Abstract
DNA methylation has a crucial role in cancer biology. In the present study, a genome-wide analysis of DNA methylation in hepatoblastoma (HB) tissues was performed to verify differential methylation levels between HB and normal tissues. As alpha-fetoprotein (AFP) has a critical role in HB, AFP methylation levels were also detected using pyrosequencing. Normal and HB liver tissue samples (frozen tissue) were obtained from patients with HB. Genome-wide analysis of DNA methylation in these tissues was performed using an Infinium HumanMethylation450 BeadChip, and the results were confirmed with reverse transcription-quantitative polymerase chain reaction. The Infinium HumanMethylation450 BeadChip demonstrated distinctively less methylation in HB tissues than in non-tumor tissues. In addition, methylation enrichment was observed in positions near the transcription start site of AFP, which exhibited lower methylation levels in HB tissues than in non-tumor liver tissues. Lastly, a significant negative correlation was observed between AFP messenger RNA expression and DNA methylation percentage, using linear Pearson's R correlation coefficients. The present results demonstrate differential methylation levels between HB and normal tissues, and imply that aberrant methylation of AFP in HB could reflect HB development. Expansion of these findings could provide useful insight into HB biology.
Keywords: epigenetics, liver, cancer, pediatrics, AFP
Introduction
Hepatoblastoma (HB) is an uncommon liver malignancy in infants and children. It accounts for just over 1% of pediatric cancers (1), but exhibits increasing incidence in North America and Europe (2). This disease is most commonly diagnosed during a child's first 3 years of life (3); only 5% of new HB cases are diagnosed in children older than 4 years (4). The disease occurs significantly more frequently in boys than in girls, for reasons that remain to be determined (4).
HB originates from immature liver precursor cells (2). Histologically, HB can be divided into epithelial or mixed epithelial/mesenchymal tissues (5). The majority of HB cases are epithelial in origin, and consist of a mixture of embryonal and fetal cell types (5). Approximately 5% of HB cases are of the small cell undifferentiated subtype (5). Currently, surgical resection, adjuvant chemotherapy and liver transplantation are the only options to treat HB (2). Thus, novel strategies are required to improve the current understanding of HB biology and to provide targets for therapy or early detection of the disease.
Methylation of DNA is the only known genetically programmed DNA modification process in mammals; it regulates several biological processes, including gene transcription, X chromosome inactivation, genomic imprinting and chromatin modification (6–8). Methylation changes in CpG islands (CGIs) and CpG shores (low CpG density areas within 2 kb of CGIs), as well as CpG shelves (low CpG density areas within 2–4 kb of CGIs) (9), affect gene expression. Hypomethylation of promoter regions of crucial genes can activate relevant gene expression and may contribute to tumorigenesis (10–12).
Several genome-wide analyses had been conducted in different types of tumors. For instance, Revill et al revealed sphingomyelin phosphodiesterase 3 as a potent tumor suppressor gene, which is hypermethylated in primary hepatocellular carcinoma (13). The present study aimed to determine whether changes in methylation status could affect HB by analyzing HB tissue samples and paired distant noncancerous tissues to profile differentially methylated genes in the disease.
Materials and methods
HB specimens and clinical information
Matched pairs of HB and adjacent non-tumor tissues were obtained from the surgical specimen archives of the Children's Hospital of Fudan University (Shanghai, China) from 10 patients who underwent partial hepatectomy, and none of them had received radiation therapy or chemotherapy prior to tumour biopsy or resection; their tissue pathology results confirmed HB with >80% viable tumor cells. The patients did not undergo any prior chemotherapy or other forms of therapy, and underwent standardized treatment post-operation. Clinical data were obtained retrospectively from clinical files (Table I). In total, 6 patients were of the mixed embryonal/fetal subtype, and the remaining 4 patients were of the epithelial subtype. The parents of the patients from whom these samples were obtained provided their written informed consent to participate in the study, and the Ethics Committee of the Children's Hospital of Fudan University approved the study as well as the consent procedure.
Table I.
