Summary
According to the hygiene hypothesis, the increasing incidence of autoimmune diseases in western countries may be explained by changes in early microbial exposure, leading to altered immune maturation. We followed gut microbiome development from birth until age three in 222 infants in Northern Europe, where early-onset autoimmune diseases are common in Finland and Estonia but less prevalent in Russia. We found that Bacteroides species are lowly abundant in Russians but dominate in Finnish and Estonian infants. Therefore their Lipopolysaccharide (LPS) exposures arose primarily from Bacteroides rather than from Escherichia coli which is a potent innate immune activator. We show that Bacteroides LPS is structurally distinct from E. coli LPS and inhibits innate immune signaling and endotoxin tolerance; furthermore, unlike LPS from E. coli, B. dorei LPS does not decrease incidence of autoimmune diabetes in non-obese diabetic mice. Early colonization by immunologically silencing microbiota may thus preclude aspects of immune education.
Introduction
According to the hygiene hypothesis, early exposure to specific microorganisms and parasites in infancy benefits immune system development and confers protection against allergic and autoimmune diseases. Indeed, several studies have demonstrated a global gradient in the incidence of type 1 diabetes (T1D), multiple sclerosis, and other autoimmune diseases in association with improved sanitation and decreased incidence of early childhood infections (reviewed in (Bach, 2002; Bach and Chatenoud, 2012)). Similarly, rates of asthma and allergy are reduced in children exposed to a farm environment (von Mutius and Vercelli, 2010). One explanation for this effect posits that hygienic measures meant to prevent infectious disease by removing microbes from individuals’ living environments may in turn alter the indigenous intestinal microbiota, eliminating microbes important for the education of the immune system (Bach, 2002; von Mutius and Vercelli, 2010). Accordingly, studies in mouse models have shown that early colonization with a protective microbiota can diminish the risk of autoimmune diabetes development in genetically susceptible animals (Markle et al., 2013). Likewise, the composition of the microbiota can protect mice from allergies (Stefka et al., 2014). However, the distinction between beneficial and harmful microbial communities and the functional mechanisms underlying their effects are still poorly understood.
A microcosm of the global gradient in immune disease incidence occurs at the border between Finland and Russian Karelia, where there is a 2–6-fold higher incidence of allergies (Seiskari et al., 2007) and a 5–6-fold higher incidence of T1D and other autoimmune disorders (Kondrashova et al., 2008a; Kondrashova et al., 2008b) in Finland relative to Russian Karelia. In nearby Estonia, coinciding with economic development and improvement in living standards, the incidence of T1D and atopy has been transitioning in recent decades from rates similar to those of Russian Karelia toward those of Finland (Teeaar et al., 2010; Voor et al., 2005). Using these three populations as a “living laboratory,” the DIABIMMUNE study (http://www.diabimmune.org/) recruited a total of ~1,000 infants from Espoo (Finland), Petrozavodsk (Russia), and Tartu (Estonia). The infants were followed from birth to three years of age by monthly stool sampling along with collection of extensive clinical metadata. The cohort thus provides the largest longitudinal functional profile of the infant gut microbiome in relation to immune-mediated diseases to date, representing an unprecedented opportunity to understand the microbial ecology and molecular mechanisms potentially underlying the hygiene hypothesis (Peet et al., 2012).
To characterize host-microbe immune interactions contributing to autoimmunity and allergy, we performed a longitudinal metagenomic characterization in 785 gut microbial communities from infants in the DIABIMMUNE cohort selected for this study (Figures 1A and 1B). Using strain-level microbial identification, we uncovered substantial differences in the composition, diversity, and stability of the early gut microbiome in Russian, Finnish, and Estonian children. We further quantified the functional potential of these microbial communities, stratifying gene families and pathways across their contributing organisms. This extensive dataset constitutes a valuable resource for infant gut microbiome investigations and is accessible through the DIABIMMNUNE Microbiome website at http://pubs.broadinstitute.org/diabimmune.
Figure 1. DIABIMMUNE Cohort.
(A) Locations of cities and countries where DIABIMMUNE infants were screened and samples were collected.
(B) Selected within-cohort statistics and stool sample collection schedule (monthly stool sampling until three years of age). Numbers next to stool samples reflect the number of samples collected in 6-month time windows. Within-cohort distribution of HLA conferred risk for autoimmunity is shown (see Table S1 for corresponding HLA allele identities), as well as prevalence of T1D-associated autoantibody seropositivity, egg allergy, and milk allergy at year 2. For T1D autoantibody seropositivity, n = 291 serum samples from 73 infants for Finns, n = 235 serum samples from 72 infants for Estonians, and n = 118 serum samples from 54 infants for Russians. For egg allergy, n = 72 for Finns, n = 51 for Estonians, and n = 24 for Russians. For milk allergy, n = 72 for Finns, n = 46 for Estonians, and n = 24 for Russians.
(C) Analysis workflow highlighting important steps in metagenomic data analysis and mechanistic experiments.
In this work, we bridge deep human longitudinal metagenomic analysis with the identification of novel molecular immune mechanisms (Figure 1C). Upon extensive analysis of this dataset, we discovered that Finnish and Estonian infants harbored both a greater proportion of Bacteroides species and an enrichment in lipopolysaccharide (LPS) biosynthesis-encoding genes, when compared to Russian infants. Our investigations revealed that these Bacteroides species produced a structurally and functionally distinct form of LPS; this LPS differed from the dominating form of LPS in the early Russian microbiome, which was almost exclusively derived from E. coli. We further demonstrated experimentally that LPS from Bacteroides dorei, previously associated with T1D pathogenesis (Davis-Richardson et al., 2014), harbored tetra-and penta-acylated lipid A structures, as opposed to the hexa-acylated lipid A seen in E. coli. Furthermore, B. dorei LPS inhibited immune stimulation and inflammatory cytokine responses to E. coli LPS in human cells. These findings suggest that differences in microbiota-derived LPS may preclude aspects of immune education in Finnish and Estonian children, uncovering one potential mechanism linking the human gut microbiome to susceptibility to immune diseases.
Results
Study Cohort
A subcohort of 74 infants from each country were selected on the basis of similar HLA risk class distribution and matching gender (Figures 1A and 1B and Table S1). For each infant, three years of monthly stool samples, questionnaires regarding breastfeeding, diet, allergies, infections, family history, use of drugs, clinical examinations, and laboratory assays were collected. In accordance with the recruitment criteria for the DIABIMMUNE cohort, all subjects had increased HLA-conferred susceptibility to autoimmunity (Figure 1B) (Larizza et al., 2012; Sollid and Thorsby, 1993). Although these children were followed only until three years of age and it was therefore unlikely to see indications of allergic disease or autoimmunity, laboratory examinations revealed a high prevalence of allergen-specific sensitization and seropositivity for T1D-associated antibodies in Finnish and Estonian infants (Figure 1B, bottom).
We also observed a gradient in T1D autoantibody seropositivity within the cohort with higher prevalence of T1D autoantibodies in Finland. The number of infants that tested seropositive for one or more T1D-associated autoantibodies were 16 for Finland, 14 for Estonia, and 4 for Russia. Other studies in an older population (7–15 years of age) have shown that children in Russian Karelia display a substantially higher microbial exposure than their Finnish peers, as denoted by higher prevalence of antibodies against Helicobacter pylori (15-fold), Toxoplasma gondii (5-fold), and hepatitis A virus (12-fold) (Seiskari et al., 2007). This increased pathogen exposure in older children suggests an overall higher exposure rate to diverse microorganisms, possibly due to higher hygiene levels in urban Finland.
