Abstract
The present study aimed to screen potential genes associated with metastatic prostate cancer (PCa), in order to improve the understanding of the mechanisms underlying PCa metastasis. The GSE3325 microarray dataset, which was downloaded from the Gene Expression Omnibus database, consists of seven clinically localized PCa samples, six hormone-refractory metastatic PCa samples and six benign prostate tissue samples. The Linear Models for Microarray Data package was used to identify differentially-expressed genes (DEGs) and a hierarchical cluster analysis for DEGs was performed with the pheatmap package. Furthermore, potential functions for the DEGs were predicted by a functional enrichment analysis. Subsequently, microRNAs (miRNAs) potentially involved in the regulation of PCa metastasis were identified by WebGestalt software, and the miRNA-DEG regulatory network was visualized using Cytoscape. In addition, a pathway enrichment analysis for DEGs in the regulatory network was performed. A total of 306 and 2,073 genes were differentially expressed in the clinically localized PCa and the metastatic PCa groups, respectively, as compared with the benign prostate group, of which 174 were differentially expressed in both groups. A number of the DEGs, including CAMK2D and SH3BP4, were significantly enriched in the cell cycle, and others, such as MAF, were associated with the regulation of cell proliferation. Furthermore, some DEGs (CAMK2D and PCDH17) were observed to be regulated by miR-30, whereas others (ADCY2, MAF, SH3BP4 and PCDH17) were modulated by miR-182. Additionally, ADCY2 and CAMK2D were distinctly enriched in the calcium signaling pathway. The present study identified novel DEGs, including ADCY2, CAMK2D, MAF, SH3BP4 and PCDH17, that may be involved in the metastasis of PCa.
Keywords: metastatic prostate cancer, differentially expressed genes, microRNA, regulatory network, pathway
Introduction
Prostate cancer (PCa) is the most common cancer among European and American men, and accounts for 27% (233,000) of cancer incidences in men in the USA (1). It has a high mortality rate as a result of its high propensity for metastasis (2,3). PCa has been shown to preferentially metastasize to the bone marrow stroma of the axial skeleton (4); however, the precise mechanism underlying PCa metastasis is currently unclear. Therefore, the identification of specific metastasis biomarkers and novel diagnostic targets is required in order to improve the prognosis and treatment of the disease.
Previous studies have made considerable progress in identifying the key regulators in the PCa metastatic process. E-cadherin, which is attached to the actin cytoskeleton via intracellular catenin, has been implicated in the process of PCa metastasis; in primary PCa, reduced E-cadherin expression was associated with bone metastasis and a poor prognosis (5). In addition, the expression of the DLC1 tumor-suppressor gene in metastatic PCa cells has been shown to upregulate the expression of E-cadherin, resulting in the suppression of highly metastatic PCa cell invasion by inhibiting the activity of RhoA-GTP and RhoC-GTP (6). The activation of Rho GTPases is dependent on the downstream Ras protein, which has a major influence on cell signaling (7). Members of the Rho GTPase family are involved in cancer cell motility by regulating actin dynamics and controlling morphological changes (8). A previous study demonstrated that the suppression of the farnesyl and geranyl-geranyl prenylation pathways markedly reduced the migration and motility of PCa cells by inhibiting Ras prenylation and concurrent Rho activation (9). Furthermore, activation of the phosphoinositide 3-kinase/protein kinase B (AKT) signaling pathway has been more frequently observed in resistant and metastatic PCa compared with primary PCa, and thus targeting this signaling pathway may improve the outcome of patients with aggressive PCa (10). Previous studies have reported various genes able to promote PCa tumorigenesis and metastasis, including CCL2 (11), SERPINB5 (12), SRC (13), TMPRSS2-ERG gene fusion and PCA3 (14). In addition, microRNAs (miRNAs), which are considered to be important regulators of gene expression, have been associated with the development of metastatic PCa. For instance, miR-203 (15), miR-16 (16), miR-205 (17), miR-24 (18), miR-29a (19) and miR-145 (16) have all been implicated in PCa metastasis.
Varambally et al (20) performed an integrative genomic and proteomic analysis of benign prostate and metastatic PCa; they reported 48–64% concordance between protein and transcript levels and demonstrated that proteomic alterations between metastatic and clinically localized PCa, which map concordantly to gene transcripts, can serve as predictors of clinical outcome in PCa as well as other solid tumors. However, to the best of our knowledge, the potential miRNAs involved in metastatic PCa, and the interactions of differentially-expressed genes (DEGs) targeted by miRNAs, have yet to be investigated. Therefore, the present study aimed to further elucidate the molecular mechanisms underlying the metastasis of PCa by analyzing the microarray data of benign prostate, clinically localized and metastatic PCa deposited by Varambally et al (20) in the Gene Expression Omnibus (GEO) database. Initially a hierarchical cluster analysis for DEGs was performed, followed by a Gene Ontology (GO) functional enrichment analysis. Furthermore, potential miRNAs in metastatic PCa were identified and a miRNA-DEG regulatory network was constructed. Finally, a pathway enrichment analysis for DEGs in the regulatory network was performed. The results of this bioinformatics analysis may shed light on the molecular mechanisms underlying the metastasis of PCa and provide novel diagnostic biomarkers.
Materials and methods
Affymetrix microarray data
The GSE3325 gene expression profile data (20) was downloaded from the GEO (http://www.ncbi.nlm.nih.gov/geo/) and was based on the GPL570 [HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array platform. A total of 19 human prostate tissue samples were available for further analysis, including seven clinically localized PCa samples, six hormone-refractory metastatic PCa samples and six benign prostate tissue samples.
CEL and probe annotation files were downloaded from GEO, and the gene expression data for all samples were preprocessed via Robust Multichip Averaging background correction, quantile normalization and probe summarization (21) in the affy software package (version 1.34.0; http://bioconductor.org/packages/release/bioc/html/affy.html), as described previously (22).
DEGs screening
The Linear Models for Microarray Data package of R (https://bioconductor.org/packages/release/bioc/html/limma.html) was used to identify genes that were differentially expressed in the primary PCa and metastatic PCa groups, as compared with the benign prostate group, as described previously (23). The raw P-value was adjusted according to the false discovery rate (FDR) using the Benjamin and Hochberg method (24). Only genes with a cut-off criteria of |log2fold change| >1 and FDR<0.01 were considered to be differentially expressed.
Hierarchical cluster analysis for DEGs
Hierarchical clustering is a common method used to determine clusters of similar data points in a multidimensional space (25). The pheatmap package (version 1.0.2; https://cran.r-project.org/web/packages/pheatmap/index.html) was used to perform hierarchical clustering of the DEGs via joint between-within distances, as described previously (26). Expression values from multiple clones or probe sets mapping to the same Unigene Cluster ID were averaged.
GO functional enrichment analysis for DEGs
The Database for Annotation, Visualization and Integrated Discovery (DAVID; https://david.ncifcrf.gov/) provides a comprehensive set of novel and powerful tools for assigning biological meaning to a set of genes (27). FDR<0.05 was used as the cut-off criterion for GO functional enrichment analysis by DAVID.
Integrated miRNA-DEG regulatory network construction
The common miRNAs in Gene set B, as predicted by the databases of miRecords (http://c1.accurascience.com/miRecords/), TarBase (http://diana.imis.athena-innovation.gr/DianaTools/index.php?r=tarbase/index) and TargetScan (http://www.targetscan.org/), were selected using WEB-based GEne SeT AnaLysis Toolkit software (update 2013; http://bioinfo.vanderbilt.edu/webgestalt/), and P<0.05 was used as the cut-off criterion. Subsequently, the Search Tool for the Retrieval of Interacting Genes (http://string-db.org/) was used to analyze the interactions between the DEGs targeted by miRNAs by calculating their combined score; a score of >0.4 was set as the cut-off criterion. Finally, the integrated miRNA-DEG regulatory network was visualized using Cytoscape (http://cytoscape.org/).
Pathway enrichment analysis for DEGs in the regulatory network
Pathway enrichment analysis was conducted as described previously (28) to identify significant metabolic pathways for the DEGs. P<0.05 was used as the cut-off criterion for the Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis using DAVID.
Results
Identification of DEGs
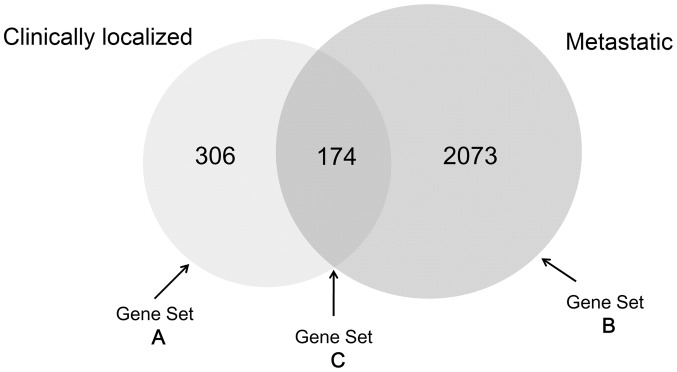
Based on the cut-off criteria, 2,727 DEGs were identified for the clinically localized PCa and metastatic PCa groups, of which 306 were differentially expressed in the clinically localized PCa group only (Gene set A). A total of 2,073 genes were differentially expressed in the metastatic PCa group only (Gene set B) and 174 genes were differentially expressed in both groups (Gene set C; Fig. 1), as compared with the benign prostate group.
Figure 1.
Venn diagram for the differentially expressed genes in the clinically localized and metastatic prostate cancer groups. Gene set A represents the genes only differentially expressed in the clinically localized prostate cancer group; Gene set B represents the genes only differentially expressed in the metastatic prostate cancer group; Gene set C represents the genes differentially expressed in both groups.
Hierarchical cluster analysis
An unsupervised hierarchical cluster analysis of the data revealed that the DEGs could be used to accurately classify prostate samples as benign, clinically localized prostate cancer or metastatic disease (Fig. 2).
Figure 2.
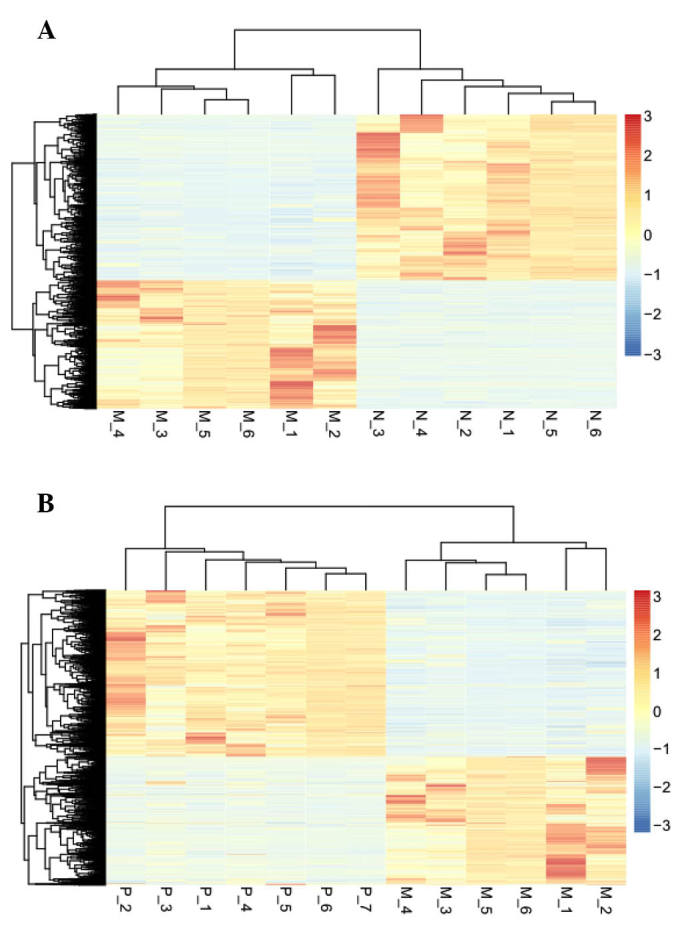
Hierarchical cluster analysis for the genes differentially expressed between the (A) metastatic prostate cancer and the benign prostate groups, and (B) the metastatic and clinically localized prostate cancer groups. Each row represents a single gene; each column represents a tissue sample. M represents the metastatic prostate cancer group; N represents the benign prostate group; P represents the clinically localized prostate cancer group. The gradual color change from orange to blue represents the changing process from upregulation to downregulation.
GO functional enrichment analysis for Gene sets A, B and C
In Gene set A, DLX2, DLX1, HOXD10 and HOXD11 DEGs were associated with proximal/distal pattern formation (FDR=3.55E-04), whereas RBP4, PDE3B and PPARG were implicated in the response to insulin (FDR=7.8400), homeostatic processes (FDR=9.6200), chemical homeostasis (FDR=0.0019) and responses to peptide hormones (FDR=0.0023) and organic substances (FDR=0.0029) (Table I).
Table I.
Enriched terms for Gene sets A, B and C.
| Category | Term | No. of genes | FDR | Genes |
|---|---|---|---|---|
| Gene set A | GO:0009954~proximal/distal pattern formation | 5 | 3.5500 | DLX2, DLX1, GREM1, HOXD10, HOXD11 |
| GO:0032868~response to insulin stimulus | 8 | 7.8400 | RBP4, EIF4EBP1, FADS1, PPARG, PDE3B, STXBP4, GAL, VLDLR | |
| GO:0042592~homeostatic process | 24 | 9.6200 | RBP4, SLC12A2, PPARG, F2RL1, PRDX4, PDE3B, CACNG2, ITPR3, PPARGC1A, MUC6… | |
| GO:0001501~skeletal system development | 14 | 0.0011 | RBP4, HOXD10, HOXD11, MSX2, DLX2, DLX1, COL9A2, BCL2, CLEC3A, NAB1… | |
| GO:0048878~chemical homeostasis | 18 | 0.0019 | RBP4, F2RL1, NOX1, PPARG, PDE3B, PPARGC1A, PRKCB, CCL11, MALL, ATP7B… | |
| GO:0021877~forebrain neuron fate commitment | 3 | 0.0022 | DLX2, DLX1, LHX6 | |
| GO:0043434~response to peptide hormone stimulus | 9 | 0.0023 | RBP4, EIF4EBP1, FADS1, BCL2, PPARG, PDE3B, STXBP4, GAL, VLDLR | |
| GO:0010033~response to organic substance | 22 | 0.0029 | RBP4, ADCY1, GNRH1, FADS1, LOC646626, PPARG, PTGS1, PDE3B, COLEC12, STXBP4… | |
| GO:0009725~response to hormone stimulus | 14 | 0.0038 | RBP4, ADCY1, GNRH1, FADS1, PTGS1, PPARG, PDE3B, STXBP4, GAL, EIF4EBP1… | |
| GO:0034637~cellular carbohydrate biosynthetic process | 6 | 60.0038 | RBP4, ISYNA1, UAP1, GNE, PPARGC1A, ACN9 | |
| Gene set B | GO:0022402~cell cycle process | 102 | 5.2300 | PRC1, ZAK, AIF1, BTRC, CDCA8, CDC6, CENPF, PTTG1, AURKB, TGFB2… |
| GO:0051726~regulation of cell cycle | 68 | 1.1100 | E2F2, PTGS2, ZAK, FAM175A, PKMYT1, PDCD4, PTEN, GTSE1, TGFB2, MYC… | |
| GO:0007049~cell cycle | 128 | 1.8200 | ZAK, PRC1, AIF1, BTRC, PKMYT1, RBM7, AURKA, AURKB, PTTG1, TGFB2… | |
| GO:0051301~cell division | 61 | 4.6100 | PRC1, PTTG1, CCNE1, CDCA2, CDC6, CABLES2, CDCA5, CCNA2, ASPM, CDK1… | |
| GO:0022403~cell cycle phase | 78 | 4.7700 | E2F1, PRC1, PKMYT1, RBM7, AURKA, AURKB, PTTG1, GTSE1, CCNE1, CDCA8… | |
| GO:0010035~response to inorganic substance | 47 | 6.4600 | CAV1, GCLC, PTGS2, PDGFA, SNCA, TPM1, PTEN, KCNMB1, FOS, GSN… | |
| GO:0007346~regulation of mitotic cell cycle | 38 | 0.0011 | CAV2, HOXA13, PML, PKMYT1, ASNS, ANLN, ZNF655, RCC1, SCRIB, MYC… | |
| GO:0042127~regulation of cell proliferation | 126 | 0.0012 | HRAS, CD38, IL6ST, PDGFA, TP63, MAF, TGFB3, STRN, PNP, TGFB2… | |
| GO:0030030~cell projection organization | 70 | 0.0015 | CAV2, HOXA13, PML, PKMYT1, ANLN, ZNF655, RCC1, SCRIB, GTSE1, MYC… | |
| GO:0000278~mitotic cell cycle | 70 | 0.0018 | E2F1, PRC1, BTRC, PKMYT1, AURKA, AURKB, PTTG1, GTSE1, CCNE1, NDE1… | |
| Gene set C | GO:0022402~cell cycle process | 21 | 3.3000 | MKI67, DLGAP5, SGOL1, NUSAP1, BIRC5, PBK, CDKN3, CCNB1, CENPA, CAMK2D… |
| GO:0022403~cell cycle phase | 18 | 3.5700 | MKI67, DLGAP5, SGOL1, NUSAP1, TTK, BIRC5, PBK, CCNB1, CAMK2D, ID4… | |
| GO:0000278~mitotic cell cycle | 17 | 4.0700 | DLGAP5, SGOL1, NUSAP1, TTK, BIRC5, PBK, CDKN3, CCNB1, CENPA, CAMK2D… | |
| GO:0000279~M phase | 15 | 2.7400 | MKI67, DLGAP5, SGOL1, NUSAP1, TTK, BIRC5, PBK, UBE2C, CCNB1, KIF2C… | |
| GO:0007049~cell cycle | 22 | 1.1800 | DLGAP5, SGOL1, NUSAP1, BIRC5, PBK, CCNB1, SH3BP4, KIF2C, CENPA, CAMK2D… | |
| GO:0000280~nuclear division | 11 | 4.5000 | CCNB1, KIF2C, CCNB2, DLGAP5, SGOL1, NUSAP1, BIRC5, PBK, UBE2C, ERCC6L… | |
| GO:0007067~mitosis | 11 | 4.5000 | CCNB1, KIF2C, CCNB2, DLGAP5, SGOL1, NUSAP1, BIRC5, PBK, UBE2C, ERCC6L… | |
| GO:0000087~M phase of mitotic cell cycle | 11 | 5.2300 | CCNB1, KIF2C, CCNB2, DLGAP5, SGOL1, NUSAP1, BIRC5, PBK, UBE2C, ERCC6L… | |
| GO:0048285~organelle fission | 11 | 6.3000 | CCNB1, KIF2C, CCNB2, DLGAP5, SGOL1, NUSAP1, BIRC5, PBK, UBE2C, ERCC6L… | |
| GO:0007346~regulation of mitotic cell cycle | 9 | 9.6800 | DLGAP5, CAMK2D, NUSAP1, TTK, BIRC5, AFAP1L2, GAS1, UBE2C, MYC |
Gene set A represents the genes only differentially expressed in the clinically localized prostate cancer group; Gene set B represents the genes only differentially expressed in the metastatic prostate cancer group; Gene set C represents the genes differentially expressed in both groups. FDR, false discovery rate.
In Gene set B, the DEGs were predominantly associated with the cell cycle: PRC1, ZAK, PTTG1, TGFB2, CDCA8, CDC6 and CENPF were associated with the cell cycle process (FDR=5.2300); PRC1, PTTG1, CCNE1, CDCA2 and CDC6 were involved in cell division (FDR=4.6100); and HRAS, CD38, IL6ST, PDGFA, TP63, MAF and TGFB3 were associated with the regulation of cell proliferation (FDR=0.0012) (Table I).
In Gene set C, the DEGs were also predominantly associated with the cell cycle. DLGAP5, SGOL1, NUSAP1, PBK, BIRC5 and CCNB1 were associated with the cell cycle process (FDR=3.3000), M phase (FDR=2.7400), mitosis (FDR=4.5000) and organelle fission (FDR=6.3000), whereas SH3BP4, KIF2C, CCNB2, CENPA and CAMK2D were associated with the cell cycle only (FDR=1.1800) (Table I).
Analysis of the miRNA-DEG regulatory network
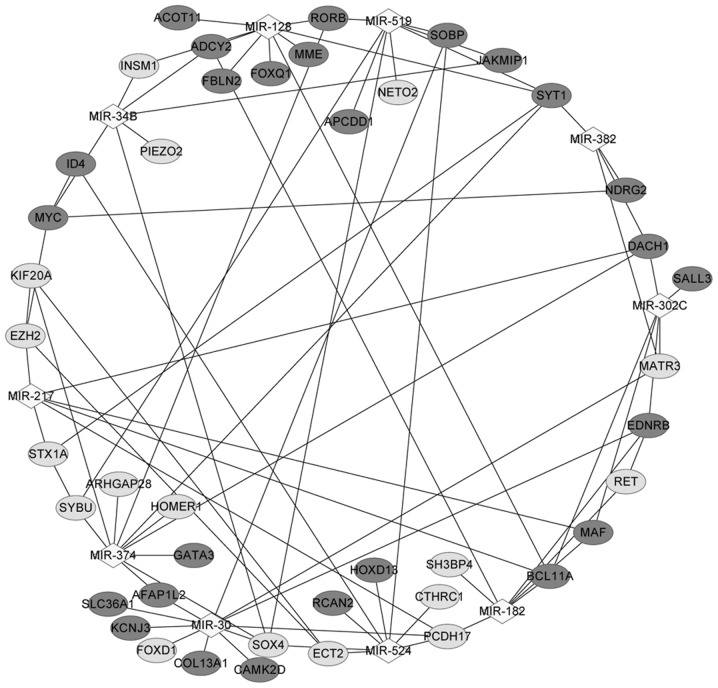
A total of 10 miRNAs were identified in Gene set B, including miR-374, miR-128, miR-182, miR-30, miR-302c and miR-524. Notably, miR-30 targeted the majority of the DEGs (11 DEGs, including CAMK2D, PCDH17, EDNRB, KCNJ3 and SOX4), and miR-182 targeted seven DEGs, including EDNRB, MAF, ADCY2, PCDH17, RET, SH3BP4 and BCL11A (Table II).