Distribution of study subjects with HB and their serum AFP levels.
| Case | Age (months)a | Gender | Diagnosis HB type | AFP (ng/ml)b | Viral hepatitis | Chip |
|---|---|---|---|---|---|---|
| 1 | 7 | Male | Mixed embryonal/fetal subtype | 68,490 | None | 1 |
| 2 | 23 | Male | Mixed embryonal/fetal subtype | >121,000 | None | 2 |
| 3 | 11 | Female | Mixed embryonal/fetal subtype | >121,000 | None | 3 |
| 4 | 10 | Female | Mixed embryonal/fetal subtype | >121,000 | None | – |
| 5 | 20 | Female | Mixed embryonal/fetal subtype | >121,000 | None | – |
| 6 | 7 | Male | Mixed embryonal/fetal subtype | >121,000 | None | – |
| 7 | 30 | Male | Epithelial type | >121,000 | None | – |
| 8 | 14 | Male | Epithelial type | >121,000 | None | – |
| 9 | 19 | Male | Epithelial type | >121,000 | None | – |
| 10 | 7 | Female | Epithelial type | >121,000 | None | – |
Age on the day that the serum sample was collected.
AFP normal range, ≤25 ng/ml. AFP, alpha-fetoprotein; HB, hepatoblastoma.
Genomic DNA was extracted from 10 HB primary tumors (fresh frozen tissue) and adjacent non-tumor tissues, of which, 3 were subjected to Infinium HumanMethylation450 BeadChip analysis (Table I). Total RNA was extracted from 10 matched HB tumor and non-tumor pairs, reverse transcribed to complementary DNA (cDNA) and then subjected to quantitative polymerase chain reaction (qPCR) analysis of alpha-fetoprotein (AFP) messenger RNA (mRNA) expression.
Infinium HumanMethylation450 BeadChip
Samples from 3 patients were analyzed with Infinium HumanMethylation450 BeadChip (Illumina Inc., San Diego, CA, USA), which is a cost-effective approach to quickly analyze each individual's methylome (14). The DNA was treated with bisulfite and hybridized to arrays according to the manufacturer's protocol. Raw data were adjusted for dye bias and quantile-normalized with data derived from Infinium I or Infinium II assay chemistry probes considered separately (Illumina Inc., San Diego, CA, USA). This high-throughput platform enabled quantitative evaluation of methylation levels with single-nucleotide resolution. The β-values were used to reflect methylation status, ranging from 0 to 1, where 0 is unmethylated and 1 is fully methylated.
Gene function analysis
The predicted target genes were input into the Database for Annotation, Visualization and Integrated Discovery (http://david.abcc.ncifcrf.gov/), which uses Gene Ontology (GO) to identify the molecular function of the profiled genes (15). The Kyoto Encyclopedia of Genes and Genomes (KEGG) database (http://www.genome.ad.jp/kegg/) was used to analyze the potential functions of these target genes in the pathways. P<0.05 was used as the cut-off value.
Reverse transcription (RT)-qPCR
Total cellular RNA was isolated from HB and normal tissues using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) and then reverse transcribed using a PrimeScript RT reagent kit (Perfect Real Time) with gDNA Eraser (Takara Biotechnology Co., Ltd., Dalian, China) following the manufacturer's protocol. The expression of AFP was analyzed by qPCR with a SYBRGreen PCR kit (Takara Biotechnology Co., Ltd.). The AFP primers used for q-PCR were: Forward, 5′-GTGGTCAGTTTGCAGCATTC-3′ and reverse, 5′-AGAGGAGATGTGCTGGATTG-3′ (length, 110 bp). Glyceraldehyde 3-phosphate dehydrogenase mRNA was used as the internal control, with forward 5′-AAAGCCCACTCCAGCATC-3′ and reverse 5′-TAGCGAGCAGCCCAAAGA-3′ primers. The PCR cycling conditions were as follows: 95°C for 15 sec, followed by 95°C for 5 sec and 60°C for 31 sec, for 40 cycles. For quantitative results, AFP expression was represented as fold-change by the 2−ΔΔCq method and statistically analyzed (16).