Regional Trends in the Gut Microbiota
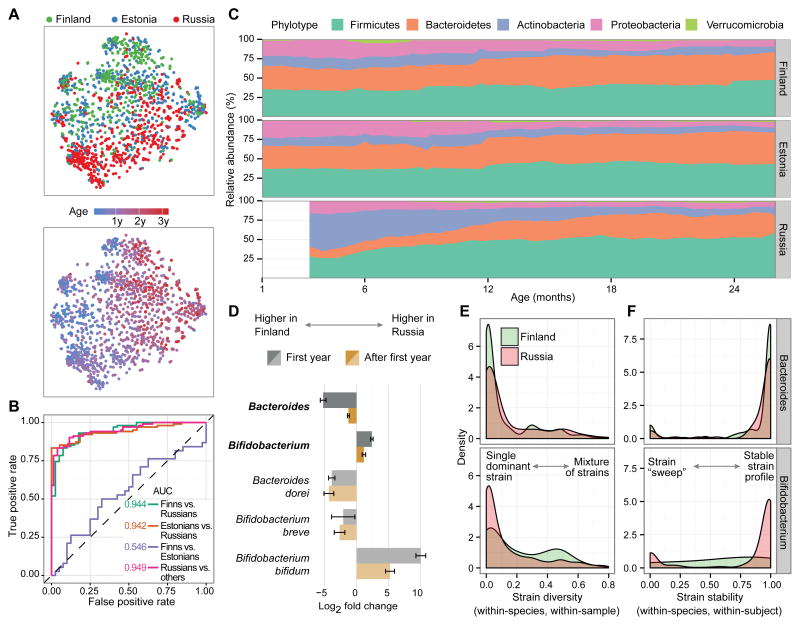
To generate an overview of the composition of the gut microbiota throughout the first three years of life, we sequenced the V4 region of the 16S rRNA gene from 1,584 samples (Figure S1) and observed several strong high-level trends within this cohort. Principal coordinate plots (Figure 2A) showed that, besides age, country was the major source of variation, particularly during the first year of life. To further confirm separability between countries, we trained a set of random forest classifiers using genus-level data from samples collected between 170 and 260 days of age. The classifier was able to predict country with high classification accuracy (AUC = 0.944 for Finns versus Russians) (Figure 2B). Classification was least accurate between Finns and Estonians (AUC = 0.546) suggesting that early microbial profiles were fairly homogenous in these two countries. Differences between the Russian samples compared to Finnish and Estonian samples were already evident at phylum-level composition (Figure 2C), represented by two distinct hallmarks. First, Finnish and Estonian children had higher levels of Bacteroidetes throughout the three-year period (FDR corrected p = 5.4 × 10−15, see Supplemental Experimental Procedures). Second, Russians had higher levels of the phylum Actinobacteria during the first year of life (FDR corrected p = 0.014). The latter difference dissipated over time and was no longer significant after two years of age. The abundance of the phylum Bacteroidetes correlated with serum insulin autoantibody (IAA) levels both within Finland (p = 0.017) and cohort-wide (p = 0.0020; Figure S2 and Supplemental Experimental Procedures). We conducted extensive testing of associations between the metadata and taxonomic groups using MaAsLin, a linear modeling tool adapted for microbial community data (Morgan et al., 2012). Hence, all reported country-level differences were corrected for all major confounding effects, such as birth mode, breastfeeding and other dietary factors, antibiotics use, and age (see Supplemental Experimental Procedures). Table 1 highlights selected associations between the metadata and microbiota. A comprehensive list of results including taxonomic differences between countries, taxonomic alterations associated with allergen-specific IgEs, and microbial changes associated with other collected metadata can be accessed at http://pubs.broadinstitute.org/diabimmune.
Figure 2. Differences in Microbial Ecology between Countries in Early Infancy.
(A) Principal coordinate analysis plots of DIABIMMUNE 16S samples, colored by country (top) and age (bottom). Each circle represents an individual stool sample (n = 1,584).
(B) ROC curves for pairwise random forest classifiers predicting country based on 16S genus data using samples collected between 170 and 260 days of age.
(C) Average phylum-level composition of DIABIMMUNE samples during the first two years of age.
(D) Genus-level (darker colors) and species-level (lighter colors) bootstrapped mean log2 fold changes and their standard deviations between Finnish and Russian gut microbiota during the first year and after.
(E and F) Strain-level diversity (E) and stability (F) in Bacteroides and Bifidobacterium species. Diversity and stability distributions for Bifidobacterium species are significantly different between the Finnish and Russian populations (two-sample Kolmogorov-Smirnov test; p = 5.0 × 10−4 and p = 1.5 × 10−6, respectively).
See also Figures S2, S3, and S4.
Table 1.
Associations between Metadata and Microbiota
| Increased | Decreased | |
|---|---|---|
|
|
||
| T1D AAB seropositivity | Rothia (g) | Bilophila (g) * |
| Gemellaceae (f) * | Sutterella (g) * | |
|
| ||
| Cesarean section | Firmicutes (p) ** | Bacteroidetes (p) ** |
| Eubacterium (g) ** | Bacteroides (g) ** | |
| Ruminococcus (g) * | Faecalibacterium prausnitzii * | |
|
| ||
| Antibiotics | Deltaproteobacteria (c) * | Gammaproteobacteria (c) ** |
| Bilophila (g) * | Clostridium (g) ** | |
| Haemophilus (g) ** | ||
|
| ||
| Breast feeding | Actinobacteria (p) ** | Blautia (g) ** |
| Bifidobacterium (g) ** | Oscillospira (g) ** | |
| Lactobacillus (g) ** | ||
|
| ||
| Baby formula | Citrobacter (g) ** | Streptococcus (g) |
| Veillonella (g) | ||
|
| ||
| Cow’s milk | Lactococcus (g) ** | Staphylococcus (g) |
| Collinsella (g) ** | ||
| Lactococcus lactis | ||
|
| ||
| Wheat | Bifidobacterium pseudocatenulatum ** | Staphylococcus (g) ** |
|
| ||
| Barley | Betaproteobacteria (c) ** | |
| Sutterella (g) ** | ||
|
| ||
| Oat | Lachnospiraceae (f) ** | Enterobacteriales (o) ** |
| Clostridium bartlettii * | ||
|
| ||
| Corn | Blautia (g) ** | Ruminococcaceae (f) |
|
| ||
| Rice | Epsilonproteobacteria (c) ** | |
| Prevotella (g) * | ||
|
| ||
| Eggs | Cyanobacteria (p) * | |
| Ruminococcus (g) ** | ||
| Lactococcus (g) | ||
|
| ||
| Vegetables | Clostridia (c) | Holdemania (g) ** |
| Lachnospiraceae (f) ** | ||
|
| ||
| Root vegetables | Ruminococcus (g) ** | Betaproteobacteria (c) |
| Coprococcus catus ** | Bacteroides (g) | |
|
| ||
| Meat | Proteobacteria (p) | Firmicutes (p) ** |
| Erysipelotrichaceae (f) ** | Coprocuccus (g) ** | |
| Bacteroides (g) * | ||
|
| ||
| Fish | Parabacteroides (g) | |
|
| ||
| Soy | Alphaproteobacteria (c) ** | Holdemania (g) ** |
| Lachnospira (g) * | ||
| Clostridium clostridioforme | ||
Microbial taxa that are associated with T1D autoantibody seropositivity, caesarean section, intake of antibiotics, breastfeeding, and other dietary compounds. Left column shows taxa that are increased and right column shows taxa that are decreased in each association. All findings have FDR-corrected p < 0.1.
p < 0.01,
p < 0.001, p = phylum, c = class, o = order, f = family, g = genus.
