Abstract
The present study aimed to investigate the associations between three distinct osteoprotegerin (OPG) gene polymorphisms and the risk of intervertebral disc degeneration (IDD). A total of 200 IDD patients and 200 healthy controls were recruited from the Department of Spine Surgery at the First Affiliated Hospital of the University of South China (Hengyang, China) between January 2013 and May 2014. The allele, genotype and haplotype frequency distributions of three OPG polymorphisms in the study and control populations were analyzed by polymerase chain reaction prior to restriction fragment length polymorphism or high resolution melting assays. In addition, serum OPG levels were measured via an ELISA. The genotype and allele frequencies of the OPG rs2073617 polymorphisms were significantly higher in the IDD patients, as compared with the control group (P<0.05). Furthermore, carriers of the C allele exhibited a higher risk of IDD, as compared with carriers of the T allele (P<0.001). Conversely, the genotype and allele frequencies of the two other gene polymorphisms, rs2073618 and rs3102735, showed no significant differences between the patients and controls (P>0.05). The serum OPG levels were significantly higher in IDD patients with TT, TC and CC genotypes at the OPG rs2073617 polymorphism, as compared with the control group (P<0.05). Logistic-regression analysis suggested that high serum levels of OPG were positively correlated with IDD risk, whereas the T-C-A, T-G-A and T-G-G haplotypes were negatively correlated with IDD risk (P<0.05). Furthermore, the G-T-G haplotype was associated with protection against IDD (P=0.008), whereas the G-C-G haplotype was associated with an elevated susceptibility to IDD (P=0.007). The results of the present study suggested that OPG rs2073617 polymorphisms and upregulated serum levels of OPG were associated with an increased risk of IDD, whereas the T-C-A, T-G-A and T-G-G haplotypes were protective factors for IDD. The results of the present study suggested that the OPG gene polymorphism may have an important role in the progression of IDD, and its serum level may function as a valuable predictive indicator of the severity of degenerative disc diseases.
Keywords: osteoprotegerin, genetic polymorphisms, intervertebral disc degeneration, susceptibility, genotypes, haplotype distributions, polymerase chain reaction-restriction fragment length polymorphism, high-resolution melting
Introduction
Intervertebral Disc Degeneration (IDD), which is also called degenerative disc disease, is the major cause of degenerative spinal disease and one of the most common ailments severely affecting quality of life in elderly populations (1). IDD is characterized by lower back pain, intervertebral disc herniation and spinal canal stenosis (2). Degeneration of the nucleus pulposus in spinal discs may reduce the production and secretion of extracellular matrix components, leading to changes in disc structure and resulting in the dysfunction of the discs (3–5). The incidence of IDD has been shown to be influenced by gender and age, with a prevalence in adolescents and young adults of <10%, which increases to 30–50% in middle adulthood (6). The exact pathophysiology underlying IDD is not well understood; however, genetic and environmental factors are thought to be involved (7–9). Previous studies have demonstrated that smoking, excessive biomechanical load, obesity, gender, age, decreased nutrition and other environmental factors are associated with a high risk of developing IDD (10–13). Furthermore, pro-inflammatory signaling pathways, tumor necrosis factor (TNF) and matrix degrading enzymes may have pivotal roles in the pathogenesis of IDD (2,14–16), and it has been suggested that osteoprotegerin (OPG), a member of the TNF receptor superfamily, has a crucial role in the etiology of IDD (17,18).
OPG, which is also called osteoclastogenesis inhibitory factor or tumor necrosis factor receptor superfamily member 11B, is a glycoprotein and cytokine receptor containing 401 amino acid residues (19,20). OPG is primarily synthesized by osteoblasts; however, it is also expressed in various organs and tissues, including the heart, vessel walls, lungs, kidneys and bone (21,22). The main role of OPG is its function as a decoy receptor for receptor activator for nuclear factor-κB ligand (RANKL) (23–25); by competitively binding to RANKL, OPG prevents RANK/RANKL association and activation of RANK, thereby modulating bone resorption, decreasing the number of osteoclasts and acting as a regulator of osteoblast-osteoclast cross-talk in bone homeostasis (26,27). The RANK-RANKL-OPG system regulates the balance between osteoblasts and osteoclasts, and has a crucial role in bone formation and absorption (28). Previous studies have suggested that a deficiency of the OPG gene enhances osteoclastogenesis and secondary hyperactive osteoblasts in long bones and vertebral bones (17,29). Therefore, it has been hypothesized that factors affecting OPG gene regulation may be major genetic factors influencing bone mass and increasing the risk of fractures, osteoporosis and osteoarthritis (30,31). The human OPG gene is located on chromosome 8q23-24, has a length of ~29 kb and consists of 5 exons and 4 introns (32). Previous studies have demonstrated that changes in the OPG serum level are influenced by genetic polymorphisms, including 950T/C at the rs2073617 locus, 1181 G/C at the rs2073618 locus and 1181 G/C at the rs2073618 locus (33–35). The association between genetic polymorphisms in the OPG gene and bone mineral density has been the focus of previous investigations (36,37). Previous studies have implicated increased bone mineral density in the etiology of IDD (38,39); however, few studies have focused on a direct association between OPG genetic polymorphisms and the risk of IDD. Therefore, the present study aimed to investigate the associations between OPG genetic polymorphisms, serum OPG levels and IDD risk.
Materials and methods
Ethical approval and patient consent
The present study was approved by the Ethical Committee of the First Affiliated Hospital of the University of South China. Written informed consent was acquired from all study participants at the time of hospitalization. In addition, the present study was performed in accordance with the guidelines and principles of the Declaration of Helsinki (40).
Subjects
Between January 2013 and May 2014, a total of 200 patients with IDD, including 100 females and 100 males, at the Department of Spine Surgery of the First Affiliated Hospital of the University of South China (Hengyang, China) were enrolled in the present study. The average age of the patients was 52.6±6.6 years (age range, 40–62 years). All patients were diagnosed with IDD according to magnetic resonance imaging (MRI) results and IDD was confirmed by postoperative pathological analyses, according to previous studies (41,42). All patients presented with the typical clinical and physical symptoms of IDD, including: i) Chronic lower back pain with radiation to the lower limb; ii) spasticity and atrophy of the paravertebral and lower limb muscles; iii) limited activity; and iv) positive results in a nerve traction test. In addition, the patients showed obvious disc degeneration in the postoperative pathological analysis. Patients were excluded from the present study if they suffered from lumbar spinal stenosis. In addition, 200 age- and gender-matched healthy subjects from our hospital, including 100 men and 100 women aged between 42 and 62-years (average age, 52.1±5.4 years), were recruited. The MRI findings of the healthy controls showed normal intervertebral disc tissue (http://www.uscspine.com/conditions/back-degenerative-disc.cfm). No significant differences in age, gender, body mass index and smoking history were observed between the IDD patients and controls. The present study was approved by the Ethics Committee of the First Affiliated Hospital of the University of South China.
