Abstract
In cells, RNAs likely adopt numerous intermediate conformations prior to formation of functional RNA-protein complexes. We used single-nucleotide resolution SHAPE to probe the structure of E. coli 16S ribosomal RNA (rRNA) in healthy growing bacteria. SHAPE-directed modeling indicated that the predominant steady-state RNA conformational ensemble in dividing cells had a base paired structure different from that expected based on comparative sequence analysis and high-resolution studies of the 30S ribosomal subunit. We identified the major cause of these differences by stopping ongoing in-cell transcription (in essence, an in-cell RNA structure pulse-chase experiment) which caused the RNA to chase into a structure that closely resembled the expected one. Most helices that formed alternate RNA conformations under growth conditions interact directly with tertiary-binding ribosomal proteins and form a C-shape that surrounds the mRNA channel and decoding site. These in-cell experiments lead to a model in which ribosome assembly factors function as molecular struts to pre-organize this intermediate and emphasize that the final stages of ribonucleoprotein assembly involve extensive protein-facilitated RNA conformational changes.
Graphical abstract
Introduction
After transcription, RNA molecules undergo complex processing, folding, and protein and ligand association reactions essential for cellular function. The conformations of many RNAs have been well defined by biochemical and high-resolution structural determination approaches; however, RNA folding and RNA-protein assembly reactions have only recently been visualized at high resolution in living cells.1–4 Under optimal growth conditions, cells spend significant energy to assemble the ribosomes that synthesize the proteome necessary for all phases of cellular function.5 In bacteria, the ribosome is composed of 30S and 50S subunits that ultimately assemble to form the 70S ribosome. The 1542-nucleotide 16S ribosomal RNA (rRNA) of E. coli is the largest component of the 30S subunit and comprises a large fraction of the total RNA in a bacterium.6
Biogenesis of the 30S ribosomal subunit involves a complex series of interrelated events: The precursor 16S rRNA molecule is transcribed then processed via endonucleolytic cleavage to the mature full-length form and covalently modified at specific, conserved sites. The RNA folds and ribosomal proteins bind in a semi-ordered fashion to form the 30S subunit. Upon association with initiation factors and the 50S subunit, 30S subunits become part of the 70S ribosome. This large complex associates with elongation factors, tRNA, and mRNA to carry out protein synthesis. Ultimately, ribosomal subunits are disassembled and constituent RNAs are degraded.6–9
Significant strides have been made toward understanding ribosome assembly through in vitro experiments,10–12 and recent work has explored the protein and RNA compositions of assembly intermediates in cells.2–4 In vitro and in-cell assembly mechanisms differ. First, in vitro assembly experiments are generally performed using full-length ribosomal RNA, whereas cellular assembly occurs co-transcriptionally. Second, the rates of 30S assembly and cell-free translation in vitro are dramatically slower than the rates in vivo.6,13 Third, 30S assembly in vitro requires a sharp temperature increase6 and specialized buffer systems.14 Despite their importance, the relationships between in-cell RNA conformation and extensive characterizations in vitro are not well understood.
Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) quantitatively measures local nucleotide flexibility at all four nucleotides in an RNA.15,16 The most useful and structurally informative SHAPE reagent developed to date is 1M7. The reactivity of 1M7 is strongly correlated with model-free measures of molecular motion17 and incorporation of 1M7 reactivities as a pseudo-free energy term in RNA structure prediction algorithms yields accurate RNA secondary structure models.18,19 1M7 is a small, slightly hydrophobic molecule that readily crosses cell membranes.1,20 SHAPE reagents are initially suspended in a non-reactive organic solvent, such as DMSO, to prevent premature hydrolysis.21 Bacterial cells grown in 2 to 4% DMSO achieve growth rates and final cell densities comparable to cells grown in the absence of DMSO.1 Thus, SHAPE can be performed in vivo without impacting bacterial cell viability.
In this work, we use SHAPE to visualize the structure of 16S rRNA in ribosome assembly intermediates in healthy growing bacterial cells. We discovered that widespread conformational changes occur in the RNA during late stages of ribosome assembly. Together with other recent work, in-cell SHAPE analysis supports a simple "molecular strut" model for how late-acting ribosome assembly factors facilitate 30S assembly.
