Cancer Research. 2004;10:5349–5354. doi: 10.1158/1078-0432.CCR-04-0555. [DOI] [PubMed] [Google Scholar]
- Prasad et al. (2008).Prasad CP, Mirza S, Sharma G, Prashad R, DattaGupta S, Rath G, Ralhan R. Epigenetic alterations of CDH1 and APC genes: relationship with activation of Wnt/beta-catenin pathway in invasive ductal carcinoma of breast. Life Sciences. 2008;83:318–325. doi: 10.1016/j.lfs.2008.06.019. [DOI] [PubMed] [Google Scholar]
- Rykova et al. (2004).Rykova EY, Skvortsova TE, Laktionov PP, Tamkovich SN, Bryzgunova OE, Starikov AV, Kuznetsova NP, Kolomiets SA, Sevostianova NV, Vlassov VV. Investigation of tumor-derived extracellular DNA in blood of cancer patients by methylation-specific PCR. Nucleosides Nucleotides Nucleic Acids. 2004;23:855–859. doi: 10.1081/NCN-200026031. [DOI] [PubMed] [Google Scholar]
- Shinozaki et al. (2005).Shinozaki M, Hoon DS, Giuliano AE, Hansen NM, Wang HJ, Turner R, Taback B. Distinct hypermethylation profile of primary breast cancer is associated with sentinel lymph node metastasis. Clinical Cancer Research. 2005;11:2156–2162. doi: 10.1158/1078-0432.CCR-04-1810. [DOI] [PubMed] [Google Scholar]
- Sparks et al. (1998).Sparks AB, Morin PJ, Vogelstein B, Kinzler KW. Mutational analysis of the APC/beta-catenin/Tcf pathway in colorectal cancer. Cancer Research. 1998;58:1130–1134. [PubMed] [Google Scholar]
- Swellam et al. (2015).Swellam M, Abdelmaksoud MD, Sayed Mahmoud M, Ramadan A, Abdel-Moneem W, Hefny MM. Aberrant methylation of APC and RARbeta2 genes in breast cancer patients. IUBMB Life. 2015;67:61–68. doi: 10.1002/iub.1346. [DOI] [PubMed] [Google Scholar]
- Taback et al. (2006).Taback B, Giuliano AE, Lai R, Hansen N, Singer FR, Pantel K, Hoon DS. Epigenetic analysis of body fluids and tumor tissues: application of a comprehensive molecular assessment for early-stage breast cancer patients. Annals of the New York Academy of Sciences. 2006;1075:211–221. doi: 10.1196/annals.1368.029. [DOI] [PubMed] [Google Scholar]
- Torre et al. (2015).Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA: A Cancer Journal for Clinicians. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- Van der Auwera et al. (2009a).Van der Auwera I, Bovie C, Svensson C, Limame R, Trinh XB, Van Dam P, Van Laere SJ, Marck EV, Vermeulen PB, Dirix LY. Quantitative assessment of DNA hypermethylation in the inflammatory and non-inflammatory breast cancer phenotypes. Cancer Biology & Therapy. 2009a;8:2252–2259. doi: 10.4161/cbt.8.23.10133. [DOI] [PubMed] [Google Scholar]
- Van der Auwera et al. (2009b).Van der Auwera I, Elst HJ, Van Laere SJ, Maes H, Huget P, Van Dam P, Van Marck EA, Vermeulen PB, Dirix LY. The presence of circulating total DNA and methylated genes is associated with circulating tumour cells in blood from breast cancer patients. British Journal of Cancer. 2009b;100:1277–1286. doi: 10.1038/sj.bjc.6605013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Auwera et al. (2008).Van der Auwera I, Van Laere SJ, Van den Bosch SM, Van den Eynden GG, Trinh BX, Van Dam PA, Colpaert CG, Van Engeland M, Van Marck EA, Vermeulen PB, Dirix LY. Aberrant methylation of the Adenomatous Polyposis Coli (APC) gene promoter is associated with the inflammatory breast cancer phenotype. British Journal of Cancer. 2008;99:1735–1742. doi: 10.1038/sj.bjc.6604705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virmani et al. (2001).Virmani AK, Rathi A, Sathyanarayana UG, Padar A, Huang CX, Cunnigham HT, Farinas AJ, Milchgrub S, Euhus DM, Gilcrease M, Herman J, Minna JD, Gazdar AF. Aberrant methylation of the adenomatous polyposis coli (APC) gene promoter 1A in breast and lung carcinomas. Clinical Cancer Research. 2001;7:1998–2004. [PubMed] [Google Scholar]
- Wojdacz et al. (2011a).Wojdacz TK, Thestrup BB, Cold S, Overgaard J, Hansen LL. No difference in the frequency of locus-specific methylation in the peripheral blood DNA of women diagnosed with breast cancer and age-matched controls. Future Oncol. 2011a;7:1451–1455. doi: 10.2217/fon.11.123. [DOI] [PubMed] [Google Scholar]
- Wojdacz et al. (2011b).Wojdacz TK, Thestrup BB, Overgaard J, Hansen LL. Methylation of cancer related genes in tumor and peripheral blood DNA from the same breast cancer patient as two independent events. Diagn Pathol. 2011b;6 doi: 10.1186/1746-1596-6-116. Article 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Li & Lang (2015).Zhang DP, Li XW, Lang JH. Prognostic value of beta-catenin expression in breast cancer patients: a meta-analysis. Asian Pacific Journal of Cancer Prevention. 2015;16:5625–5633. doi: 10.7314/APJCP.2015.16.14.5625. [DOI] [PubMed] [Google Scholar]
- Zhang et al. (2007).Zhang JJ, Ouyang T, Wan WH, Xu GW, Deng GR. Detection and significance of APC gene promoter hypermethylation in serum of breast cancer patients. Ai Zheng. 2007;26:44–47. [PubMed] [Google Scholar]
- Zmetakova et al. (2013).Zmetakova I, Danihel L, Smolkova B, Mego M, Kajabova V, Krivulcik T, Rusnak I, Rychly B, Danis D, Repiska V, Blasko P, Karaba M, Benca J, Pechan J, Fridrichova I. Evaluation of protein expression and DNA methylation profiles detected by pyrosequencing in invasive breast cancer. Neoplasma. 2013;60:635–646. doi: 10.4149/neo_2013_082. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The following information was supplied regarding data availability: