Abstract
RNA SHAPE chemistry exploits the discovery that conformationally dynamic nucleotides preferentially adopt conformations that facilitate reaction between the 2′-OH group and a hydroxyl-selective electrophile, such as benzoyl cyanide (BzCN), to form a 2′-O-adduct. BzCN is ideally suited for quantitative, time-resolved analysis of RNA folding and RNP assembly mechanisms because this reagent both reacts with flexible RNA nucleotides and also undergoes auto-inactivating hydrolysis with a half-life of 0.25 s at 37 °C. RNA folding is initiated by addition of Mg2+ or protein, or other change in solution conditions, and nucleotide resolution structural images are obtained by adding aliquots of the evolving reaction to BzCN and then “waiting” for 1 sec. Sites of 2′-O-adduct formation are subsequently scored as stops to primer extension using reverse transcriptase. This time resolved SHAPE protocol makes it possible to obtain 1 sec snapshots in time-resolved kinetic studies for RNAs of arbitrary length and complexity in a straightforward and concise experiment.
INTRODUCTION
RNA structural transitions are central to the ability of RNA to function inside the cell1-3. A full understanding of RNA function therefore requires a nucleotide resolution view of the time-resolved mechanisms by which RNA molecules fold, interconvert between distinct states, and function in higher-order assemblies. RNA folding events occur on a wide range of time-scales, ranging from μs to minutes but many rate-determining steps occur on timescales spanning tens of seconds to minutes4-7. These critical events might, in principle, be conveniently monitored using simple bench top kinetics.
Current, well-established and highly useful, approaches for probing the kinetics of RNA folding at nucleotide resolution include dimethyl sulfate (DMS) and hydroxyl radical footprinting8-11. In practice, both time-resolved DMS and hydroxyl radical footprinting are experimentally challenging to perform and require significant experiment-specific optimization. DMS reactivity probes a subset of nucleotides, largely adenosine, and requires that the reagent be quenched in a separate inactivation step8. Hydroxyl radical footprinting is a powerful approach for probing the solvent accessibility of the RNA backbone but either requires the use of a synchotron10 or adding a separate quench step12. To date, these experimental challenges have limited the interest in evaluating folding states and the associated folding pathways for large RNAs at nucleotide resolution. Time-resolved RNA SHAPE chemistry addresses these limitations and makes possible experimentally straightforward single-nucleotide resolution and simultaneous analysis of local structure at nearly every position in an RNA with one-second time resolution13.
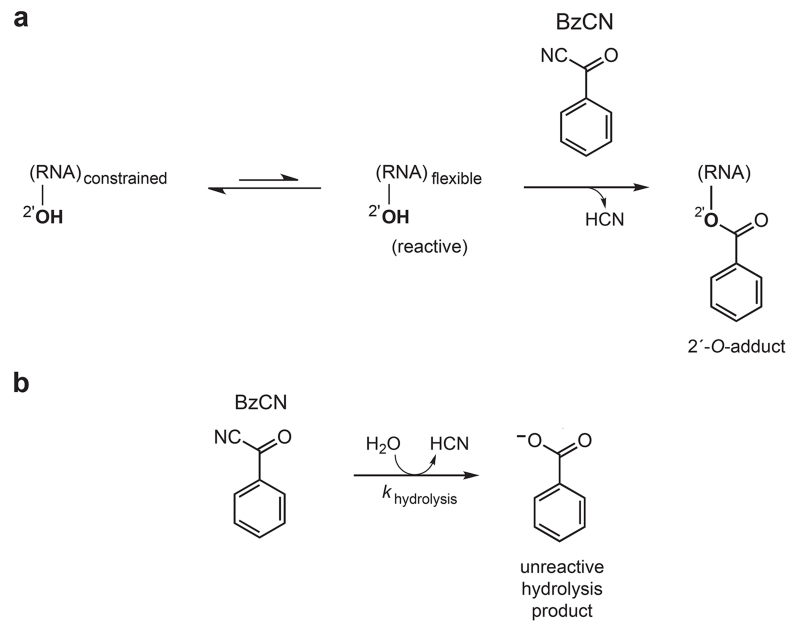
SHAPE chemistry exploits the discovery that the nucleophilic reactivity of the ribose 2′-hydroxyl position is strongly gated by the underlying local nucleotide flexibility (Figure 1a)14-16. Flexible nucleotides preferentially adopt conformations that react with a hydroxyl-selective electrophile, such as benzoyl cyanide (BzCN), to form a 2′-O-adduct. In contrast, base-paired or otherwise conformationally constrained nucleotides are unreactive. Because almost all ribonucleotides possess a free 2′-hydroxyl, local nucleotide flexibility at each position in an RNA is interrogated simultaneously in a single experiment14,17. Because RNA folding and ribonucleoprotein assembly reactions typically involve large changes in the local environment at many nucleotides, SHAPE represents an ideal approach for monitoring these changes, comprehensively and at single nucleotide resolution.
Figure 1.
Mechanism of RNA SHAPE chemistry with BzCN. (a) BzCN reacts with 2′-hydroxyl groups at conformationally flexible positions to form a 2′-O-adduct. (b) Parallel BzCN inactivation by hydrolysis.
During the RNA structure-probing reaction, SHAPE electrophiles are concurrently inactivated by hydrolysis. Thus, a specific quench step is not required, provided the adduct-forming reaction with RNA is allowed to continue until all reagent is consumed (Figure 1b). BzCN undergoes hydrolysis with a half-life of 0.25 s at 37 °C: the reaction between BzCN and RNA is therefore complete in ~1 s.
Once the reaction is complete, sites of 2′-O-adduct formation are identified by annealing a 5′-labeled DNA primer to the modified RNA and scoring stops to primer extension by reverse transcriptase. The length and amount of a cDNA transcript correlates with the position and degree of modification at each position in the RNA. The length of each cDNA is assigned by comparison with a dideoxy sequencing reaction13,18.