Methods
1M7 lifetime in growth media
Hydrolysis of 1M7 was followed by addition of 2.0 mM 1M7 in 100 µL DMSO to 900 µL Luria broth (LB) (pH 6.5 or 7.0) pre-equilibrated at 37 °C in a cuvette. Pseudo-first-order rate constants were obtained by monitoring the absorbance of the hydrolysis product (2-methylamino-4-nitrobenzoate) at 430 nm.1,21
SHAPE analysis of 16S rRNA structure in vivo
DH5α cells (500 mL) were grown in LB to an OD600 of ~0.6 starting from 5 mL of an overnight culture. RNA modification was initiated by adding an aliquot of cell culture (14 mL) to 433 µL of 167 mM 1M7 in DMSO (5 mM 1M7 final). The control cell sample was treated with neat DMSO (3% vol/vol). Samples were allowed to react with shaking for 5 min at 37 °C. An untreated aliquot of cell culture was obtained for dideoxy sequencing. Cells were collected by centrifugation (4,000 ×g for 20 min, 4 °C). Cell pellets were resuspended in 200 µL TE buffer containing 30 mg/mL lysozyme and incubated on ice for 5 min. RNA was purified by affinity chromatography (RNeasy Mini column; Qiagen). RNA concentrations were determined by absorbance at 260 nm. For all samples, the purified RNA contained total cellular RNA. For experiments in which transcription was inhibited, cell growth was initiated as above and, at an OD600 of ~0.5, 50 mL of cells were added to either 5.55 mL rifampicin in H2O (final concentration 18.75 µg/mL) or 5.55 mL H2O. Cells were incubated with vigorous shaking for 20 min and in-cell SHAPE was performed as described above.
Primer extension
Five primer sets were used to analyze the 16S rRNA with the 3′ positions of the primers complementary to 16S rRNA positions 323, 559, 947, 1112, and 1492.16 We were ultimately able to examine 94% of the nucleotides in the 16S rRNA, but did not analyze ~65 nts at the very 3' end of the RNA. Primer extension was performed as described22 except that modified total cellular RNA (2.1 µg) and primer (2 pmol) were mixed and diluted to 13 µL with water prior to the primer-annealing step. Dideoxy sequencing ladders were produced using unlabeled, unmodified total cellular RNA (in 12 µL) and 1 µL 2′,3′-dideoxycytidine (5 mM) triphosphate. cDNA fragments were separated by capillary electrophoresis using an Applied Biosystems 3500 capillary electrophoresis instrument.
Data analysis
Capillary electrophoresis traces were processed using QuShape.23 Datasets were normalized using model-free statistics to a scale of 0 to ~2. Nucleotides with normalized SHAPE reactivities 0–0.4, 0.4–0.85, and >0.85 correspond to unreactive, moderately reactive, and highly reactive positions, respectively. Each experiment was performed at least twice, and data were reproducible.
RNA structure modeling
Alternative secondary structure models were generated using RNAstructure24 in conjunction with SHAPE pseudo-free energy change constraints.18,19
Solvent accessibility
Solvent accessibility surface area (SASA) of the 2′-hydroxyl group was calculated with NACCESS 2.1.1 (http://www.bioinf.machester.ac.uk/naccess/) using a probe size of 2.8 Å and the atomic coordinates from 3i1m (30S) and 3i1n (50S).25 Nucleotides were taken to be surface nucleotides if the SASA was greater than 5 Å2. Nucleotides at the subunit interface of the 30S were defined as those that became inaccessible when the 50S subunit was added. Nucleotides in the 16S rRNA in direct contact with proteins were defined as those that became inaccessible when 30S proteins were added. Nucleotides in the 16S rRNA involved in RNA-RNA domain interactions were defined as nucleotides that became inaccessible when the other domains were included. Domains of the 16S rRNA were defined as: 5′, nts 1–566; central, nts 567–912; 3′-major, nts 913–1396; and 3′-minor, nts 1397–1542.
