Abstract
Background
Neurodevelopment is orchestrated by a wide range of genes, and the genetic causes of neurodevelopmental disorders are thus heterogeneous. We applied whole exome sequencing (WES) for molecular diagnosis and in silico analysis to identify novel disease gene candidates in a cohort from Saudi Arabia with primarily Mendelian neurologic diseases.
Methods
We performed WES in 31 mostly consanguineous Arab families and analyzed both single nucleotide and copy number variants (CNVs) from WES data. Interaction/expression network and pathway analyses, as well as paralog studies were utilized to investigate potential pathogenicity and disease association of novel candidate genes. Additional cases for candidate genes were identified through the clinical WES database at Baylor Miraca Genetics Laboratories and GeneMatcher.
Results
We found known pathogenic or novel variants in known disease genes with phenotypic expansion in 6 families, disease-associated CNVs in 2 families, and 12 novel disease gene candidates in 11 families, including KIF5B, GRM7, FOXP4, MLLT1, and KDM2B. Overall, a potential molecular diagnosis was provided by variants in known disease genes in 17 families (54.8 %) and by novel candidate disease genes in an additional 11 families, making the potential molecular diagnostic rate ~90 %.
Conclusions
Molecular diagnostic rate from WES is improved by exome-predicted CNVs. Novel candidate disease gene discovery is facilitated by paralog studies and through the use of informatics tools and available databases to identify additional evidence for pathogenicity.
Trial registration
Not applicable.
Electronic supplementary material
The online version of this article (doi:10.1186/s12920-016-0208-3) contains supplementary material, which is available to authorized users.
Keywords: Whole exome sequencing (WES), Copy Number Variants (CNV), Neurodevelopment, Developmental Delay/Intellectual Disability (DD/ID), GRM7
Background
Neurodevelopmental disorders reflect the biology and underlying complexity of nervous system developmental processes, its constituent cellular and anatomical structure, and neurophysiological functions. The human brain cortex consists of nearly 20 billion neurons, each expressing over 80 % of the roughly 25,000 known genes [1] and participating in thousands of synaptic connections [2]. Deleterious gene variants can give rise to a wide range of clinical presentations, which may be divided broadly into either functional (i.e. intellectual disability) or structural (i.e. malformations of cortical development) abnormalities [3]. The study of developmental brain disorders is further challenged by the high degree of genetic heterogeneity observed, even within clinically well-defined disorders, such as pontocerebellar hypoplasia [4]. These observations underscore both the necessity for and power of comprehensive molecular studies in this field [5]. The ultimate goal of these studies is to identify novel neurodevelopmental disease genes and establish potential genotype/phenotype correlations, but the identities of many of these genes remain elusive. Functional studies in animal models and cell lines play a fundamental role in the establishment of genotype/phenotype correlations; however, the need for basic infrastructure and time can be a bottleneck in processing the tremendous amount of data generated by the extensive and widespread use of new genomic tools [6]. The study of Mendelian phenotypes, including neurodevelopmental disorders, provides perhaps the most direct route to identify the link between gene function and resultant phenotype, and a foundation for investigating the underlying biology [7].
In this study, we used whole exome sequencing (WES) to identify potentially deleterious gene variants in a mostly consanguineous cohort from Saudi Arabia with either structural or functional brain abnormalities. CNVs predicted from WES data provide some molecular diagnoses. In addition, investigation of paralogs associated with similar phenotypes also facilitates the identification of novel candidates. Moreover, cross-database gene mining can provide further support for novel candidate disease genes.
Methods
Patients
This cohort consists of 31 probands (13 males and 18 females) and their siblings and parents from families with brain malformation and/or developmental delay/intellectual disability. Additional file 1: Table S1 and Additional file 2: Figure S1 provide the pedigree structures and clinical information. All patients were evaluated by clinical geneticists and neurologists.
Whole exome sequencing (WES) analysis
For the eight multiple-affected families, two members in each family were studied by WES. For the 23 singleton families, four were studied by WES as trios (i.e. affected child plus both parents), and both proband and unaffected twin sibling were studied in 1 family. In the remaining families, only the proband was studied and the DNA samples of other family members were used in segregation analysis. DNA samples from selected individuals were sequenced by WES at the BCM Human Genome Sequencing Center (HGSC) through the Baylor-Hopkins Center for Mendelian Genomics (BHCMG) research initiative. All experiments and analyses were performed as previously described [8], and WES performed in this study had 80X average depth-of-coverage, with 93 % of base positions at a minimum of 20X.
PCR confirmation for WES data
We used standard PCR to confirm the variants identified from WES and to perform segregation analysis. PCR products were analyzed by Sanger sequencing (DNA Sequencing Core Facility at BCM).
Absence of heterozygosity (AOH) analysis
Calculated B-allele frequency information was determined from WES as the ratio of variant reads to total reads in WES data and then processed using the Circular Binary Segmentation algorithm implemented in the DNAcopy R bioconductor package [9]. We subtracted 0.5 from the calculated B-allele frequency. Segments with the mean signal > 0.45 and size >1 kb were detected as AOH regions. In the summary statistics in Additional file 1: Table S4, for each proband, we provided the number, the maximum/minimum length, the median length, the mean length, and the total length of potential AOH regions (> = 0.5 Mb).
CNV analysis from WES data
We used CoNIFER [10], CoNVex (Sanger Centre, unpublished), and HMZDelFinder (https://github.com/BCM-Lupskilab/HMZDelFinder) to predict CNVs from WES data. HMZDelFinder is an in-house-developed algorithm implemented in the R programming language (R Core Team 2014, http://www.R-project.org). WES data were transformed into per-exon read depth, RPKM. Homozygous/hemizygous deletions of exons were defined as meeting three criteria: exons with RPKM <0.5, frequency <0.5 % in the whole cohort, and located within an AOH region (>1 kb), were called together with all the exomes in BHCMG database.
Evaluation of candidate genes/variants
To assess the candidacy of novel disease genes, we ranked each candidate gene in the cohort from most likely (red) to less (green) according to the following criteria [7]: Genes found in multiple families with similar phenotypes (3 points); Interactors/paralogs of the gene showing overlapping features in human (2 points); Animal studies of the gene or its interactor/paralogs showing overlapping features (2 points); Genomic region associated with the phenotypes (1 point); Loss-of-function variant (2 points); Predicted deleterious by multiple programs (1 point); rare variants in multiple databases (1 point); Expressed in the tissues/organs being affected (1 point).
We further classified variants for all known and novel disease genes using the ACMG/AMP guidelines for variant interpretation in Mendelian disorders (2015) [11] which recommends classification of variant pathogenicity into 5 levels: pathogenic, likely pathogenic, variant of uncertain significance (VUS), likely benign, and benign (Additional file 1: Table S1). As these guidelines provide for additional sub-classification of VUS, we used the term “VUS-favoring likely pathogenic” to indicate variants for which there is evidence of pathogenicity that does not meet criteria for consideration as a likely pathogenic variant. In addition, as the ACMG/AMP guidelines were developed for clinical use and are not designed to address pathogenicity of variants in genes not yet established to be disease-causing, i.e. candidate disease genes in gene discovery efforts, we have categorized all variants in novel candidate disease genes as VUS (Additional file 1: Table S1), while still providing evidence for variant pathogenicity as appropriate.
Arrays and droplet PCR confirmation for CNVs
Deletion in GRID2 was confirmed using BCM chromosomal microarray version 10.2 [12]. Droplet digital (dd) PCR for RPS6KC1 was performed using the QX200™ AutoDG™ Droplet Digital™ PCR System (Bio-Rad) following the manufacturer’s protocols and analyzed in QuantaSoft™. Detailed conditions and primers are listed in Additional file 3: Supplementary Materials and Methods.
Results
This genomics study includes 31 Arab families exhibiting neurological disease phenotypes, among which 28 were reported to be consanguineous or from the same tribe (Additional file 2: Figure S1). There were 23 (74 %) families with a single affected subject and 8 (26 %) with multiple affected family members. Nineteen (61 %) probands exhibited various structural brain malformations and 12 (39 %) probands presented developmental delay/intellectual disability (DD/ID) without brain malformations (Fig. 1a). We hypothesize that disease causative genes in the first group mainly affect brain development whereas those in the second group mainly affect neuronal functions. The cohort was further clinically assigned into phenotypic subgroups including syndromic brain malformation, corpus callosum abnormalities, cortical dysgenesis, microcephaly, hindbrain malformation, white matter abnormalities, as well as syndromic and non-syndromic DD/ID (Fig. 1b and Additional file 1: Table S2). Individual families could be assigned to multiple subgroups if they had multiple neurodevelopmental phenotypes.
Fig. 1.
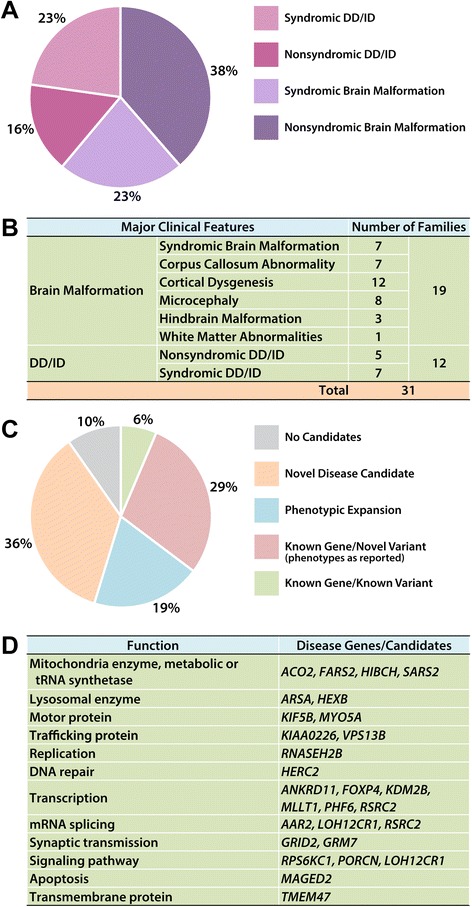
Clinical categories in the cohort and summary of WES findings. a Based on the brain structural findings and accompanying features, this cohort is grouped into nonsyndromic brain malformations (38 %), syndromic brain malformations (23 %), nonsyndromic DD/ID (16 %), and syndromic DD/ID (23 %). b Based on the brain structural defects, families can be further grouped into corpus callosum abnormalities, cortical dysgenesis, microcephaly, hindbrain malformations, and white matter changes; families may be counted in more than one of these groups. c Summary of WES findings (including CNVs): We identified a known variant in a known gene in 2 families (6 %), 10 novel variants in known genes in 9 families presenting reported clinical features (29 %), 7 novel variants in 6 known genes with phenotypic expansion in 6 families (19 %), and 12 novel disease candidates in 11 families (36 %). d The disease genes/candidates identified in this study can be grouped into several biological processes, including mitochondrial enzymes (metabolic or tRNA synthetase), lysosomal enzymes, motor proteins, trafficking proteins, DNA replication, DNA repair, transcription, mRNA splicing, synaptic transmission, signaling pathways, apoptosis, and transmembrane proteins
We analyzed both SNVs and CNVs derived from the exome data (Additional file 2: Figure S2). SNVs (including small indels) were computationally filtered using the allele frequency data from different exome databases and bioinformatic predictions of damaging effects on the corresponding protein. In addition to the public exome variant databases, we also searched the variants and genes in our internal BHCMG database with documented clinical features as annotated in PhenoDB [13, 14]. Sanger sequencing of all candidate variants was performed for each proband and all available relatives for confirmation and to assess co-segregation with the phenotype. Only variants that segregated with the phenotype are reported. Additional analyses included an assessment of any previous functional or disease studies of the gene, its orthologs in other model organisms, and its paralogs or known interactors in humans.
We identified rare genetic variants potentially contributing to the phenotypes in ~90 % (28/31) of families studied, including 54.8 % for which a molecular diagnosis involved variants in known disease genes (Fig. 1c-d and Additional file 1: Table S3). Known or suspected consanguinity was reported in 28/31 families and analysis of absence of heterozygosity (AOH) demonstrated one or more regions of AOH > 0.5 Mb in all families with one or more variants in known or candidate disease genes (Additional file 1: Tables S2 and S4). Total personal genome AOH regions ranged from 86.5 Mb to 507.5 Mb in the 28/31 families with historical evidence for consanguinity and from 97.1 Mb to 118.8 Mb in the 3 families in which there was no historical evidence for consanguinity. Most of the variants in potential candidate disease genes that we identified map to the AOH regions and are thus homozygous recessive. In total, we identified 4 de novo variants and 1 case with compound heterozygous alleles in candidate genes (Additional file 1: Table S1). One of the de novo variants and the one case with compound heterozygous alleles, were found in consanguineous families (Additional file 1: Tables S1 and S2).
Variants in known disease genes included previously described pathogenic variants in 2 families (FARS2, PORCN) and novel variants in 15 families (ACO2, RNASEH2B, SARS2, MLC1, ARSA, HEXB, HIBCH, HERC2, PHF6, GRID2, MYO5A, ANKRD11, KIAA0226, C12ORF57, VPS13B, ATP2B3) (Additional file 1: Table S1), 6 of which are clinically observed to likely represent phenotypic expansion from the previously reported associated traits (Table 1 and Additional file 1: Table S1). Moreover, we propose and provide evidenced-based ranking for 12 novel candidate disease genes (GRM7, FOXP4, KIF5B, KDM2B, MLLT1, RPS6KC1, CDK20, MAGED2, TMEM47, AAR2, LOH12CR1, RSRC2) in 11 families (Table 2 and Additional file 1: Table S1).
Table 1.
Known disease genes with phenotypic expansion
| Family | ID | Gene | Variant | Zyg | Clinical features of patients | MIM | Phenotypic expansion |
|---|---|---|---|---|---|---|---|
| 037 | BAB6843 | MYO5A | NM_000259.3:c.4200C > G:p.Ser1400Arg | Hom | DD, seizures, cerebellar atrophy, nystagmus, dystonia | 214450 | Lacks classical Griscelli syndrome phenotype of skin hypopigmentation and silver-grey hair |
| 019 | BAB6938 | ANKRD11 | NM_013275.5:c.5317G > T:p.E1773* | Het | DD/ID, hypotonia, esotropia, hyperopia, astigmatism, broad nasal bridge, hypertelorism, epicanthal folds, retrognathia, cryptorchidism | 148050 | Hypotonia, esotropia, hyperopia, astigmatism |
| 022 | BAB6787 | KIAA0226 | NM_014687.2:c.1642A > G:p.Thr548Ala | Hom | DD/ID, hypotonia, diffuse cortical hypomyelination, contractures of elbows and knees, nystagmus | 615705 | Hypotonia, contractures, lacks cerebellar atrophy |
| NM_014687.2:c.319G > A:p.Glu107Lys | Hom | ||||||
| 024 | BAB6793 | C12ORF57 | NM_138425.3:c.53-2A > G | Hom | DD/ID, ADHD, bilateral hydronephrosis and VUR, atrophic non-functioning left kidney | 218340 | Vesicoureteric reflux and bilateral hydronephrosis, small non-functioning left kidney |
| 034 | BAB6835 | VPS13B | NM_152564.4:c.1219C > T:p.Gln407* | Hom | DD/ID, ADHD, microcephaly, hypotonia, joint hyperlaxity, unsteady gait, severe dental caries | 216550 | Autistic behavior, unsteady gait, severe dental caries |
| 026 | BAB7039 | ATP2B3 | NM_001001344.2:c.3594G > T:p.Lys1198Asn | Hem | DD/ID, microcephaly, abnormal cortical pattern, hypotonia and muscle atrophy, oropharyngeal dysphagia, dental caries | 302500 | Microcephaly, abnormal cortical pattern, muscle atrophy, oropharyngeal dysphagia |
DD/ID developmental delay/intellectual disability, ADHD attention deficit hyperactivity disorder, VUR Vesicoureteral Reflux, Hom homozygous, Het heterozygous, Hem hemizygous; * stopgain
Table 2.
Ranking for the candidate disease genes

The novel candidate genes identified in this cohort were ranked from most likely (red) to less (green) based on the scores using the following criteria: a. Genes found in multiple families with similar phenotypes (3 points); b. Interactors/paralogs of the gene showing overlapping features in human (2 points); c. Animal studies of the gene or its interactor/paralogs showing overlapping features (2 points); d. Genomic region associated with the phenotypes (1 point); e. Loss-of-function variant (2 points); f. Predicted deleterious by multiple programs (1 point); g. Rare variants in multiple databases (1 point); h. Expressed in the tissues/organs being affected (1 point). DD/ID: Developmental Delay/Intellectual Disability; ADHD: attention deficit hyperactivity disorder; PFO: Patent foramen ovale; VSD: Ventricular Septal Defect; VUR: Vesicoureteral Reflux; Hom: homozygous; Het: heterozygous; cHet: compound heterozygous; Hem: hemizygous
CNVs predicted from WES facilitate establishing a potential etiologic molecular diagnosis
We used CoNIFER [10], CoNVex, and HMZDelFinder to predict potential heterozygous and homozygous CNVs based on exome data. Although many subjects have been pre-screened with karyotyping and array CGH, this bioinformatics approach enables higher resolution evaluation of genomic regions poorly covered by clinical arrays, including homozygous/hemizygous single exon dropout alleles. Among the 31 cases, we found compound heterozygous variant alleles consisting of a SNV and a CNV in a novel candidate gene RPS6KC1 (Figure 2 and Additional file 1: Table S1) as well as a homozygous intragenic deletion in GRID2 (Additional file 2: Figure S3).
Fig. 2.

A combination of SNV and CNV in RPS6KC1 in BAB6797. a CoNVex reveals a 42 kb heterozygous deletion (chr1:g.213403839_213445978del) in RPS6KC1 in BAB6797 (family 025). There is a SNV (NM_001287219.1:c.2074G > A:p.Gly692Ser) uncovered by this deletion. Gray dotted line: the read depth of other samples in the cohort; red line: the read depth for BAB6797; pink line: the predicted deletion region. b Droplet Digital PCR (ddPCR) detects heterozygous deletion of RPS6KC1 in proband BAB6797 and father BAB6799 but not in mother BAB6798; primer pair targeting RPS6KC1 around chr1:g.213415529[hg19] and three control primer pairs targeting copy-number neutral regions were used to perform ddPCR. Absolute positive droplet concentrations (copies/ul) are plotted from ddPCR results for each primer pair in each sample. Positive droplet concentrations of BAB6797 and BAB6799 for the RPS6KC1 primer pair (around 300 bp) are approximately half of the value in BAB6798 and the results from all control primers (around 600 bp), indicating heterozygous deletion of the RPS6KC1 gene in BAB6797 and BAB6799, but not in BAB6798. Corresponding raw data of ddPCR and primer sequences are shown in Table S5 . Ctrl, control. c This deletion is also confirmed by a customized array. The combination of SNV and CNV in RPS6KC1 segregates in the family. d Breakpoint junction is mapped to chr1:g.213396378-213592823[hg19] by long-range PCR and Sanger sequencing of the junction
The combined variants in RPS6KC1 were identified in the proband (BAB6797) in family 025 with structural brain malformation, including cortical dysgenesis and corpus callosum abnormality (Fig. 2; Additional file 1: Table S1). The SNV in RPS6KC1 was initially annotated as homozygous (A/A) in the proband; segregation of this variant in the parents demonstrated a heterozygous (G/A) mother and a wild-type (G/G) father, suggesting the possibility of a paternally inherited deletion uncovering the locus of the maternally inherited SNV. Indeed, a heterozygous deletion was predicted by CoNVex (chr1:g.213,403,839-213,445,978[hg19]; 42 kb, including the last six exons of RPS6KC1) (Fig. 2a) and CoNIFER (chr1:g.213,341,200-214,171,606[hg19]; 83 kb, including the last seven exons of RPS6KC1 and the first two exons of the adjacent gene, data not shown). Since the clinical array utilized has suboptimal coverage in this genomic interval, this suspected deletion CNV was confirmed in both father and the proband by droplet digital PCR (Fig. 2b and Additional file 1: Table S5) and a customized high density array CGH (Fig. 2c), and was demonstrated to be 196,446 bp (chr1:g.213396378-213592823[hg19]; including the last 7 exons of this gene) through breakpoint junction PCR (Fig. 2d). RPS6KC1 mRNA is expressed in the brain (Additional file 2: Figure S4a) and may regulate sphingosine-1 phosphate signaling [15], which is essential for neurogenesis and proper embryonic development [16], suggesting RPS6KC1 as a candidate disease gene in this family.
The homozygous deletion in GRID2 was predicted by all three programs in the personal genome WES from proband (BAB6883) in family 041 who presented with hindbrain malformation (Additional file 2: Figure S3a and Additional file 1: Table S1). The deletion predicted by CoNIFER (chr4:g.92007100-94138063[hg19]; 2.1 Mb, data not shown) is much larger than that predicted by HMZDelFinder and CoNVex (chr4:94006145-94032105[hg19]; 25 kb, data not shown), as well as the size experimentally verified by BCM clinical array (chr4:g.93985826-94074965[hg19]; 89 kb) (Additional file 2: Figure S3b) [12]. It was further fine-mapped to chr4:93978239-94078203[hg19] by breakpoint junction PCR (99,965 bp) (Additional file 2: Figure S3c). This homozygous deletion includes exons 3 and 4 of this 15 exon gene. GRID2 is the receptor for excitatory glutamate neurotransmitter in the cerebellum [17] and homozygous deletions, ranging from 37 kb–335 kb in size, were reported to cause cerebellar ataxia and atrophy (MIM#616204).
Sub-network analysis and cross-database mining provides supporting evidence for candidate genes
In addition to RPS6KC1, we identified 11 other potential novel candidate disease genes (Additional file 1: Table S1). The supporting evidence for pathogenicity of these variants is listed in Additional file 1: Table S1 and ranked evidence supporting disease gene candidacy is listed in Table 2. The protein network analysis for individual candidate genes provides potential functional correlation with the phenotypes, such as observed for CDK20, LOH12CR1, and RSRC2 (Additional file 1: Table S1). We performed additional bioinformatic analyses of mRNA expression in the brain at different developmental stages, biological functional annotation, and network connectivity based on protein-protein interactions to investigate the novel and known genes as a cohort (Additional file 2: Figure S4 and Figure S5). There is no obvious correlation between the phenotypes and the temporal mRNA expression in the brain. Although the network analysis does not show significant enrichment, which likely reflects the small cohort size as well as heterogeneous phenotypes and genetic causes (Fig. 1d and Additional file 1: Table S1), we observed some sub-networks among a subset of genes, such as MLLT1 and KDM2B (Additional file 2: Figure S5a).
MLLT1 is an essential gene during development [18] that co-activates SWI/SNF complexes and regulates histone H3K79 demethylation [19]. The proband (BAB6950) in family 058 with a potentially deleterious homozygous variant in MLLT1 presents with developmental delay, hypotonia, infantile spasm, and cortical dysgenesis (Additional file 1: Table S1 and Fig. 3a). KDM2B is a histone demethylase for H3K4 and H3K36 [20, 21]. The homozygous variant is identified in two affected siblings (BAB6693 and BAB6694) in family 009 presenting developmental delay, hypotonia, and infantile spasms (Fig. 3b and Additional file 1: Table S1). Variants in the genes involved in chromatin remodeling and histone modification are known to be associated with brain malformation and DD/ID [7].
Fig. 3.