The diversity of the microbiota within individual samples (alpha diversity) increased with age (Figure S3A) as the microbiota developed toward an adult composition (Koenig et al., 2011). However, Russians displayed a significantly less diverse microbiota compared to Finns and Estonians during the first year (Figure S3B). This difference could be explained by the two-fold overrepresentation of the phylum Actinobacteria and the genus Bifidobacterium in the Russians over that time period (Figures 2C and 2D). Lastly, we also uncovered differences in stability within taxa between the countries. These differences were particularly evident when comparing samples collected during early and late time windows (Figure S3C and S3D). Russians had an overall more plastic microbiota during the first three years of life, with the exception of the most dominant genus Bifidobacterium in the early time window. In contrast, the phylum Bacteroidetes and the genus Bacteroides were more stable in Finns and Estonians throughout the entire observation period. Taken together, we uncovered strong global differences between the Russian versus Finnish and Estonian microbiota, with the largest differences occurring in the first year and dissipating during the second and third years.
Species- and Strain-Level Microbial Dynamics
To obtain a more complete and higher-resolution taxonomic view of the infants’ microbiome, we performed deep whole-genome shotgun metagenomic sequencing on a representative subset of 785 samples (Figure S1). We first investigated the metagenomic reads for their detailed taxonomic composition down to the species level using MetaPhlAn 2.2 (Metagenomic Phylogenetic Analysis 2.2) (Truong et al., 2015) and observed multiple differentially abundant species in the Bacteroides and Bifidobacterium genera between Finland and Russia (Figure 2D). Notably, the Bacteroides species with the largest fold change between Finns and Russians was B. dorei, which has been previously associated with T1D pathogenesis (Davis-Richardson et al., 2014). We confirmed the validity of the metagenomics data by running quantitative PCR (qPCR) on DNA from 85 stool samples. Interpolated absolute abundances of B. dorei and E. coli species were in good agreement with absolute abundances predicted by the metagenomics data when total bacterial mass was estimated using universal 16S primers (Figure S3E and Supplemental Experimental Procedures).
We next analyzed the metagenomics data at the strain level using ConStrains, a recently developed strain haplotyping tool, and evaluated the diversity and stability of the infant microbiota (Luo et al., 2015). In 60% of all strain profiles, communities were composed of species with a single dominant strain (> 90% within-species abundance), as reflected in low within-species, within-sample haplotype diversity scores (Figure S4A). However, species in some genera, such as Faecalibacterium and Veillonella, had bimodal haplotype diversity distributions, indicative of more complex strain compositions. Moreover, strain diversity had a tendency to increase with age (Figure S4B). Analysis of the strain stability over time revealed that species tended to either (i) remain stable, maintaining their single dominant strain over time, or (ii) experience a strain “sweep,” in which the original dominant strain was replaced by a new dominant strain (Figure S4C). We observed an inverse correlation between the longitudinal distance of the samples and the corresponding strain stability (Figure S4D). When comparing strain stability with average diversity of the compared samples, we saw an inverse correlation, indicating that less diverse strain profiles (i.e., single dominant strain behavior) tended to be more stable compared to more diverse strain profiles (Figure S4E). Within the genera of interest, we observed that Bifidobacterium species failed to establish stable single-strain communities in Finnish children, as shown by a more evenly distributed strain diversity and stability compared to Russians (Figures 2E and 2F). In contrast, Bacteroides species (when present) tended to establish stable, single-strain compositions in both Finns and Russians (Figures 2E and 2F).
Differential Microbial Functions between Populations
To survey the functional and metabolic consequences of the taxonomic differences between countries, we next analyzed the metagenomic sequences for their genomic functional potential using HUMAnN2 (Abubucker et al., 2012) and linked quantified gene abundances (reads per kilobase per million reads, RPKMs) to gene ontology (GO) terms. As observed for microbial diversity, functional diversity of the microbiome also started with a less complex composition in Russians but developed to reach greater diversity by the end of the three-year period (Figure S5A). We identified multiple GO categories with significantly different abundances between Finns and Russians in both the early (first year) and late (after first year) time windows (Figure 3A). For instance, siderophore-related functions, which include iron scavenging as well as virulence-related functionalities, were higher in Finnish infants, possibly reflecting an increase in pathobiont organisms in Finland. A comprehensive list of differential categories between the two countries is shown in Figure S5B and Table S2.
Figure 3. Functional Differences, HMO Utilization, and Lipid A Biosynthesis.
(A) Bootstrapped mean log2 fold changes and their standard deviations in the functional categories with the largest differences between Finnish and Russian children.
(B) Mean human milk oligosaccharide utilization gene abundance across the three countries within the first year of life, stratified by taxonomic origin of each gene (“conserved” genes were too highly conserved to confidently assign to a unique genus).
(C) Lipid A biosynthesis pathway normalized read counts (RPKM) per sample (n = 785) and a linear fit per country. (D and E) Mean relative abundances of 15 species with the largest contributions to lipid A biosynthesis signal
(D), and their relative contributions (E) to the signal in all samples within each country.
Glycolytic functions were differentially abundant between the two populations (Figure 3A), which led us to computationally investigate differences in milk oligosaccharide metabolism. The gut microbiome composition within the first year is largely shaped by milk, the sole nutrient source available to infants, whether from breast- or bottle-feeding (reviewed in (Sela and Mills, 2010)). The Bifidobacterium and Bacteroides genera are the two main groups of human milk oligosaccharide (HMO)-metabolizing bacteria (Marcobal et al., 2011). Within Bifidobacterium, B. bifidum and B. longum (predominant in Russians) are capable of metabolizing HMOs, whereas B. breve (present in Finns) is incapable of metabolizing intact HMOs, though it readily utilizes monosaccharides liberated from HMOs (Locascio et al., 2009). This observation led us to hypothesize that HMO metabolism in Finnish and Estonian children is performed by Bacteroides species, whereas it is performed by B. bifidum and B. longum in Russians. Indeed, by analyzing the taxonomic origin of genes belonging to a bona fide HMO gene cluster (Sela et al., 2008), we showed that although the average abundance of HMO utilization genes is approximately equal across the three countries (mean ± SD in RPKM: Finland 460 ± 372, Estonia 462 ± 331, Russia 504 ± 469), most of the genes are conferred by Bifidobacterium in Russians and Bacteroides in Finns and Estonians (Figures 3B and S5C). We note that the higher abundance of B. bifidum in Russians is not a result of increased breastfeeding; Finnish infants were breastfed for a longer period than Russians on average (mean ± SD breastfeeding / days: Finland 268 ± 149, Estonia 307 ± 217, Russia 199 ± 165).
Most significantly, we found that GO terms related to LPS functions, LPS biosynthetic process (GO:0009103) and lipid A biosynthetic process (GO:0009245), showed a striking difference in abundance between countries (Figures 3A and 3C), indicating that microbial communities in Finnish and Estonian subjects produced more LPS. This molecule is of particular interest because it elicits a strong immune response in mammalian cells (Cullen et al., 2015). When deconvoluting the species contributing to biosynthesis of lipid A, the subunit responsible for the immunostimulatory properties of LPS, we made two key observations. In all three countries, E. coli was a major contributor to lipid A biosynthesis, but in Finland and Estonia a number of other bacterial species contributed to the lipid A biosynthesis potential, many of which belong to the genus Bacteroides (Figures 3D and 3E). LPS subtypes derived from Bacteroides species have been shown to exhibit lower endotoxicity relative to LPS isolated from other enteric bacteria (Hofstad et al., 1977). This finding prompted us to examine whether there was a difference in immunogenicity of the LPS subtypes derived from the predominant species of the three populations.