Results
Cell viability, time-scale, and quality of in vivo SHAPE
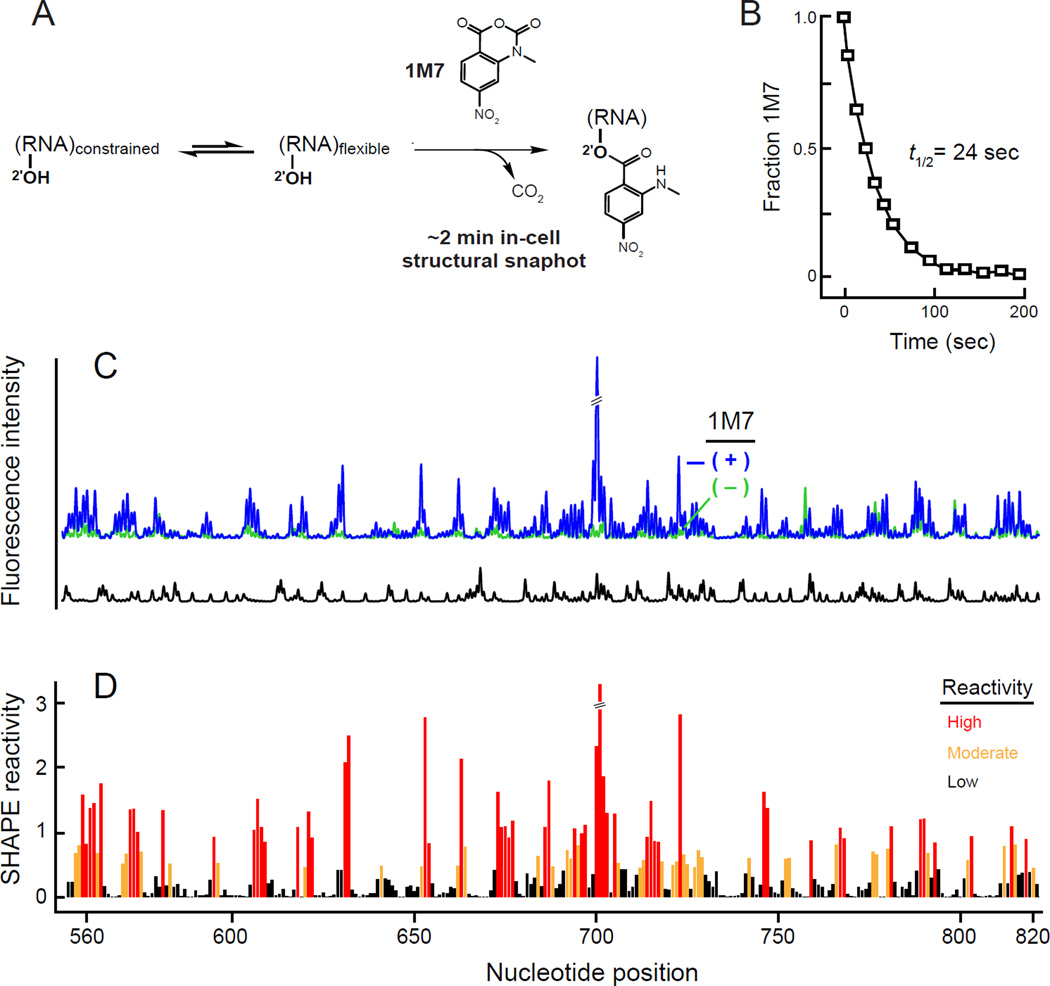
The hydrolysis half-life of 1M7 in standard growth media during mid-log phase of cell growth is 24 sec (Fig. 1A, B); under these conditions, the SHAPE reaction will be complete in approximately 2 minutes (5 hydrolysis half-lives). This is an ideal timescale because the 1M7 reagent is sufficiently stable to afford straightforward experimental use but makes rapid and facile in-cell structure analysis possible.
Figure 1. In vivo SHAPE of 16S ribosomal RNA.
(A) Schematic for using 1M7 to obtain ~2 min structural snapshots in vivo. (B) Pseudo-first order hydrolysis of 1M7 in Luria Broth (37 °C) at pH 7.0, corresponding to the mid-log phase of cell growth. This panel was adapted from prior work.1 (C) Electropherogram resulting from in-cell SHAPE probing of 16S rRNA. Region shown lies in the central domain. Reactions performed in the presence and absence of 1M7 are indicated by blue and green, respectively; dideoxy sequencing (ddC) trace is black. (D) Histogram of normalized SHAPE reactivities.23
In vivo SHAPE was performed by growing E. coli cell cultures to mid-log phase (OD600 = 0.6) and adding an aliquot of cells directly to 1M7 in DMSO (3% vol/vol, final). After 2 minutes, cells were pelleted and lysed, the cellular debris and proteins removed, and total E. coli cellular RNA purified. Sites of in vivo SHAPE adduct formation in the 1542-nucleotide 16S rRNA were detected by primer extension and resolved by capillary gel electrophoresis using five sets of DNA primers.16 Quantitative SHAPE reactivity data were obtained for 1,450 nucleotides (94%) of the 16S rRNA. The quality of the single-nucleotide resolution SHAPE data was very high as evidenced by a low level of background in the no-reagent control reaction and by the perfect agreement between the RNA-based sequencing reaction and the known 16S rRNA sequence (Fig. 1C,D). The 1M7 SHAPE reagent thus readily crossed the complex double membrane of the Gram-negative E. coli bacterial cell to yield an ~2-minute-resolution snapshot of the conformationally dynamics of almost all nucleotides in the 16S RNA.