Statistical analyses
Statistical analyses and graphical depiction of data were conducted with GraphPad Prism 5.0 (GraphPad Software, Inc., La Jolla, CA, USA). Results were presented as the mean ± standard error of the mean and were evaluated with Student's t-test (two-tailed) unless otherwise specified (paired t-test or Pearson's correlation). Certain statistical calculations were performed using SPSS version 19.0 (IBM SPSS, Armonk, NY, USA). P<0.05 was considered to indicate a statistically significant difference.
Results
Hierarchical clustering analysis by Infinium HumanMethylation450 BeadChip and functional annotations of differentially methylated CpG sites
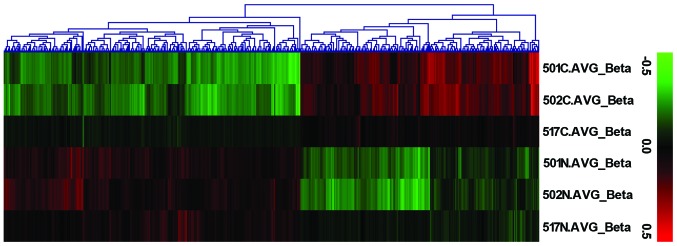
The Infinium HumanMethylation450 BeadChip was used to obtain data from an independent cohort of 3 HB-non-tumor tissue pairs (probe call rate, >99% for all samples). Principal component analysis revealed separation of HB-non-tumor pairs and demonstrated the methylation levels of HB tissues to be significantly lower than those of non-tumor tissues (Fig. 1).
Figure 1.
Infinium HumanMethylation450 BeadChip analysis revealed an unsupervised hierarchical clustering of samples based on their β-values from the CpG loci. Rows represent the samples (three cancer tissues and three normal tissues) and columns represent CpGs. Green β-values indicate hypomethylation and red hypermethylation. C.AVG_Beta, cancer tissue denomination; N.AVG_Beta, non-cancer/normal tissue denomination.
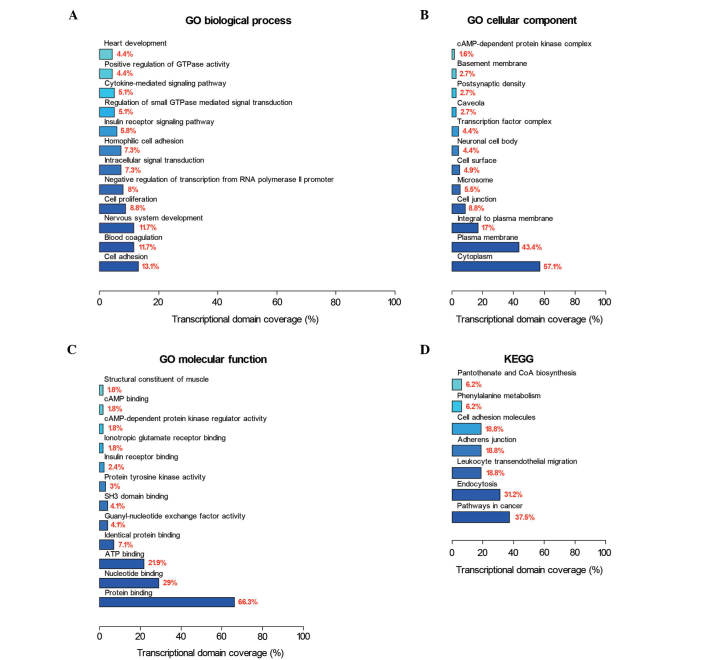
Next, GO and KEGG pathway database categories were used to analyze the 524 differential methylated genes identified. Cell adhesion, blood coagulation and nervous system development were observed to be the three most affected biological processes; while cytoplasm, plasma membrane and integral plasma membrane structures were observed to be the three most affected cellular components; and protein binding, nucleotide binding and adenosine triphosphate binding were observed to be the three most affected molecular functions (Fig. 2A-C). Pathway-based analyses revealed significant enrichment for genes in cancer pathways (Fig. 2D).
Figure 2.
GO enrichment and KEGG pathway-based analyses of differentially methylated sites-targets. GO analysis of differentially methylated sites-target genes according to (A) biological process, (B) cellular component and (C) molecular function. (D) KEGG pathway-based analysis of differentially methylated sites-target genes. GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; CoA, coenzyme A; GTP, guanosine-5′-triphosphate; cAMP, cyclic adenosine 3′,5′-monophosphate; ATP, adenosine triphosphate; SH3, SRC homology 3.