Contrasting Immunogenicity of LPS Subtypes
Inter-species differences in LPS structure are associated with alterations in their capacity to elicit an innate immune response (Whitfield and Trent, 2014). Specifically, the lipid A domain of LPS is responsible for immune signaling through recognition and activation of the TLR4 complex (Kim et al., 2007); as such, structural changes in lipid A impact recognition by TLR4 and influence multiple facets of microbial ecology (Cullen et al., 2015; Whitfield and Trent, 2014). We purified LPS (see below) and used matrix-assisted laser desorption/ionization-time of flight mass spectrometry to examine the structure of the lipid A domain of two bacterial species (type strains): E. coli as a representative of the most common immunostimulatory lipid A structure, and B. dorei which was the most differentially abundant Bacteroides species between the countries (Figures 2D and 3D). Lipid A extracted from E. coli produced a predominant peak at a mass-to-charge ratio (m/z) of 1798.3, consistent with the published [M-H]− ion structure of E. coli lipid A (Needham et al., 2013) carrying two phosphate groups and six acyl chains (predicted exact mass: 1797.2 m/z) (Figure 4A). Lipid A extracted from B. dorei produced two predominant peaks at m/z 1690.9 and 1436.2, consistent with the [M-H]− ion structures with one phosphate group and four and five acyl chains, respectively (predicted exact mass: 1689.2 and 1435.0 m/z) (Figure 4B).
Figure 4. Structures of LPS Molecules and Impact on Tolerogenic Function.
MALDI-TOF MS analysis of lipid A purified from E. coli (A) and B. dorei (B). Representative structures are shown as insets with predicted exact mass.
See also Figure S6.
In order to ensure that LPS from our B. dorei type strain was representative of clinical samples, we isolated B. dorei strains from stool samples of six healthy donors for comparative LPS structural analysis. These data revealed identical LPS structural features across all B. dorei isolates (Figures S6A and S6B). Thus, our findings regarding B. dorei LPS structure and function are likely to be recapitulated in patients.
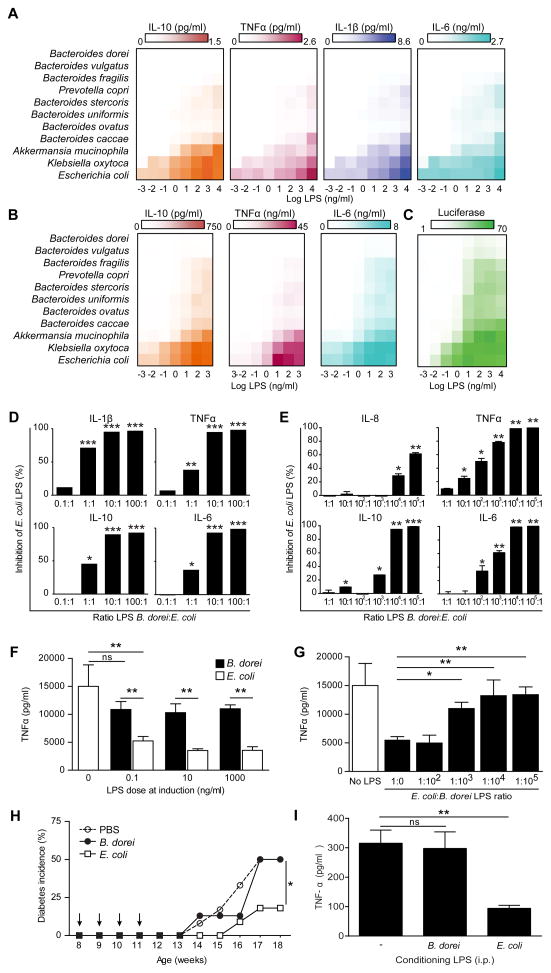
Extensive lipid A structure-function studies have shown that the number of acyl chains is a strong determinant of immune activation by LPS (Hajjar et al., 2002; Needham et al., 2013), and that penta- and tetra-acylated lipid A structures elicit reduced TLR4 responses (Herath et al., 2011). In order to understand the consequences of the structural differences between the LPS subtypes, we assessed the immunogenicity of LPS derived from the bacterial species contributing to the LPS load in our samples (see Fig. 3E). Of the 15 strongest contributors, we were able to purify LPS from 11 type strains listed in Table S3. We first used the LPS purified from these strains to stimulate primary human peripheral blood mononuclear cells (PBMCs), which contain LPS-responsive cell types similar to those present in the gut and are thus a common proxy for mucosal leukocytes (Ardeshir et al., 2014; Sokol et al., 2008). LPS derived from E. coli produced a strong response as measured by the production of the NFκB-dependent cytokines IL-10, TNFα, IL-1β, and IL-6 in primary PBMCs (Figures 5A and S7A), whereas LPS derived from B. dorei failed to elicit any response regardless of dose. Notably, LPS derived from all analyzed members of the phylum Bacteroidetes (Bacteroides species and Prevotella copri) also showed a severely impaired capacity to elicit the production of these inflammatory cytokines. We then measured cytokine production in human monocyte-derived dendritic cells upon stimulation with LPS from these same strains and obtained similar results (Figures 5B and S7B). Consistent with assays in primary cells, E. coli-derived LPS elicited high levels of luciferase activity in TLR4-NFκB reporter cells, whereas Bacteroides species failed to induce an inflammatory signal in these cells (Figures 5C and S7C).
Figure 5. Immunostimulatory Properties of LPS from Different Bacterial Strains.
(A) Mean cytokine production in PBMCs stimulated with indicated doses of LPS as assessed by cytokine bead array.
(B) Mean cytokine production in monocyte-derived dendritic cells stimulated with indicated doses of LPS.
(C) Reporter cells expressing human TLR4 were stimulated with LPS from indicated bacterial strains for 6 h and NFκB activity was measured by luciferase activity. Activity is expressed as percent of maximum luciferase signal.
(D and E) Inhibition of E. coli LPS-induced PBMC (D) or monocyte-derived dendritic cells (E) cytokine production by additional doses of LPS from B. dorei. Inhibition of the cytokine production is expressed as measured upon stimulation with E. coli LPS alone.
(F) Induction of endotoxin tolerance by LPS from E. coli or B. dorei in primary human monocytes as assessed by cytokine bead array. Bars show TNFα concentration in monocyte supernatants upon 24 h restimulation with zymosan as assessed by cytokine bead array.
(G) Inhibition of E. coli-driven endotoxin tolerance induction in human monocytes by B. dorei LPS.
(H) Impact of E. coli- or B. dorei-derived LPS exposure on diabetes incidence in NOD mice. Mice were injected i.p. once a week (arrows) with LPS from E. coli (n = 9 mice) or B. dorei (n = 12 mice). Blood glucose was monitored weekly.
(I) Induction of endotoxin tolerance in NOD mice. The mice (n = 5 per group) were injected i.p. with LPS purified from E. coli or B. dorei. The splenocytes were isolated after 24 h and restimulated in vitro with zymosan. Bars show TNFα concentration assessed by cytokine bead array after 24 h. In vitro data are representative of three or more independent experiments and are presented as mean (and SD) of triplicate assessments. *p < 0.05, **p < 0.005 by Student’s t-test compared to E. coli stimulation (D and E), E. coli LPS treatment alone (F and G) or PBS treatment (I), or by ANOVA for diabetes incidence (H).