16S rRNA structure in rapidly growing cells differs from the accepted structure
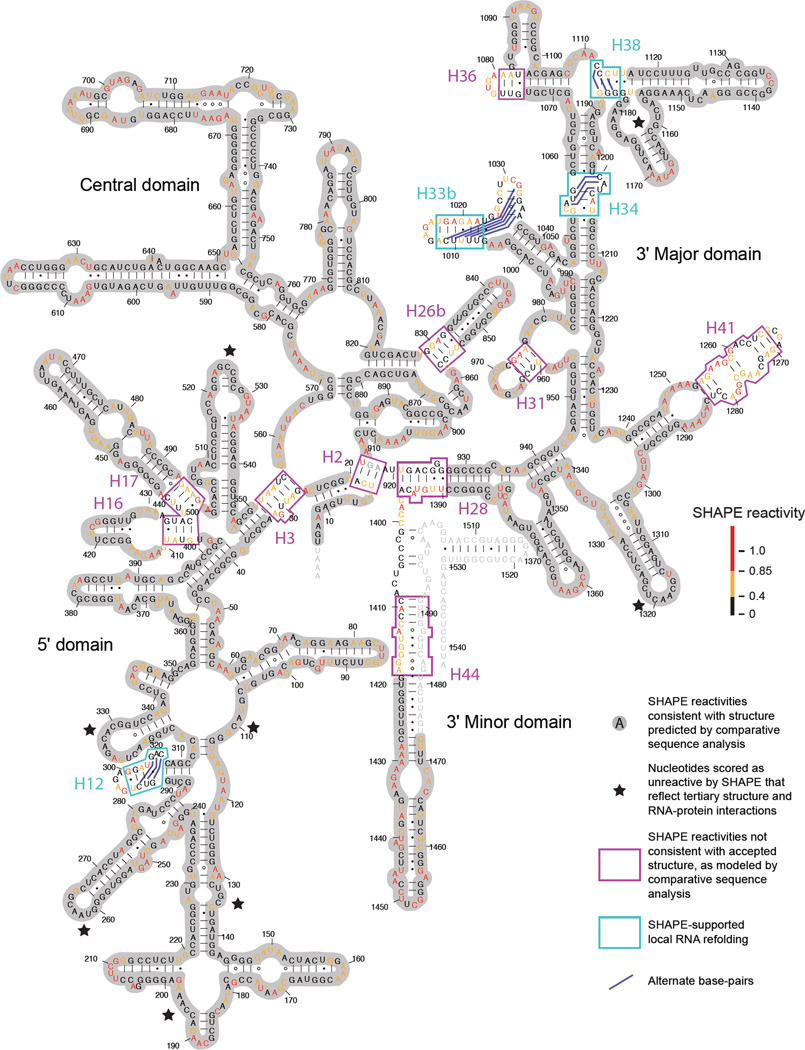
When the in vivo SHAPE reactivities were superimposed on the accepted secondary structure model26,27 of the E. coli 16S rRNA, the majority of reactivities showed excellent agreement with the accepted secondary structure (Fig. 2, grey highlights). In general, single-stranded nucleotides in the accepted structure showed medium or high reactivities, whereas most nucleotides expected to be base paired were unreactive by SHAPE.
Figure 2. In vivo SHAPE reactivities superimposed on the E. coli 16S secondary structure model determined by comparative sequence analysis.
Regions in which SHAPE data are consistent with the phylogenetically26 and crystallographically25 supported structures are shaded in gray. Helices where SHAPE data are not consistent with the secondary structure model are boxed. Alternate base pair pairs, supported by SHAPE probing, are emphasized with dark blue lines.