Specimen collection
Blood samples (10 ml) were drawn from the patient's elbow vein in the morning following an overnight fast, and 3 ml blood was added to ethylenediamine tetraacetic acid (EDTA) tubes (Sigma-Aldrich, St. Louis, MO, USA) for anticoagulation. Genomic DNA was extracted from the 3 ml blood samples using a Whole Blood Genomic DNA Extraction kit (cat. no. OSR-M102/M104; Tiangen Biotech, Co., Ltd., Beijing, China). The remaining blood (without EDTA) was allowed to clot for 1 h at room temperature, followed by centrifugation at 1006.2 × g for 10 min at 37°C to obtain the serum, which was stored at −80°C until further use.
High resolution melting (HRM) analysis
The primers used for HRM genotyping were designed by LightScanner® Primer Design software, version 1.0 (Idaho Technology, Inc., Salt Lake City, UT USA) and are presented in Table I. The polymerase chain reaction (PCR)-HRM analysis was performed using a final reaction volume of 11 µl, consisting of 1 µl genomic DNA (20 ng/l), 0.2 µl each of the forward and reverse primers (each 10 pmol), 5 µl 2X Taq PCR Master mix (containing Taq DNA polymerase, Mg2+ and dNTPs; Tiangen Biotech, Co., Ltd.), 3.6 µl sterilizing ultrapure water and 1 µl 10X LCGreen Plus dye (Idaho Technology, Inc.). The PCR reaction was conducted using the Type-it HRM PCR kit (cat. no. 206542; Qiagen GmbH, Hilden, Germany), according to the manufacturer's protocol. The PCR reaction conditions were as follows: Initial denaturation at 96°C for 5 min, followed by 40 cycles of 96°C for 20 sec, 57°C for 20 sec and 72°C for 20 sec. The HRM analysis was synchronized with the PCR reaction, as follows: 95°C for 5 min, 40°C for 2 min, 60°C for 1 min and then heating from 72–95°C at 0.1°C/sec, during which time the melting curve data was collected. Each HRM analysis detected the standard three genotypes already known from each locus and the genomic DNA of study subjects. LightScanner software (Idaho Technology, Inc.) was used in the melting curve analysis to determine the genotypes.
Table I.
Sequences of the primers used for polymerase chain reaction-high resolution melting genotyping of the osteoprotegerin gene.
| Locus | Primer sequence | Product size (bp) |
|---|---|---|
| rs2073617 | F: 5′-CTGGTAGGACAAATATTGG-3′ | 88 |
| R: 5′-ACTTACCATTTGCGATCACC-3′ | ||
| rs2073618 | F: 5′-CGTTGTCTTGAGAAGGTTGA-3′ | 141 |
| R: 5′-TCCTCAAAAACATGTCAGTGTG-3′ | ||
| rs3102735 | F: 5′- TCCCACTATCATGATTATTTCCC-3′ | 163 |
| R: 5′-ATTATAGGTTTTTAAGTAATTTGT-3′ |
F, forward; R, reverse.
PCR-restriction fragment length polymorphism (PCR-RFLP) assays
PCR primers were designed using Primer Premier 5.0 software (Premier Biosoft International, Palo Alto, CA, USA) and were synthesized by Sangon Biotech Co., Ltd. (Shanghai, China). The sequences and lengths of the primers are shown in Table II. The PCR reaction system consisted of 100 ng genomic DNA, 125 ng each of the upstream and downstream primers, 12.5 µl 2X Taq PCR Master mix and 9.5 µl double-distilled H2O, in a final volume of 25 µl. The cycling conditions were as follows: Initial denaturation at 95°C for 5 min, followed by 30 cycles at 94°C for 30 sec, 62°C for 45 sec and 72°C for 60 sec, and a final extension step at 72°C for 10 min. HincII, XspI and AseI restriction enzymes (Takara Biotechnology Co., Ltd., Dalian, China) were added to the PCR products and then 10% agarose gel electrophoresis was used to assess PCR fragment patterns. An automated DNA sequencer (model 370; Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA) was used to determine the genotypes of the rs2073617, rs2073618 and rs3102735 loci.
Table II.
Sequences of the primers used for polymerase chain reaction-restriction fragment length polymorphism analysis of the three loci of the osteoprotegerin gene.
| Locus | Primer sequence | Product size (bp) | Endonucleases |
|---|---|---|---|
| rs2073617 | F: 5′-GAAGTGAAGGGGTCAGGCAGC-3′ | 342 | HincII |
| R: 5′-GTCTGTCTCTCTCTTGCTGTCTTCC-3′ | |||
| rs2073618 | F: 5′-GTCTGTCTCTCTCTTGCTGTCTTCC-3′ | 340 | XspI |
| R: 5′-GAGATGAAGACAGAAGGTTAATGAC-3′ | |||
| rs3102735 | F: 5′-TTCCTTCCCTTGAATCTGGTG-3′ | 300 | AseI |
| R: 5′-CTAAAGCCCGTGCTATTCTGC-3′ |
F, forward; R, reverse.
Measurement of OPG protein levels in serum
Serum levels of OPG were measured using an OPG ELISA kit (cat. no. K1011; Immundiagnostik AG, Bensheim, Germany), according to the manufacturer's protocol. Optical density (OD) values were measured at 450 nm using a microplate reader. The standard curve was drawn by setting standard concentrations as the vertical axis and OD values as the horizontal axis. All samples were measured twice and averaged, and the detection met laboratory quality control standards.
Construction of haplotypes
OPG rs2073617 and rs3102735 are located in the promoter region of the OPG gene, whereas rs2073618 is located in the exon 1 (43). Haplotype construction of these three genotypes of polymorphic loci was performed using the individual-driven Bayesian method implemented in PHASE 2.1 software (http://stephenslab.uchicago.edu/phase/download.html), as previously described (44).
Statistical analysis
SPSS software, version 18.0 (SPSS, Inc., Chicago, IL, USA) was used to perform data analyses. Hardy-Weinberg equilibrium was used to assess and confirm the representation of the selected samples in the population. Measurement data are presented as the mean ± standard deviation. One-way analysis of variance was used for comparisons between the three groups and Student's t-test was used to compare the measurement data between the groups. The non-parametric Wilcoxon signed-rank test was used to calculate the significance of comparisons when the data did not meet the normal distribution. The χ2 test was applied to compare the distribution frequencies of the genotypes, alleles and haplotypes between the two groups. Logistic regression analysis was conducted to calculate the odds ratios (OR), and 95% confidence intervals (CI) represented the relative risk. All statistical tests were two-sided probability tests. P<0.05 was considered to indicate a statistically significant difference.
Results
Genotyping of the OPG gene by HRM analysis
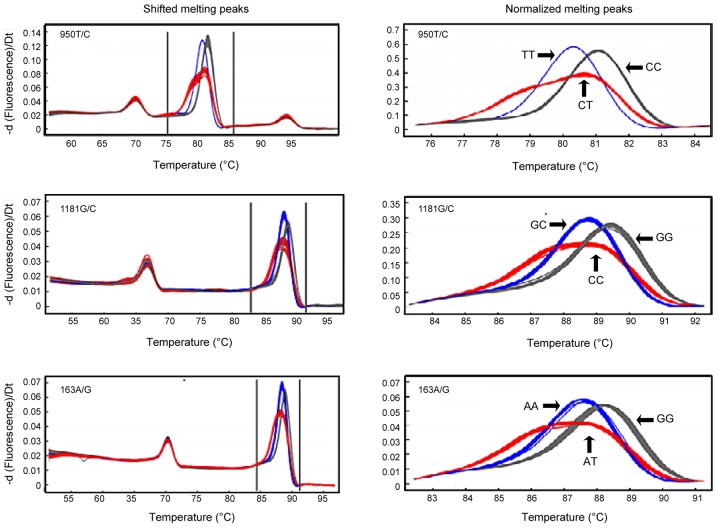
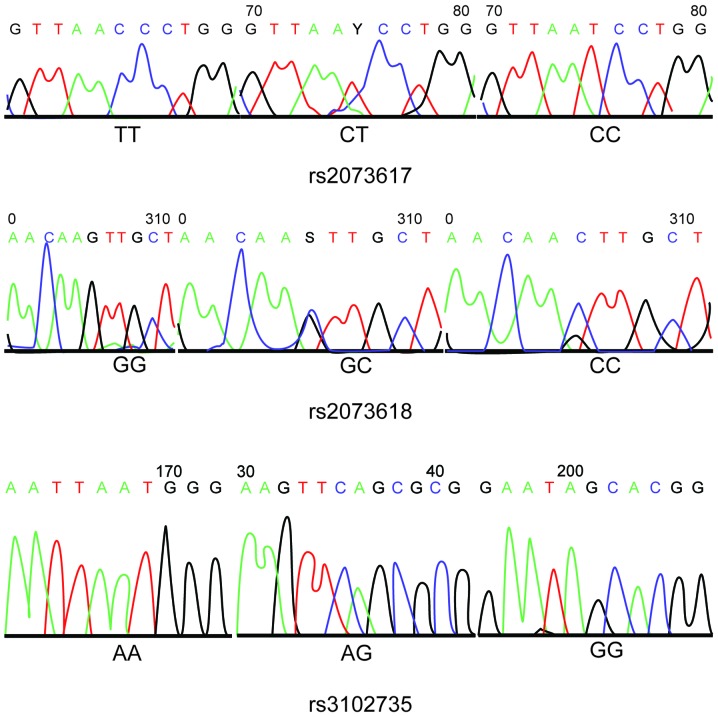
DNA fragments containing polymorphic loci were amplified by PCR and analyzed by HRM. The OPG gene has some common, functionally important genetic polymorphisms that have been associated with various human diseases. The most important polymorphisms of the OPG gene are rs2073617 (950T/C) and rs3102735 (163A/G), which are located in the promoter region, and rs2073618 (1181G/C), which is located in exon 1 (44). The HRM analysis demonstrated that the 950T/C (rs2073617), 1181G/C (rs2073618) and 163A/G (rs3102735) OPG gene polymorphisms were associated with three genotypes (Fig. 1). The HRM genotyping results were consistent with the sequences of the polymorphisms, as determined by PCR and DNA sequencing (Fig. 2).
Figure 1.
Melting curves of OPG genotyping by high resolution melting analysis. There were three different genotypes for the OPG rs2073617, rs2073618 and rs3102735 loci in the study population. OPG, osteoprotegerin.
Figure 2.
Results of the sequencing of 950T/C (rs2073617), 1181G/C (rs2073618) and 163A/G (rs3102735) polymorphisms of the osteoprotegerin gene.
OPG genotyping
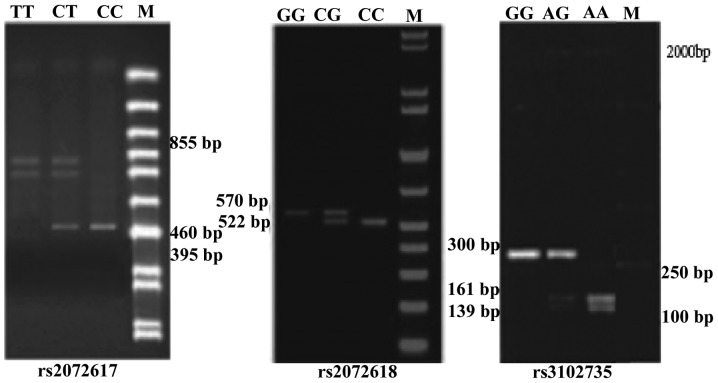
PCR-RFLP analysis of the OPG rs2073617 locus detected a single band of 855 bp corresponding to the homozygous wild-type genotype (TT), three fragments of 855 bp, 460 bp and 395 bp corresponding to the heterozygous mutated genotype (TC), and two fragments of 395 bp and 460 bp corresponding to the homozygous mutated genotype (CC). Similarly, for rs2073618, an undigested and single band of 570 bp corresponded to the homozygous wild-type GG genotype, three fragments of 570 bp, 522 bp and 48 bp corresponded to the heterozygous mutated GC genotype, and two bands of 522 bp and 48 bp corresponded to the homozygous mutant CC genotype, although the 48 bp fragment was too small to be visible. Furthermore, the OPG rs3102735 locus showed three genotypes: Homozygous wild-type (A/A) with 161 bp and 139 bp fragments; heterozygous mutated genotype (G/A) with 300 bp, 161 bp and 139 bp fragments, and homozygous mutated genotype (G/G) with a 300 bp fragment. The results of DNA sequencing confirmed the existence of these genotypes for the rs2073617, rs2073618 and rs3102735 loci (Figs. 2 and 3).
Figure 3.
Agarose gel electrophoresis of the various genotypes of the osteoprotegerin rs2073617, rs2073618 and rs3102735 loci, following restriction enzyme digestion.
