Abstract
Hepatocellular carcinoma (HCC) is one of the most common cancers worldwide and has an extremely poor prognosis. Surgical resection is always inapplicable to HCC patients diagnosed at an advanced tumor stage. The mechanisms underlying HCC cell proliferation remain obscure. In the present study, SWItch/sucrose nonfermentable catalytic subunit SNF2 (SNF2H) expression was tested in HCC tissues and Wnt/β-catenin pathway activation upon overexpression of SNF2H or knockdown of SNF2H expression was investigated in cultured HCC cells. It was demonstrated that SNF2H is a vital factor for HCC growth. The SNF2H expression level is increased in HCC tissues compared with paratumoral liver tissues. SNF2H promotes HCC cell proliferation and colony formation ability in vitro. SNF2H may increase the protein level of β-catenin and enhance its nuclear accumulation in HCC cells, thereby leading to the activation of the Wnt/β-catenin signaling pathway. In conclusion, the present results indicate that SNF2H plays a vital role in HCC cell growth, suggesting that SNF2H may be a promising therapeutic target for HCC treatment.
Keywords: hepatocellular carcinoma, proliferation, SNF2H, Wnt/β-catenin
Introduction
Hepatocellular carcinoma (HCC) is the fifth most common tumor in men and the seventh most common tumor in women. It is the third leading cause of tumor-associated mortality worldwide (1). HCC has an extremely poor prognosis, with a 5-year survival rate of 40–50%. Patients with HCC are usually diagnosed at an advanced tumor stage, which renders surgical resection inapplicable (2). The molecular mechanism underlying HCC cell proliferation is not well understood, and the clarification of this mechanism may facilitate the prevention and treatment of patients with HCC.
Approximately one-half of HCC patients exhibit aberrant activation of the Wnt/β-catenin pathway, which indicates the importance of this signaling cascade in hepatocarcinogenesis (3–5). The Wnt/β-catenin signaling pathway is comprehensively involved in cell proliferation (6). The Wnt/β-catenin pathway is activated upon the binding of the Wnt ligands to Frizzled receptors, which is followed by cytosolic accumulation of β-catenin via interruption of destruction complex-mediated protein degradation of β-catenin. Stabilized β-catenin may translocate into the nucleus to induce the transcription of Wnt/β-catenin pathway target genes, such as cyclin D1 and cellular v-myc avian myelocytomatosis viral oncogene homolog (c-MYC) (4,7). The Wnt/β-catenin signaling pathway plays a vital role in hepatocarcinogenesis. Although several factors, such as activator protein 1, human zinc finger protein 191, prospero homeobox protein 1 and T-cell factor (TCF)/lymphocyte enhancer factor (LEF), have been reported to regulate β-catenin expression (8–11), the molecular mechanism of the aberrant expression of β-catenin remains unknown in HCC.
SWItch/sucrose nonfermentable (SWI/SNF) catalytic subunit SNF2 (SNF2H) is a member of the SWI/SNF chromatin remodeling family with ATPase activity (12). SNF2H plays a vital role in gene transcription, DNA repair and DNA replication (12,13). SWI/SNF complexes are associated with malignant transformation (14). Previously, several studies have reported aberrant high expression of SNF2H in breast cancer, acute leukemia, ovarian cancer and gastric cancer (15–17). These studies suggest that SNF2H has an important role in cancer progression. However, whether SNF2H plays a vital role in HCC progression has not been investigated.
In the present study, it was demonstrated that SNF2H plays a vital role in HCC cell growth. The SNF2H expression level was increased in HCC tissues compared with the paratumoral liver tissues. SNF2H promotes the cell proliferation and colony formation ability of HCC cells in vitro. SNF2H promotes the growth of HCC cells by activating the Wnt/β-catenin pathway, which increases β-catenin protein levels and enhances the nuclear accumulation β-catenin. These results indicate that SNF2H plays a vital role in hepatocarcinogenesis.
Materials and methods
Clinical samples
Tissue specimens were collected from 30 patients with HCC during surgical resection. The collection of the tissue specimens and associated information from patients was approved by the Ethics Committee of Renji Hospital, Shanghai Jiaotong University School of Medicine (Shanghai, China). All patients enrolled in the present study provided written informed consent prior to their inclusion.
Plasmid constructs
For lentivirus-mediated expression of SNF2H, FLAG-tagged full-length SNF2H ORF was cloned in pWPI.1 (plasmid no. 12254; Addgene, Inc., Cambridge, MA, USA). For RNA interference of SNF2H expression, DNA fragments encoding the hairpin precursors for shSNF2H#1 (5′-CGTCGAATTAAGGCTGATGTT-3′) and shSNF2H#2 (5′-CGACTGCTGATGTAGTAATTT-3′) were inserted into the pLKO.1-TRC cloning vector. A scrambled small interfering RNA precursor (Scr) of similar GC-content to shSNF2H#1 and shSNF2H#2, but without sequence identity to SNF2H cDNA, was used as the control.