Our metagenomics analyses revealed that E. coli and B. dorei LPS often co-occur in the gut of Finnish and Estonian infants. In order to study possible interactions between these LPS subtypes, we used a base dose of E. coli LPS while co-treating human primary immune cells with B. dorei LPS at increasing ratios. We then measured changes in the production of inflammatory cytokines with respect to baseline E. coli LPS stimulation. Cytokine production was inhibited by B. dorei LPS in primary human PBMCs (Figure 5D) and in monocyte-derived dendritic cells (Figure 5E). Notably, we observed maximal inhibition in PBMCs at a ratio of 10:1 B. dorei:E. coli LPS, which corresponds to the computational prediction of the ratio between inhibitory and stimulatory LPS typical for IAA-seropositive infants (Figure S2). Similar to cytokine production in PBMCs, NFκB-luciferase activity was inhibited by B. dorei LPS in a dose-dependent manner (Figure S7D). We also obtained similar results when examining cord blood mononuclear cells (Figure S7E and S7F), suggesting that our observations reflect the reaction of the naïve immune system of infants. Our results show that B. dorei LPS acts as an inhibitor of immune stimulation by E. coli-derived LPS, with a potency that is concordant with ratios of the LPS subtypes observed in vivo.
It has been proposed that lipid A phosphorylation pattern contributes to LPS immunogenicity. However, LPS from a mutant strain of B. thetaiotaomicron (ΔLpxF), whose lipid A structure is identical to that of B. dorei but harbors two phosphate groups similar to E. coli (Cullen et al., 2015), did not increase LPS immunogenicity or alter its inhibitory capacity (Figures S7G and S7H). This suggests that lipid A phosphorylation status is unlikely to be the underlying mechanism of our observations.
Stimulation of immune cells with LPS induces a temporary refractory state to a repeated immune challenge, a phenomenon known as endotoxin tolerance (Watson and Kim, 1963). This mechanism of immunosuppression was originally described in sepsis, but is thought to underlie multiple other physiological contexts of innate immune unresponsiveness, such as the immune protective effect conferred by microbial exposure suggested by the hygiene hypothesis (Biswas and Lopez-Collazo, 2009). We assessed the potency of E. coli and B. dorei LPS subtypes to induce endotoxin tolerance in primary human monocytes. Initial exposure to E. coli LPS prevented TNFα production upon restimulation at all conditioning doses tested (Figure 5F). In contrast, B. dorei LPS conditioning did not abrogate cytokine production in these cells even at the highest concentrations, corresponding to a potency at least 4 orders of magnitude lower than E. coli LPS. Hence, the LPS produced by B. dorei failed to induce protective endotoxin tolerance. Finally, the addition of B. dorei LPS to E. coli LPS during the endotoxin tolerance induction phase prevented the establishment of endotoxin tolerance by E. coli LPS in a dose-dependent manner (Figure 5G), suggesting that the presence of B. dorei in the infant gut could prevent the establishment of protective immune tolerance by E. coli LPS.
To demonstrate the relevance of LPS-driven immune suppression to the development of autoimmunity in vivo, we assessed the impact of different LPS subtypes on diabetes development in the Non-Obese Diabetic (NOD) mouse model of T1D. Intraperitoneal (i.p.) injection of E. coli LPS resulted in a delayed onset and reduced overall incidence of disease, as measured by blood glucose levels (Figure 5H). In contrast, B. dorei LPS did not delay the onset of diabetes, nor did it decrease incidence compared to the mock-injected group (PBS). Interestingly, as shown in Figure 5I, we also found that splenocytes isolated from NOD mice 24 hours after i.p. E. coli LPS injection were hyporesponsive to further in vitro innate immune stimulation, whereas B. dorei LPS did not alter the response. This shows that E. coli LPS, but not B. dorei LPS, can induce endotoxin tolerance in vivo in NOD mice. These results suggest that exposure to immunostimulatory LPS can contribute to the protection from immune-mediated diseases by modulating the immune system responsiveness.
Discussion
A growing body of evidence suggests that the gut microbiome may be a key factor in influencing predisposition to autoimmunity and allergic diseases. Here, we characterized infant gut microbiome development between three environmentally disparate populations and identified marked differences between these populations in the prevalence of important intestinal microbes, such as Bifidobacterium and LPS-producing Bacteroides species (Figure 6). Subsequently, we identified lipid A biosynthesis as one of the most differentially abundant functional pathways between the populations, suggesting that early microbial communities in Finnish and Estonian subjects produce more LPS compared to their Russian counterparts. Additionally, we uncovered functional and structural differences in the dominant microbial LPS subtypes. Notably, we showed that LPS produced by different constituents of the human gut microbiome could either stimulate or actively inhibit TLR4, NFκB activation and endotoxin tolerance. Hence, rather than the mere amount of LPS, the nature and composition of different LPS subtypes seem to determine the level of immune activation triggered by the microbe-derived LPS cocktail. Importantly, we show that injection of an immunogenic subtype of LPS from E. coli can both elicit endotoxin tolerance in vivo in NOD mice, and decrease the incidence of diabetes in these mice. These effects were not observed with LPS from B. dorei. Our observations suggest that microbiome-derived LPS could impact long-term immunosuppressive mechanisms in more complex ways than previously appreciated.
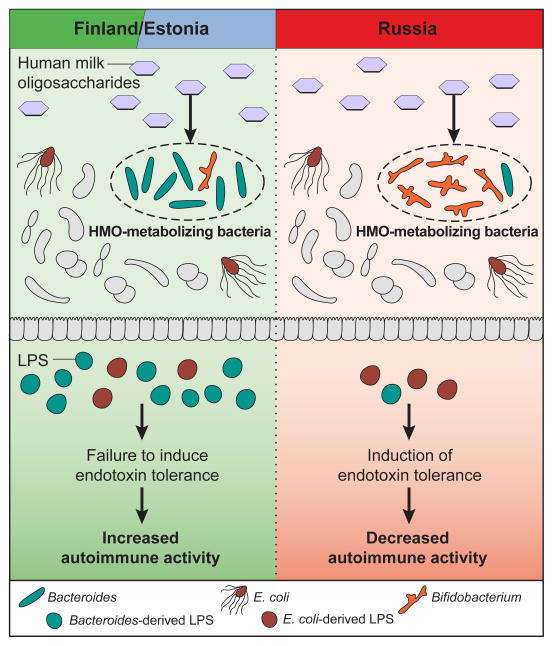
Figure 6. Differences in HMO-Utilizing Bacteria Provide a Route to Differences in Immune Education.
Human milk oligosaccharides can be metabolized by different prevalent microbes in Russia (primarily Bifidobacterium species) versus Finland and Estonia (primarily Bacteroides species). Potentially as a result of these population differences, Bacteroides-derived lipopolysaccharide (LPS) constitutes the major portion of LPS produced by microbes in Finnish and Estonian infants, whereas LPS in Russian infants is mostly derived from E. coli. Bacteroides-derived LPS is of an immunoinhibitory subtype, thus leading to differential immune education by means of endotoxin tolerance or other routes.
In the absence of specific biomarkers, we cannot determine what impact the differences in early LPS exposure has had in Russian and Finnish infants in our cohort. However, the proposition of a direct impact of LPS on the pathophysiology of T1D is further supported by previous studies in mice. For example, Wen et al. (2008) have demonstrated that components of the microbiome modulate immune system activity, resulting in altered disease development in NOD mice. In addition, LPS has a direct impact on T1D progression in NOD mice by i.p. injection (Aumeunier et al., 2010) and oral gavage (Sai and Rivereau, 1996). The impact of the microbiome on T1D onset and development in NOD mice is dependent on TLR4 and MyD88, critical components of the LPS/TLR4 signal transduction pathway (Gulden et al., 2013; Wen et al., 2008). Taken together, these studies support a model whereby exposure to different LPS subtypes produced by the gut microbiome can contribute to immune modulation and alter the course of autoimmunity.