There are two potential limitations of time-resolved SHAPE as described in this protocol. SHAPE primarily measures local nucleotide flexibility. In general, this information gives good coverage of both secondary and tertiary structure interactions during an RNA folding or ribonucleoprotein assembly reaction15,19-21. However, there may be some instances in which analysis of backbone solvent accessibility is essential or can provide critical additional information. In these cases, fast hydroxyl radical cleavage is a good alternative10,11. The method outlined here emphasizes using time-resolved SHAPE in an experimentally straightforward bench top approach that ultimately yields ~1 second structural images of RNA structure, taken 5-15 seconds apart. SHAPE can be adapted to faster timescales, in a more complex experiment, by use of a rapid mixing device and quenching the BzCN reagent with dithiothreitol13.
Time-resolved SHAPE with BzCN was initially used to study the time-resolved folding of the specificity domain of the RNase P RNA13. We anticipate that further application of this approach will make possible facile kinetic studies of many RNA in ~1 s snapshots. Time-resolved SHAPE holds broad potential for understanding structural biogenesis and the conformational interconversions essential to the function of complex RNA molecules at single nucleotide resolution.
EXPERIMENTAL DESIGN
Time-resolved SHAPE takes advantage of the intrinsic feature of BzCN that it degrades completely in water in ~1 sec. Thus, the experiment is as simple as adding a solution of an RNA folding reaction to a small aliquot of BzCN, “waiting” for 1 sec, and then analyzing the results by primer extension at a later point. Experimental conditions should be adjusted so that roughly 1 in 100-300 nts are modified prior to BzCN inactivation. It is not necessary that exactly one modification occur per RNA, only that the level of modification is sufficiently sparse that sites of 2′-O-adduct formation are uncorrelated. Higher levels of modification produce stronger peak signals; whereas, lower reagent concentrations yield longer read lengths for analysis by capillary electrophoresis. Because BzCN reacts rapidly with trace amounts of water, it is important to take care in setting up the experiment to work with dry BzCN and DMSO. The concentration of buffer (for examples Hepes) should be higher than the final concentration of BzCN to prevent acidification of the solution upon reagent hydrolysis (Figure 1b). The reaction is largely insensitive to presence of other solution components that might react with BzCN, including amines, carbohydrates, and proteins.
RNA folding
RNA folding reactions can be initiated in many ways including changes in temperature, ionic strength, or addition of a protein or small molecule ligand. This protocol focuses on analyzing the formation of RNA tertiary structure initiated by addition of Mg2+ to a solution that otherwise contains all components necessary to stabilize the native structure. However, this approach is readily applied to most other methods of initiating the RNA folding or ribonucleoprotein assembly reaction and then makes it possible to analyze the resulting RNA conformational changes on the second time scale.
Primer Extension
Sites of time-dependent 2′-O-adduct formation can be analyzed using DNA primers containing either a 5′-radiolabel or a 5′-fluorescent tag. This protocol focuses on quantifying sites of 2′-O-adduct formation in a high-throughput way using fluorescently labeled primers in the primer extension step and then rapidly resolving and quantifying cDNA products by capillary electrophoresis13,18. To use 5′-[32P]-labeled DNA primers and conventional sequencing gel electrophoresis, the procedure below should be followed up to step 17. An alternate protocol for analyzing radiolabeled cDNAs by conventional gel electrophoresis has been described previously (and replaces steps 18-24 below)17. In general, the quality of the quantitative nucleotide reactivity information is far superior using capillary electrophoresis; however, use of radiolabeled primers can be more sensitive in cases where RNA is limiting. Using either primer extension approach, no reactivity information is obtained at the sequence to which the primer binds, nor 20-50 nts 5′ of from the reverse transcriptase start site14,17. Thus, the DNA primer binding site can either be a natural sequence 3′ of the RNA sequence of interest or an appended ‘structure cassette’ sequence14 that allows the entire structure of the RNA to be evaluated.
Analysis of time-progress curves
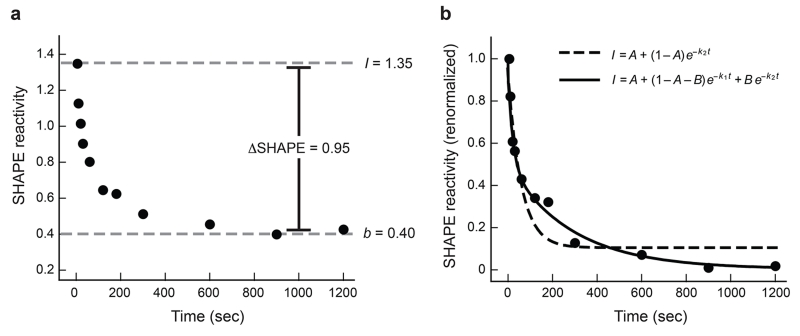
We typically normalize SHAPE data to a scale that spans 0 to ~2. Zero corresponds to an unreactive position and 1.0 is defined as the average intensity of highly reactive nucleotides13,15,18. On this scale, time-dependent changes in reactivity are generally significant and quantifiable if they are 0.2 SHAPE units or greater (Figure 2a). The quality and reproducibility of the time-resolved SHAPE profiles are typically quite high and it is often possible to distinguish processes characterized by either a single
| (1) |
or double
| (2) |
exponential (Figure 2b)13. Or in terms of two irreversible consecutive steps4:
| (3) |
More complex kinetic schemes can also be handled by other approaches22.
Figure 2.
Analysis of time-progress curves. Nucleotide G217 of the RNase P specificity domain is shown. (a) Representative change in SHAPE reactivity as a function of time. Y-axis data are shown using absolute SHAPE reactivities that have been normalized to a scale that spans 0 to ~1.5, where 1.0 is defined as the average intensity of highly reactive positions. The absolute change in SHAPE reactivity at this nucleotide is 0.95, which means the time-progress curve can be quantified at a high level of accuracy and significance. (b) Quantitative analysis of time-resolved changes in local nucleotide flexibility. Data from panel A have been renormalized on the y-axis such that reactivity at time = 0 is defined as 1.0 and is obtained by dividing by the initial maximum intensity (I = 1.35), after first subtracting the plateau SHAPE reactivity (b = 0.40). Most nucleotides tend to follow simple kinetic behavior and are well fit using straightforward kinetic equations. A double exponential (solid line, Eqn. 2) is the best choice for these data.