Segregation results and AOH plots of patients with variants in MLLT1, KDM2B, FOXP4, and KIF5B. In each panel, the segregation results are shown to the left and AOH plots on the corresponding chromosomes are shown to the right. a In family 058, the proband (BAB6950) has developmental delay, hypotonia, and infantile spasms with diffuse brain atrophy, delay and hypomyelination, as well as malformation of cortical development. A homozygous variant in MLLT1 (NM_005934.3:c.1418G > A:p.Arg473Gln) is identified and segregates with the phenotypes in the family. b In family 009, the proband (BAB6693) has developmental delay, hypotonia, and infantile spasms. A homozygous variant in KDM2B (NM_032590.4:c.3050G > A:p.Arg1017His) is identified and segregates with the phenotypes in the family. c In family 028, the proband (BAB6800) exhibits developmental delay, laryngeal hypoplasia, and ventricular septal defect (VSD). A homozygous variant in FOXP4 (NM_001012426.1:c.815delT:p.Lys272fs) is identified and segregates in the family. d In family 015, the proband (BAB6712) has developmental delay, seizures, increased reflexes in upper and lower extremities, short stature with mild diffuse brain atrophy, and thinning of the corpus callosum. The younger brother (BAB6713) is mildly affected with speech delay. A homozygous variant in KIF5B (NM_004521.2:c.2252A > G:p.His751Arg) is identified and segregates with the phenotypes in the family, present in a homozygous state in both brothers
We previously reported that Drosophila genes with multiple human homologs are more likely to be associated with human disorders [8]. We investigated paralogs which have been associated with neurological disorders by adding them into the interaction network and observed sub-network enrichment (Additional file 2: Figure S4b and S5b-d; Additional file 1: Table S6). Paralogs of FOXP4, KIF5B, and GRM7, have been associated with disorders presenting related phenotypes, which provide supporting evidence for these novel candidate disease genes (Fig. 3 and Additional file 1: Table S1).
FOXP4 may play a role in multiple systems affected in the proband (BAB6800) in family 028, such as the larynx and heart [22–24]. Moreover, FOXP4 and its paralogs, FOXP1 and FOXP2, function together in several developmental processes [25, 26]. Variants in FOXP1 and FOXP2 can lead to mental retardation with language impairment (MIM#613670) and Speech-language disorder-1 (MIM#602081), respectively. Genetic abnormalities in FOXP1 are also associated with congenital heart defects [27]. Therefore, FOXP4 may work together with FOXP1/FOXP2 in brain function and heart development and contribute to the phenotypes observed (Fig. 3c, Additional file 2: Figure S5b, Additional file 1: Table S1).
A homozygous KIF5B variant was identified in the proband (BAB6712) and his mildly affected brother (BAB6713) in family 015 (Fig. 3d and Additional file 1: Table S1). KIF5B and its paralogs, KIF5A and KIF5C (Additional file 2: Figure S5c and Additional file 1: Table S6), can transport RNA granules to the neuronal dendrites for protein synthesis and translational silencing [28]. Loss of KIF5B is embryonic lethal with severe growth retardation in mice [29]. Moreover, variants in KIF5A and KIF5C lead to spastic paraplegia (MIM#604187) and cortical dysplasia (MIM#615282), respectively. The severity of the phenotypes in both disorders is variable, which may potentially explain the different severity in the clinical features between the affected siblings. The variability may also be potentially explained by additional unknown genetic variant(s) that exacerbate or suppress clinical severity [30].
GRM6 and GRM8, the paralogs of GRM7, have been implicated in neurodevelopmental diseases such as attention deficit hyperactivity disorder (ADHD) and autism spectrum disorders (ASD) [31, 32]. Accordingly, recent genomic studies also revealed an association between GRM7 and ASD [33, 34]. Another related member, GRM1 (Additional file 1: Table S6), causes autosomal recessive spinocerebellar ataxia type 13 (MIM#614831) which may include cerebellar atrophy. A recent study indicates GRM7 regulates neurogenesis in early developing mouse brain [35]. Together, these data support GRM7 as a disease candidate in the two affected siblings (BAB6708 and BAB6709) in family 014 presenting with hypotonia, brain malformation including cortical atrophy, very thin and shortened corpus callosum, and mild cerebellar volume loss (Fig. 4; Additional file 1: Table S1). These examples illustrate how expanding analyses to include paralogs may help identify potential candidate disease genes.
Fig. 4.

Comparison of the two families carrying GRM7 variants. a Homozygous variant in GRM7 (NM_000844.3:c.461 T > C:p.Ile154Thr) identified in family 014 with two affected children. b Compound heterozygous variants in GRM7 (NM_000844.3:c.1972C > T:p.Arg658Trp and NM_000844.3:c.2024C > A:p.Thr675Lys) segregates in the family with two affected children identified in the BMGL database. c MRI images of BAB6708 and BAB6709. d MRI images of BAB8506 and her brother. e Clinical features comparison among four patients
To date, the candidate gene list submitted to GeneMatcher has not resulted in the identification of additional families with phenotypes consistent with those observed in our cohort. However, a similar analysis of candidate genes in clinical WES data from the Baylor-Miraca Genetics Laboratory revealed a family with compound heterozygous GRM7 variants in two affected siblings presenting similar clinical features, including cortical atrophy, very thin and short corpus callosum, and hypotonia in the affected members of family 014 (Fig. 4). This further supports our findings that GRM7 is a causative gene for both families.
Discussion
We performed genomic studies in 31 mostly consanguineous Arab families with either brain malformation or DD/ID and used bioinformatics analyses to identify candidate disease genes and candidate variants likely contributing to the observed clinical neurodevelopmental phenotypes. By considering minor allele frequency, variant properties and predicted effects on protein structure and function, previous functional/disease studies of the gene and its paralogs/interactors, as well as studying both SNVs and CNVs, we identified known disease genes in 55 % and candidate disease genes in 35 % of families (Table 2 and Fig. 1c). The overall potential molecular diagnostic rate considering both known and candidate disease genes is around 90 %. This relatively high rate likely reflects the fact that many neurodevelopmental disease genes have been reported previously and that families included are mostly consanguineous. In this small Arab cohort, there are no shared causative/candidate genes among the families even for the known disease genes. In addition, these genes function in a variety of biological processes, including chromatin modification, transcriptional regulation, signal transduction, intracellular transport, autophagy, mitosis, and apoptosis (Fig. 1d). The diversity of genes found speaks to the heterogeneity of the genetic causes and cellular processes underlying neurological disorders.
Highlights of a genomic study in a cohort with a high consanguinity rate
We performed WES on two probands in each of the eight families with multiple affected children (Additional file 2: Figure S1). A molecular diagnosis was not achieved in families 006 and 018 since none of the candidate variants assessed segregate with the phenotype among the family members. This may result from shared variants either not being detected or potentially being filtered out by the exome analysis pipeline. Alternatively, an inheritance model that is different than that assumed by pedigree analysis may apply. Although in most of the cases variation at a single locus may explain the clinical features, one must consider the possibility that multiple genetic causes may contribute in a given family and different genes may be involved in different affected family members.
When affected siblings are observed to exhibit variable clinical features in a family, there may still be a shared genetic cause. In family 015 with one severely affected proband and one mildly affected brother, we identified a shared homozygous variant in KIF5B. Its paralogs, KIF5A and KIF5C, are responsible for neurological disorders presenting a variable severity of clinical features, which may also be the case in this family. We cannot exclude the possibility that other gene variants in the proband may contribute to the severity or that other variants in the mildly affected brother may have a protective effect (Additional file 1: Table S1) [30]. Nevertheless, the related functional studies support the molecular diagnosis and KIF5B as the major contributor, i.e. the molecular driver variant, for the observed phenotypes (Additional file 1: Table S1).