One limitation of mouse studies is that LPS subtypes that are antagonistic or silent in humans are instead recognized as stimulatory in mice due to differences between the mouse and human LPS co-receptor MD2 (Hajjar et al., 2002). However, our observation that mice react to E. coli LPS but not to B. dorei LPS suggests that the non-immunogenicity of B. dorei LPS is possibly independent of MD2 in both species. The specific mechanism of the antagonism mediated by B. dorei LPS in humans remains to be explored.
This study achieved a deep-level understanding of microbial community establishment in three different infant populations. Our analyses revealed a lengthy list of associations between microbial taxa and the rich metadata collected in the DIABIMMUNE study, such as dietary information, mode of delivery, and the use of antibiotics (see Table 1). A comprehensive list of our findings, including microbial alterations relative to allergen-specific IgEs and T1D autoantibody seropositivity, containing numerous potentially interesting features of microbiota, can be found at http://pubs.broadinstitute.org/diabimmune.
Our analysis more broadly exercises a generalizable discovery and validation process for identifying and characterizing bioactive microbial products from the microbiome (Figure 1C). We began by identifying differentially abundant microbial processes between phenotypically distinct populations, assigned them to specific microbes, and ultimately identified structural differences within these pathways (e.g., LPS) that induced distinct immune responses in vitro. We targeted LPS biosynthesis for initial mechanistic follow-up because it was among the strongest signals and has a well-established connection to immune activation. This can be expanded in the future, since our study population included many additional functional differences in the gut, ranging from microbial metabolism (e.g., glycolysis) to iron uptake.
We found that HMO metabolism was a potential factor in establishing and/or maintaining a Bifidobacterium-dominant versus Bacteroides-dominant gut microbiota in the first year of life, likely because the two genera compete for HMOs as a common energy source (Marcobal et al., 2011). A significant role for HMO metabolism in determining microbial community composition is consistent with the hygiene hypothesis, given that mothers are also under environmental stress and can transfer these effects to their infants. In vaginal births, most of the early infant gut colonizers are derived from the mother’s gut (Backhed et al., 2015) and possibly consolidated by the microbiome in breast milk (Hunt et al., 2011). Transcriptomic analysis of cord blood from the infants in this cohort revealed a signal resembling the response to LPS exposure, suggesting that these infants are exposed to environmental stresses even before birth (Kallionpaa et al., 2014).
The effects of the hygiene hypothesis are most likely mediated through not just one mechanism but rather a complex interplay of environmental factors. These likely include immune responses to multiple different parasites, helminths, microbes, and viruses. Here, we have identified one potential contributing factor, namely immunogenicity of early colonizing symbiotic bacteria. Understanding how the different members of our microbiota contribute to the development of our immune system alone and in combination will be a key step in the development of probiotic interventions that may alter the increasing trends of autoimmune diseases in countries such as Finland.
Experimental Procedures
Study cohort
The international DIABIMMUNE study recruited 832 families in Finland (Espoo), Estonia (Tartu) and Russia (Petrozavodsk) with infants carrying HLA alleles that conferred risk for autoimmunity (Larizza et al., 2012; Sollid and Thorsby, 1993). The newborns were followed by monthly stool sampling, periodic laboratory assays and questionnaires regarding breastfeeding, diet, allergies, infections, family history, use of drugs and clinical examinations. For this study, data from 74 infants per country were selected to be analyzed based on similar HLA risk class distribution and matching gender between the countries.
Stool sample collection and DNA extraction
Stool samples were collected by the participants’ parents and stored in the household freezer (−20°C) until the next visit to the local study center; samples were then shipped on dry ice to the DIABIMMUNE Core Laboratory in Helsinki. The samples were then stored at −80°C until shipping to the Broad Institute for DNA extraction. DNA extractions from stool were carried out using the QIAamp DNA Stool Mini Kit (QIAGEN).
Library construction, sequencing, and analysis of the 16S rRNA gene and shotgun metagenomics
16S rRNA gene libraries were constructed as previously described in Kostic et al. (2015). Metagenomic libraries were prepared using Nextera XT DNA Library Preparation kit (Illumina). 16S and metagenomic libraries were sequenced on the Illumina HiSeq 2500 platform. 16S data was processed using QIIME and taxonomy was assigned according to Greengenes taxonomy map. Metagenomic data was analyzed using MetaPhlAn 2.2 (Truong et al., 2015) for taxonomic profiling and HUMAnN2 (http://huttenhower.sph.harvard.edu/humann2) for functional profiling. Associations with metadata were analyzed using MaAsLin, a linear modeling system adapted for microbial community data (http://huttenhower.sph.harvard.edu/maaslin). Metagenomic samples were additionally analyzed using ConStrains (Luo et al., 2015), which conducts within-species strain haplotyping by deconvoluting SNP patterns detected from mapping reads to species core genes across samples. See Supplemental Experimental Procedures for detailed methodology.
Bacterial strains and LPS purification
The bacterial strains used in the study are summarized in Table S3. LPS purification was performed by hot phenol-water method (Hirschfeld et al., 2000).
Human immune stimulation assays
Primary human PBMCs, in vitro-differentiated monocyte-derived dendritic cells or HEK293-NFκB reporter cells expressing hTLR4 were stimulated with indicated doses of LPS purified from various strains (Table S3). In primary cells, cytokine concentrations in the supernatant after 24h were measured by cytokine bead array analysis (BD biosciences). In HEK-293 cells, stimulation was measured by Luciferase (BrightGlo, Promega.
Endotoxin Tolerance Assays
Primary monocytes were isolated from human PBMCs and incubated in the presence of LPS purified from B. dorei or E. coli. at indicated doses for 18–20 h. Cells were then washed and cultured in cRPMI for 3 days. Monocytes were challenged with a standard dose of 5 μg/ml of zymosan. Supernatants were collected after 20 h and analyzed using the cytokine bead array human inflammation kit (BD Biosciences) according to the manufacturer’s instructions.
Diabetes incidence in NOD mice
All animal studies were conducted under protocols approved by the Institutional Animal Care and Use Committee (IACUC) at NIBR. NOD/ShiLTj mice were purchased from Jackson Laboratory. Groups of 9 to 12, 8-week old mice were injected intraperitoneally with 10 μg LPS purified from either E. coli or B. dorei once a week for 4 consecutive weeks. Non-fasting blood glucose was monitored weekly. The experimenter was blinded to the nature of the treatment for each group. Animals with either one reading above 300 mg/dL or two consecutive readings above 250 mg/dL were deemed diabetic.
Endotoxin tolerance in NOD mice
Groups of 5 mice were injected i.p. with 10 μg LPS purified from either E. coli or B. dorei. After 24 h, the splenocytes were isolated and restimulated in vitro with zymosan (2.5 μg/ml). TNFα concentration was assessed by cytokine bead array after 24 h.