Table II.
Enriched microRNAs in Gene set B.
| microRNA | P-value | Count | Genes targeted by microRNA |
|---|---|---|---|
| hsa_TATTATA, MIR-374 | 2.1100 | 10 | RORB, HOMER1, KIF20A, SYBU, DACH1, GATA3, ARHGAP28, AFAP1L2, SOX4, SYT1 |
| hsa_CACTGTG, MIR-128 | 0.0003 | 9 | RORB, FBLN2, ADCY2, ACOT11, INSM1, SYT1, FOXQ1, MME, BCL11A |
| hsa_ATGCAGT, MIR-217 | 0.0003 | 6 | STX1A, MAF, PCDH17, DACH1, EZH2, BCL11A |
| hsa_TGTTTAC, MIR-30 | 0.0005 | 11 | SOBP, CAMK2D, COL13A1, SLC36A1, PCDH17, AFAP1L2, EDNRB, KCNJ3, SOX4, MATR3…… |
| hsa_ACAACTT, MIR-382 | 0.0032 | 4 | NDRG2, SYT1, MATR3, DACH1 |
| hsa_ACTGCCT, MIR-34B | 0.0032 | 6 | INSM1, SOX4, MYC, ADCY2, PIEZO2, JAKMIP1 |
| hsa_TTGCCAA, MIR-182 | 0.0036 | 7 | EDNRB, MAF, ADCY2, PCDH17, RET, SH3BP4, BCL11A |
| hsa_CTTTGTA, MIR-524 | 0.0036 | 8 | SOBP, CTHRC1, PCDH17, ECT2, ID4, RCAN2, HOXD13, SOX4 |
| hsa_ATGTTAA, MIR-302C | 0.0038 | 6 | EDNRB, SALL3, MAF, MATR3, DACH1, BCL11A |
| hsa_TGCACTT, MIR-519 | 0.0038 | 8 | SOBP, RORB, SYBU, NETO2, SOX4, SYT1, APCDD1, JAKMIP1 |
Count represents the number of differentially-expressed genes targeted by microRNA. Gene set B represents the genes only differentially expressed in the metastatic prostate cancer group.
The miRNA-DEG regulatory network in Fig. 3 contained 10 miRNAs and 43 corresponding DEGs. ADCY2 was regulated by miR-128, miR-34B and miR-182; EDNRB was regulated by miR-30, miR-182 and miR-302C; CAMK2D was regulated by miR-30; PCDH17 was modulated by miR-217, miR-30, miR-182 and miR-524; SH3BP4 was modulated by miR-182; and MAF interacted with miR-182, miR-302c and BCL11A.
Figure 3.
Regulatory network containing microRNAs and their corresponding differentially expressed genes for metastatic prostate cancer. Dark grey nodes represent upregulated genes; light grey nodes represent downregulated genes; and diamonds represent microRNAs.
Pathway enrichment analysis for the DEGs in the regulatory network
The DEGs in the regulatory network were enriched in two pathways, including the calcium signaling pathway (EDNRB, ADCY2 and CAMK2D), and thyroid cancer (RET and MYC; Table III).
Table III.
Enriched pathways for the differentially-expressed genes in the regulatory network.
| Term | Description | Count | P-value | Genes |
|---|---|---|---|---|
| hsa04020 | Calcium signaling pathway | 3 | 0.02231 | EDNRB, ADCY2, CAMK2D |
| hsa05216 | Thyroid cancer | 2 | 0.03927 | RET, MYC |
Discussion
The present study identified 306 and 2,073 genes that were differentially expressed in the clinically localized PCa group and the metastatic PCa group, respectively, as compared with the benign prostate group. Of these, 174 genes were differentially expressed in both the clinically localized PCa and metastatic PCa groups.
ADCY2, which encodes adenylate cyclase 2, and CAMK2D, which encodes calcium/calmodulin-dependent protein kinase II δ (29,30), were shown to be enriched in the calcium signaling pathway. Metastasis is the predominant cause of mortality in patients with PCa, and Ca2+ is a crucial regulator of cell migration (31). Elevated intracellular concentrations of Ca2+ may facilitate the metastasis of PCa by triggering the activation of the Akt signaling pathway and promoting PCa cell (PC3) attachment (32). CAMK2D encodes components of the Wnt/β-catenin-signaling pathway, the inhibition of which delays metastatic PCa cell cycle progression and proliferation (33). In the present study, CAMK2D was associated with the cell cycle, which is known to be a critical event in tumor growth and metastasis (34). Furthermore, CAMK2D was observed to be regulated by miR-30. As a tumor suppressor, miR-30 has been shown to be downregulated by oncogenic signals, such as hepatocyte growth factor and epidermal growth factor, in PCa samples (35), and overexpression of miR-30 in PCa cells was reported to suppress the epithelial-to-mesenchymal transition and inhibit cell migration and invasion (36).