Cell lines, transfection and lentivirus preparation
The embryo kidney HEK293T and human HCC SMCC-7721, PLC, BEL-7402 and Huh7 cell lines were acquired from the Cell Bank of Shanghai Institutes of Biological Sciences, Chinese Academy of Sciences (Shanghai, China). The cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Thermo Fisher Scientific, Inc., Waltham, MA, USA) with 10% fetal bovine serum (Thermo Fisher Scientific, Inc.) at 37°C in a 5% CO2 atmosphere. After culturing cells for 24 h, cells underwent transfection.
The lentivirus plasmid-mediated SNF2H expression vectors pWPI.1-Flag-SNF2H, pLKO.1-shSNF2H#1 and pLKO.1-shSNF2H#2 were co-transfected with the helper plasmids pMD2.G and psPAX2 into HEK293T cell to package recombinant lentivirus. In total, 24 h subsequent to cell transfection, medium was replaced with fresh DMEM medium to continue cell culture. After 48 h, the lentivirus supernatants from HEK293T cells were used for the infection of BEL-7402 and Huh7 cells using Lipofectamine 2000 (Thermo Fisher Scientific, Inc.).
Western blot analysis
Total tissue and cell proteins were obtained using radioimmunoprecipitation assay buffer lysis buffer with proteinase inhibitors (protease inhibitor cocktail; Sigma-Aldrich, St. Louis, MO, USA). Equivalent amounts of proteins (30 µg) were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto nitrocellulose membranes (Invitrogen; Thermo Fisher Scientific, Inc.). Subsequent to blocking with 5% skimmed milk for 30 min, the membranes were incubated with a rabbit polyclonal anti-SNF2H primary antibody (dilution, 1:500; catalog no., ab3749; Abcam, Cambridge, MA, USA) at 4°C overnight. A goat anti-rabbit IgG horseradish peroxidase-conjugated secondary antibody (dilution, 1:3,000; catalog no., 7074; Cell Signaling Technology, Inc., Danvers, MA, USA) was used for the subsequent incubation at room temperature for 1 h. In addition, a mouse monoclonal β-actin horseradish peroxidase-conjugated antibody (dilution, 1:5,000; catalog no., ab20272; Abcam) was used as a loading control. The expression of SNF2H was detected on X-ray film using the ECL detection system (Pierce; Thermo Fisher Scientific, Inc.).
RNA isolation and reverse transcription-quantitative PCR (RT-qPCR)
Total RNA of the sample was extracted using TRIzol reagent (Thermo Fisher Scientific, Inc.), and RT was performed using the PrimeScript RT kit (Takara, Otsu, Japan). For the RT-qPCR assay, aliquots of cDNA were amplified using SYBR Premix Ex Taq (Takara). PCR reactions were performed on MXP3000 (Stratagene, La Jolla, CA, USA). The primer sequences were as follows: SFN2H forward, CCTTTGAAGATGAAACCAGGGCGC and reverse, CTGTTAATAGCTCTTCATCCTCCTC; and β-actin forward, TCCCTGGAGAAGAGCTACG and reverse, GTAGTTTCGTGGATGCCACA.
Cell proliferation
Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc., Kumamoto, Japan) to measure the cell proliferation. BEL-7402-Mock, BEL-7402-SNF2H, Huh7-scr, Huh7-shSNF2H#1 and Huh7-shSNF2H#2 cell lines were seeded into 96-well plates at a density of 2×103 cells/well with DMEM medium containing 10% FBS. At the indicated time points (24, 48, 72 or 96 h), the cell absorbance was detected at 450 nm using xMark Microplate Spectrophotometer (Bio-Rad Laboratories, Inc., Hercules, CA, USA). The CCK-8 assay procedure was performed according to the manufacturer's instructions.
Colony formation assay
BEL-7402-Mock, BEL-7402-SNF2H, Huh7-scr, Huh7-shSNF2H#1 and Huh7-shSNF2H#2 cell lines were separately seeded into 6-well plates at a density of 3×102 cells/well using DMEM medium containing 10% FBS, and the cells were cultured at 37°C in a 5% CO2 atmosphere. After 10–14 days of culturing, the cells were washed with PBS and fixed using 10% formaldehyde for 30 min. Finally, the cells were stained with Giemsa stain for 60 min.
Statistical Analysis
Pearson's χ2 test or Fisher's exact test were used to compare SNF2H expression levels between tissue samples and the Student's t-test was used for quantitative variables. P<0.05 was considered to indicate a statistically significant difference.