Supplementary Material
Acknowledgments
We thank Tiffany Poon and Scott Steelman (Broad Institute) for help in sequence production and sample management, Leon Murphy (Novartis) for help with experimental design, Katriina Koski and Matti Koski (University of Helsinki) for the coordination and database work in the DIABIMMUNE study, Chengwei Luo (Broad Institute) for help in the strain analysis, and Natalia Nedelsky (Massachusetts General Hospital) for editorial help in writing and figure generation. T.V. was supported by funding from JDRF and Hecse (Helsinki Doctoral Programme in Computer Science). H.L. and T.V. were supported by funding from The Academy of Finland Center of Excellence in Systems Immunology and Physiology Research. A.D.K received support as the Merck Fellow of the Helen Hay Whitney Foundation and as the Lawrence H. Summers Fellow of the Broad Institute. M.K. was supported by the European Union Seventh Framework Programme FP7/2007–2013 (grant 202063) and the Academy of Finland Centre of Excellence in Molecular Systems Immunology and Physiology Research (decision 250114). R.J.X. was supported by funding from JDRF, National Institutes of Health (NIH) grants U54 DK102557, R01 DK092405, and P30 DK043351, the Leona M. and Harry B. Helmsley Charitable Trust, and the Center for Microbiome Informatics and Therapeutics at MIT.
Footnotes
Author Contributions
T.V. and A.D.K. performed 16S and metagenomics data analysis. A.D.K. and T.D.A. performed qPCR to validate metagenomics data. E.H. and T.W.C. performed LPS purification and immunological assays and analyzed the data. T.V., A.D.K., E.H., T.W.C., E.A.F., H.V., C.H., D.G., and R.J.X. assembled and wrote the paper. T.V., A.D.K., E.H., T.W.C., V.T., S.M., D.G., M.K., and R.J.X. served as project leaders. H.S., A.-M.H., A.P., R.U., N.D., S.M.V., and M.K. designed the cohort study. A.D.K., E.H., T.W.C., J.A.P., and S.J.S. designed the LPS study. T.V., A.D.K., and D.G. designed the DNA sequencing experiments and sample management pipelines. T.V., E.A.F., M.Y., R.K., J.I., C.H., and D.G. led the method and research development. A.-M.H., A.P., V.T., R.U., S.M., N.D., J.I., and S.M.V. collected clinical samples. H.L., C.H., D.G., T.W.C., M.K., and R.J.X. served as principal investigators. The NCBI BioProject ID for these data containing all publicly deposited sequencing data is PRJNA290380. The authors report no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abubucker S, Segata N, Goll J, Schubert AM, Izard J, Cantarel BL, Rodriguez-Mueller B, Zucker J, Thiagarajan M, Henrissat B, et al. Metabolic reconstruction for metagenomic data and its application to the human microbiome. PLoS computational biology. 2012;8:e1002358. doi: 10.1371/journal.pcbi.1002358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardeshir A, Narayan NR, Mendez-Lagares G, Lu D, Rauch M, Huang Y, Van Rompay KK, Lynch SV, Hartigan-O’Connor DJ. Breast-fed and bottle-fed infant rhesus macaques develop distinct gut microbiotas and immune systems. Sci Transl Med. 2014;6:252ra120. doi: 10.1126/scitranslmed.3008791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aumeunier A, Grela F, Ramadan A, Pham Van L, Bardel E, Gomez Alcala A, Jeannin P, Akira S, Bach JF, Thieblemont N. Systemic Toll-like receptor stimulation suppresses experimental allergic asthma and autoimmune diabetes in NOD mice. PloS one. 2010;5:e11484. doi: 10.1371/journal.pone.0011484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach JF. The effect of infections on susceptibility to autoimmune and allergic diseases. The New England journal of medicine. 2002;347:911–920. doi: 10.1056/NEJMra020100. [DOI] [PubMed] [Google Scholar]
- Bach JF, Chatenoud L. The hygiene hypothesis: an explanation for the increased frequency of insulin-dependent diabetes. Cold Spring Harbor perspectives in medicine. 2012;2:a007799. doi: 10.1101/cshperspect.a007799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, Li Y, Xia Y, Xie H, Zhong H, et al. Dynamics and Stabilization of the Human Gut Microbiome during the First Year of Life. Cell host & microbe. 2015;17:690–703. doi: 10.1016/j.chom.2015.04.004. [DOI] [PubMed] [Google Scholar]
- Biswas SK, Lopez-Collazo E. Endotoxin tolerance: new mechanisms, molecules and clinical significance. Trends in immunology. 2009;30:475–487. doi: 10.1016/j.it.2009.07.009. [DOI] [PubMed] [Google Scholar]
- Cullen TW, Schofield WB, Barry NA, Putnam EE, Rundell EA, Trent MS, Degnan PH, Booth CJ, Yu H, Goodman AL. Gut microbiota. Antimicrobial peptide resistance mediates resilience of prominent gut commensals during inflammation. Science. 2015;347:170–175. doi: 10.1126/science.1260580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis-Richardson AG, Ardissone AN, Dias R, Simell V, Leonard MT, Kemppainen KM, Drew JC, Schatz D, Atkinson MA, Kolaczkowski B, et al. Bacteroides dorei dominates gut microbiome prior to autoimmunity in Finnish children at high risk for type 1 diabetes. Frontiers in microbiology. 2014;5:678. doi: 10.3389/fmicb.2014.00678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulden E, Ihira M, Ohashi A, Reinbeck AL, Freudenberg MA, Kolb H, Burkart V. Toll-like receptor 4 deficiency accelerates the development of insulin-deficient diabetes in non-obese diabetic mice. PloS one. 2013;8:e75385. doi: 10.1371/journal.pone.0075385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajjar AM, Ernst RK, Tsai JH, Wilson CB, Miller SI. Human Toll-like receptor 4 recognizes host-specific LPS modifications. Nature immunology. 2002;3:354–359. doi: 10.1038/ni777. [DOI] [PubMed] [Google Scholar]
- Herath TD, Wang Y, Seneviratne CJ, Lu Q, Darveau RP, Wang CY, Jin L. Porphyromonas gingivalis lipopolysaccharide lipid A heterogeneity differentially modulates the expression of IL-6 and IL-8 in human gingival fibroblasts. Journal of clinical periodontology. 2011;38:694–701. doi: 10.1111/j.1600-051X.2011.01741.x. [DOI] [PubMed] [Google Scholar]
- Hirschfeld M, Ma Y, Weis JH, Vogel SN, Weis JJ. Cutting edge: repurification of lipopolysaccharide eliminates signaling through both human and murine toll-like receptor 2. Journal of immunology. 2000;165:618–622. doi: 10.4049/jimmunol.165.2.618. [DOI] [PubMed] [Google Scholar]
- Hofstad T, Sveen K, Dahlen G. Chemical composition, serological reactivity and endotoxicity of lipopolysaccharides extracted in different ways from Bacteroides fragilis, Bacteroides melaninogenicus and Bacteroides oralis. Acta Pathologica et Microbiologica Scandinavica Section B, Microbiology. 1977;85:262–270. doi: 10.1111/j.1699-0463.1977.tb01972.x. [DOI] [PubMed] [Google Scholar]
- Hunt KM, Foster JA, Forney LJ, Schutte UM, Beck DL, Abdo Z, Fox LK, Williams JE, McGuire MK, McGuire MA. Characterization of the diversity and temporal stability of bacterial communities in human milk. PloS one. 2011;6:e21313. doi: 10.1371/journal.pone.0021313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallionpaa H, Laajala E, Oling V, Harkonen T, Tillmann V, Dorshakova NV, Ilonen J, Lahdesmaki H, Knip M, Lahesmaa R, et al. Standard of hygiene and immune adaptation in newborn infants. Clinical immunology. 2014;155:136–147. doi: 10.1016/j.clim.2014.09.009. [DOI] [PubMed] [Google Scholar]
- Kim HM, Park BS, Kim JI, Kim SE, Lee J, Oh SC, Enkhbayar P, Matsushima N, Lee H, Yoo OJ, et al. Crystal structure of the TLR4-MD-2 complex with bound endotoxin antagonist Eritoran. Cell. 2007;130:906–917. doi: 10.1016/j.cell.2007.08.002. [DOI] [PubMed] [Google Scholar]
- Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, Angenent LT, Ley RE. Succession of microbial consortia in the developing infant gut microbiome. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(Suppl 1):4578–4585. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashova A, Mustalahti K, Kaukinen K, Viskari H, Volodicheva V, Haapala AM, Ilonen J, Knip M, Maki M, Hyoty H, et al. Lower economic status and inferior hygienic environment may protect against celiac disease. Annals of medicine. 2008a;40:223–231. doi: 10.1080/07853890701678689. [DOI] [PubMed] [Google Scholar]
- Kondrashova A, Viskari H, Haapala AM, Seiskari T, Kulmala P, Ilonen J, Knip M, Hyoty H. Serological evidence of thyroid autoimmunity among schoolchildren in two different socioeconomic environments. The Journal of clinical endocrinology and metabolism. 2008b;93:729–734. doi: 10.1210/jc.2007-1644. [DOI] [PubMed] [Google Scholar]
- Kostic AD, Gevers D, Siljander H, Vatanen T, Hyotylainen T, Hamalainen AM, Peet A, Tillmann V, Poho P, Mattila I, et al. The dynamics of the human infant gut microbiome in development and in progression toward type 1 diabetes. Cell host & microbe. 2015;17:260–273. doi: 10.1016/j.chom.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larizza D, Calcaterra V, Klersy C, Badulli C, Caramagna C, Ricci A, Brambilla P, Salvaneschi L, Martinetti M. Common immunogenetic profile in children with multiple autoimmune diseases: the signature of HLA-DQ pleiotropic genes. Autoimmunity. 2012;45:470–475. doi: 10.3109/08916934.2012.697594. [DOI] [PubMed] [Google Scholar]
- Locascio RG, Ninonuevo MR, Kronewitter SR, Freeman SL, German JB, Lebrilla CB, Mills DA. A versatile and scalable strategy for glycoprofiling bifidobacterial consumption of human milk oligosaccharides. Microbial biotechnology. 2009;2:333–342. doi: 10.1111/j.1751-7915.2008.00072.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo C, Knight R, Siljander H, Knip M, Xavier RJ, Gevers D. ConStrains identifies microbial strains in metagenomic datasets. Nature biotechnology. 2015;33:1045–1052. doi: 10.1038/nbt.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcobal A, Barboza M, Sonnenburg ED, Pudlo N, Martens EC, Desai P, Lebrilla CB, Weimer BC, Mills DA, German JB, et al. Bacteroides in the infant gut consume milk oligosaccharides via mucus-utilization pathways. Cell host & microbe. 2011;10:507–514. doi: 10.1016/j.chom.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markle JG, Frank DN, Mortin-Toth S, Robertson CE, Feazel LM, Rolle-Kampczyk U, von Bergen M, McCoy KD, Macpherson AJ, Danska JS. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science. 2013;339:1084–1088. doi: 10.1126/science.1233521. [DOI] [PubMed] [Google Scholar]
- Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV, Reyes JA, Shah SA, LeLeiko N, Snapper SB, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome biology. 2012;13:R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needham BD, Carroll SM, Giles DK, Georgiou G, Whiteley M, Trent MS. Modulating the innate immune response by combinatorial engineering of endotoxin. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:1464–1469. doi: 10.1073/pnas.1218080110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peet A, Kool P, Ilonen J, Knip M, Tillmann V, Group DS. Birth weight in newborn infants with different diabetes-associated HLA genotypes in three neighbouring countries: Finland, Estonia and Russian Karelia. Diabetes/metabolism research and reviews. 2012;28:455–461. doi: 10.1002/dmrr.2303. [DOI] [PubMed] [Google Scholar]
- Sai P, Rivereau AS. Prevention of diabetes in the nonobese diabetic mouse by oral immunological treatments. Comparative efficiency of human insulin and two bacterial antigens, lipopolysacharide from Escherichia coli and glycoprotein extract from Klebsiella pneumoniae. Diabetes Metab. 1996;22:341–348. [PubMed] [Google Scholar]
- Seiskari T, Kondrashova A, Viskari H, Kaila M, Haapala AM, Aittoniemi J, Virta M, Hurme M, Uibo R, Knip M, et al. Allergic sensitization and microbial load--a comparison between Finland and Russian Karelia. Clinical and experimental immunology. 2007;148:47–52. doi: 10.1111/j.1365-2249.2007.03333.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sela DA, Chapman J, Adeuya A, Kim JH, Chen F, Whitehead TR, Lapidus A, Rokhsar DS, Lebrilla CB, German JB, et al. The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:18964–18969. doi: 10.1073/pnas.0809584105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sela DA, Mills DA. Nursing our microbiota: molecular linkages between bifidobacteria and milk oligosaccharides. Trends in microbiology. 2010;18:298–307. doi: 10.1016/j.tim.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermudez-Humaran LG, Gratadoux JJ, Blugeon S, Bridonneau C, Furet JP, Corthier G, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollid LM, Thorsby E. HLA susceptibility genes in celiac disease: genetic mapping and role in pathogenesis. Gastroenterology. 1993;105:910–922. doi: 10.1016/0016-5085(93)90912-v. [DOI] [PubMed] [Google Scholar]
- Stefka AT, Feehley T, Tripathi P, Qiu J, McCoy K, Mazmanian SK, Tjota MY, Seo GY, Cao S, Theriault BR, et al. Commensal bacteria protect against food allergen sensitization. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:13145–13150. doi: 10.1073/pnas.1412008111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teeaar T, Liivak N, Heilman K, Kool P, Sor R, Paal M, Einberg U, Tillmann V. Increasing incidence of childhood-onset type 1 diabetes mellitus among Estonian children in 1999–2006. Time trend analysis 1983–2006. Pediatric diabetes. 2010;11:107–110. doi: 10.1111/j.1399-5448.2009.00535.x. [DOI] [PubMed] [Google Scholar]
- Truong DT, Franzosa EA, Tickle TL, Scholz M, Weingart G, Pasolli E, Tett A, Huttenhower C, Segata N. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nature methods. 2015;12:902–903. doi: 10.1038/nmeth.3589. [DOI] [PubMed] [Google Scholar]
- von Mutius E, Vercelli D. Farm living: effects on childhood asthma and allergy. Nature reviews Immunology. 2010;10:861–868. doi: 10.1038/nri2871. [DOI] [PubMed] [Google Scholar]
- Voor T, Julge K, Bottcher MF, Jenmalm MC, Duchen K, Bjorksten B. Atopic sensitization and atopic dermatitis in Estonian and Swedish infants. Clinical and experimental allergy : journal of the British Society for Allergy and Clinical Immunology. 2005;35:153–159. doi: 10.1111/j.1365-2222.2005.02157.x. [DOI] [PubMed] [Google Scholar]
- Watson DW, Kim YB. Modification of Host Responses to Bacterial Endotoxins. I. Specificity of Pyrogenic Tolerance and the Role of Hypersensitivity in Pyrogenicity, Lethality, and Skin Reactivity. The Journal of experimental medicine. 1963;118:425–446. doi: 10.1084/jem.118.3.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen L, Ley RE, Volchkov PY, Stranges PB, Avanesyan L, Stonebraker AC, Hu C, Wong FS, Szot GL, Bluestone JA, et al. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. 2008;455:1109–1113. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield C, Trent MS. Biosynthesis and export of bacterial lipopolysaccharides. Annual review of biochemistry. 2014;83:99–128. doi: 10.1146/annurev-biochem-060713-035600. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.