Allele and genotype frequency distributions of OPG polymorphisms
The genotype and allele frequencies of OPG rs2073617, rs2073618 and rs3102735 polymorphisms were in accordance with the Hardy-Weinberg equilibrium, thus suggesting that each polymorphism had reached equilibrium and that the selected sample was representative of the population (P>0.05). There were significant differences in the allele and genotype frequencies of the OPG rs2073617 polymorphisms between the case and control groups (P<0.05). In addition, patients carrying the C allele exhibited an increased risk of IDD, as compared with carriers of the other alleles (OR=1.79; 95% CI=1.33–2.41; P<0.001). However, no significant differences were observed in the allele and genotype frequencies of the rs2073618 and rs3102735 polymorphisms between the case and control groups (P>0.05; Table III).
Table III.
Genotype and allele frequency distributions of osteoprotegerin genetic polymorphisms and their associations with intervertebral disc degeneration risk.
| Locus | Genotype/Allele | Control Group [n (%)] | Case group [n (%)] | P1 | OR (95% CI) | P2 |
|---|---|---|---|---|---|---|
| rs2073617 | TT | 112 (56.0) | 84 (41.8) | 0.018 | Ref. | |
| TC | 55 (32.8) | 70 (35.2) | 1.69 (1.07–2.67) | 0.021 | ||
| CC | 33 (11.2) | 46 (23.0) | 1.19 (1.10–3.16) | 0.020 | ||
| TC+CC | 88 (46.0) | 116 (58.2) | 1.76 (1.18–2.61) | 0.005 | ||
| T | 290 (72.4) | 238 (59.4) | <0.001 | Ref. | ||
| C | 110 (27.6) | 162 (40.6) | 1.79 (1.33–2.41) | <0.001 | ||
| rs2073618 | GG | 107 (53.5) | 111 (55.2) | 0.890 | Ref. | |
| CG | 77 (38.5) | 75 (37.8) | 0.94 (0.62–1.42) | 0.765 | ||
| CC | 16 (8.0) | 14 (7.0) | 0.84 (0.39–1.81) | 0.662 | ||
| CC+CG | 93 (46.5) | 89 (44.8) | 1.04 (0.70–1.53) | 0.845 | ||
| G | 291 (72.8) | 304 (74.1) | 0.292 | Ref. | ||
| C | 109 (27.3) | 96 (25.9) | 1.07 (0.78–1.47) | 0.682 | ||
| rs3102735 | AA | 116 (58.4) | 113 (56.6) | 0.945 | Ref. | |
| AG | 54 (26.4) | 55 (27.0) | 1.05 (0.87–1.17) | 0.848 | ||
| GG | 30 (15.2) | 32 (16.4) | 1.10 (0.62–1.92) | 0.781 | ||
| AG+GG | 84 (41.6) | 87 (43.4) | 1.06 (0.71–1.58) | 0.761 | ||
| A | 287 (71.6) | 280 (70.1) | 0.586 | Ref. | ||
| G | 113 (28.4) | 120 (29.9) | 1.09 (0.80–1.47) | 0.586 |
P1, comparison of genotypes and allele frequencies between the case and control groups; P2, comparison with the control group in the risk assessment. OR, odds-ratio; CI, confidence interval; P, P-value.
Association between serum OPG levels and OPG genotype frequency distributions
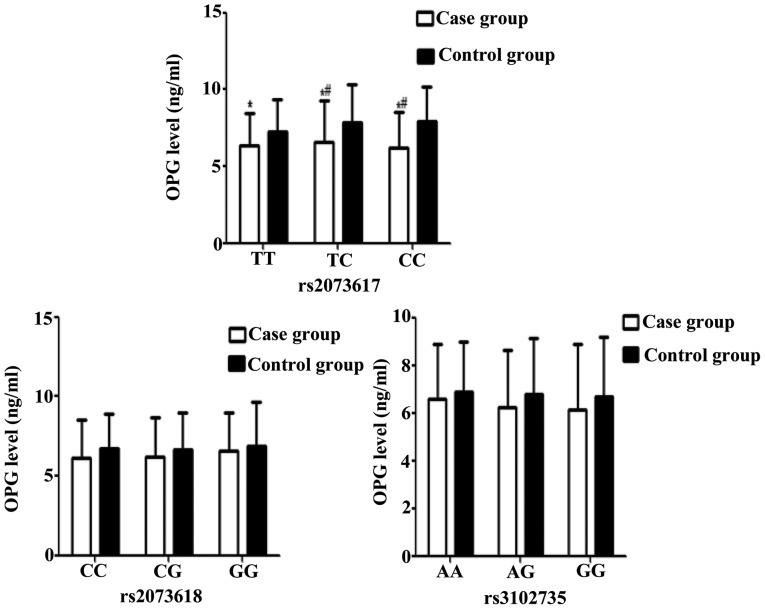
Serum levels of OPG were significantly higher in IDD patients with TT, TC or CC genotypes at the OPG rs2073617 locus, as compared with the control group (all P<0.05). In addition, OPG serum levels were significantly higher in IDD patients harboring the TC or CC mutated genotypes, as compared with those carrying the wild-type homozygous (TT) genotype (P<0.05). No significant differences were observed between the patient and control groups in the OPG serum levels associated with the various genotypes of the rs3102735 and rs2073618 loci (P>0.05; Table IV and Fig. 4). These results suggest that TT, TC and CC genotypes may contribute to the elevation of serum OPG levels in IDD patients.
Table IV.
OPG serum levels in individuals with certain genetic polymorphisms of the OPG gene.
| SNP | Genotype | Control group (ng/ml) | Case group (ng/ml) |
|---|---|---|---|
| rs2073617 | TT | 6.36±2.10 | 7.25±2.12a |
| TC | 6.56±2.45 | 7.86±2.70a,b | |
| CC | 6.20±2.23 | 7.95±2.30a,b | |
| rs2073618 | CC | 6.16±2.13 | 6.75±2.34 |
| CG | 6.21±2.35 | 6.69±2.45 | |
| GG | 6.54±2.82 | 6.84±2.41 | |
| rs3102735 | AA | 6.56±2.13 | 6.86±2.30 |
| AG | 6.21±2.16 | 6.76±2.42 | |
| GG | 6.14±2.52 | 6.69±2.74 |
Data are presented as the mean ± standard deviation.
P<0.05 vs. the control group
P<0.05 vs. the homozygous wild-type genotypes. SNP, single nucleotide polymorphism; OPG, osteoprotegerin.
Figure 4.
OPG serum levels associated with genetic polymorphisms at the rs2073617, rs2073618 and rs3102735 loci of the OPG gene in the case and control groups. Higher serum levels of OPG were detected in intervertebral disc degeneration patients harboring the TT, TC and CC genotypes of the OPG rs2073617 locus, as compared with the control group. *P<0.05 vs. the same genotype in the control group; #P<0.05 vs. the TT genotype. OPG, osteoprotegerin.