Aberrant methylation of AFP in HB
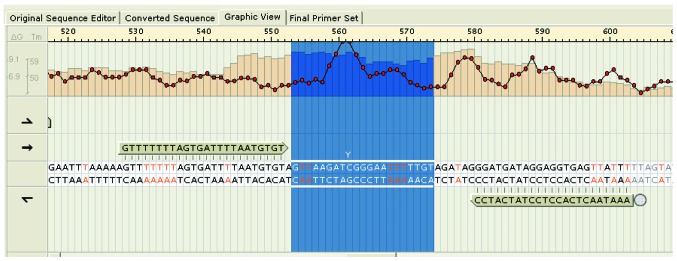
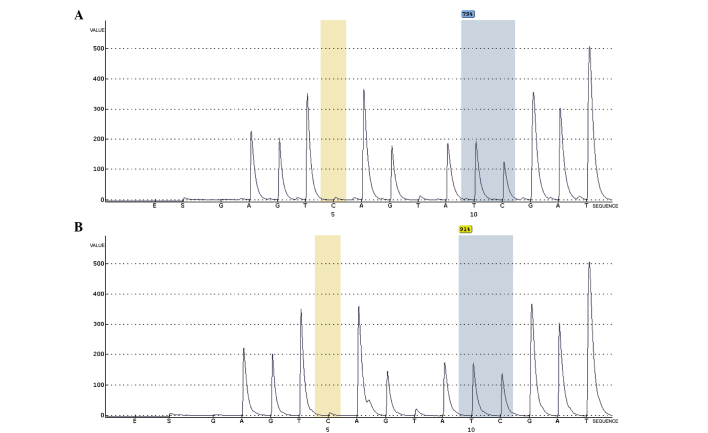
The methylation microarray indicated enriched methylation of positions near the transcription start site of AFP (Fig. 3). A representative illustration of AFP methylation levels in a matched HB-non-tumor tissue pair is shown in Fig. 4. As HB tissues had lower AFP methylation levels than non-tumor liver tissues, aberrant methylation may be a tumor-specific event in HB.
Figure 3.
Methylation position enrichment near the transcription start site of alpha-fetoprotein [cg10778295 (TSS1500)] revealed hypomethylated regions. Tm, melting temperature.
Figure 4.
Representative measurements of AFP methylation using pyrosequencing. (A) AFP locus cg10778295 methylation status in cancer tissues (79% methylation). (B) AFP locus cg10778295 methylation status in normal tissues (91% methylation). The methylation percentages are percentages of C at each CpG site after bisulfite conversion. Orange bar indicates no residual C at the non-CpG site, ensuring complete bisulfite conversion. AFP, alpha-fetoprotein.
Correlation of AFP mRNA expression and percentage of DNA methylation
The expression levels of the AFP gene in 10 HB-non-tumor liver tissue pairs were observed to be significantly higher in the HB tissues than in their non-tumor counterparts (Fig. 5A). The correlations between AFP mRNA expression and DNA methylation percentage were tested using the linear Pearson's R correlation coefficients, which were interpreted using the scale provided by Salkin (R=1-0.8, very strong; R=0.8–0.6, strong; R=0.6–0.4, moderate; R=0.4–0.2, weak; and R=0.2–0.0, very weak or not correlated) (17) (Fig. 5B), and were observed to be negatively correlated (R=0.89; P<0.05).
Figure 5.