MATERIALS
REAGENTS
RNA at a concentration of ~1 μM, ~3 pmol per time point, dissolved in 0.5× TE (5 mM Tris, 0.5 mM EDTA, pH 8.0).
Deoxyadenosine, deoxycytidine, and deoxythymidine triphosphates (dATP, dCTP, dTTP), 100 mM (Invitrogen cat. No. 55082, 55083, 55085)
Deoxyinosine triphosphate (dITP), 100 mM (Trilink Biotechnologies, cat. No. N2012. Use of dITP reduces band compression at G residues and increases resolution of primer extension products by capillary electrophoresis.
Dideoxyadenosine, dideoxycytidine, dideoxythymidine and dideoxyguanosine triphosphates (ddATP, ddCTP, ddTTP, ddGTP), (Trilink Biotechnologies, cat. No. N-4001, N-4005, N-4004, N-4002); make 10 mM solutions of ddATP, ddCTP, ddTTP and/or 0.25 mM ddGTP by dilution in sterile water.
CRITICAL: Nucleotide solutions are stable for months at −20 °C but are intolerant of freeze-thaw cycles. Maintaining small aliquots at −20 °C is recommended.
Glycogen, 20 mg/mL (Invitrogen, cat. No. 10814-010)
EDTA, 0.5 M (Ambion, 9260G)
Deionized formamide (Applied Biosystems, cat. No. 4311320)
DMSO, molecular biology grade (Sigma-Aldrich, cat. No. D8418)
CRITICAL: DMSO bottle should be stored in a desiccator at room temperature (~22 °C).
Benzoyl cyanide (BzCN) (Sigma-Aldrich, cat. No. 115959)
CRITICAL: BzCN bottle should be stored in a desiccator at room temperature.
Superscript III reverse transcriptase (Invitrogen, 18080-093)
5× SSIII FS buffer [250 mM Tris (pH 8.3), 375 mM KCl, 15 mM MgCl2] (Invitrogen cat. No. 18080-093)
0.1 M DTT (Invitrogen cat. No. 18080-093)
DNA primer: use a primer ~18-20 nt in length and that forms a 3′ G-C base pair with the target RNA
0.65 and 1.5 mL RNase-free polypropylene (Eppendorf) reaction tubes
REAGENT SET UP
The first four reagents can be stored at room temperature and are stable for many months.
Enzyme stop mix (50 mM EDTA, pH 8.0)
TE (10 mM Tris, 1 mM EDTA, pH 8.0)
3.3× no-Mg2+ RNA folding solution (333 mM HEPES, pH 8.0, 333 mM NaCl). These are the conditions under which the RNA is equilibrated before initiating the folding reaction. Buffer, ions, and ionic strength can be varied. In general, reaction with BzCN is tolerant of wide variations in experimental conditions. The primary critical requirement is that the final concentration of buffer should be greater than the BzCN concentration during the modification reaction.
10× MgCl2 (100 mM MgCl2). Any concentration of MgCl2 sufficient to yield the biologically active or target RNA structure can be used.
10× BzCN in DMSO The optimal concentration can vary with RNA length. The useful range is 100-800 mM. For longer RNAs, use the lower end of these BzCN concentrations. A good starting concentration is 400 mM BzCN (40 mM, final). The BzCN solution should be prepared fresh for every experiment and can be stored at room temperature for one day if kept in a desiccator.
Superscript enzyme mix (250 mM KCl, 161 mM Tris-HCl, pH 8.3, 1.61 mM each dNTP, 11 mM DTT, 10 mM MgCl2). The enzyme mix can be prepared by combining 4 parts SSIII FS buffer, one part 0.1 M DTT, and one part 10 mM dNTP mix (10 mM in each deoxynucleotide). The enzyme mix must be stored at −20 °C and is sensitive to freeze thaw cycles. Maintaining small aliquots is recommended.
5′-fluorescently labeled primers These can be ordered from several companies including Applied Biosystems, IDT, and Trilink Biotechnologies. At least three, preferentially four, DNA primers, each labeled with a different fluorophore, are needed. Compatible dye sets depend on the sequencing instrument used. Fluorescent labeled primers must be stored at −20 °C away from light and are sensitive to freeze thaw cycles. Maintaining small aliquots is recommended.
EQUIPMENT
−80 °C and −20 °C freezers
microfuge for 1.5 ml reaction tubes at 4 ° C
automated capillary electrophoresis instrument. We use instruments typically sold for DNA sequencing applications and have obtained excellent results with instruments from both Beckman15,18 and Applied Biosystems (ABI)13,23. It is easier to obtain fluorescently labeled DNA primers for ABI instruments.
Programmable incubator or heat block. We recommend an incubator with a heated top of the type typically used for performing PCR.
ShapeFinder software system (freely available to academic researchers at http://bioinfo.unc.edu/Downloads).
PROCEDURE
RNA folding
Add 78 pmol RNA [3 pmol per each (+) and (−) reagent time point] in 144 μL sterile H2O to a 0.65 mL reaction tube. This procedure yields 13 total data points, including a no-Mg2+ reference.
Heat the RNA to 95 °C for 2 min; then place the RNA on ice for 1 min.
Add 72 μL 3.3× no-Mg2+ folding buffer and mix thoroughly.
Incubate the tube at the desired reaction temperature (25 or 37 °C) for 5-10 min.
Preincubate a 0.65 mL reaction tube containing 22 μL 10× MgCl2 solution at the same temperature as the RNA solution.
RNA modification
-
6.
Aliquot 1 μL 10× BzCN in DMSO and 1 μL neat DMSO into each of 13 0.65 mL reaction tubes; these are the (+) and (−) BzCN tubes, respectively.
-
7.
Take a no-Mg2+ initial reference time point by transferring two 9 μL aliquots of the RNA solution to reaction tubes containing 1 μL BzCN in DMSO or 1 μL DMSO. This point can also be taken as a time = 0 reference.