In family 023, the proband exhibits low activity of arylsulfatase A (ARSA), leading to metachromatic leukodystrophy (MIM#250100). Targeted sequencing for the previously reported ARSA variants showed a negative result while WES reveals a novel homozygous ARSA variant (Additional file 1: Table S1), likely explaining the low ARSA activity. In family 046, WES was performed on both affected children. Although the analysis pipeline only revealed a homozygous frameshift variant in ACO2 in the proband (BAB7017) (Additional file 1: Table S1), Sanger confirmation indicated that the affected sibling (BAB7018) also has this homozygous variant. Retrospective re-analysis of the original sequencing files showed that this frameshift in BAB7018 was filtered out in the analysis pipeline due to poor quality metrics. Similarly, frameshift variants in DVL1, a recently reported disease gene for Robinow Syndrome (MIM#616331) [36], also evaded detection or were interpreted as missense by the WES analysis pipeline. These examples point out a current challenge in calling indels in WES pipelines. Filtering criteria may improve through utilization of a ‘training set’ of cases with known indels. Alternatively, multiple calling algorithms may be necessary for optimized indel detection.
One challenge in the study of consanguineous families is the presence of multiple rare homozygous variants within extended genomic intervals of AOH that are distributed throughout the genome (Additional file 1: Table S4). This is exemplified by family 001 which is categorized as unsolved because there are multiple candidate variants in different genes that co-segregate with the phenotypes. Other prioritization filters to identify the potential disease contributing variants among rare homozygous variants (Additional file 2: Figure S2 and Table 2) include: i) detecting loss-of-function variants, ii) analyzing potential deleteriousness of missense variants using multiple prediction programs, iii) examining whether the gene is expressed in disease related tissues, iv) assessing available functional studies of the gene or its interactor/paralogs with overlapping phenotypes, and v) finding the gene is a candidate disease gene in multiple families with similar features. In this study, there were 4 de novo and 2 compound heterozygous variants found for the potential molecular diagnoses rendered; however, the majority of the disease or candidate variants identified map within AOH regions (Fig. 3 and Additional file 1: Table S4). Therefore, in some families provided with candidate disease genes, there may be additional contributors to the phenotypes (Additional file 1: Table S1).
Estimating the potential inheritance mode based on the pedigree structure before analysis facilitates the speed of analysis. However, all potential inheritance models should be investigated. In family 060, there are eight children with only the youngest affected suggesting a de novo event in the proband; therefore trio WES was performed with analysis initially focused on de novo variant identification. However, the molecular analysis identified homozygous variants in RNASEH2B (NM_001142279.2:c.356A > G:p.Asp119Gly) using a presumed recessive model (Additional file 1: Table S1). Notably, one of the four de novo and the compound heterozygous variants are found in consanguineous families (Additional file 1: Tables S1 and S2), indicating the importance of considering all inheritance modes during analysis.
Expanded phenotypes of the known disease-associated genes and multifunction genes
In family 022, the proband with KIAA0226 variants exhibits microcephaly without cortical dysgenesis, hindbrain malformation, or corpus callosum abnormality, clinically distinct from the reported spinocerebellar ataxia (MIM#615705). The reported case is a consanguineous Saudi Arabia family with homozygous variants in the same gene, suggesting phenotypic expansion potentially related to intrinsic properties of the identified variants. In family 037 with hindbrain malformation, we identified a homozygous variant in MYO5A, which is responsible for Griscelli syndrome (MIM#214450). MYO5A, an actin-based motor protein, regulates many different cellular processes, including melanosome transport in pigment cells [37] and ER transport in Purkinje cells [38]. Different from previously reported truncating variants in Griscelli syndrome, the missense variant in this family seems mainly to affect cerebellar function.
Further empowering the analysis of the WES data by CNV-detection tools
Predicting CNVs from exome data allows a more complete analysis of possible pathogenic genomic variation, which may result from a variety of mutational mechanisms. The combination of a SNV and a CNV in RPS6KC1 provides a possible explanation for the proband’s phenotypes in family 025 (Fig. 2, Table 2 and Additional file 1: Table S1). This indicates that by using the read depth information in BAM files and B-allele frequency in VCF files, one can detect intragenic CNV. We anticipate that using next generation sequencing for both CNV and SNV analysis may lead to more molecular diagnoses.
Paralogs with functional studies and disease association provide additional supportive evidence
Previously, by combining the phenotypic information from a large scale mutagenesis screen in fruit flies with human exome and associated phenotype data from the BHCMG database, we observed that fly genes with multiple human orthologs are more likely to be associated with human disorders [8]. This observation led us to hypothesize that functional or genetic studies of paralogs can facilitate candidate gene identification, as shown in the cases of FOXP4, KIF5B, and GRM7, of which paralogs are closely linked in the interaction network (Additional file 1: Table S6 and Additional file 2: Figure S5). Moreover, utilizing data from clinical sequencing and computational programs for gene matching such as GeneMatcher can provide additional families to support our candidate genes [13].
WES research study as a building block toward disease gene discovery
In this research-based WES study, we identified several potential candidate disease genes and provide supporting evidence for each gene’s proposed association with disease [7] (Table 2 and Additional file 1: Table S1). We also applied the ACMG variant classification criteria to objectively demonstrate the evidence for pathogenicity for these variants; notably, because the ACMG criteria were designed for the clinical classification of variants in known disease genes, we have classified all variants associated with novel candidate disease genes as VUS [11] (Additional file 1: Table S1). These variant classifications are meant to form a foundation for further study. Nevertheless, this study provides information that may serve as a building block for future disease-gene discovery efforts.
Conclusions
In summary, by combining traditional WES analysis with WES-derived CNV detection, database mining and bioinformatic analyses of paralog, and interactome studies, we attained a potential molecular diagnosis rate of ~90 % of families in this cohort. We also identified biallelic variants representing recessive disease in a novel disease gene GRM7 in two families with decreased cerebral and cerebellar volume. Our data suggest that this combined strategy for WES analysis can provide a high potential molecular diagnostic rate in this genetically heterogeneous cohort exhibiting neurologic disease.
Abbreviations
ADHD, attention deficit hyperactivity disorder; AOH, absence of heterozygosity; ASD, autism spectrum disorders; BHCMG, Baylor-Hopkins Center for Mendelian Genomics; CNV, copy number variant; DD/ID, developmental delay/intellectual disability; ddPCR, droplet digital PCR; HGSC, human genome sequencing center; PFO, patent foramen ovale; VSD, ventricular septal defect; VUR, vesicoureteral reflux; WES, whole exome sequencing
Acknowledgements
We thank all the collaborators and families involved in this study.
Funding
This work is supported by the US National Human Genome Research Institute (NHGRI) National Heart Lung and Blood Institute (NHBLI) grant U54HG006542 to the Baylor-Hopkins Center for Mendelian Genomics, NINDS grant RO1 NS058529 to JRL and NHGRI 5U54HG003273 to RAG. W-LC is supported by The Cancer Prevention & Research Institute of Texas (CPRIT) training Program RP140102. JEP is supported by the NIH/NIGMS T32 GM07526 Medical Genetics Research Fellowship Program.
Availability of data and materials
Variants described in this manuscript (Additional file 1: Table S1) have been submitted to ClinVar (accessions SCV000282188 through SCV000282221). The ClinVar entries will be publicly released after publication. In addition, the genomic data have been submitted to dbGaP and are currently under review by dbGaP. Once the submission has been approved, the data will be submitted to the Baylor Hopkins dbGaP Study Accession number phs000711, which can be found at the following link: http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000711.v3.p1.
Authors’ contributions
W-LC participated in the design of the study, performed the majority of the analyses/experiments, and prepared the initial draft of the manuscript. EK participated in the design of the study, performed analyses, and prepared the initial draft of the manuscript. ZCA, TG contributed to statistical and bioinformatic analysis of the genomics and phenotype data, and critically revised the manuscript. MMA participated in the acquisition of data and helped prepare the initial draft of the manuscript. SG participated in the acquisition of data and critically revised the manuscript. JEP participated in the interpretation of clinical phenotype and molecular evidence as well as critically revised the manuscript. SNJ, HD, JH participated in the design of the study, generated the whole exome sequencing (WES) data, and critically revised the manuscript. DM participated in the design of the study, the generation and quality control/assessment of the WES data, and critically revised the manuscript. EB, RAG participated in the design of the study, overview of WES informatics pipeline, and critically revised the manuscript. JAR, KM contributed to the interpretation of clinical phenotype data, and critically revised the manuscript. HC, FX contributed to the acquisition, analysis, and interpretation of the molecular data, and critically revised the manuscript. YY contributed to the acquisition of the molecular and clinical data and critically revised the manuscript. EAF, AAA, MAMS, AWE-H contributed to the acquisition of the clinical data, and critically revised the manuscript. JRL participated in the design of the study, analyses of all molecular and clinical data, data interpretation, oversaw manuscript preparation, and critically revised the manuscript. All authors approved the final submitted version of the manuscript, and are accountable for the accuracy and integrity of the work.