Results
SNF2H is highly expressed in HCC
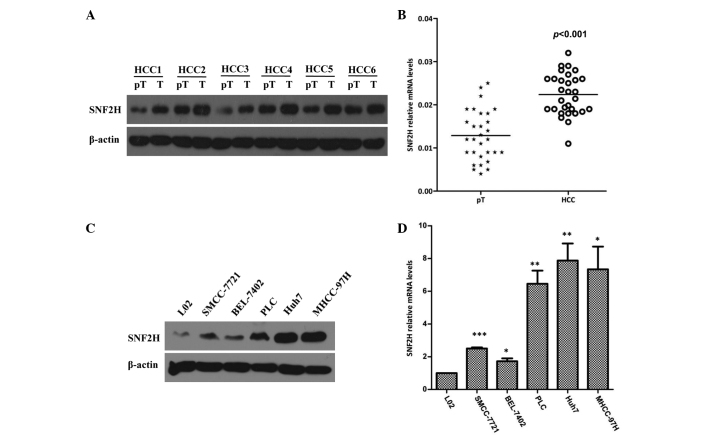
The expression of SNF2H was first assessed in HCC and paired paratumoral liver tissues. The SNF2H protein level was significantly increased by 1.7-fold in HCC tissues compared with the matched paratumoral liver tissues (P=0.018; Fig. 1A). To confirm these results, the expression of SNF2H in 30 HCC specimens and the paired paratumoral liver tissues was evaluated. It was found that the expression of SNF2H in HCC specimens was increased 1.9-fold compared with the paired paratumoral liver tissues (P<0.001; Fig. 1B). Next, the expression of SNF2H in the normal liver cell and HCC SMCC-7721, BEL-7402, PLC, Huh7 and MHCC-97H cell lines was examined. Western blotting showed that the HCC Huh7 and MHCC97H cell lines had increased SNF2H expression compared with the normal liver L02 cell line (Fig. 1C). It was also found that the mRNA level of SNF2H in the HCC SMCC-7721, BEL-7402, PLC, Huh7 and MHCC-97H cell lines was increased 2.5-fold, 1.7-fold, 6.34-fold, 7.72-fold and 7.09-fold, respectively, compared with the mRNA level in the L02 cell line (P=0.0018, P=0.0160, P=0.0025, P=0.0027 and P=0.0102, respectively; Fig. 1D). Overall, these results indicate that the high expression of SNF2H may improve the development of HCC.
Figure 1.
SNF2H is highly expressed in HCC. (A) Western blot assays showed that SNF2H expression in HCC was increased compared with the corresponding paratumoral tissues. (B) Reverse transcription-quantitative polymerase chain reaction analysis of SNF2H expression in HCC and corresponding paratumoral tissues. The relative expression of SNF2H mRNA in HCC tissue samples was increased compared with the paired paratumoral tissues. (C) SNF2H protein levels in the normal liver cell were decreased compared with several HCC cell lines. Western blotting showed that the HCC cell lines Huh7 and MHCC97H express more SNF2H than the normal liver L02 cell lines. (D) The SNF2H mRNA level in the HCC cell lines was increased compared with the mRNA level in the L02 cell line. Significant differences were determined by Student's t-test. *P<0.05; **P<0.01; ***P<0.001. SNF2H, SWItch/sucrose nonfermentable catalytic subunit SNF2; HCC, hepatocellular carcinoma; mRNA, messenger RNA; pT, paratumoral; T, tumor.
Overexpression of SNF2H improves HCC cell growth
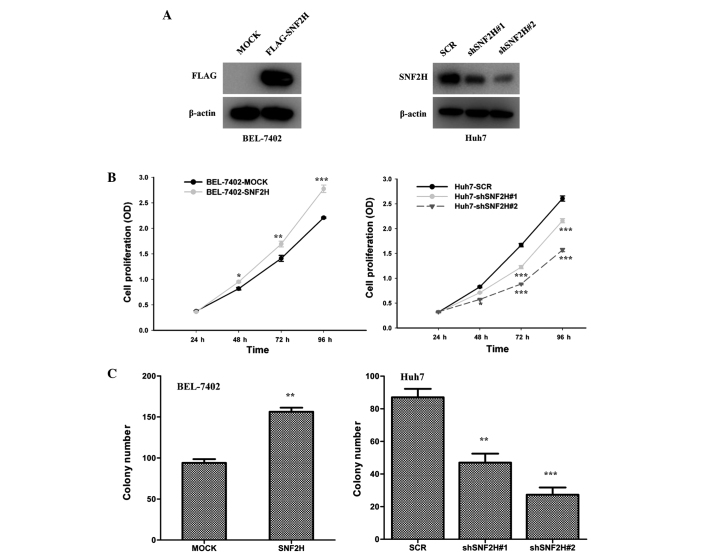
BEL-7402 and Huh7 cells were used as model HCC cell lines for the following investigations. Lentivirus-mediated overexpression of SNF2H in the BEL-7402 cells and knockdown of SNF2H expression of SNF2H in the Huh7 cells were used to evaluated the function of SNF2H in HCC cell growth. Overexpression of SNF2H in BEL-7402 cells and knockdown of the expression of SNF2H in the Huh7 cells were performed (Fig. 2A). Overexpression of SNF2H in BEL-7402 cells improved the cell growth over 96 h (24 h, P=0.1070; 48 h, P=0.0015; 72 h, P<0.001; 96 h, P<0.001), but knockdown of SNF2H expression in Huh7 cells inhibited cell growth (shSNF2H#1 vs. SCR: 24 h, P=0.97; 48 h, P=0.0054; 72 h, P<0.001; and 96 h, P<0.001) (shSNF2H#2 vs. SCR: 24 h, P=0.345; 48 h, P=0.001; 72 h, P<0.001; and 96 h, P<0.001) (Fig. 2B). Overexpression of SNF2H also increased the number of BEL-7402 cell colonies by 1.6-fold (P<0.001) (Fig. 2C). However, knockdown of SNF2H expression reduced the number of Huh7 cell colonies by 50 and 70%, respectively (shSNF2H#1 vs. SCR, P=0.006; shSNF2H#2 vs. SCR, P<0.001) (Fig. 2C). These results indicate that SNF2H improves HCC cell growth.