-
8.
Initiate RNA folding by adding the remaining 198 μL solution of re-folded RNA to the tube containing 10× MgCl2 (~220 μL total final volume). Mix thoroughly and start timing.
-
9.
At desired time points (up to 12 total) remove 9 μL and add to a 0.65 mL reaction tube containing 1 μL pre-aliquoted 10× BzCN in DMSO. Mix thoroughly by rapid pipetting 2-3 times. Repeat by adding 9 μL to a tube containing 1 μL neat DMSO and mix thoroughly (each time point requires 18 μL total of the folded RNA solution). Reaction with BzCN is complete in 3 and 1 sec at 25 and 37 °C, respectively; no explicit quench step is required. Obtaining time points at 15 sec intervals is straightforward. With practice, time points at 5-7 second intervals are possible.
-
10.
At the end of the time course, when all time points have been obtained, recover the modified RNA by ethanol precipitation. To each tube (26 total) add 90 μL sterile H2O, 5 μL 4 M NaCl, 1 μL 20 mg/mL glycogen, 400 μL 100% ethanol; mix; and incubate at −80 °C for 30 min. Sediment the RNA by spinning at ≥ 10K rpm (≥ 9000 × g) speed in a microfuge at 4 °C for 30 min.
-
11.
Remove supernatant and redissolve RNA in 10 μL 0.5× TE.
PAUSE POINT. Modified RNA can be stored at −20 °C overnight.
Primer extension and RNA sequencing
-
12.
Add 3 μL 0.3 μM fluorescently or radiolabeled primers to the (+) and (−) BzCN reactions.
-
13.
For each required sequencing reaction, add 3 μL 0.3 μM fluorescently or radiolabeled primer to 3 pmol of RNA in 8 μL 0.5× TE. Each time point requires its own sequencing reaction, so multiply by the number of time points (one reference, plus 12 time points, as described here)
CRITICAL: For resolution by capillary electrophoresis instrument, each reaction will be monitored using a DNA primer labeled with a different color-coded fluorophore. Several compatible fluorescent dye sets are possible. For resolution on an ABI instrument, we typically use dye sets consisting of FAM, VIC, NED and PET (ABI G5 dye set); FAM, TET, HEX, and NED; or FAM, JOE, TAMRA, and ROX.
-
14.
Anneal the primer to the RNA by heating at 65 °C for 5 min and placing on ice for 1 min.
-
15.
Add 6 μL of superscript enzyme mix to the (+) and (−) BzCN reactions and to the sequencing reactions. To each sequencing reaction, also add 1 μL ddNTP solution.
-
16.
Add 1 μL of Superscript III to each tube. Mix well and place at 45 °C.
-
17.
Incubate at 45 °C for 1 min, 52 °C for 25 min, 65 °C for 5 min, and then place on ice.
CRITICAL: Omitting the 45 °C step can lead to poor primer extension, especially for longer RNAs.
For radiolabeled primers, omit steps 18-24 below and replace with steps 17-23 from ref. 17.
-
18.
Add 4 μL 50 mM EDTA (pH 8.0) to each tube to quench the extension reaction and place on ice.
-
19.
For each time point, combine the (+) and (−) BzCN reactions and a sequencing reaction (22 μL each, 66-88 μL total ) into a 1.5 mL reaction tube and recover the cDNAs by ethanol precipitation: add 240 μL 100 % ethanol to each tube (12 total) and incubate at −80 °C for 15 min. Sediment the cDNA by spinning at ≥10K rpm (≥9000 × g) in a microfuge at 4 °C for 15 min.
-
20.
Remove supernatant and add 800 μL 70% ethanol. Invert the tube to dislodge pellet and spin at maximum speed in a microfuge at 4 °C for 2 min.
-
21.
Repeat step 19.
CRITICAL: Omitting this step leaves residual salt in the primer extension reaction solutions and can lead to poor resolution during capillary electrophoresis.
-
22.
Dry the pellet by vacuum for 10 min and resuspend in 10 μL deionized formamide (this volume will vary depending on the capillary electrophoresis instrument used).
PAUSE POINT. The resuspended pellets can be stored at −20 °C away from light for an extended period of time. For best results, run samples immediately.
cDNA analysis by capillary electrophoresis
-
23.
Load each 10 μL sample into separate wells on a capillary electrophoresis DNA sequencing instrument and run.
-
24.
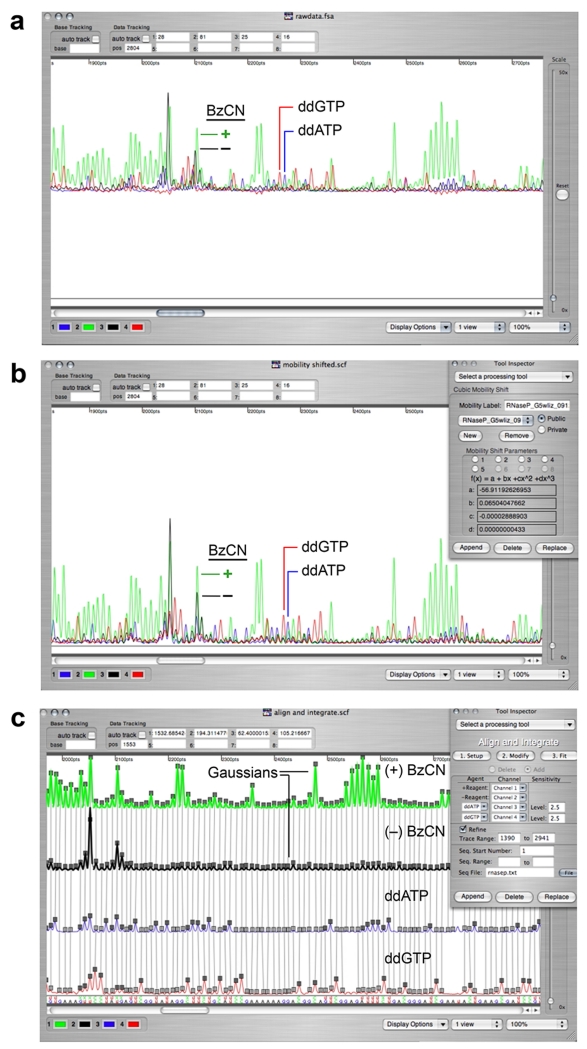
Export the resulting raw traces into ShapeFinder24 (Figure 3a), process the data by applying a: (i) baseline adjustment, (ii) mobility shift appropriate for the dye set used24, and (iii) a multiplicative scaling factor, if necessary, to the (+) and (−) BzCN traces (Figures 3b & 4a; see Troubleshooting table and reference 24 for details). Quantify the intensity of all peaks in the (+) and (−) BzCN reactions by whole-trace Gaussian integration using the Align and Integrate tool in ShapeFinder (Figure 3c). ShapeFinder outputs a text file containing the integrated areas for each peak in the electropherogram, the net reactivity after subtraction of the (−) reagent background, and the normalized SHAPE reactivities using a scale in which 1.0 is defined as the average intensity of highly reactive positions (Figure 4b). These steps can be fine tuned in a spreadsheet application.