Competing interests
J.R.L. has stock ownership in 23andMe and Lasergen, is a paid consultant for Regeneron, and a coinventor on multiple United States and European patents related to molecular diagnostics for inherited neuropathies, eye diseases and bacterial genomic fingerprinting. The Department of Molecular and Human Genetics at Baylor College of Medicine derives revenue from the clinical exome sequencing offered in the Baylor Miraca Genetics Laboratory (BMGL; http://www.bmgl.com/BMGL/); RAG served as Chief Scientific Officer and JRL is on the Scientific Advisory Board of BMGL. Other authors declare that they have no competing interests.
Consent for publication
The informed consent to publish identifiable information, including images, genetic pedigrees and clinical information was obtained for all adult participants and parental or legal guardian consent for all participants under the age of 16.
Ethics approval and consent to participate
This study has been approved by the Institutional Review Board at Baylor College of Medicine (protocol H-29697 granted by National Human Genome Research Institute [NHGRI]) and also by the Institutional Review Board of King Fadad Medical City, Riyadh, Saudi Arabia; and the National Committee of Biomedical Ethics, King Abdulaziz City for Science and Technology, Riyadh, Saudi Arabia. All participants provided informed consent prior to participation in the study.
Additional files
Summary of clinical features and disease/candidate variants identified. The major clinical features and the disease/candidate variants as well as the prediction scores and classifications for damaging effect of the variants are listed. For Polyphen2 (Pph2), D for probably damaging, P for possibly damaging, and B for benign. For LRT, D for deleterious, N for neutral, and U for unknown. For MutationTaster (MT), A for disease causing automatic, D for disease causing, N for polymorphism, and P for polymorphism automatic. Moreover, this table includes the ranking and ACMG criteria for each gene, the supporting evidence and the discussion of other variants, as well as the frequency/number of the variants in our internal CMG database, Atherosclerosis Risk in Communities Study (ARIC), ExAC database, Thousand Genome project, and NHLBI GO Exome Sequencing Project (ESP). pLI: probability of loss-of-function (LoF) intolerance. Table S2. Categorization of families based on major clinical features. Y: the family has this clinical feature; N: the family does not have this clinical feature. Families with brain malformations were not counted in the ID/DD groups, even if this feature was present. Percentages of families with each feature are shown at the bottom of the table. Table S3. Categorization of disease genes/candidates by major clinical features. Table S4. AOH metrics for the probands carrying known or candidate disease genes. Table S5. Raw data of ddPCR in RPS6KC1 in family 025. Table S6. Homologs of disease genes/candidates between human and fruit fly. The HCOP website (http://www.genenames.org/cgi-bin/hcop) was used to identify the fly homologs of the identified disease/candidate genes, listed in the upper panel. These fly homologs are then used to search for additional human homologs to find paralogs of the original human genes, as shown in the bottom part of the list. (XLS 165 kb)
Thirty one Arab family pedigrees in this study. Family numbers are given as numeric series and individual IDs as BAB series. They are arranged into: known variant in known gene (green), novel variant in known gene (light red), phenotypic expansion (blue), novel disease candidates (light orange), and unsolved cases (gray). Gray shading indicates a minor phenotype. Figure S2. Workflow of WES analysis. The SNVs predicted to have (probably) damaging effects are filtered with the allele frequency data from Atherosclerosis Risk in Communities Study (ARIC) (http://drupal.cscc.unc.edu/aric/), NHLBI GO Exome Sequencing Project (ESP), Seattle, WA (http://evs.gs.washington.edu/EVS/), 1000 Genomes Project (http://www.1000genomes.org), and Exome Aggregation Consortium (ExAC) Cambridge, MA (http://exac.broadinstitute.org) [Oct 2014] and Baylor-Hopkins Center for Mendelian Genomics (BHCMG) database with more than 5,000 exomes. WES data are also used to predict CNVs. The information of the paralogs of candidate genes provides supporting evidence for the findings. Moreover, cross-database gene mining identifies additional cases. Figure S3. CNV prediction identifies a homozygous deletion in GRID2 in BAB6883. a HMZDelFinder reveals a homozygous deletion in GRID2 (chr4:g.94006145_94032105del). The lower threshold for RPKM is 0.5 and upper threshold is 1. b This deletion is confirmed by clinical array (chr4:g.93,985,826_94,074,965del) and segregates with phenotypes in the family. c Breakpoint junction maps to chr4:g.93,978,239_94,078,203del. Figure S4. Network analysis of known and candidate genes. a Temporal RNA expression in brain. b The protein-protein interaction network: Interaction STRING scores are displayed in an upper triangular heatmap. Low and high values are represented in blue and red, respectively. Figure S5. Protein interaction network of the candidate genes. a KDM2B and MLLT1 are in the chromatin remodeling sub-network. b FOXP4 and its paralogs. c KIF5B and its paralogs. d GRM7 and its related members. (ZIP 4859 kb)
Suplementary Materials and Methods. (DOCX 16 kb)
Contributor Information
Wu-Lin Charng, Email: charng@alumni.bcm.edu.
Ender Karaca, Email: karaca@bcm.edu.
Zeynep Coban Akdemir, Email: Zeynep.CobanAkdemir@bcm.edu.
Tomasz Gambin, Email: Tomasz.Gambin@bcm.edu.
Mehmed M. Atik, Email: Mehmed.Atik@bcm.edu
Shen Gu, Email: Shen.Gu@bcm.edu.
Jennifer E. Posey, Email: Jennifer.Posey@bcm.edu
Shalini N. Jhangiani, Email: jhangian@bcm.edu
Donna M. Muzny, Email: donnam@bcm.edu
Harsha Doddapaneni, Email: doddapan@bcm.edu.
Jianhong Hu, Email: Jianhong.Hu@bcm.edu.
Eric Boerwinkle, Email: Eric.Boerwinkle@uth.tmc.edu.
Richard A. Gibbs, Email: agibbs@bcm.edu
Jill A. Rosenfeld, Email: Jill.Mokry@bcm.edu
Hong Cui, Email: Hongcuicn@gmail.com.
Fan Xia, Email: fxia@bcm.edu.
Kandamurugu Manickam, Email: kmanickam@geisinger.edu.
Yaping Yang, Email: yapingy@bcm.edu.
Eissa A. Faqeih, Email: efaqeih@kfmc.med.sa
Ali Al Asmari, Email: aalasmari@kfmc.med.sa.