ADCY2 was observed to be modulated by miR-182. A previous study demonstrated that ectopic expression of miR-182 in PC3 significantly reduced protein expression levels of GNA13, GNA13-3′-untranslated region (UTR)-reporter activity and extracorporeal invasion of these cells (37). In addition, aberrant overexpression of miR-182 was shown to promote the proliferation, increase the invasion, facilitate the G1/S cell cycle transition and reduce early apoptosis of PC3 cells; and, miR-182 was able to suppress the expression of the NDRG1 tumor suppressor gene by directly targeting the NDRG1 3′-UTR (38). Therefore, CAMK2D and ADCY2 may be involved in the metastasis of PCa via calcium signaling and regulation by miR-30 and miR-182, respectively.
MAF, which was also modulated by miR-182 in the present study, was associated with the regulation of cell proliferation. MAF acts as a macrophage-activating factor and is generated from a precursor protein termed the Gc protein (39). Deglycosylation of the Gc protein prevents its conversion to MAF, inhibiting macrophage activation and resulting in immunosuppression (40). In a previous study, patients with metastatic PCa were administered Gc protein with MAF precursor activity (100 ng/week), and were shown to have serum activity levels of Nagalase equivalent to those of healthy controls, thus suggesting that these patients were tumor-free (41). Furthermore, MAF expression has been associated with the receptor tyrosine kinase, platelet-derived growth factor receptor (PDGFR)-β status (42). In the miRNA-DEG regulatory network, MAF was also modulated by miR-302c, and it has been reported that miR-302c is downregulated in clinical PCa samples (43). In addition, MAF interacted with BCL11A, which was observed to be upregulated in PC3 holoclones (44). Therefore, MAF may have an important role in the metastasis of PCa by interacting with miR-182, miR-302c and BCL11A.
In the present study, the downregulated DEG SH3BP4 was shown to be associated with the cell cycle and was also regulated by miR-182. SH3BP4 encodes SH3-domain binding protein 4 (45). SH3 domains are found in a variety of proteins, including tyrosine kinases, such as Abl and Src, and are involved in endocytosis, intracellular sorting and the cell cycle (46). Another downregulated DEG PCDH17, which encodes protocadherin 17, was shown to interact with miR-182 and miR-30. PCDH17 methylation is a common tumor-specific event in PCa and has been associated with a shorter biochemical recurrence-free survival rate and a reduced overall survival rate of patients with PCa following a radical prostatectomy (47). Therefore, SH3BP4 and PCDH17 may be responsible for the metastasis of PCa via their interactions with miR-182 and/or miR-30. Furthermore, miR-374 was significantly enriched in Gene set B. Previous studies have reported that miR-374 is markedly downregulated in PCa (48,49). Furthermore, miR-374b, which is a subtype of miR-374, has been shown to be downregulated in prostate fluid or serum samples from prostate cancer patients, and thus may serve as a PCa biomarker in clinical diagnosis (50).
In conclusion, the present study identified numerous important DEGs, including ADCY2, CAMK2D, MAF, SH3BP4 and PCDH17, that may be involved in the metastasis of PCa. However, the results of the present study require validation by further experiments, and the molecular mechanisms underlying metastatic PCa require further investigation.
Acknowledgements
The present study was supported by the National Science Foundation of China (grant no. 81470923).
References
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Parkin DM, Steliarova-Foucher E. Estimates of cancer incidence and mortality in Europe in 2008. Eur J Cancer. 2010;46:765–781. doi: 10.1016/j.ejca.2009.12.014. [DOI] [PubMed] [Google Scholar]
- 3.Watahiki A, Wang Y, Morris J, Dennis K, O'Dwyer HM, Gleave M, Gout PW, Wang Y. MicroRNAs associated with metastatic prostate cancer. PLoS One. 2011;6:e24950. doi: 10.1371/journal.pone.0024950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun YX, Schneider A, Jung Y, Wang J, Dai J, Wang J, Cook K, Osman NI, Koh-Paige AJ, Shim H, et al. Skeletal localization and neutralization of the SDF-1 (CXCL12)/CXCR4 axis blocks prostate cancer metastasis and growth in osseous sites in vivo. J Bone Miner Res. 2005;20:318–329. doi: 10.1359/JBMR.041109. [DOI] [PubMed] [Google Scholar]
- 5.Cheng L, Nagabhushan M, Pretlow TP, Amini SB, Pretlow TG. Expression of E-cadherin in primary and metastatic prostate cancer. Am J Pathol. 1996;148:1375–1380. [PMC free article] [PubMed] [Google Scholar]
- 6.Tripathi V, Popescu NC, Zimonjic DB. DLC1 induces expression of E-cadherin in prostate cancer cells through Rho pathway and suppresses invasion. Oncogene. 2013;33:724–733. doi: 10.1038/onc.2013.7. [DOI] [PubMed] [Google Scholar]