Figure 3.
Electropherogram analysis using ShapeFinder24. (a) Unprocessed capillary electrophoresis electropherogram exported from an ABI sequencer (no matrixing step required). (b) Resulting trace after application of the Fitted Baseline Adjust and Mobility Shift:Cubic tools. (c) Application of the Align and Integrate tool to quantify all peaks in the (+) and (−) reagent channels by whole-trace Gaussian integration. Channels are split for clarity and ease of analysis; the aligned RNA sequence is at the bottom of the window.
Figure 4.
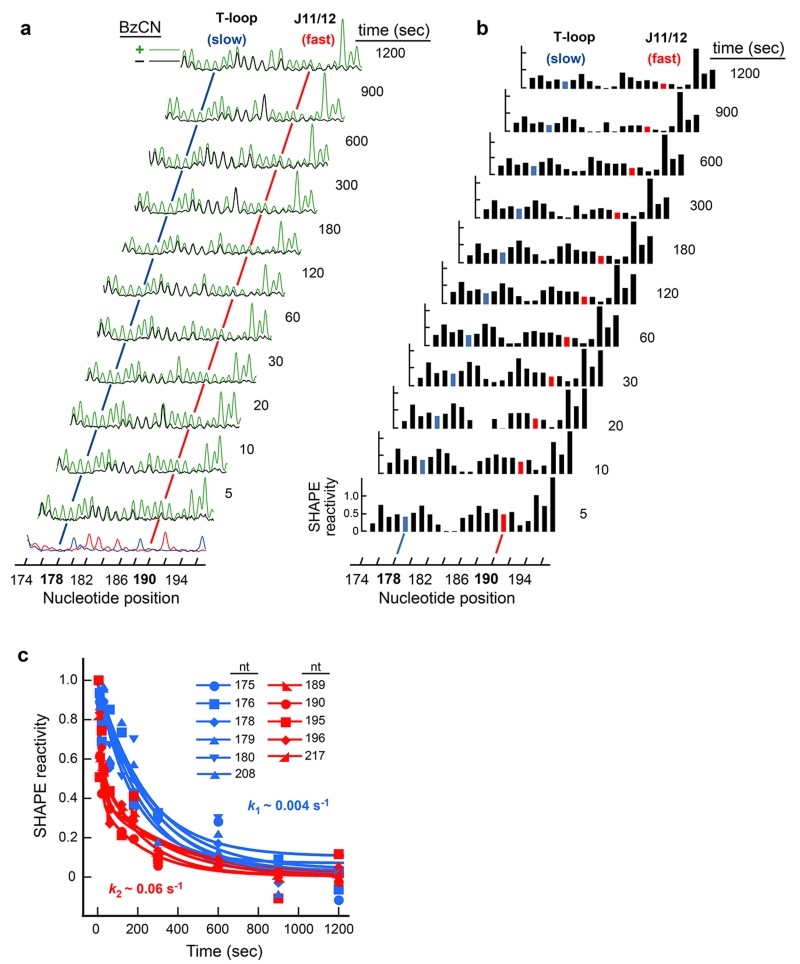
Representative time-resolved SHAPE analysis of the RNase P specificity domain RNA. (a) Processed capillary electrophoresis traces obtained for the reaction of BzCN with the RNase P specificity domain for folding times spanning 5 s to 20 min. RNA folding was initiated by addition of Mg2+ to RNA pre-equilibrated in buffer and NaCl. (+) and (−) reagent channels are shown in green and black; sequencing channels (at bottom) were used to assign peak position in ShapeFinder24. The difference in peak intensity between the (+) and (−) BzCN channels reports the extent of 2′-O-adduct formation at each point. Nucleotides involved in tertiary interactions show a decrease in reactivity over time. For example, nucleotide 178 becomes protected from reaction with BzCN on a slow time scale, while nucleotide 190 undergoes this transition more rapidly (blue and red lines, respectively). (b) Absolute SHAPE reactivities as a function of nucleotide position and time. (c) Representative time-progress curves monitored by time-resolved SHAPE. For the RNase P RNA, all nucleotides show a decrease in SHAPE reactivity as a function of time, indicating a significant increase in structural constraints as the RNA forms tertiary interactions. Each time-dependent profile was fit individually to either a single or double exponential (Equations 1 or 2 in the main text). All curves found to fit a single exponential (in blue) were characterized by the same rate constant, at 0.004 sec−1. Nucleotides best fit by a double exponential (in red) were characterized by a fast phase at 0.06 sec−1 and a slow phase at 0.004 sec−1. Adapted in part from the J. Am. Chem. Soc. 130:16178 (Copyright 2008 American Chemical Society).
Data analysis
-
25.