Mohammed A. M. Saleh, Email: msaleh@kfmc.med.sa
Ayman W. El-Hattab, Email: elhattabaw@yahoo.com
James R. Lupski, Phone: (713) 798-3723, Email: jlupski@bcm.edu
References
- 1.Hawrylycz MJ, Lein ES, Guillozet-Bongaarts AL, Shen EH, Ng L, Miller JA, van de Lagemaat LN, Smith KA, Ebbert A, Riley ZL, Abajian C, Beckmann CF, Bernard A, Bertagnolli D, Boe AF, Cartagena PM, Chakravarty MM, Chapin M, Chong J, Dalley RA, Daly BD, Dang C, Datta S, Dee N, Dolbeare TA, Faber V, Feng D, Fowler DR, Goldy J, Gregor BW, Haradon Z, Haynor DR, Hohmann JG, Horvath S, Howard RE, Jeromin A, Jochim JM, Kinnunen M, Lau C, Lazarz ET, Lee C, Lemon TA, Li L, Li Y, Morris JA, Overly CC, Parker PD, Parry SE, Reding M, Royall JJ, Schulkin J, Sequeira PA, Slaughterbeck CR, Smith SC, Sodt AJ, Sunkin SM, Swanson BE, Vawter MP, Williams D, Wohnoutka P, Zielke HR, Geschwind DH, Hof PR, Smith SM, Koch C, Grant SG, Jones AR. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 2012;489(7416):391–9. doi: 10.1038/nature11405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Drachman DA. Do we have brain to spare? Neurology. 2005;64(12):2004–5. doi: 10.1212/01.WNL.0000166914.38327.BB. [DOI] [PubMed] [Google Scholar]
- 3.Guerrini R, Dobyns WB. Malformations of cortical development: clinical features and genetic causes. Lancet Neurol. 2014;13(7):710–26. doi: 10.1016/S1474-4422(14)70040-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rudnik-Schoneborn S, Barth PG, Zerres K. Pontocerebellar hypoplasia. Am J Med Genet C Semin Med Genet. 2014;166C(2):173–83. doi: 10.1002/ajmg.c.31403. [DOI] [PubMed] [Google Scholar]
- 5.Alazami AM, Patel N, Shamseldin HE, Anazi S, Al-Dosari MS, Alzahrani F, Hijazi H, Alshammari M, Aldahmesh MA, Salih MA, Faqeih E, Alhashem A, Bashiri FA, Al-Owain M, Kentab AY, Sogaty S, Al Tala S, Temsah MH, Tulbah M, Aljelaify RF, Alshahwan SA, Seidahmed MZ, Alhadid AA, Aldhalaan H, AlQallaf F, Kurdi W, Alfadhel M, Babay Z, Alsogheer M, Kaya N, Al-Hassnan ZN, Abdel-Salam GM, Al-Sannaa N, Al Mutairi F, El Khashab HY, Bohlega S, Jia X, Nguyen HC, Hammami R, Adly N, Mohamed JY, Abdulwahab F, Ibrahim N, Naim EA, Al-Younes B, Meyer BF, Hashem M, Shaheen R, Xiong Y, Abouelhoda M, Aldeeri AA, Monies DM, Alkuraya FS. Accelerating novel candidate gene discovery in neurogenetic disorders via whole-exome sequencing of prescreened multiplex consanguineous families. Cell Rep. 2015;10(2):148–61. [DOI] [PubMed]
- 6.Chong JX, Buckingham KJ, Jhangiani SN, Boehm C, Sobreira N, Smith JD, Harrell TM, McMillin MJ, Wiszniewski W, Gambin T, Coban Akdemir ZH, Doheny K, Scott AF, Avramopoulos D, Chakravarti A, Hoover-Fong J, Mathews D, Witmer PD, Ling H, Hetrick K, Watkins L, Patterson KE, Reinier F, Blue E, Muzny D, Kircher M, Bilguvar K, López-Giráldez F, Sutton VR, Tabor HK, Leal SM, Gunel M, Mane S, Gibbs RA, Boerwinkle E, Hamosh A, Shendure J, Lupski JR, Lifton RP, Valle D, Nickerson DA, Centers for Mendelian Genomics. Bamshad MJ. The Genetic Basis of Mendelian Phenotypes: Discoveries, Challenges, and Opportunities. Am J Hum Genet. 2015;97(2):199–215. doi: 10.1016/j.ajhg.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karaca E, Harel T, Pehlivan D, Jhangiani SN, Gambin T, Coban Akdemir Z, Gonzaga-Jauregui C, Erdin S, Bayram Y, Campbell IM, Hunter JV, Atik MM, Van Esch H, Yuan B, Wiszniewski W, Isikay S, Yesil G, Yuregir OO, Tug Bozdogan S, Aslan H, Aydin H, Tos T, Aksoy A, De Vivo DC, Jain P, Geckinli BB, Sezer O, Gul D, Durmaz B, Cogulu O, Ozkinay F, Topcu V, Candan S, Cebi AH, Ikbal M, Yilmaz Gulec E, Gezdirici A, Koparir E, Ekici F, Coskun S, Cicek S, Karaer K, Koparir A, Duz MB, Kirat E, Fenercioglu E, Ulucan H, Seven M, Guran T, Elcioglu N, Yildirim MS, Aktas D, Alikaşifoğlu M, Ture M, Yakut T, Overton JD, Yuksel A, Ozen M, Muzny DM, Adams DR, Boerwinkle E, Chung WK, Gibbs RA, Lupski JR. Genes that Affect Brain Structure and Function Identified by Rare Variant Analyses of Mendelian Neurologic Disease. Neuron. 2015;88(3):499–513. [DOI] [PMC free article] [PubMed]
- 8.Yamamoto S, Jaiswal M, Charng WL, Gambin T, Karaca E, Mirzaa G, Wiszniewski W, Sandoval H, Haelterman NA, Xiong B, Zhang K, Bayat V, David G, Li T, Chen K, Gala U, Harel T, Pehlivan D, Penney S, Vissers LE, de Ligt J, Jhangiani SN, Xie Y, Tsang SH, Parman Y, Sivaci M, Battaloglu E, Muzny D, Wan YW, Liu Z, Lin-Moore AT, Clark RD, Curry CJ, Link N, Schulze KL, Boerwinkle E, Dobyns WB, Allikmets R, Gibbs RA, Chen R, Lupski JR, Wangler MF, Bellen HJ. A drosophila genetic resource of mutants to study mechanisms underlying human genetic diseases. Cell. 2014;159(1):200–14. doi: 10.1016/j.cell.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5(4):557–72. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 10.Krumm N, Sudmant PH, Ko A, O’Roak BJ, Malig M, Coe BP. Copy number variation detection and genotyping from exome sequence data. Genome Res. 2012;22(8):1525–32. doi: 10.1101/gr.138115.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL, ACMG Laboratory Quality Assurance Committee Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–24. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wiszniewska J, Bi W, Shaw C, Stankiewicz P, Kang SH, Pursley AN, Lalani S, Hixson P, Gambin T, Tsai CH, Bock HG, Descartes M, Probst FJ, Scaglia F, Beaudet AL, Lupski JR, Eng C, Cheung SW, Bacino C, Patel A. Combined array CGH plus SNP genome analyses in a single assay for optimized clinical testing. Eur J Hum Genet. 2014;22(1):79–87. doi: 10.1038/ejhg.2013.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sobreira N, Schiettecatte F, Boehm C, Valle D, Hamosh A. New tools for Mendelian disease gene identification: PhenoDB variant analysis module; and GeneMatcher, a web-based tool for linking investigators with an interest in the same gene. Hum Mutat. 2015;36(4):425–31. doi: 10.1002/humu.22769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamosh A, Sobreira N, Hoover-Fong J, et al. PhenoDB: a new web-based tool for the collection, storage, and analysis of phenotypic features. Hum Mutat. 2013;34(4):566–71. doi: 10.1002/humu.22283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hayashi S, Okada T, Igarashi N, Fujita T, Jahangeer S, Nakamura S. Identification and characterization of RPK118, a novel sphingosine kinase-1-binding protein. J Biol Chem. 2002;277(36):33319–24. doi: 10.1074/jbc.M201442200. [DOI] [PubMed] [Google Scholar]
- 16.Mizugishi K, Yamashita T, Olivera A, Miller GF, Spiegel S, Proia RL. Essential role for sphingosine kinases in neural and vascular development. Mol Cell Biol. 2005;25(24):11113–21. doi: 10.1128/MCB.25.24.11113-11121.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takayama C, Nakagawa S, Watanabe M, Mishina M, Inoue Y. Light- and electron-microscopic localization of the glutamate receptor channel delta 2 subunit in the mouse Purkinje cell. Neurosci Lett. 1995;188(2):89–92. doi: 10.1016/0304-3940(95)11403-J. [DOI] [PubMed] [Google Scholar]