A small number of nucleotides predicted to be non-base paired in the accepted secondary structure model had low SHAPE reactivities. Since SHAPE measures molecular motion,17 it is likely these nucleotides are involved in higher-order tertiary contacts or interactions with proteins. These unreactive nucleotides in the 16S rRNA were examined in context of the three-dimensional structure of the 70S ribosome.25 Almost all of these unreactive nucleotides are indeed involved in RNA tertiary interactions or are located within or near protein binding sites (Fig. 2, black stars). For example, the unreactive nucleotides 132–135 and 260–265 interact via direct RNA-RNA contacts. Similarly, the unreactive loop nucleotides at positions 260–265, 321–323, 326–330 are all located near the protein-binding site for the S20 protein, one of the first proteins to bind during 30S assembly.6
Approximately 10% of 16S rRNA nucleotides predicted to be constrained by base pairing in the accepted structure exhibited medium or high SHAPE reactivities. These nucleotides were not spread uniformly throughout the RNA but were clustered in a small number of helices, most of which were located in irregular or multi-helix junctions (Fig. 2, magenta and cyan boxes). Of the 14 helices in which these nucleotides are located, ten have reactive nucleotides on both strands of the helix (H2, H3, H16/H17, H26b, H28, H31, H36, H41, and H44), making it unlikely that these helices are stably formed in the steady-state 16S rRNA structure in vivo (Fig. 2, magenta boxes). In four of the 14 regions (H12, H33b, H34, and H38), SHAPE-reactive nucleotides are adjacent to unreactive nucleotides that are single-stranded in the accepted secondary structure. In these regions, the predominant pattern of base pairing in vivo likely differs from that in the accepted secondary structure model.
We identified plausible alternative structures in these discordant regions by using the in-cell SHAPE data to model local RNA folding18,19 (Fig. 2, cyan boxes, alternate base pairs shown with dark blue lines). For example, in helix 34 in the accepted secondary structure, nucleotides 1055 and 1200–1202 are bulged; these nucleotides were expected to be reactive to SHAPE reagent but instead are unreactive in vivo. In contrast, nucleotides 1053, 1204, and 1205 are base paired in the accepted structure, but are reactive in vivo. In the alternate SHAPE-directed structure model, nucleotides 1200–1202 pair with nucleotides 1057–1055 (Fig. 2, dark blue lines in H34), and nucleotides 1053, 1054, and 1203–1205 are bulged. In-cell SHAPE also suggests local base pairing rearrangements relative to the accepted structure in helices H12, H33, and H38 (Fig. 2, cyan boxes). In each case, the alternative base-pairing patterns are more consistent with the SHAPE-measured in vivo nucleotide dynamics than is the accepted structural model.
At mid-log phase, E. coli cells are dividing actively, and levels of ribosomal RNA synthesis are high. Ribosome components, including the rRNA, are present at multiple stages of biogenesis. The experimental SHAPE reactivities measured in cells grown to mid-log phase may therefore reflect reactivities of 16S rRNA molecules that have not been fully assembled into 30S subunits. Differences observed between the SHAPE-directed model and the accepted secondary structure likely reflect the fact that the snapshot of the 16S rRNA taken by SHAPE in rapidly growing cells reflects the conformations of intermediates in ribosome assembly.
Halting transcription converts 16S rRNA structure to roughly the accepted one
Halting transcription will stop production of new 16S rRNA transcripts and should allow the16S rRNA to be incorporated into 30S subunits and 70S ribosomes. To evaluate the effect of transcription inhibition on 16S rRNA structure, we performed a kinetic "chase" experiment at nucleotide resolution in vivo. We added the antibiotic rifampicin, which inhibits transcription,28 to cells at mid-log phase (OD600 = 0.5). Rifampicin inhibits transcription by binding to the bacterial RNA polymerase, but does not bind to the ribosome. Changes in SHAPE reactivity of the 16S rRNA will thus reflect the effects of arrested transcription rather than direct effects on ribosomal RNA structure. Rifampicin halts transcription initiation within 70 seconds of addition. It requires approximately 2.2 minutes to transcribe an rRNA operon29 and an additional ~2 minutes for the 16S rRNAs to be processed and incorporated into 30S subunits.13 When cells were incubated with rifampicin for either 20 or 60 minutes prior modification with 1M7, SHAPE reactivity profiles were essentially identical. Therefore, we analyzed SHAPE reactivity profiles for the 16S rRNA 20 minutes after rifampicin addition (~5 half lives for rRNA synthesis and assembly).
During rRNA maturation, RNase III cleaves the primary 16S rRNA transcript to generate a form of the 16S rRNA precursor (the 17S rRNA) that contains an additional 115 and 33 nucleotides at the 5′ and 3′ ends, respectively, relative to the mature form.6 The amount of 17S precursor relative to mature 16S rRNA was quantified by primer extension analysis across the 5′ terminus of the mature rRNA.30 In log-phase cells without addition of antibiotic, 16% of the total 16S rRNA contained an unprocessed 5′ end. After incubation with rifampicin for 20 minutes, the percentage of RNA with an unprocessed 5′ end decreased to 2.5%. Thus, mid-log phase E. coli cells contain a significant fraction of incompletely processed 16S rRNA, and rifampicin addition promotes nearly complete conversion to the fully processed rRNA.