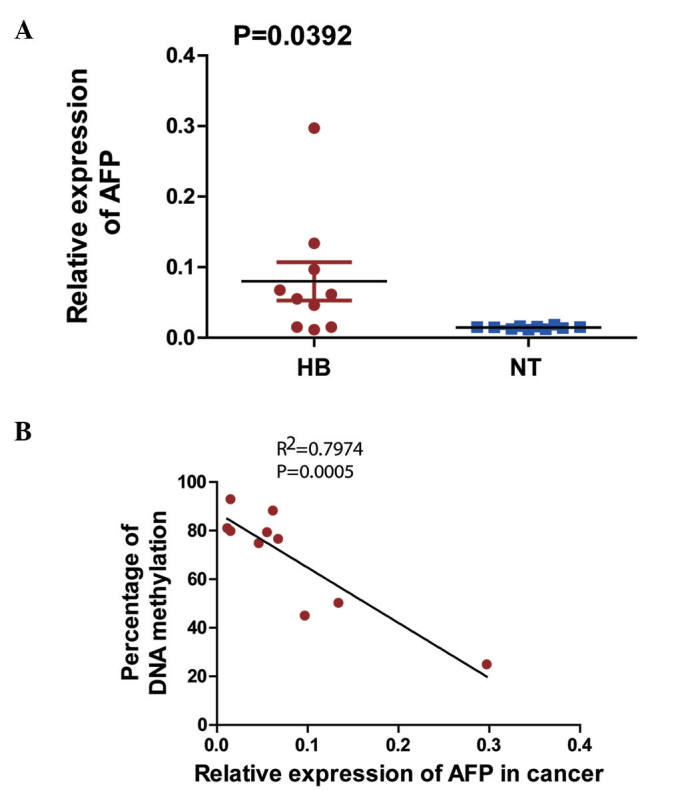
(A) AFP mRNA expression by reverse transcription-quantitative polymerase chain reaction in an independent cohort of 10 paired HBs and adjacent non-tumor tissues with complementary DNA (P<0.01; paired Student's t-test). (B) Correlation analysis of AFP mRNA expression and percentage of DNA methylation in 10 HB primary tissues, analyzed by two-tailed Pearson's correlation test. AFP, alpha-fetoprotein; HB, hepatoblastoma; NT, non-tumor; mRNA, messenger RNA.
Discussion
Although HB is an uncommon cancer in the general population, it is the most common liver tumor in children (4). Its etiology is still unknown, but it has been associated with familial adenomatous polyposis (18), low and high birth weights (19) and constitutional trisomy 18 (20). Mutations in numerous components of the wingless-related integration site/β-catenin signaling pathway are also often present in HB tissues (21).
Although the mechanisms responsible for HB development have been extensively studied for the last two decades, the pathogenesis of this disease is still vague, and the majority of its altered gene expression and regulation remain to be delineated. The present authors recently sequenced the HB exome and identified a novel oncogene (caprin family member 2) and three tumor suppressors (speckle-type POZ protein, olfactory receptor family 5 subfamily I member 1 and cell division cycle 20B) that influence HB cell growth (22). A link between long non-coding RNA and HB was also detected (23).
Hypermethylation or hypomethylation of gene promoter regions can result in their transcriptional silencing or activation (24). Although genome-wide copy number alterations in HB have been studied, little is known about genome-wide methylation changes (4). The present study is the first to explore whole-genome DNA methylation status in HB. In the present study, Infinium HumanMethylation450 BeadChip, a new platform for high-throughput DNA methylation analysis, was used (25). The cluster analysis revealed markedly less methylation in HB tissues than in normal liver tissues in the three sets of paired tissues analyzed. HB tissues are relatively low in genome-wide DNA methylation with preponderant CpG sites, and highly methylated genes may also have more numerous CpG sites (26). The present results revealed the DNA in HB tissues to be less methylated than in normal liver tissues. This may explain how the disease begins: A number of oncogenes are activated by demethylation, resulting in rapid tumor growth.
Serum AFP screening for HB has been a clinical practice for a long time (27). Additionally, high mRNA levels of AFP are reportedly associated with HB (27). To further understand the association between HB and AFP methylation, a methylation microarray was used in the present study to specifically detect AFP methylation status. A methylation-enriched position near the AFP transcription start site [g10778295 (TSS1500)] was identified, which was hypomethylated in HB. Pyrosequencing demonstrated the methylation level of the AFP locus cg10778295 to be lower in cancer tissues (79%) than in normal liver tissues (91%). Finally, RT-qPCR demonstrated AFP mRNA expression in HB to be much higher than in normal liver tissue in an independent cohort of 10 adjacent HB-non-tumor tissues pairs. The expression of AFP mRNA was significantly negatively correlated with its methylation status.