Figure 2.
SNF2H promotes HCC cell proliferation in vitro. (A) SNF2H expression in HCC cells infected with lentivirus. The left panel shows overexpression of SNF2H in BEL-7402 cells. The right panel shows knockdown of SNF2H expression in Huh7 cells. (B) The Cell Counting Kit-8 assay revealed that SNF2H overexpression significantly improved the cell growth rate (left panel). Knockdown of SNF2H expression in Huh7 cells significantly reduced the cell growth rate. The mean ± SD from three independent experiments is presented. (C) SNF2H promotes HCC cell colony formation. The left panel shows that overexpression of SNF2H expression promoted cell colony-forming capacity in BEL-7402 cells. The right panel shows that knockdown of SNF2H expression inhibited colony formation in Huh7 cells. The mean ± SD from three independent experiments is presented. Significant differences were determined by Student's t-test. *P<0.05; **P<0.01; ***P<0.001. SNF2H, SWItch/sucrose nonfermentable catalytic subunit SNF2; HCC, hepatocellular carcinoma; SD, standard deviation; SCR, scrambled small interfering RNA precursor; OD, optical density.
SNF2H promotes the protein levels of β-catenin and enhances its nuclear accumulation
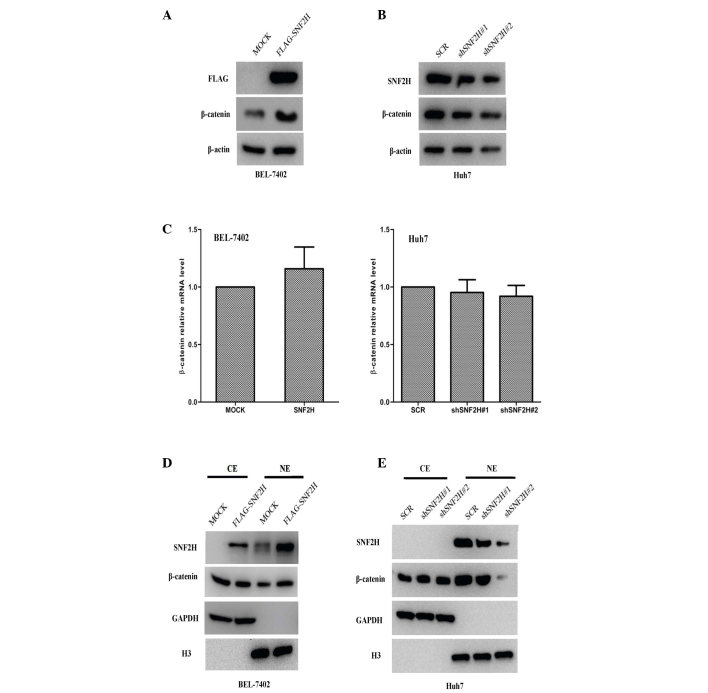
The Wnt/β-catenin pathway is always aberrantly activated during the development of HCC, and the β-catenin expression level is often increased in HCC (3–5,18,19). Therefore, the present study investigated whether SNF2H enhances β-catenin expression to promote tumor growth. It was found that the β-catenin protein level in BEL-7402 cells was upregulated 2.6-fold by the exogenous expression of SNF2H (P=0.0012; Fig. 3A), but the β-catenin protein level was downregulated by 30% (shSNF2H#1 vs. SCR, P=0.008) and 70% (shSNF2H#2 vs. SCR, P=0.001) in Huh7 cells by SNF2H knockdown (Fig. 3B). The increase in β-catenin protein levels may result in induced β-catenin transcription or improved β-catenin protein stability. The present study investigated whether SNF2H used this mechanism to upregulate β-catenin protein levels. Subsequently, it was observed that neither SNF2H overexpression in BEL-7402 cells nor SNF2H knockdown in Huh7 cells resulted in different β-catenin mRNA levels compared with the control BEL-7402-MOCK (increase of 1.1-fold; P=0.158) and Huh7-Scr cells (shSNF2H#1 vs. SCR, decrease of 7%, P=0.62; shSNF2H#2 vs. SCR, increase of 8%, P=0.574) (Fig. 3C). In addition, there were no differences in the cytoplasmic level of β-catenin. However, the nuclear level of β-catenin was markedly upregulated 2-fold in the SNF2H overexpression BEL-7402 cells compared with the control group (P=0.003) (Fig. 3D). Knockdown of SNF2H did not change the cytoplasmic level of β-catenin, but the nuclear level of β-catenin was markedly downregulated by 20 and 70%, respectively, compared with the control group in the Huh7 cells (shSNF2H#1 vs. SCR, P=0.023; shSNF2H#2 vs. SCR, P=0.002) (Fig. 3E). These results indicate that SNF2H may promote β-catenin protein levels and enhance the nuclear accumulation of β-catenin in HCC cells.