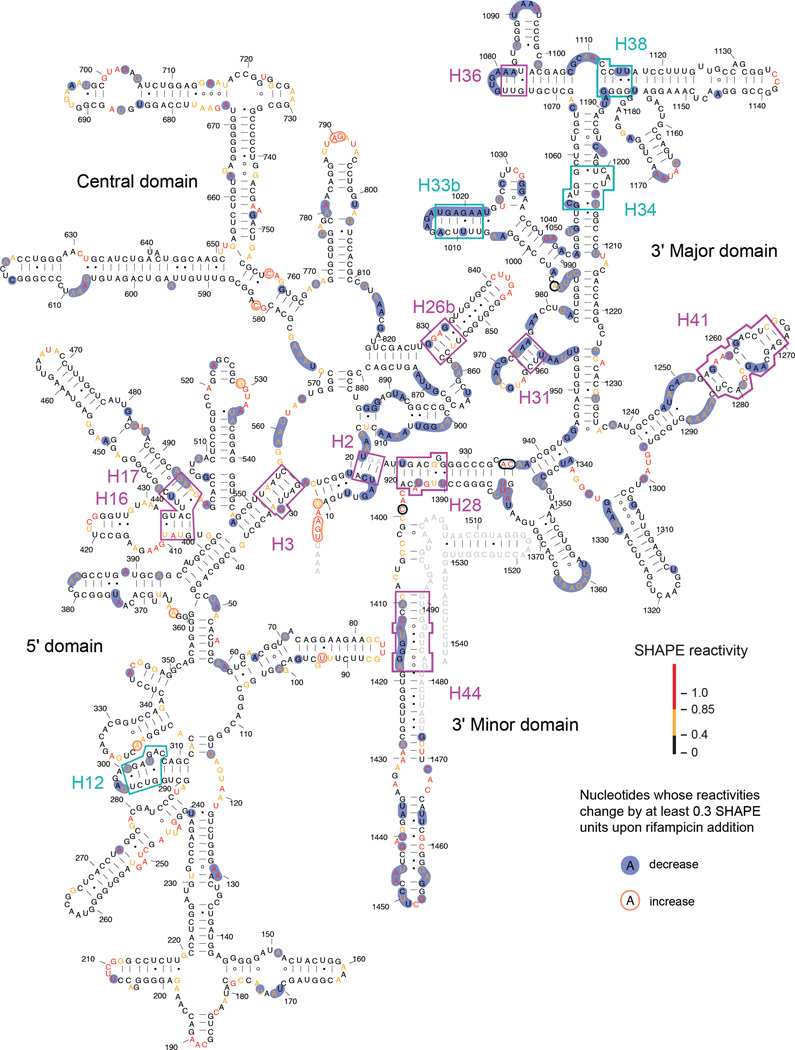
After inhibition of transcription, SHAPE reactivities for many nucleotides in the 16S rRNA differed from those measured in the absence of rifampicin. Of the nucleotides with moderate or high reactivity in the absence of rifampicin (defined as ≥ 0.4), more than half (273/509) exhibited a significant decrease (greater than 0.3 SHAPE units) in reactivity upon rifampicin addition (Fig. 3, highlighted in blue). Overall, the 16S rRNA becomes much more structured after transcription arrest. Of the 273 nucleotides that decreased by more than 0.3 SHAPE reactivity units upon inhibition of transcription, 58 (~20%) changed in a way that resolved discrepancies between the SHAPE-directed model and accepted secondary structure models (Fig. 3, magenta and cyan boxes). For example, the mean in vivo SHAPE reactivity of nucleotides in the H33b helix decreased from 0.5 to 0.2 when transcription was halted, consistent with formation of the base pairs observed in the accepted secondary structure. Transcription arrest ultimately yielded an in-cell SHAPE profile that was ~99% consistent with the accepted secondary structure. A small number (17 or ~1%) of 16S rRNA nucleotides increased by more than 0.3 SHAPE reactivity units upon rifampicin addition (Fig. 3, circled in orange). All of these nucleotides are in single-stranded or weakly paired regions, suggestive of changes in loop dynamics that accompanied formation of higher levels of overall structure in the 16S rRNA.16,17
Figure 3. In vivo SHAPE reactivities for the 16S rRNA after transcription inhibition.