In conclusion, the current study of methylation in HB tissues is a proof-of-principle that methylation status probably affects HB development and progression. HB is an uncommon malignant liver neoplasm of children, and its etiology, pathophysiology and molecular mechanisms are largely unknown. Further studies are required to fully understand and control this disease. Although the current study demonstrated only that high AFP expression is associated with epigenetic modification, this topic warrants further investigation.
Acknowledgements
The present study received financial support from the National Natural Science Foundation of China (Beijing, China; grant nos. 81370472 and 81300517), the Shanghai City Health Bureau for Youth Scientific Fund Project (Shanghai, China; grant no. 20134y100), the Science Foundation of Shanghai (Shanghai, China; grant nos. 11JC1401300, 13ZR1451800 and 15ZR1404200) and the State Key Laboratory of Oncogenes and Related Genes, Shanghai Cancer Institute (Shanghai, China; grant no. SKLORG #90-12-01).
References
- 1.Herzog CE, Andrassy RJ, Eftekhari F. Childhood cancers: Hepatoblastoma. Oncologist. 2000;5:445–453. doi: 10.1634/theoncologist.5-6-445. [DOI] [PubMed] [Google Scholar]
- 2.von Schweinitz D. Hepatoblastoma: Recent developments in research and treatment. Semin Pediatr Surg. 2012;21:21–30. doi: 10.1053/j.sempedsurg.2011.10.011. [DOI] [PubMed] [Google Scholar]
- 3.De Ioris M, Brugieres L, Zimmermann A, Keeling J, Brock P, Maibach R, Pritchard J, Shafford L, Zsiros J, Czaudzerna P, Perilongo G. Hepatoblastoma with a low serum alpha-fetoprotein level at diagnosis: The SIOPEL group experience. Eur J Cancer. 2008;44:545–550. doi: 10.1016/j.ejca.2007.11.022. [DOI] [PubMed] [Google Scholar]
- 4.Litten JB, Tomlinson GE. Liver tumors in children. Oncologist. 2008;13:812–820. doi: 10.1634/theoncologist.2008-0011. [DOI] [PubMed] [Google Scholar]
- 5.Haas JE, Feusner JH, Finegold MJ. Small cell undifferentiated histology in hepatoblastoma may be unfavorable. Cancer. 2001;92:3130–3134. doi: 10.1002/1097-0142(20011215)92:12<3130::AID-CNCR10115>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 6.Geiman TM, Robertson KD. Chromatin remodeling, histone modifications and DNA methylation-how does it all fit together? J Cell Biochem. 2002;87:117–125. doi: 10.1002/jcb.10286. [DOI] [PubMed] [Google Scholar]
- 7.Reik W, Walter J. Genomic imprinting: Parental influence on the genome. Nat Rev Genet. 2001;2:21–32. doi: 10.1038/35048096. [DOI] [PubMed] [Google Scholar]
- 8.Chow J, Heard E. X inactivation and the complexities of silencing a sex chromosome. Curr Opin Cell Biol. 2009;21:359–366. doi: 10.1016/j.ceb.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 9.Bibikova M, Barnes B, Tsan C, Ho V, Klotzle B, Le JM, Delano D, Zhang L, Schroth GP, Gunderson KL, et al. High density DNA methylation array with single CpG site resolution. Genomics. 2011;98:288–295. doi: 10.1016/j.ygeno.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 10.Doi A, Park IH, Wen B, Murakami P, Aryee MJ, Irizarry R, Herb B, Ladd-Acosta C, Rho J, Loewer S, et al. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat Genet. 2009;41:1350–1353. doi: 10.1038/ng.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ogoshi K, Hashimoto S, Nakatani Y, Qu W, Oshima K, Tokunaga K, Sugano S, Hattori M, Morishita S, Matsushima K. Genome-wide profiling of DNA methylation in human cancer cells. Genomics. 2011;98:280–287. doi: 10.1016/j.ygeno.