Statistical analysis of haplotype frequencies of three polymorphic loci
The T-G-A haplotype was shown to be a potential protective factor for IDD (OR=0.62; 95% CI=0.41–0.94; P=0.02), whereas the C-G-G haplotype was a potential risk factor for IDD (OR=2.24; 95% CI=1.09–4.60; P=0.02; Table V).
Table V.
Frequency distributions of the haplotypes of osteoprotegerin genetic polymorphisms in the case and control groups.
| Haplotypes | Case group | Control group | χ2 | P-value | OR (95% CI) |
|---|---|---|---|---|---|
| T-C-A | 21 (10.5) | 28 (14.0) | 0.82 | 0.36 | 0.70 (0.40–1.32) |
| T-C-G | 9 (4.5) | 12 (6.0) | 0.19 | 0.66 | 0.73 (0.30–1.79) |
| C-C-A | 14 (7.0) | 10 (5.0) | 0.71 | 0.39 | 1.43 (0.61–3.30) |
| C-C-G | 6 (3.0) | 4 (2.0) | 0.41 | 0.52 | 1.51 (0.42–5.45) |
| T-G-A | 55 (27.5) | 76 (38.0) | 5.01 | 0.02 | 0.62 (0.41–0.94) |
| T-G-G | 26 (13.0) | 30 (15.0) | 0.33 | 0.56 | 0.84 (0.48–1.50) |
| C-G-A | 44 (22.0) | 28 (14.0) | 2.61 | 0.06 | 1.73 (1.03–2.91) |
| C-G-G | 25 (12.5) | 12 (6.0) | 5.03 | 0.02 | 2.24 (1.091–4.60) |
Data are presented as n (%). The haplotypes are presented as rs2073617-rs2073618-rs3102735. OR, odds-ratio; CI, confidence interval.
Logistic regression analysis
A multivariate stepwise logistic regression was performed to calculate the adjusted OR. The IDD was the dependent variable and the genotypes and haplotypes of OPG rs2073617, rs3102735 and rs2073618 loci, body mass index, smoking, OPG levels (>6.69 ng/ml as high levels and ≤6.69 ng/ml as low levels), age and gender, were the independent variables. The multivariate stepwise logistic regression results showed that upregulated OPG serum levels were positively correlated with IDD risk, whereas the T-C-A, T-G-A and T-G-G haplotypes were negatively correlated with IDD risk (P<0.05; Table VI).
Table VI.
Multivariate stepwise logistic regression analysis for the associated risk factors of patients with intervertebral disc degeneration.
| Variable | B | S.E. | Wald | df | P-value | Exp (B) | 95% CI |
|---|---|---|---|---|---|---|---|
| T-C-A | −1.022 | 0.455 | 5.051 | 1 | 0.025 | 0.360 | 0.148–0.878 |
| T-G-A | −1.057 | 0.393 | 7.228 | 1 | 0.007 | 0.347 | 0.161–0.751 |
| T-G-G | −0.877 | 0.442 | 3.942 | 1 | 0.047 | 0.416 | 0.175–0.989 |
| High serum level of OPG | 0.567 | 0. 141 | 7.791 | 1 | 0.005 | 1.758 | 1.183–2.611 |
The haplotypes are presented as rs2073617-rs2073618-rs3102735. High serum levels of OPG were defined as >6.69 ng/ml. B, partial regression coefficient; S.E., standard error of regression; Wald, Wald χ2; df, degree of freedom; Exp (B), adjusted odds-ratio; 95% CI, 95% confidence interval; OPG, osteoprotegerin.
Discussion
The pathogenesis of IDD is considered multifactorial, involving various genetic and environmental factors (7,9); however, the exact mechanism underlying IDD remains incompletely understood. The present study evaluated the associations between polymorphisms of the OPG gene and the risk of IDD, and demonstrated that the 950T/C variant at the rs2073617 locus was associated with an increased risk of IDD, as compared with the 950T/T wild-type genotype. Conversely, the 1181G/C (rs2073618) and 163A/G (rs3102735) SNPs were not associated with an increased susceptibility to IDD. These results suggested that the OPG gene, in particular the 950T/C (rs2073617) SNP, may serve as a potential indicator of IDD risk. Previous studies based on pathophysiological analyses have reported the involvement of various cytokines in IDD (45,46). In addition, the RANK/RANKL/OPG signaling pathway has been proposed to have a role in bone metabolism (26,47,48). Previous studies have demonstrated that disruption of the RANKL/OPG balance led to cartilage degradation due to mechanical loading and resulted in the progression of IDD or osteoarthritis (49,50). In addition, it has been reported that OPG genetic polymorphisms negatively influence bone resorption, and thereby regulate bone mineral density, resulting in the deterioration of patients with IDD (51,52). OPG gene expression has a crucial role in maintaining the integrity of endplate cartilage by preventing its resorption by osteoclasts (17).
In the present study, a haplotype analysis suggested that the G-T-G, T-C-A, T-G-A and T-G-G haplotypes were associated with protection against IDD, whereas the G-C-G haplotype was associated with an increased susceptibility to IDD. Furthermore, the serum levels of OPG were significantly higher in IDD patients with TT, TC and CC genotypes at the OPG rs2073617 locus, as compared with the control group (P<0.05), which suggested that TT, TC and CC genotypes may be significant risk factors for IDD development and that upregulated serum levels of OPG are correlated with IDD risk.
Insufficient nutrition has been shown to be involved in the degeneration of intervertebral discs (50,53,54). Metabolism and nutrient exchange at intervertebral discs is dependent on the interaction of endplate cartilage with intervertebral discs (55), and endplate cartilage has an important role in the biomechanical structure of intervertebral discs (55). OPG polymorphisms have been closely associated with the degeneration of endplate cartilage and intervertebral discs by upregulating OPG serum levels; thus OPG has already been associated with an increased risk of IDD (53,56). Furthermore, a previous study demonstrated that the TT, TC and CC genotypes of the rs2073617 locus affected the protein expression levels of OPG and were associated with elevated OPG serum levels; thus suggesting that upregulated OPG expression is a high risk factor for IDD (57). In addition, it was reported that the CC genotype of the rs2073617 locus was associated with increased serum levels of OPG and increased production of OPG by osteoblasts in order to restore disrupted bone metabolism (58). Consistent with this, the present study demonstrated that IDD patients with the TT, TC or CC genotypes of the OPG rs2073617 polymorphism had significantly higher serum levels of OPG, as compared with the healthy controls. These results suggested that IDD patients with high OPG serum levels and the 950T/C SNP at the rs2073617 locus may be at a high risk of developing impaired bone metabolism and endplate cartilage degeneration, resulting in IDD.