Transcription was halted by treatment with rifampicin.28 Nucleotides with significant decreases or increases in SHAPE reactivity (≥ 0.3) due to transcription arrest are highlighted in blue and orange, respectively. Boxed helices are those shown in Fig. 2.
Strong 5' to 3' polarity in rifampicin-induced changes in 16S rRNA structure
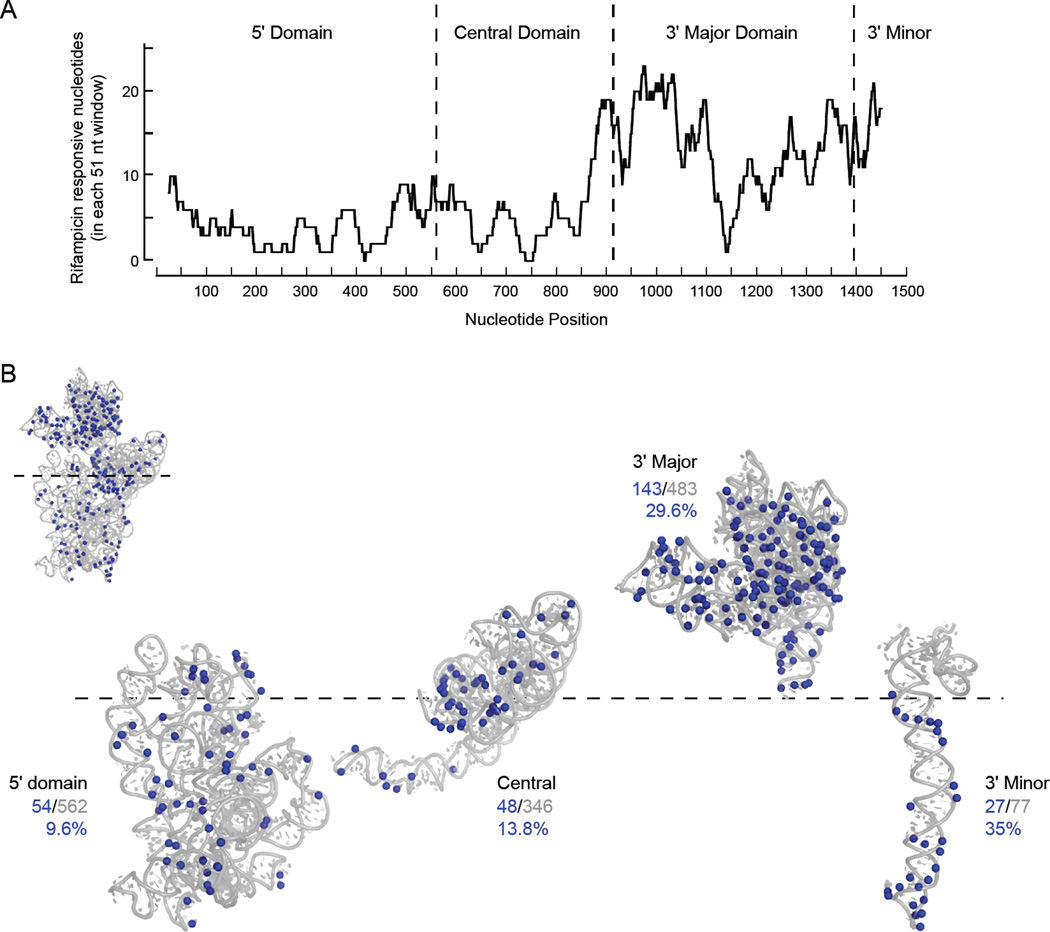
Roughly 20 times more nucleotides showed decreases in SHAPE reactivity versus increases upon rifampicin addition. There is a clear 5′ to 3′ polarity in the differences between SHAPE reactivity in the absence and the presence of rifampicin (Fig. 4A). As averaged over 51-nt windows, relatively few nucleotides in the 5' domain differed in SHAPE reactivity when reactivity profiles in the absence and presence of rifampicin are compared, but 30 to 35% of the nucleotides in the 3' major and 3' minor domains decreased significantly in reactivity in the absence of transcription. In addition, local peaks in the number of reactivity differences occur precisely at the junctions between domains of the 16S rRNA (Fig. 4A, vertical dashed lines). The positions that become less reactive to 1M7 in cells upon rifampicin treatment are superimposed on the three-dimensional structure of the 16S rRNA (Fig. 4B). The observed polarity in structural differences is consistent with an ordered ribosomal assembly process and with co-transcriptional folding and assembly into 30S subunits.