Plot the normalized SHAPE reactivities as a function of time for each nucleotide in the RNA. Inspect the individual curves for their kinetic behavior. Typically, one-half of the nucleotides in an RNA will show no change. This is the expected result because the local environment at some positions does not change with folding. At other nucleotides, the SHAPE reactivities will decrease (largest number of positions) or increase (a few positions). In both cases, a significant change in the local nucleotide flexibility during folding is one with ΔSHAPE ≥ 0.2 (Figure 2a).
-
26.
We generally interpret time-progress curves in term of net, normalized intensity plots at each nucleotide position: (i) perform an initial exponential fit to determine the plateau value (b) at long times; (ii) subtract this value from all data points, and (iii) renormalize intensities to a scale spanning 0 to 1.0 by dividing by the intensity of the first time point (I) (Figure 2a).
-
27.
Individually fit time-dependent changes in SHAPE reactivity for each nucleotide to an appropriate kinetic equation. For most nucleotides, a single or double exponential or an equation describing two consecutive steps is generally sufficient (Figure 2b and see Eqns. 1-3). For each time-progress curve, the equation that best corresponds to the time-resolved SHAPE data can be distinguished either by eye or using Pearson’s r-value. Group nucleotides according to rate constants that are less than 2-fold apart (Figure 4c).
-
28.
Superimpose kinetic data on a structural model of the RNA. Develop a model for the RNA folding or RNP assembly mechanism based on the clustering of nucleotides that fold with similar rates. Use the number of observed rate constants to estimate the number of kinetically significant structural intermediates (Figure 5).
Figure 5.
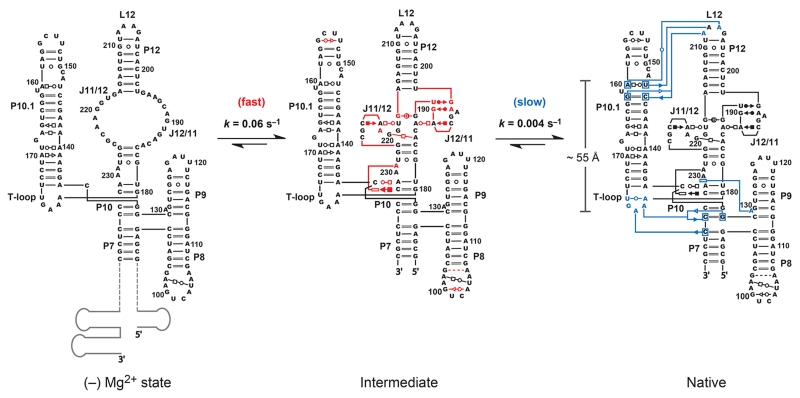
Mechanism for folding of the RNase P specificity domain determined by time-resolved SHAPE. Secondary structure is drawn to approximate arrangement of RNA helices and tertiary interactions as they occur in three dimensions2. Fast-forming interactions are emphasized in red. These are interpreted as reflecting formation of a distinct folding intermediate. Interactions that form in the slow, rate-determining, step are blue. Structure cassette sequences are shown schematically in gray. Reprinted with permission from the J. Am. Chem. Soc. 130:16178 (Copyright 2008 American Chemical Society).
Timing:
RNA folding (Steps 1-5): 20 min
RNA modification (Steps 6-11): Length of time course plus 1 h 30 min
Primer extension and RNA sequencing (Steps 12-22): 2 h
cDNA analysis by capillary electrophoresis (Step 23): 45 min
Data analysis (Steps 24-28): 1 or more days depending on the nature and depth of analysis.
TROUBLE SHOOTING
| Step | Problem | Reason | Solution |
|---|---|---|---|
| 1-21 | Intense peaks present in the (−) BzCN capillary electrophoresis trace. |
RNase contamination; this is, by far, the most common problem encountered for new practitioners of SHAPE technology. |
Identify contaminated solution by running mock SHAPE experiments with 5′ fluorescently or [32P]- labeled RNA. |
| 12-17 | Structure-induced pausing by the reverse transcriptase enzyme. |
Heat RNA in 0.5× TE to 95 °C for 3 min, cool on ice for 3 min before adding primer. Increase the time of extension at both 52 °C and 65 °C. |
|
| 9 | Very low signal in the (+) BzCN channel but an intense full length product is observed. |
Insufficient modification of RNA. |
Perform modification using a 2-fold higher concentration of BzCN. |
| 9 | No signal in the (+) BzCN trace. |
No modification of RNA. |
BzCN is very sensitive to trace amounts of water. Make sure BzCN/DMSO stock is kept dry and use fresh solutions for each experiment. |
| 9 | No full length product in the (+) BzCN trace or intense bands that disappear rapidly with read length. |
Excessive modification of RNA. |
Perform modification using a 2-fold lower concentration of BzCN. |
| 12-17 | No full length product in any channel in the capillary electrophoresis electropherogram. |
Poor or incomplete primer extension. |
The reverse transcriptase enzyme is very sensitive to MgCl2 concentration. Make sure final solution conditions are 3 mM in MgCl2. Enzyme is also sensitive to freezing. Retry the experiment with fresh enzyme. |
| 23 | Poor resolution or broad peaks in the electropherogram |
Too much residual salt present in sample when loaded onto the capillary electrophoresis instrument. |
Perform additional 70% ethanol wash before drying and resuspending pellet in formamide. |
| 15 | No sequencing bands in ddNTP sequencing trace (s). |
ddNTPs were incorporated in the wrong proportion. |
Adjust the ddNTP concentrations upwards or downwards by 2-fold. |
| 1,12 | Low fluorescence signal in electropherogram. |
Not enough primer or RNA in primer extension reactions. |
Increase amount of primer and/or RNA (to 5-8 pmol). |
| 23,24 | The faintest bands in the (+) BzCN channel have significantly different intensity as compared to the corresponding peaks in the (−) BzCN channel. |
Random error involved with volume measurement; difference in quantum yield of different fluorophores. |
Renormalize traces using the Scale Factor tool in ShapeFinder. Run primers labeled with each fluorophore to determine the multiplicative factor that yields equal intensities across fluorophores. |
| 23,24 | Sharp or negative peaks are present in the unprocessed or processed electropherogram. |
Improper automatic matrixing of dyes. |
Make sure dye overall dye intensity is roughly the same in all lanes. Use primer concentrations that yield similar intensities for each fluorescent signal. |
| 25-27 | Time-progress curves do not clearly fit a simple exponential equation. |
Multiple RNA folding populations or transitions. |
Ensure that the RNA is in a single defined conformation upon initiation of the folding reaction. Perform equilibrium SHAPE experiments on the RNA under initial folding conditions and evaluate whether data are consistent with a single conformation. |
Portions of this table are adapted from ref. 17.