Figure 3.
SNF2H upregulates β-catenin protein levels and promotes the nuclear accumulation of β-catenin. (A) Overexpression of SNF2H in BEL-7402 cells increased β-catenin protein levels. (B) Knockdown of SNF2H expression in Huh7 cells reduced β-catenin protein levels. (C) SNF2H did not change β-catenin mRNA expression. Neither SNF2H-overexpression BEL-7402 cells nor SNF2H-knockdown Huh7 cells altered β-catenin mRNA levels compared with the control BEL-7402 and Huh7 cells. β-catenin mRNA was measured using reverse transcription-quantitative polymerase chain reaction. The mean ± standard deviation from three independent experiments is presented as a relative ratio to the control, whose value was taken as 1.0. (D) Cell fractionation assays revealed that overexpression of SNF2H increased the levels of nuclear β-catenin protein in BEL-7402 cells. (E) Knockdown of SNF2H expression reduced the levels of nuclear β-catenin protein in Huh7 cells. CE, cytoplasmic extract; NE, nuclear extract; SNF2H, SWItch/sucrose nonfermentable catalytic subunit SNF2; HCC, hepatocellular carcinoma; SCR, scrambled small interfering RNA precursor; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; H3, histone H3.
SNF2H activates the Wnt/β-catenin signaling pathway
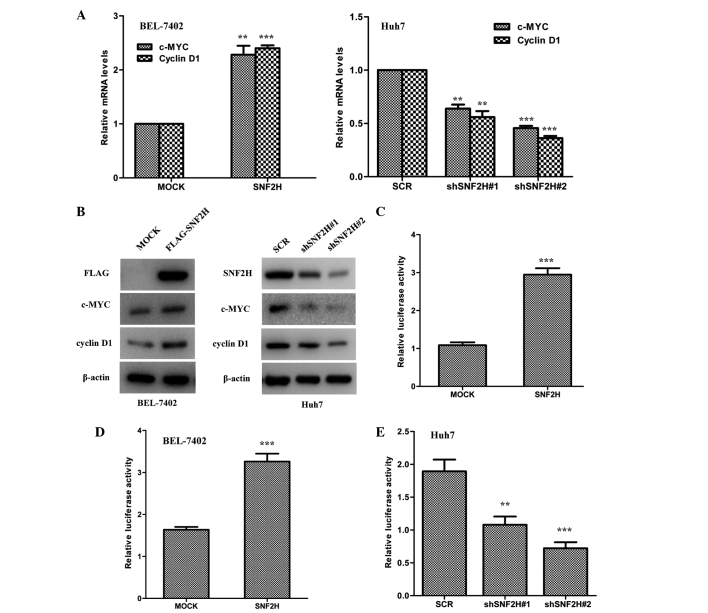
SNF2H increases the protein level of β-catenin and enhance its nuclear translocation in HCC cells. These findings indicate that SNF2H may drive the Wnt/β-catenin signaling pathway in HCC cells. To test this hypothesis, it was investigated whether overexpression of SNF2H in BEL-7402 cells could regulate the transcription of the Wnt/β-catenin target genes, such as cyclin D1 and c-MYC. SNF2H overexpression in BEL-7402 cells significantly upregulated c-MYC and cyclin D1 mRNA by 2.5-fold (P=0.0056 vs. MOCK) and 2.2-fold (P<0.001 vs. MOCK), respectively, and protein levels (Fig. 4A and B). However, knockdown of the expression of SNF2H decreased the mRNA expression of cyclin D1 by 60% (shSNF2H#1 vs. SCR, P<0.001; shSNF2H#2 vs. SCR, P=0.0053) and c-MYC by 50% (shSNF2H#1 vs. SCR, P=0.0037; shSNF2H#2 vs. SCR, P<0.001) and also decreased protein levels in Huh7 cells compared with the control group (Fig. 4A and B). It was investigated whether SNF2H could activate the LEF luciferase reporter that is always used as indicator of the level of Wnt/β-catenin signaling pathway activity. Indeed, exogenous expression of SNF2H in HEK-293T cells could enhance the LEF luciferase reporter activity by 3.0-fold compared with the control group (P<0.001) (Fig. 4C). Next, it was investigated whether SNF2H could also activate the LEF luciferase reporter activity in the HCC cells. Overexpression of SNF2H in BEL-7402 cells enhanced LEF luciferase reporter activity by 2.0-fold compared with the control group (P=0.0031) (Fig. 4D). However, knockdown of SNF2H expression in Huh7 cells decreased LEF luciferase reporter activity by 50% (shSNF2H#1 vs. SCR, P=0.0166) and 60% (shSNF2H#2 vs. SCR, P=0.0018) compared with the control group (Fig. 4E). These results indicate that SNF2H activates the Wnt/β-catenin signaling pathway to promote HCC cell growth.