In conclusion, the present study demonstrated that the OPG genetic polymorphism, 950T/C (rs2073617), was associated with an increased risk of IDD, and that the C allele may be a high risk factor for elevated IDD risk by promoting increased serum levels of OPG. In addition, the G-T-G haplotype was associated with protection against IDD, whereas the G-C-G haplotype was associated with an elevated risk of IDD. Furthermore, an elevated OPG serum level was positively correlated with IDD risk, whereas the T-C-A, T-G-A and T-G-G haplotypes were negatively correlated with IDD risk. These results suggested that genetic polymorphisms of the OPG gene may influence susceptibility to IDD by altering the protein serum levels of OPG. With continued efforts, the OPG rs2073617 polymorphism may emerge as a biomarker for the diagnosis of IDD, and targeting OPG may represent a novel and promising therapeutic strategy for biologically-induced disc repair in early stage disc degeneration.
References
- 1.Sakai D. Future perspectives of cell-based therapy for intervertebral disc disease. Eur Spine J. 2008;17(Suppl 4):452–458. doi: 10.1007/s00586-008-0743-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sudo H, Yamada K, Iwasaki K, Higashi H, Ito M, Minami A, Iwasaki N. Global identification of genes related to nutrient deficiency in intervertebral disc cells in an experimental nutrient deprivation model. PLoS One. 2013;8:e58806. doi: 10.1371/journal.pone.0058806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Richardson SM, Walker RV, Parker S, Rhodes NP, Hunt JA, Freemont AJ, Hoyland JA. Intervertebral disc cell-mediated mesenchymal stem cell differentiation. Stem Cells. 2006;24:707–716. doi: 10.1634/stemcells.2005-0205. [DOI] [PubMed] [Google Scholar]
- 4.Sobajima S, Vadala G, Shimer A, Kim JS, Gilbertson LG, Kang JD. Feasibility of a stem cell therapy for intervertebral disc degeneration. Spine J. 2008;8:888–896. doi: 10.1016/j.spinee.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 5.Song YQ, Karasugi T, Cheung KM, Chiba K, Ho DW, Miyake A, Kao PY, Sze KL, Yee A, Takahashi A, et al. Lumbar disc degeneration is linked to a carbohydrate sulfotransferase 3 variant. J Clin Invest. 2013;123:4909–4917. doi: 10.1172/JCI69277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang YX, Griffith JF, Zeng XJ, Deng M, Kwok AW, Leung JC, Ahuja AT, Kwok T, Leung PC. Prevalence and sex difference of lumbar disc space narrowing in elderly chinese men and women: Osteoporotic fractures in men (Hong Kong) and osteoporotic fractures in women (Hong Kong) studies. Arthritis Rheum. 2013;65:1004–1010. doi: 10.1002/art.37857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hangai M, Kaneoka K, Kuno S, Hinotsu S, Sakane M, Mamizuka N, Sakai S, Ochiai N. Factors associated with lumbar intervertebral disc degeneration in the elderly. Spine J. 2008;8:732–740. doi: 10.1016/j.spinee.2007.07.392. [DOI] [PubMed] [Google Scholar]
- 8.Liu G, Cao P, Chen H, Yuan W, Wang J, Tang X. MiR-27a regulates apoptosis in nucleus pulposus cells by targeting PI3K. PLoS One. 2013;8:e75251. doi: 10.1371/journal.pone.0075251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kalichman L, Hunter DJ. The genetics of intervertebral disc degeneration. Familial predisposition and heritability estimation. Joint Bone Spine. 2008;75:383–387. doi: 10.1016/j.jbspin.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 10.Guehring T, Wilde G, Sumner M, Grünhagen T, Karney GB, Tirlapur UK, Urban JP. Notochordal intervertebral disc cells: Sensitivity to nutrient deprivation. Arthritis Rheum. 2009;60:1026–1034. doi: 10.1002/art.24407. [DOI] [PubMed] [Google Scholar]
- 11.Smith LJ, Nerurkar NL, Choi KS, Harfe BD, Elliott DM. Degeneration and regeneration of the intervertebral disc: Lessons from development. Dis Model Mech. 2011;4:31–41. doi: 10.1242/dmm.006403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takatalo J, Karppinen J, Taimela S, Niinimäki J, Laitinen J, Blanco Sequeiros R, Paananen M, Remes J, Näyhä S, Tammelin T, et al. Body mass index is associated with lumbar disc degeneration in young Finnish males: Subsample of Northern Finland birth cohort study 1986. BMC Musculoskelet Disord. 2013;14:87. doi: 10.1186/1471-2474-14-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang YX, Griffith JF. Menopause causes vertebral endplate degeneration and decrease in nutrient diffusion to the intervertebral discs. Med Hypotheses. 2011;77:18–20. doi: 10.1016/j.mehy.2011.03.014. [DOI] [PubMed] [Google Scholar]
- 14.Omair A, Holden M, Lie BA, Reikeras O, Brox JI. Treatment outcome of chronic low back pain and radiographic lumbar disc degeneration are associated with inflammatory and matrix degrading gene variants: A prospective genetic association study. BMC Musculoskelet Disord. 2013;14:105. doi: 10.1186/1471-2474-14-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kalb S, Martirosyan NL, Kalani MY, Broc GG, Theodore N. Genetics of the degenerated intervertebral disc. World Neurosurg. 2012;77:491–501. doi: 10.1016/j.wneu.2011.07.014. [DOI] [PubMed] [Google Scholar]
- 16.Purmessur D, Walter BA, Roughley PJ, Laudier DM, Hecht AC, Iatridis J. A role for TNF-α in intervertebral disc degeneration: A non-recoverable catabolic shift. Biochem Biophys Res Commun. 2013;433:151–156. doi: 10.1016/j.bbrc.2013.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liang QQ, Li XF, Zhou Q, Xing L, Cheng SD, Ding DF, Xu LQ, Tang DZ, Bian Q, Xi ZJ, et al. The expression of osteoprotegerin is required for maintaining the intervertebral disc endplate of aged mice. Bone. 2011;48:1362–1369. doi: 10.1016/j.bone.2011.03.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ciacli C, Puşchiţă M. RANKL/RANK/OPG molecular complex-control factors in bone remodeling in psoriatic arthritis. Rev Med Chir Soc Med Nat Iasi. 2011;115:354–360. [PubMed] [Google Scholar]