- 18.Doty RT, Vanasse GJ, Disteche CM, Willerford DM. The leukemia-associated gene Mllt1/ENL: characterization of a murine homolog and demonstration of an essential role in embryonic development. Blood Cells Mol Dis. 2002;28(3):407–17. doi: 10.1006/bcmd.2002.0525. [DOI] [PubMed] [Google Scholar]
- 19.Mueller D, Bach C, Zeisig D, Garcia-Cuellar MP, Monroe S, Sreekumar A, Zhou R, Nesvizhskii A, Chinnaiyan A, Hess JL, Slany RK. A role for the MLL fusion partner ENL in transcriptional elongation and chromatin modification. Blood. 2007;110(13):4445–54. doi: 10.1182/blood-2007-05-090514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frescas D, Guardavaccaro D, Bassermann F, Koyama-Nasu R, Pagano M. JHDM1B/FBXL10 is a nucleolar protein that represses transcription of ribosomal RNA genes. Nature. 2007;450(7167):309–13. doi: 10.1038/nature06255. [DOI] [PubMed] [Google Scholar]
- 21.He J, Kallin EM, Tsukada Y, Zhang Y. The H3K36 demethylase Jhdm1b/Kdm2b regulates cell proliferation and senescence through p15(Ink4b) Nat Struct Mol Biol. 2008;15(11):1169–75. doi: 10.1038/nsmb.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hannenhalli S, Putt ME, Gilmore JM, Wang J, Parmacek MS, Epstein JA, Morrisey EE, Margulies KB, Cappola TP. Transcriptional genomics associates FOX transcription factors with human heart failure. Circulation. 2006;114(12):1269–76. doi: 10.1161/CIRCULATIONAHA.106.632430. [DOI] [PubMed] [Google Scholar]
- 23.Li S, Zhou D, Lu MM, Morrisey EE. Advanced cardiac morphogenesis does not require heart tube fusion. Science. 2004;305(5690):1619–22. doi: 10.1126/science.1098674. [DOI] [PubMed] [Google Scholar]
- 24.Teufel A, Wong EA, Mukhopadhyay M, Malik N, Westphal H. FoxP4, a novel forkhead transcription factor. Biochim Biophys Acta. 2003;1627(2–3):147–52. doi: 10.1016/S0167-4781(03)00074-5. [DOI] [PubMed] [Google Scholar]
- 25.Spaeth JM, Hunter CS, Bonatakis L, Guo M, French CA, Slack I, Hara M, Fisher SE, Ferrer J, Morrisey EE, Stanger BZ, Stein R. The FOXP1, FOXP2 and FOXP4 transcription factors are required for islet alpha cell proliferation and function in mice. Diabetologia. 2015;58(8):1836–44. doi: 10.1007/s00125-015-3635-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao H, Zhou W, Yao Z, Wan Y, Cao J, Zhang L, Zhao J, Li H, Zhou R, Li B, Wei G, Zhang Z, French CA, Dekker JD, Yang Y, Fisher SE, Tucker HO, Guo X. Foxp1/2/4 regulate endochondral ossification as a suppresser complex. Dev Biol. 2015;398(2):242–54. doi: 10.1016/j.ydbio.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang SW, Mislankar M, Misra C, Huang N, Dajusta DG, Harrison SM, McBride KL, Baker LA, Garg V. Genetic abnormalities in FOXP1 are associated with congenital heart defects. Hum Mutat. 2013;34(9):1226–30. doi: 10.1002/humu.22366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kanai Y, Dohmae N, Hirokawa N. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron. 2004;43(4):513–25. doi: 10.1016/j.neuron.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 29.Tanaka Y, Kanai Y, Okada Y, Nonaka S, Takeda S, Harada A, Hirokawa N. Targeted disruption of mouse conventional kinesin heavy chain, kif5B, results in abnormal perinuclear clustering of mitochondria. Cell. 1998;93(7):1147–58. doi: 10.1016/S0092-8674(00)81459-2. [DOI] [PubMed] [Google Scholar]
- 30.Gonzaga-Jauregui C, Harel T, Gambin T, Kousi M, Griffin LB, Francescatto L, Ozes B, Karaca E, Jhangiani SN, Bainbridge MN, Lawson KS, Pehlivan D, Okamoto Y, Withers M, Mancias P, Slavotinek A, Reitnauer PJ, Goksungur MT, Shy M, Crawford TO, Koenig M, Willer J, Flores BN, Pediaditrakis I, Us O, Wiszniewski W, Parman Y, Antonellis A, Muzny DM, Baylor-Hopkins Center for Mendelian Genomics. Katsanis N, Battaloglu E, Boerwinkle E, Gibbs RA, Lupski JR. Exome Sequence Analysis Suggests that Genetic Burden Contributes to Phenotypic Variability and Complex Neuropathy. Cell Rep. 2015;12(7):1169–83. doi: 10.1016/j.celrep.2015.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carlsson ML. Hypothesis: is infantile autism a hypoglutamatergic disorder? Relevance of glutamate - serotonin interactions for pharmacotherapy. J Neural Transm. 1998;105(4–5):525–35. doi: 10.1007/s007020050076. [DOI] [PubMed] [Google Scholar]
- 32.Jamain S, Betancur C, Quach H, Philippe A, Fellous M, Giros B, Gillberg C, Leboyer M, Bourgeron T. Linkage and association of the glutamate receptor 6 gene with autism. Mol Psychiatry. 2002;7(3):302–10. doi: 10.1038/sj.mp.4000979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Y, Zhang Y, Zhao D, Dong R, Yang X, Tammimies K, Uddin M, Scherer SW, Gai Z. Rare de novo deletion of metabotropic glutamate receptor 7 (GRM7) gene in a patient with autism spectrum disorder. Am J Med Genet B Neuropsychiatr Genet. 2015;168B(4):258–64. doi: 10.1002/ajmg.b.32306. [DOI] [PubMed] [Google Scholar]
- 34.Yang Y, Pan C. Role of metabotropic glutamate receptor 7 in autism spectrum disorders: a pilot study. Life Sci. 2013;92(2):149–53. doi: 10.1016/j.lfs.2012.11.010. [DOI] [PubMed] [Google Scholar]
- 35.Xia W, Liu Y, Jiao J. GRM7 Regulates Embryonic Neurogenesis via CREB and YAP. Stem Cell Reports. 2015;4(5):795–810. doi: 10.1016/j.stemcr.2015.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.White J, Mazzeu JF, Hoischen A, Jhangiani SN, Gambin T, Alcino MC, Penney S, Saraiva JM, Hove H, Skovby F, Kayserili H, Estrella E, Vulto-van Silfhout AT, Steehouwer M, Muzny DM, Sutton VR, Gibbs RA, Baylor-Hopkins Center for Mendelian Genomics. Lupski JR, Brunner HG, van Bon BW, Carvalho CM. DVL1 frameshift mutations clustering in the penultimate exon cause autosomal-dominant Robinow syndrome. Am J Hum Genet. 2015;96(4):612–22. doi: 10.1016/j.ajhg.2015.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fukuda M, Kuroda TS, Mikoshiba K. Slac2-a/melanophilin, the missing link between Rab27 and myosin Va: implications of a tripartite protein complex for melanosome transport. J Biol Chem. 2002;277(14):12432–6. doi: 10.1074/jbc.C200005200. [DOI] [PubMed] [Google Scholar]
- 38.Wagner W, Brenowitz SD, Hammer JA., 3rd Myosin-Va transports the endoplasmic reticulum into the dendritic spines of Purkinje neurons. Nat Cell Biol. 2011;13(1):40–8. doi: 10.1038/ncb2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Variants described in this manuscript (Additional file 1: Table S1) have been submitted to ClinVar (accessions SCV000282188 through SCV000282221). The ClinVar entries will be publicly released after publication. In addition, the genomic data have been submitted to dbGaP and are currently under review by dbGaP. Once the submission has been approved, the data will be submitted to the Baylor Hopkins dbGaP Study Accession number phs000711, which can be found at the following link: http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000711.v3.p1.