- 7.Hu L, Shi Y, Hsu JH, Gera J, Van Ness B, Lichtenstein A. Downstream effectors of oncogenic ras in multiple myeloma cells. Blood. 2003;101:3126–3135. doi: 10.1182/blood-2002-08-2640. [DOI] [PubMed] [Google Scholar]
- 8.Hall A. Rho family GTPases. Biochem Soc Trans. 2012;40:1378–1382. doi: 10.1042/BST20120103. [DOI] [PubMed] [Google Scholar]
- 9.Khafagy R, Stephens T, Hart C, Ramani V, Brown M, Clarke N. In vitro effects of the prenyl transferase inhibitor AZD3409 on prostate cancer epithelial cells. J Clin Oncol. 2004;22:4744. [Google Scholar]
- 10.Toren P, Zoubeidi A. Targeting the PI3K/Akt pathway in prostate cancer: Challenges and opportunities (review) Int J Oncol. 2014;45:1793–1801. doi: 10.3892/ijo.2014.2601. [DOI] [PubMed] [Google Scholar]
- 11.Zhang J, Patel L, Pienta KJ. CC chemokine ligand 2 (CCL2) promotes prostate cancer tumorigenesis and metastasis. Cytokine Growth Factor Rev. 2010;21:41–48. doi: 10.1016/j.cytogfr.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luo JL, Tan W, Ricono JM, Korchynskyi O, Zhang M, Gonias SL, Cheresh DA, Karin M. Nuclear cytokine-activated IKKalpha controls prostate cancer metastasis by repressing Maspin. Nature. 2007;446:690–694. doi: 10.1038/nature05656. [DOI] [PubMed] [Google Scholar]
- 13.Cai H, Smith DA, Memarzadeh S, Lowell CA, Cooper JA, Witte ON. Differential transformation capacity of Src family kinases during the initiation of prostate cancer. Proc Natl Acad Sci USA. 2011;108:6579–6584. doi: 10.1073/pnas.1103904108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salagierski M, Schalken JA. Molecular diagnosis of prostate cancer: PCA3 and TMPRSS2: ERG gene fusion. J Urol. 2012;187:795–801. doi: 10.1016/j.juro.2011.10.133. [DOI] [PubMed] [Google Scholar]
- 15.Saini S, Majid S, Yamamura S, Tabatabai L, Suh SO, Shahryari V, Chen Y, Deng G, Tanaka Y, Dahiya R. Regulatory role of miR-203 in prostate cancer progression and metastasis. Clin Cancer Res. 2011;17:5287–5298. doi: 10.1158/1078-0432.CCR-10-2619. [DOI] [PubMed] [Google Scholar]
- 16.Schaefer A, Jung M, Mollenkopf HJ, Wagner I, Stephan C, Jentzmik F, Miller K, Lein M, Kristiansen G, Jung K. Diagnostic and prognostic implications of microRNA profiling in prostate carcinoma. Int J Cancer. 2010;126:1166–1176. doi: 10.1002/ijc.24827. [DOI] [PubMed] [Google Scholar]
- 17.Gandellini P, Folini M, Zaffaroni N. Towards the definition of prostate cancer-related microRNAs: Where are we now? Trends Mol Med. 2009;15:381–390. doi: 10.1016/j.molmed.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 18.Szczyrba J, Löprich E, Wach S, Jung V, Unteregger G, Barth S, Grobholz R, Wieland W, Stöhr R, Hartmann A, et al. The microRNA profile of prostate carcinoma obtained by deep sequencing. Mol Cancer Res. 2010;8:529–538. doi: 10.1158/1541-7786.MCR-09-0443. [DOI] [PubMed] [Google Scholar]
- 19.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Varambally S, Yu J, Laxman B, Rhodes DR, Mehra R, Tomlins SA, Shah RB, Chandran U, Monzon FA, Becich MJ, et al. Integrative genomic and proteomic analysis of prostate cancer reveals signatures of metastatic progression. Cancer Cell. 2005;8:393–406. doi: 10.1016/j.ccr.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 21.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 22.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy-analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 23.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 24.Verhoeven KJ, Simonsen KL, McIntyre LM. Implementing false discovery rate control: Increasing your power. Oikos. 2005;108:643–647. doi: 10.1111/j.0030-1299.2005.13727.x. [DOI] [Google Scholar]
- 25.Olson CF. Parallel algorithms for hierarchical clustering. Parallel Computing. 1995;21:1313–1325. doi: 10.1016/0167-8191(95)00017-I. [DOI] [Google Scholar]
- 26.Kolde R. Pheatmap: Pretty Heatmaps. Journal. 2012 R package version 0.7. 7. [Google Scholar]
- 27.Huang DW, Sherman BT, Tan Q, Collins JR, Alvord G, Roayaei J, Stephens R, Baseler MW, Lane HC, Lempicki RA. The DAVID Gene functional classification tool: A novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007;8:R183. doi: 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Visel A, Alvarez-Bolado G, Thaller C, Eichele G. Comprehensive analysis of the expression patterns of the adenylate cyclase gene family in the developing and adult mouse brain. J Comp Neurol. 2006;496:684–679. doi: 10.1002/cne.20953. [DOI] [PubMed] [Google Scholar]
- 30.Hagemann D, Bohlender J, Hoch B, Krause EG, Karczewski P. Expression of Ca2+/calmodulin-dependent protein kinase II delta-subunit isoforms in rats with hypertensive cardiac hypertrophy. Mol Cell Biochem. 2001;220:69–76. doi: 10.1023/A:1010899724222. [DOI] [PubMed] [Google Scholar]
- 31.Prevarskaya N, Skryma R, Shuba Y. Calcium in tumour metastasis: New roles for known actors. Nat Rev Cancer. 2011;11:609–618. doi: 10.1038/nrc3105. [DOI] [PubMed] [Google Scholar]