- 19.Chollet ME, Brouland JP, Bal Dit Sollier C, Bauduer F, Drouet L, Bellucci S. Evidence of a colocalisation of osteoprotegerin (OPG) with von Willebrand factor (VWF) in platelets and megakaryocytes alpha granules. Studies from normal and grey platelets. Br J Haematol. 2010;148:805–807. doi: 10.1111/j.1365-2141.2009.07989.x. [DOI] [PubMed] [Google Scholar]
- 20.Pobeha P, Petrasova D, Tkacova R, Joppa P. Circulatory osteoprotegerin is related to osteoporosis of the hip in patients with COPD. Respir Med. 2014;108:621–627. doi: 10.1016/j.rmed.2013.12.011. [DOI] [PubMed] [Google Scholar]
- 21.Jayakumar P, Di Silvio L. Osteoblasts in bone tissue engineering. Proc Inst Mech Eng H. 2010;224:1415–1440. doi: 10.1243/09544119JEIM821. [DOI] [PubMed] [Google Scholar]
- 22.Tousoulis D, Siasos G, Maniatis K, Oikonomou E, Kioufis S, Zaromitidou M, Paraskevopoulos T, Michalea S, Kollia C, Miliou A, et al. Serum osteoprotegerin and osteopontin levels are associated with arterial stiffness and the presence and severity of coronary artery disease. Int J Cardiol. 2013;167:1924–1928. doi: 10.1016/j.ijcard.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 23.Trouvin AP, Goeb V. Receptor activator of nuclear factor-κB ligand and osteoprotegerin: Maintaining the balance to prevent bone loss. Clin Interv Aging. 2010;5:345–354. doi: 10.2147/CIA.S10153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohmori R, Momiyama Y, Taniguchi H, Tanaka N, Kato R, Nakamura H, Ohsuzu F, Nagano M, Egashira T. Association between osteoprotegerin gene polymorphism and coronary artery disease in Japanese men. Atherosclerosis. 2006;187:215–217. doi: 10.1016/j.atherosclerosis.2006.03.026. [DOI] [PubMed] [Google Scholar]
- 25.Granchi D, Pellacani A, Spina M, Cenni E, Savarino LM, Baldini N, Giunti A. Serum levels of osteoprotegerin and receptor activator of nuclear factor-kappaB ligand as markers of periprosthetic osteolysis. J Bone Joint Surg Am. 2006;88:1501–1509. doi: 10.2106/JBJS.E.01038. [DOI] [PubMed] [Google Scholar]
- 26.Vega D, Maalouf NM, Sakhaee K. CLINICAL Review #: The role of receptor activator of nuclear factor-kappaB (RANK)/RANK ligand/osteoprotegerin: Clinical implications. J Clin Endocrinol Metab. 2007;92:4514–4521. doi: 10.1210/jc.2007-0646. [DOI] [PubMed] [Google Scholar]
- 27.Grimaud E, Soubigou L, Couillaud S, Coipeau P, Moreau A, Passuti N, Gouin F, Redini F, Heymann D. Receptor activator of nuclear factor kappaB ligand (RANKL)/osteoprotegerin (OPG) ratio is increased in severe osteolysis. Am J Pathol. 2003;163:2021–2031. doi: 10.1016/S0002-9440(10)63560-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Silva I, Branco JC. Rank/Rankl/opg: Literature review. Acta Reumatol Port. 2011;36:209–218. [PubMed] [Google Scholar]
- 29.Nakamura M, Udagawa N, Matsuura S, Mogi M, Nakamura H, Horiuchi H, Saito N, Hiraoka BY, Kobayashi Y, Takaoka K, et al. Osteoprotegerin regulates bone formation through a coupling mechanism with bone resorption. Endocrinology. 2003;144:5441–5449. doi: 10.1210/en.2003-0717. [DOI] [PubMed] [Google Scholar]
- 30.Jurado S, Nogués X, Agueda L, Garcia-Giralt N, Urreizti R, Yoskovitz G, Pérez-Edo L, Saló G, Carreras R, Mellibovsky L, et al. Polymorphisms and haplotypes across the osteoprotegerin gene associated with bone mineral density and osteoporotic fractures. Osteoporos Int. 2010;21:287–296. doi: 10.1007/s00198-009-0956-4. [DOI] [PubMed] [Google Scholar]
- 31.Sun T, Chen M, Lin X, Yu R, Zhao Y, Wang J. The influence of osteoprotegerin genetic polymorphisms on bone mineral density and osteoporosis in Chinese postmenopausal women. Int Immunopharmacol. 2014;22:200–203. doi: 10.1016/j.intimp.2014.06.023. [DOI] [PubMed] [Google Scholar]
- 32.Morinaga T, Nakagawa N, Yasuda H, Tsuda E, Higashio K. Cloning and characterization of the gene encoding human osteoprotegerin/osteoclastogenesis-inhibitory factor. Eur J Biochem. 1998;254:685–691. doi: 10.1046/j.1432-1327.1998.2540685.x. [DOI] [PubMed] [Google Scholar]
- 33.García-Unzueta MT, Riancho JA, Zarrabeitia MT, Sañudo C, Berja A, Valero C, Pesquera C, Paule B, González-Macías J, Amado JA. Association of the 163A/G and 1181G/C osteoprotegerin polymorphism with bone mineral density. Horm Metab Res. 2008;40:219–224. doi: 10.1055/s-2008-1046793. [DOI] [PubMed] [Google Scholar]
- 34.Vidal C, Formosa R, Xuereb-Anastasi A. Functional polymorphisms within the TNFRSF11B (osteoprotegerin) gene increase the risk for low bone mineral density. J Mol Endocrinol. 2011;47:327–333. doi: 10.1530/JME-11-0067. [DOI] [PubMed] [Google Scholar]
- 35.Kim JG, Kim JH, Kim JY, Ku SY, Jee BC, Suh CS, Kim SH, Choi YM. Association between osteoprotegerin (OPG), receptor activator of nuclear factor-kappaB (RANK) and RANK ligand (RANKL) gene polymorphisms and circulating OPG, soluble RANKL levels and bone mineral density in Korean postmenopausal women. Menopause. 2007;14:913–918. doi: 10.1097/gme.0b013e31802d976f. [DOI] [PubMed] [Google Scholar]
- 36.Liu S, Yi Z, Ling M, Shi J. Association between g.19163A>G and g.23298T>C genetic variants of the osteoprotegerin gene and bone mineral density in Chinese women. Hormones (Athens) 2013;12:578–583. doi: 10.14310/horm.2002.1446. [DOI] [PubMed] [Google Scholar]
- 37.Wang Q, Chen Z, Huang Y, Li Q, Zhu L, Cai X, He G, Xie Y, Liu Q. The relationship between osteoprotegerin gene polymorphisms and bone mineral density in Chinese postmenopausal women. Int Immunopharmacol. 2013;17:404–407. doi: 10.1016/j.intimp.2013.06.031. [DOI] [PubMed] [Google Scholar]