ANTICIPATED RESULTS
Time-resolved SHAPE provides 1 s snapshots of RNA structure for RNAs of nearly arbitrary complexity over time periods spanning a few seconds to many minutes. The fluorescently encoded data for each individual time point are resolved in three or four channels in a single capillary from a capillary electrophoresis instrument (Figure 3a). Time-progress curves can also be monitored using 32P-labeled DNA primers and resolved on sequencing gels17, but data are generally of much higher quality using capillary electrophoresis.
The representative experiment outlined here was performed using an in vitro transcript corresponding to the Bacillus subtilis RNase P specificity domain25 embedded within a structure cassette that facilitates analysis of the very 5′ and 3′ ends of the RNA14. Tertiary folding of the RNase P RNA was initiated by addition of Mg2+ (to 10 mM) to a pre-equilibrated solution containing RNA, buffer, and 100 mM NaCl. Each capillary electrophoresis electropherogram (Figure 4a) contains reactivity data for the entire RNase P RNA (~150 nts). Peaks in the processed capillary electrophoresis traces are quantified by subtracting background intensities in the no-reaction control from the (+) BzCN reaction. After normalization, absolute SHAPE reactivities were obtained at almost every position within the RNA over a dense sampling of time points (Figure 4b). Nucleotides that show significant changes in SHAPE reactivity typically fall naturally into a smaller set of kinetically distinct behaviors. In the RNase P RNA, nucleotides partition into two distinct categories, both in terms of the observed rate constant and in terms of whether each curve was best fit by a single or double exponential. Approximately half the nucleotides in the RNase P RNA folded slowly (k1 = 0.004 s−1) in a transition characterized by a single exponential (in blue, Figure 4c).
The remaining nucleotides exhibiting significant time-dependent change in SHAPE reactivity were well described by fitting to a double exponential (in red, Figure 4c). The slower rate constant, 0.004 s−1, was identical to that observed for the positions characterized by a single exponential, while the faster rate constant, at 0.06 s−1, represents an additional kinetically significant step.
Time-resolved SHAPE data then make possible modeling of kinetic intermediates in the folding pathway for an RNA at nucleotide resolution. For the RNase P specificity domain RNA, time-resolved SHAPE clearly identifies two kinetically significant steps. A good first step is typically to superimpose the observed kinetic behavior on a secondary structure model for the RNA. Consistent with the two categories of reactivity changes, these changes cluster within two secondary (and tertiary) structure regions of the RNA. The fast folding step involves nucleotides in the tertiary structure module involving the J11/12 and J12/11 loops (residues 185-196 and 217-225) and also in the interaction of A229 with C134 and C232 (in red, Figure 5). Nucleotides characterized by the slower folding step involve the formation of three distinct sets of long-range interactions: docking of the T-loop (U175-A179) at P7-P10, stacking of A230 on A130 in P9, and the docking of the GAAA tetraloop (P12) into its receptor in P10.1 (in blue, Figure 5).
A comprehensive folding mechanism for the RNase P specificity domain was also developed prior to our work using equilibrium and kinetic approaches26,27. In general, nucleotide resolution information was obtained for folding intermediates under equilibrium conditions using chemical mapping experiments26 and time-resolved information was obtained for the overall folding of the whole RNA domain, including for relatively fast processes that are not accessible by bench top time-resolved SHAPE27. These approaches yielded a model that shares major features with that determined by time-resolved SHAPE, including that short-range tertiary interactions form prior to longer-range tertiary interactions and that the tetraloop-receptor and T-loop interactions form simultaneously in the rate determining step (Figure 5). A useful feature of time-resolved SHAPE was that sufficient information required to create a nucleotide-resolution model for the predominant RNA folding pathway could be obtained in a single, concise, set of experiments. Time-resolved SHAPE also revealed the distinct rates for each folding transition and the specific nucleotides involved in each step.
We envision that time-resolved SHAPE with BzCN will make possible facile analysis of the pathways for formation of individual sets of tertiary interactions in complex RNAs and RNP complexes on timescales as short as a few seconds using very simple bench top kinetics.
Acknowledgements
This work was supported by a grant from the National Science Foundation (MCB-0416941 to K.M.W.)
Footnotes
Author contributions: S.A.M. and K.M.W. collaborated on all aspects of the conception, design, presentation, and writing of this manuscript.
The authors declare that they have no competing financial interests.