Figure 4.
SNF2H activates the Wnt/β-catenin pathway. (A) Left, SNF2H overexpression in BEL-7402 cells upregulated c-MYC and cyclin D1 mRNA levels. Right, SNF2H knockdown in Huh7 cells downregulated c-MYC and cyclin D1 mRNA levels. The mean ± SD from three independent experiments is presented as a relative ratio to the control, which was assigned a value of 1.0. (B) Left, overexpression of SNF2H increased c-MYC and cyclin D1 protein levels in BEL-7402 cells. Right, knockdown of SNF2H decreased c-MYC and cyclin D1 protein levels in Huh7 cells. (C) SNF2H activated the LEF reporter. HEK293T cells were co-transfected with the SNF2H and the LEF reporter expression constructs. (D) Overexpression of SNF2H upregulated LEF reporter activity in BEL-7402 cells. (E) Knockdown of SNF2H expression downregulated LEF reporter activity in Huh7 cells. The mean ± SD of normalized luciferase activity from three independent experiments is presented. Significant differences were determined using Student's t-test. **P<0.01; ***P<0.001. SNF2H, SWItch/sucrose nonfermentable catalytic subunit SNF2; c-MYC, cellular v-myc avian myelocytomatosis viral oncogene homolog; mRNA, messenger RNA; LEF, lymphocyte enhancer factor; SD, standard deviation.
Discussion
HCC is the fifth most common cancer and the third most common cause of cancer-associated mortality worldwide. The incidence and mortality of HCC has been increasing rapidly worldwide in previous decades. China alone accounts for >50% of cases of HCC worldwide (20,21). Due to the poor prognosis of HCC patients, the molecular mechanism of HCC development urgently requires clarification. Although numerous genes associated with the development of HCC have been confirmed (22–26), the mechanism of HCC development has not been well elucidated. In the present study, the expression of SNF2H in HCC specimens and the paired paratumoral liver tissues was assessed. It was found that the expression of SNF2H in HCC specimens was increased compared with the paired paratumoral liver tissues. The present findings suggest that SNF2H may be a novel intervention target for the treatment of HCC.
The extremely poor prognosis of HCC is mainly due to tumor growth and vascular invasion (27–30). The molecular mechanism of HCC cell growth is not well known. Therefore, a thorough knowledge of the molecular mechanisms of HCC cell growth is extremely important for the therapeutics for HCC patients. The present results showed that SNF2H plays a vital role in promoting HCC cell growth and colony formation ability, indicating that SNF2H is a vital factor for the development of HCC.
The Wnt/β-catenin signaling pathway is aberrantly activated during the development of HCC (3–5). When binding with Wnt ligands, the destruction complex, which includes adenomatous polyposis coli protein, casein kinase 1, Axin and glycogen synthase kinase 3, may not induce the degradation of β-catenin. Further, stabilized β-catenin may induce the expression of Wnt/β-catenin target genes by interacting with TCF/LEF transcription factors. Previous data show that certain Wnt target genes, such as cyclin D1 and c-MYC, regulate tumor cell growth (6,31). However, the association between SNF2H and the Wnt/β-catenin pathway in HCC remains unknown. In the present study, SNF2H was shown to activate the Wnt/β-catenin pathway in HCC cells by upregulating β-catenin protein levels and enhancing β-catenin nuclear translocation. The mechanism for SNF2H-mediated upregulation of the expression of β-catenin requires elucidation. Neither SNF2H-overexpression BEL-7402 cells nor SNF2H-knockdown Huh7 cells regulated β-catenin mRNA levels. Thus, it is possible that SNF2H may interfere with the degradation machinery of β-catenin. The mechanism of SNF2H enhancement of the nuclear translocation of β-catenin is a vital focus for additional investigation, which may indicate a novel target for the treatment of HCC patients.
SNF2H plays an important role in cell growth and viability (32,33). Stopka et al (13) reported that knockout of SNF2H reduced cell proliferation by the inhibition of mitosis. The present results showed that SNF2H also promotes cell proliferation in HCC. The role of SNF2H in carcinogenesis is well established. Mohamed et al (12) reported that SNF2H showed higher expression in prostatic neoplasia compared with benign prostatic hyperphasia. The results of this study suggest that SNF2H is important in prostate carcinogenesis (12). Previously, another study observed increased expression of SNF2H in gastric cancer samples compared with normal mucosa (16). However, no study has reported an association between SNF2H and HCC. The present study demonstrated that SNF2H expression was markedly upregulated in HCC tissues compared with the paired paratumoral liver tissues. The present results indicate that SNF2H may be a promising therapeutic target strategy for HCC patients.