- 38.Wang Y, Battié MC, Boyd SK, Videman T. The osseous endplates in lumbar vertebrae: Thickness, bone mineral density and their associations with age and disk degeneration. Bone. 2011;48:804–809. doi: 10.1016/j.bone.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 39.Salo S, Leinonen V, Rikkonen T, Vainio P, Marttila J, Honkanen R, Tuppurainen M, Kröger H, Sirola J. Association between bone mineral density and lumbar disc degeneration. Maturitas. 2014;79:449–455. doi: 10.1016/j.maturitas.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 40.World Medical Association: World Medical Association Declaration of Helsinki: Ethical principles for medical research involving human subjects. JAMA. 2013;310:2191–2194. doi: 10.1001/jama.2013.281053. [DOI] [PubMed] [Google Scholar]
- 41.Kim NK, Shin DA, Han IB, Yoo EH, Kim SH, Chung SS. The association of aggrecan gene polymorphism with the risk of intervertebral disc degeneration. Acta Neurochir (Wien) 2011;153:129–133. doi: 10.1007/s00701-010-0831-2. [DOI] [PubMed] [Google Scholar]
- 42.Mwale F, Iatridis JC, Antoniou J. Quantitative MRI as a diagnostic tool of intervertebral disc matrix composition and integrity. Eur Spine J. 2008;17(Suppl 4):432–440. doi: 10.1007/s00586-008-0744-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hashemi M, Ebrahimi M, Amininia S, Naderi M, Eskandari-Nasab E, Taheri M. Evaluation of rs3102735 and rs2073617 Osteoprotegerin Gene Polymorphisms and the Risk of Childhood Acute lymphoblastic Leukemia in Zahedan Southeast Iran. Int J Hematol Oncol Stem Cell Res. 2014;8:39–44. [PMC free article] [PubMed] [Google Scholar]
- 44.Stephens M, Scheet P. Accounting for decay of linkage disequilibrium in haplotype inference and missing-data imputation. Am J Hum Genet. 2005;76:449–462. doi: 10.1086/428594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Risbud MV, Shapiro IM. Role of cytokines in intervertebral disc degeneration: Pain and disc content. Nat Rev Rheumatol. 2014;10:44–56. doi: 10.1038/nrrheum.2013.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shamji MF, Setton LA, Jarvis W, So S, Chen J, Jing L, Bullock R, Isaacs RE, Brown C, Richardson WJ. Proinflammatory cytokine expression profile in degenerated and herniated human intervertebral disc tissues. Arthritis Rheum. 2010;62:1974–1982. doi: 10.1002/art.27444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li WF, Hou SX, Yu B, Li MM, Férec C, Chen JM. Genetics of osteoporosis: Accelerating pace in gene identification and validation. Hum Genet. 2010;127:249–285. doi: 10.1007/s00439-009-0773-z. [DOI] [PubMed] [Google Scholar]
- 48.Zupan J, Mencej-Bedrac S, Jurković-Mlakar S, Prezelj J, Marc J. Gene-gene interactions in RANK/RANKL/OPG system influence bone mineral density in postmenopausal women. J Steroid Biochem Mol Biol. 2010;118:102–106. doi: 10.1016/j.jsbmb.2009.10.013. [DOI] [PubMed] [Google Scholar]
- 49.Shimizu S, Asou Y, Itoh S, Chung UI, Kawaguchi H, Shinomiya K, Muneta T. Prevention of cartilage destruction with intraarticular osteoclastogenesis inhibitory factor/osteoprotegerin in a murine model of osteoarthritis. Arthritis Rheum. 2007;56:3358–3365. doi: 10.1002/art.22941. [DOI] [PubMed] [Google Scholar]
- 50.Kadri A, Ea HK, Bazille C, Hannouche D, Lioté F, Cohen-Solal ME. Osteoprotegerin inhibits cartilage degradation through an effect on trabecular bone in murine experimental osteoarthritis. Arthritis Rheum. 2008;58:2379–2386. doi: 10.1002/art.23638. [DOI] [PubMed] [Google Scholar]
- 51.Styrkarsdottir U, Halldorsson BV, Gretarsdottir S, Gudbjartsson DF, Walters GB, Ingvarsson T, Jonsdottir T, Saemundsdottir J, Center JR, Nguyen TV, et al. Multiple genetic loci for bone mineral density and fractures. N Engl J Med. 2008;358:2355–2365. doi: 10.1056/NEJMoa0801197. [DOI] [PubMed] [Google Scholar]
- 52.Lee YH, Woo JH, Choi SJ, Ji JD, Song GG. Associations between osteoprotegerin polymorphisms and bone mineral density: A meta-analysis. Mol Biol Rep. 2010;37:227–234. doi: 10.1007/s11033-009-9637-9. [DOI] [PubMed] [Google Scholar]
- 53.Stolina M, Schett G, Dwyer D, Vonderfecht S, Middleton S, Duryea D, Pacheco E, Van G, Bolon B, Feige U, et al. RANKL inhibition by osteoprotegerin prevents bone loss without affecting local or systemic inflammation parameters in two rat arthritis models: Comparison with anti-TNFalpha or anti-IL-1 therapies. Arthritis Res Ther. 2009;11:R187. doi: 10.1186/ar2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kepler CK, Ponnappan RK, Tannoury CA, Risbud MV, Anderson DG. The molecular basis of intervertebral disc degeneration. Spine J. 2013;13:318–330. doi: 10.1016/j.spinee.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 55.Raj PP. Intervertebral disc: Anatomy-physiology-pathophysiology-treatment. Pain Pract. 2008;8:18–44. doi: 10.1111/j.1533-2500.2007.00171.x. [DOI] [PubMed] [Google Scholar]
- 56.Matzko ME, Bowen TR, Smith WR. Orthogenomics: An update. J Am Acad Orthop Surg. 2012;20:536–546. doi: 10.5435/JAAOS-20-08-536. [DOI] [PubMed] [Google Scholar]
- 57.Biscetti F, Porreca CF, Bertucci F, Straface G, Santoliquido A, Tondi P, Angelini F, Pitocco D, Santoro L, Gasbarrini A, et al. TNFRSF11B gene polymorphisms increased risk of peripheral arterial occlusive disease and critical limb ischemia in patients with type 2 diabetes. Acta Diabetol. 2014;51:1025–1032. doi: 10.1007/s00592-014-0664-1. [DOI] [PubMed] [Google Scholar]
- 58.Straface G, Biscetti F, Pitocco D, Bertoletti G, Misuraca M, Vincenzoni C, Snider F, Arena V, Stigliano E, Angelini F, et al. Assessment of the genetic effects of polymorphisms in the osteoprotegerin gene, TNFRSF11B, on serum osteoprotegerin levels and carotid plaque vulnerability. Stroke. 2011;42:3022–3028. doi: 10.1161/STROKEAHA.111.619288. [DOI] [PubMed] [Google Scholar]