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 13.Revill K, Wang T, Lachenmayer A, Kojima K, Harrington A, Li J, Hoshida Y, Llovet JM, Powers S. Genome-wide methylation analysis and epigenetic unmasking identify tumor suppressor genes in hepatocellular carcinoma. Gastroenterology. 2013;145:1424–1435. doi: 10.1053/j.gastro.2013.08.055. e1-e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sandoval J, Heyn H, Moran S, Serra-Musach J, Pujana MA, Bibikova M, Esteller M. Validation of a DNA methylation microarray for 450,000 CpG sites in the human genome. Epigenetics. 2011;6:692–702. doi: 10.4161/epi.6.6.16196. [DOI] [PubMed] [Google Scholar]
- 15.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
- 16.Johnson MR, Wang KS, Smith JB, Heslin MJ, Diasio RB. Quantitation of dihydropyrimidine dehydrogenase expression by real-time reverse transcription polymerase chain reaction. Anal Biochem. 2000;278:175–184. doi: 10.1006/abio.1999.4461. [DOI] [PubMed] [Google Scholar]
- 17.Perek B, Malinska A, Stefaniak S, Ostalska-Nowicka D, Misterski M, Zabel M, Suri A, Nowicki M. Predictive factors of late venous aortocoronary graft failure: Ultrastructural studies. PLoS One. 2013;8:e706288. doi: 10.1371/journal.pone.0070628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kingston JE, Herbert A, Draper GJ, Mann JR. Association between hepatoblastoma and polyposis coli. Arch Dis Child. 1983;58:959–962. doi: 10.1136/adc.58.12.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ross JA. Hepatoblastoma and birth weight: Too little, too big, or just right? J Pediatr. 1997;130:516–517. [PubMed] [Google Scholar]
- 20.Mamlok V, Nichols M, Lockhart L, Mamlok R. Trisomy-18 and hepatoblastoma. Am J Med Genet. 1989;33:125–126. doi: 10.1002/ajmg.1320330119. [DOI] [PubMed] [Google Scholar]
- 21.Takayasu H, Horie H, Hiyama E, Matsunaga T, Hayashi Y, Watanabe Y, Suita S, Kaneko M, Sasaki F, Hashizume K, et al. Frequent deletions and mutations of the beta-catenin gene are associated with overexpression of cyclin D1 and fibronectin and poorly differentiated histology in childhood hepatoblastoma. Clin Cancer Res. 2001;7:901–908. [PubMed] [Google Scholar]
- 22.Jia D, Dong R, Jing Y, Xu D, Wang Q, Chen L, Li Q, Huang Y, Zhang Y, Zhang Z, et al. Exome sequencing of hepatoblastoma reveals novel mutations and cancer genes in the Wnt pathway and ubiquitin ligase complex. Hepatology. 2014;60:1686–1696. doi: 10.1002/hep.27243. [DOI] [PubMed] [Google Scholar]
- 23.Dong R, Jia D, Xue P, Cui X, Li K, Zheng S, He X, Dong K. Genome-wide analysis of long noncoding RNA (lncRNA) expression in hepatoblastoma tissues. PLoS One. 2014;9:e85599. doi: 10.1371/journal.pone.0085599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baylin SB, Ohm JE. Epigenetic gene silencing in cancer-a mechanism for early oncogenic pathway addiction? Nat Rev Cancer. 2006;6:107–116. doi: 10.1038/nrc1799. [DOI] [PubMed] [Google Scholar]
- 25.Morris TJ, Butcher LM, Feber A, Teschendorff AE, Chakravarthy AR, Wojdacz TK, Beck S. ChAMP: 450k chip analysis methylation pipeline. Bioinformatics. 2014;30:428–430. doi: 10.1093/bioinformatics/btt684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tomlinson GE, Kappler R. Genetics and epigenetics of hepatoblastoma. Pediatr Blood Cancer. 2012;59:785–792. doi: 10.1002/pbc.24213. [DOI] [PubMed] [Google Scholar]
- 27.Clericuzio CL, Chen E, McNeil DE, O'Connor T, Zackai EH, Medne L, Tomlinson G, DeBaun M. Serum alpha-fetoprotein screening for hepatoblastoma in children with Beckwith-Wiedemann syndrome or isolated hemihyperplasia. J Pediatr. 2003;143:270–272. doi: 10.1067/S0022-3476(03)00306-8. [DOI] [PubMed] [Google Scholar]