Acknowledgements
The present study was sponsored by the National Natural Science Foundation of China (grant nos. 81322025 and 81371875), and the Shanghai Committee of Science and Technology, China (grant no. 14140901000) and the Foundation for Innovative Research Groups of the National Natural Science Foundation of China (grant no. 81421001).
References
- 1.Ferlay J, Shin H, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Forner A, Hessheimer AJ, Isabel Real M, Bruix J. Treatment of hepatocellular carcinoma. Crit Rev Oncol Hematol. 2006;60:89–98. doi: 10.1016/j.critrevonc.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 3.Boyault S, Rickman DS, deReyniès A, Balabaud C, Rebouissou S, Jeannot E, Hérault A, Saric J, Belghiti J, Franco D, et al. Transcriptome classification of HCC is related to gene alterations and to new therapeutic targets. Hepatology. 2007;45:42–52. doi: 10.1002/hep.21467. [DOI] [PubMed] [Google Scholar]
- 4.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 5.Takigawa Y, Brown AM. Wnt signaling in liver cancer. Curr Drug Targets. 2008;9:1013–1024. doi: 10.2174/138945008786786127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Polakis P. The many ways of Wnt in cancer. Curr Opin Genet Dev. 2007;17:45–51. doi: 10.1016/j.gde.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 7.Valenta T, Hausmann G, Basler K. The many faces and functions of β-catenin. EMBO J. 2012;31:2714–2736. doi: 10.1038/emboj.2012.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guigon CJ, Kim DW, Zhu X, Zhao L, Cheng SY. Tumor suppressor action of liganded thyroid hormone receptor beta by direct repression of beta-catenin gene expression. Endocrinology. 2010;151:5528–5536. doi: 10.1210/en.2010-0475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Q, Dashwood WM, Zhong X, Al-Fageeh, Dashwood RH. Cloning of the rat beta-catenin gene (Ctnnb1) promoter and its functional analysis compared with the Catnb and CTNNB1 promoters. Genomics. 2004;83:231–242. doi: 10.1016/j.ygeno.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 10.Liu G, Jiang S, Wang C, et al. Zinc finger transcription factor 191, directly binding to β-catenin promoter, promotes cell proliferation of hepatocellular carcinoma. Hepatology. 2012;55:1830–1839. doi: 10.1002/hep.25564. [DOI] [PubMed] [Google Scholar]
- 11.Liu Y, Ye X, Zhang JB, et al. PROX1 promotes hepatocellular carcinoma proliferation and sorafenib resistance by enhancing β-catenin expression and nuclear translocation. Oncogene. 2015;34:5524–5535. doi: 10.1038/onc.2015.7. [DOI] [PubMed] [Google Scholar]
- 12.Mohamed MA, Greif PA, Diamond J, Sharaf O, Maxwell P, Montironi R, Young RA, Hamilton PW. Epigenetic events, remodelling enzymes and their relationship to chromatin organization in prostatic intraepithelial neoplasia and prostatic adenocarcinoma. BJU Int. 2007;99:908–915. doi: 10.1111/j.1464-410X.2006.06704.x. [DOI] [PubMed] [Google Scholar]
- 13.Stopka T, Skoultchi AI. The ISWI ATPase Snf2h is required for early mouse development. Proc Natl Acad Sci USA. 2003;100:14097–14102. doi: 10.1073/pnas.2336105100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reisman D, Glaros S, Thompson EA. The SWI/SNF complex and cancer. Oncogene. 2009;28:1653–1668. doi: 10.1038/onc.2009.4. [DOI] [PubMed] [Google Scholar]
- 15.Jin Q, Mao X, Li B, Guan S, Yao F, Jin F. Overexpression of SMARCA5 correlates with cell proliferation and migration in breast cancer. Tumour Biol. 2015;36:1895–1902. doi: 10.1007/s13277-014-2791-2. [DOI] [PubMed] [Google Scholar]
- 16.Stopka T, Zakova D, Fuchs O, Kubrova O, Blafkova J, Jelinek J, Necas E, Zivny J. Chromatin remodeling gene SMARCA5 is dysregulated in primitive hematopoietic cells of acute leukemia. Leukemia. 2000;14:1247–1252. doi: 10.1038/sj.leu.2401807. [DOI] [PubMed] [Google Scholar]
- 17.Gigek CO, Lisboa LC, Leal MF, Silva PN, Lima EM, Khayat AS, Assumpção PP, Burbano RR, Smith Mde A. SMARCA5 methylation and expression in gastric cancer. Cancer Invest. 2011;29:162–166. doi: 10.3109/07357907.2010.543365. [DOI] [PubMed] [Google Scholar]