- 32.Liao J, Schneider A, Datta NS, McCauley LK. Extracellular calcium as a candidate mediator of prostate cancer skeletal metastasis. Cancer Res. 2006;66:9065–9073. doi: 10.1158/0008-5472.CAN-06-0317. [DOI] [PubMed] [Google Scholar]
- 33.Rajan P, Sudbery IM, Villasevil ME, Mui E, Fleming J, Davis M, Ahmad I, Edwards J, Sansom OJ, Sims D, et al. Next-generation sequencing of advanced prostate cancer treated with androgen-deprivation therapy. Eur Urol. 2014;66:32–39. doi: 10.1016/j.eururo.2013.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Whitfield ML, Sherlock G, Saldanha AJ, Murray JI, Ball CA, Alexander KE, Matese JC, Perou CM, Hurt MM, Brown PO, Botstein D. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol Biol Cell. 2002;13:1977–2000. doi: 10.1091/mbc.02-02-0030.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.White R, Kung H. miR-30 as a tumor suppressor connects EGF/Src signal to ERG and EMT. Oncogene. 2014;33:2495–2503. doi: 10.1038/onc.2013.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kao C, Martiniez A, Shi X, Yang J, Evans C, Dobi A, Devere White R, Kung H. miR-30 as a tumor suppressor connects EGF/Src signal to ERG and EMT. Oncogene. 2014;33:2495–2503. doi: 10.1038/onc.2013.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rasheed SA, Teo CR, Beillard EJ, Voorhoeve PM, Casey PJ. MicroRNA-182 and microRNA-200a control G-protein subunit α-13 (GNA13) expression and cell invasion synergistically in prostate cancer cells. J Biol Chem. 2013;288:7986–7995. doi: 10.1074/jbc.M112.437749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu R, Li J, Teng Z, Zhang Z, Xu Y. Overexpressed microRNA-182 promotes proliferation and invasion in prostate cancer PC-3 cells by down-regulating N-myc downstream regulated gene 1 (NDRG1) PLoS One. 2013;8:e68982. doi: 10.1371/journal.pone.0068982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nagasawa H, Uto Y, Sasaki H, Okamura N, Murakami A, Kubo S, Kirk KL, Hori H. Gc protein (vitamin D-binding protein): Gc genotyping and GcMAF precursor activity. Anticancer Res. 2005;25:3689–3695. [PubMed] [Google Scholar]
- 40.Yamamoto N, Naraparaju VR, Asbell SO. Deglycosylation of serum vitamin D3-binding protein leads to immunosuppression in cancer patients. Cancer Res. 1996;56:2827–2831. [PubMed] [Google Scholar]
- 41.Yamamoto N, Suyama H, Yamamoto N. Immunotherapy for prostate cancer with Gc protein-derived macrophage-activating factor, GcMAF. Transl Oncolo. 2008;1:65–72. doi: 10.1593/tlo.08106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sharad S, Srivastava A, Ravulapalli S, Parker P, Chen Y, Li H, Petrovics G, Dobi A. Prostate cancer gene expression signature of patients with high body mass index. Prostate Cancer Prostatic Dis. 2011;14:22–29. doi: 10.1038/pcan.2010.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Coppola V, De Maria R, Bonci D. MicroRNAs and prostate cancer. Endocr Relat Cancer. 2010;17:F1–F17. doi: 10.1677/ERC-09-0172. [DOI] [PubMed] [Google Scholar]
- 44.Zhang K, Waxman DJ. PC3 prostate tumor-initiating cells with molecular profile FAM65Bhigh/MFI2low/LEF1low increase tumor angiogenesis. Mol Cancer. 2010;9:319. doi: 10.1186/1476-4598-9-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dunlevy JR, Koppelman ED, Kolberg JB. The expression of a SH3BP4-related protein in retinal cells. Invest Ophthalmol Vis Sci. 2005;46:2996–2996. [Google Scholar]
- 46.Dunlevy JR, Berryhill BL, Vergnes JP, SundarRaj N, Hassell JR. Cloning, chromosomal localization and characterization of cDNA from a novel gene, SH3BP4, expressed by human corneal fibroblasts. Genomics. 1999;62:519–524. doi: 10.1006/geno.1999.5994. [DOI] [PubMed] [Google Scholar]
- 47.Lin YL, Xie PG, Wang L, Ma JG. Aberrant methylation of protocadherin 17 and its clinical significance in patients with prostate cancer after radical prostatectomy. Med Sci Monit. 2014;20:1376–1382. doi: 10.12659/MSM.891247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tang X, Tang X, Gal J, Kyprianou N, Zhu H, Tang G. Detection of microRNAs in prostate cancer cells by microRNA array. Methods Mol Biol. 2011;732:69–88. doi: 10.1007/978-1-61779-083-6_6. [DOI] [PubMed] [Google Scholar]
- 49.Ma S, Chan YP, Kwan PS, Lee TK, Yan M, Tang KH, Ling MT, Vielkind JR, Guan XY, Chan KW. MicroRNA-616 induces androgen-independent growth of prostate cancer cells by suppressing expression of tissue factor pathway inhibitor TFPI-2. Cancer Res. 2011;71:583–592. doi: 10.1158/1538-7445.AM2011-583. [DOI] [PubMed] [Google Scholar]
- 50.He HC, Han ZD, Dai QS, Ling XH, Fu X, Lin ZY, Deng YH, Qin GQ, Cai C, Chen JH, et al. Global analysis of the differentially expressed miRNAs of prostate cancer in Chinese patients. BMC Genomics. 2013;14:757. doi: 10.1186/1471-2164-14-757. [DOI] [PMC free article] [PubMed] [Google Scholar]