REFERENCES
- 1.Tinoco I, Jr., Bustamante C. How RNA folds. J. Mol. Biol. 1999;293:271–281. doi: 10.1006/jmbi.1999.3001. [DOI] [PubMed] [Google Scholar]
- 2.Leontis NB, Westhof E. Analysis of RNA motifs. Curr. Opin. Struct. Biol. 2003;13:300308. doi: 10.1016/s0959-440x(03)00076-9. [DOI] [PubMed] [Google Scholar]
- 3.Gesteland RF, Cech TR, Atkins JF, editors. The RNA World. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 2004. [Google Scholar]
- 4.Webb AE, Weeks KM. A collapsed state functions to self-chaperone RNA folding into a native ribonucleoprotein complex. Nat. Struct. Biol. 2001;8:135–140. doi: 10.1038/84124. [DOI] [PubMed] [Google Scholar]
- 5.Furtig B, et al. Time-resolved NMR studies of RNA folding. Biopolymers. 2007;86:360–383. doi: 10.1002/bip.20761. [DOI] [PubMed] [Google Scholar]
- 6.Williamson JR. Biophysical studies of bacterial ribosome assembly. Curr. Opin. Struct. Biol. 2008;18:299–304. doi: 10.1016/j.sbi.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Woodson SA. RNA folding and ribosome assembly. Curr. Opin. Chem. Biol. 2008;12:667673. doi: 10.1016/j.cbpa.2008.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tijerina P, Mohr S, Russell R. DMS footprinting of structured RNAs and RNA-protein complexes. Nat. Protoc. 2007;2:2608–2623. doi: 10.1038/nprot.2007.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hennelly SP, et al. A Time-resolved investigation of ribosomal subunit association. J. Mol. Biol. 2005;346:1243–1258. doi: 10.1016/j.jmb.2004.12.054. [DOI] [PubMed] [Google Scholar]
- 10.Sclavi B, Woodson S, Sullivan M, Chance M, Brenowitz M. Following the folding of RNA with time-resolved synchrotron X-ray footprinting. Methods Enzymol. 1998;295:379–402. doi: 10.1016/s0076-6879(98)95050-9. [DOI] [PubMed] [Google Scholar]
- 11.Shcherbakova I, Brenowitz M. Monitoring structural changes in nucleic acids with single residue spatial and millisecond time resolution by quantitative hydroxyl radical footprinting. Nat. Protoc. 2008;3:288–302. doi: 10.1038/nprot.2007.533. [DOI] [PubMed] [Google Scholar]
- 12.Brenowitz M, Chance MR, Dhavan G, Takamoto K. Probing the structural dynamics of nucleic acids by quantitative time-resolved and equilibirum hydroxy radical ‘footprinting’. Curr. Opin. Struct. Biol. 2002;12:648–653. doi: 10.1016/s0959-440x(02)00366-4. [DOI] [PubMed] [Google Scholar]
- 13.Mortimer SA, Weeks KM. Time-resolved RNA SHAPE chemistry. J. Am. Chem. Soc. 2008;130:16178–16180. doi: 10.1021/ja8061216. [DOI] [PubMed] [Google Scholar]
- 14.Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. RNA structure analysis at single nucleotide resolution by Selective 2′-Hydroxyl Acylation and Primer Extension (SHAPE) J. Am. Chem. Soc. 2005;127:4223–4231. doi: 10.1021/ja043822v. [DOI] [PubMed] [Google Scholar]
- 15.Mortimer SA, Weeks KM. A fast-acting reagent for accurate analysis of RNA secondary and tertiary structure by SHAPE chemistry. J. Am. Chem. Soc. 2007;129:4144–4145. doi: 10.1021/ja0704028. [DOI] [PubMed] [Google Scholar]
- 16.Gherghe CM, Shajani Z, Wilkinson KA, Varani G, Weeks KM. Strong correlation between SHAPE chemistry and the generalized NMR order parameter (S2) in RNA. J. Am. Chem. Soc. 2008;130:12244–12245. doi: 10.1021/ja804541s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilkinson KA, Merino EJ, Weeks KM. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): Quantitative RNA structure analysis at single nucleotide resolution. Nat. Protoc. 2006;1:1610–1616. doi: 10.1038/nprot.2006.249. [DOI] [PubMed] [Google Scholar]
- 18.Wilkinson KA, et al. High-throughput SHAPE analysis reveals structures in HIV-1 genomic RNA strongly conserved across distinct biological states. PLoS Biol. 2008;6:e96. doi: 10.1371/journal.pbio.0060096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilkinson KA, Merino EJ, Weeks KM. RNA SHAPE Chemistry Reveals Nonhierarchical Interactions Dominate Equilibrium Structural Transitions in tRNAAsp Transcripts. J. Am. Chem. Soc. 2005;127:4659–4667. doi: 10.1021/ja0436749. [DOI] [PubMed] [Google Scholar]
- 20.Wang B, Wilkinson KA, Weeks KM. Complex Ligand-Induced Conformational Changes in tRNAAsp Revealed by Single-Nucleotide Resolution SHAPE Chemistry. Biochemistry. 2008;47:3454–3461. doi: 10.1021/bi702372x. [DOI] [PubMed] [Google Scholar]
- 21.Duncan CDS, Weeks KM. SHAPE analysis of long-range interactions reveals extensive and thermodynamically preferred misfolding in a fragile group I intron RNA. Biochemistry. 2008;47:8504–8513. doi: 10.1021/bi800207b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leaderach A, Shcherbakova I, Liang MP, Brenowitz M, Altman RB. Local Kinetic Measures of Macromolecular Structure Reveal Partitioning among Multiple Parallel Pathways from the Earliest Steps in the Folding of a Large RNA molecule. J. Mol. Biol. 2006;358:1179–1190. doi: 10.1016/j.jmb.2006.02.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gherghe CM, Mortimer SA, Krahn JM, Thompson NL, Weeks KM. Slow conformational dynamics at C2’-endo nucleotides in RNA. J. Am. Chem. Soc. 2008;130:8884–8885. doi: 10.1021/ja802691e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vasa SM, Guex N, Wilkinson KA, Weeks KM, Giddings MC. ShapeFinder: a software system for high-throughput quantitative analysis of nucleic acid reactivity information resolved by capillary electrophoresis. RNA. 2008;14:1979–1990. doi: 10.1261/rna.1166808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krasilnikov AS, Yang X, Pan T, Mondragon A. Crystal structure of the specificity domain of ribonuclease P. Nature. 2003;421:760–764. doi: 10.1038/nature01386. [DOI] [PubMed] [Google Scholar]
- 26.Baird NJ, Westhof E, Qin H, Pan T, Sosnick TR. Structure of a Folding Intermediate Reveals the Interplay Between Core and Peripheral Elements in RNA folding. J. Mol. Biol. 2005;352:712–722. doi: 10.1016/j.jmb.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 27.Baird NJ, Fang X, Srividya N, Pan T, Sosnick TR. Folding of a universal ribozyme:the ribonuclease P RNA. Q. Rev. Biophys. 2007;40:113–161. doi: 10.1017/S0033583507004623. [DOI] [PubMed] [Google Scholar]