- 18.Cui J, Zhou X, Liu Y, Tang Z. Mutation and overexpression of the beta-catenin gene may play an important role in primary hepatocellular carcinoma among Chinese people. J Cancer Res Clin Oncol. 2001;127:577–581. doi: 10.1007/s004320100259. [DOI] [PubMed] [Google Scholar]
- 19.Lai TY, Su CC, Kuo WW, Yeh YL, Kuo WH, Tsai FJ, Tsai CH, Weng YJ, Huang CY, Chen LM. β-catenin plays a key role in metastasis of human hepatocellular carcinoma. Oncol Rep. 2011;26:415–422. doi: 10.3892/or.2011.1323. [DOI] [PubMed] [Google Scholar]
- 20.Farazi PA, DePinho RA. Hepatocellular carcinoma pathogenesis: From genes to environment. Nat Rev Cancer. 2006;6:674–687. doi: 10.1038/nrc1934. [DOI] [PubMed] [Google Scholar]
- 21.El-Serag HB, Rudolph KL. Hepatocellular carcinoma: Epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–2576. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 22.Chen L, Chan TH, Yuan YF, et al. CHD1L promotes hepatocellular carcinoma progression and metastasis in mice and is associated with these processes in human patients. J Clin Invest. 2010;120:1178–1191. doi: 10.1172/JCI40665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Y, Zhang JB, Qin Y, et al. PROX1 promotes hepatocellular carcinoma metastasis by way of up-regulating hypoxia-inducible factor 1α expression and protein stability. Hepatology. 2013;58:692–705. doi: 10.1002/hep.26398. [DOI] [PubMed] [Google Scholar]
- 24.Chuma M, Sakamoto M, Yasuda J, Fujii G, Nakanishi K, Tsuchiya A, Ohta T, Asaka M, Hirohashi S. Overexpression of cortactin is involved in motility and metastasis of hepatocellular carcinoma. J Hepatol. 2004;41:629–636. doi: 10.1016/j.jhep.2004.06.018. [DOI] [PubMed] [Google Scholar]
- 25.Lau SH, Sham JS, Xie D, Tzang CH, Tang D, Ma N, Hu L, Wang Y, Wen JM, Xiao G, Zhang WM, et al. Clusterin plays an important role in hepatocellular carcinoma metastasis. Oncogene. 2006;25:1242–1250. doi: 10.1038/sj.onc.1209141. [DOI] [PubMed] [Google Scholar]
- 26.Sun CK, Ng KT, Sun BS, Ho JW, Lee TK, Ng I, Poon RT, Lo CM, Liu CL, Man K, Fan ST. The significance of proline-rich tyrosine kinase2 (Pyk2) on hepatocellular carcinoma progression and recurrence. Br J Cancer. 2007;97:50–57. doi: 10.1038/sj.bjc.6603827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tung-Ping Poon R, Fan ST, Wong J. Risk factors, prevention, and management of postoperative recurrence after resection of hepatocellular carcinoma. Ann Surg. 2000;232:10–24. doi: 10.1097/00000658-200007000-00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang W, Lu Y, Xu Y, Xu L, Zheng W, Wu Y, Li L, Shen P. Estrogen represses hepatocellular carcinoma (HCC) growth via inhibiting alternative activation of tumor-associated macrophages (TAMs) J Biol Chem. 2012;287:40140–40149. doi: 10.1074/jbc.M112.348763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang WL, Wei L, Huang WQ, Li R, Shen WY, Liu JY, Xu JM, Li B, Qin Y. Vigilin is overexpressed in hepatocellular carcinoma and is required for HCC cell proliferation and tumor growth. Oncol Rep. 2014;31:2328–2334. doi: 10.3892/or.2014.3111. [DOI] [PubMed] [Google Scholar]
- 30.Jiang YF, Yang ZH, Hu JQ. Recurrence or metastasis of HCC: Predictors, early detection and experimental antiangiogenic therapy. World J Gastroenterol. 2000;6:61–65. doi: 10.3748/wjg.v6.i1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmidt-Ott KM, Barasch J. WNT/beta-catenin signaling in nephron progenitors and their epithelial progeny. Kidney Int. 2008;74:1004–1008. doi: 10.1038/ki.2008.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Corona DF, Längst G, Clapier CR, Bonte EJ, Ferrari S, Tamkun JW, Becker PB. ISWI is an ATP-dependent nucleosome remodeling factor. Mol Cell. 1999;3:239–245. doi: 10.1016/S1097-2765(00)80314-7. [DOI] [PubMed] [Google Scholar]
- 33.VargaWeisz PD, Becker PB. Chromatin-remodeling factors: Machines that regulate? Curr Opin Cell Biol. 1998;10:346–353. doi: 10.1016/S0955-0674(98)80010-0. [DOI] [PubMed] [Google Scholar]