Figure 4. Transcription-dependent differences in SHAPE reactivity in the 16S rRNA.
(A) Positions with significant differences in SHAPE reactivity in the absence and presence of rifampicin. The number of nucleotides with significant differences in reactivity when the two conditions were compared were summed in 51-nucleotide windows. Domain junctions are emphasized with vertical dashed lines. (B) All nucleotides with significant differences in SHAPE reactivity in the absence and presence of rifampicin visualized on the structure of the 16S rRNA (3i1m).25 Ribosomal domains are expanded along a horizontal axis corresponding to the 5' to 3' polarity of their synthesis during transcription (dashed line). Inset shows complete ribosomal RNA. Number and percentage of nucleotides in each 16S rRNA domain whose SHAPE reactivities changed relative to the number of nucleotides in the domain are given.
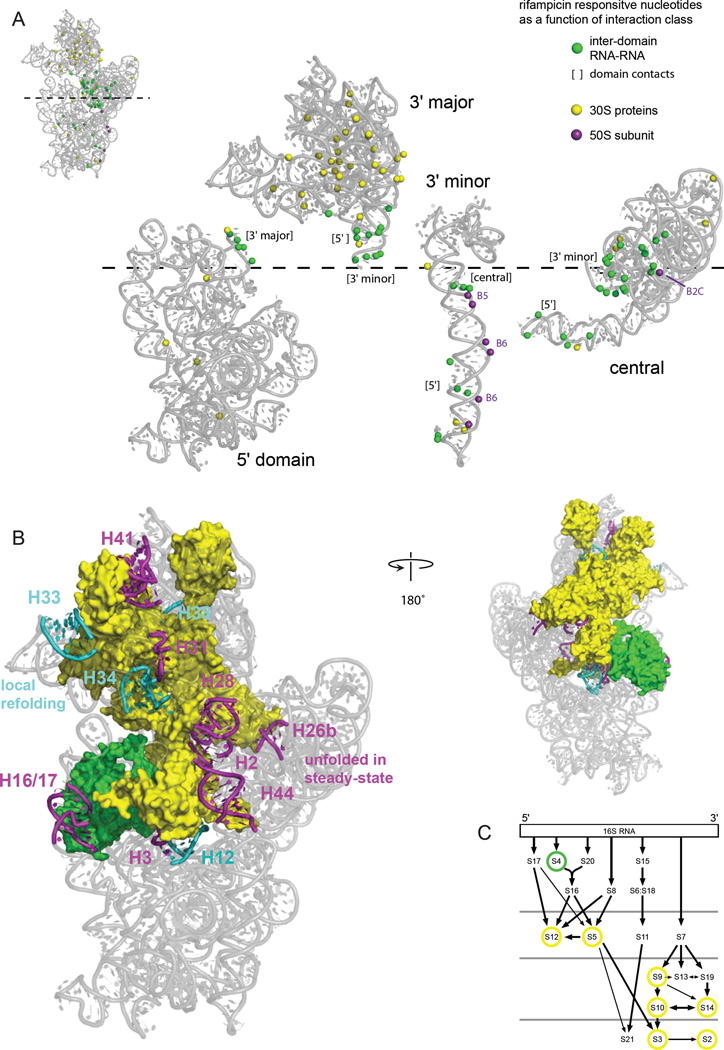
We next examined the role that higher-order interactions play in the in-cell assembly process by identifying nucleotides accessible to solvent in the context of each folded domain. SHAPE reactivities are not fundamentally governed by solvent or molecular accessibility: they are instead determined by local nucleotide dynamics.16,17 However, we used solvent accessibility as a metric to identify nucleotides located on the exterior of each domain in the 16S rRNA. We then identified the nucleotides that differed in reactivity in the presence and absence of rifampicin that became protected from solvent in the context of the crystal structure of the 70S ribosome and assigned these as predominately reflecting inter-domain RNA-RNA interactions (Fig. 5A, green spheres, 50 total), RNA-protein interactions (yellow spheres, 47 total), and interactions with the 50S subunit (purple spheres, 7 total). Most nucleotides whose protection likely reflects direct RNA-RNA domain interactions are located in the central domain and make contact with the 3′ minor domain (Fig. 5A, green spheres). Other nucleotides that differed in reactivity in the presence and absence of rifampicin are located at a compact interface between the 5' and 3' domains.
Figure 5. RNA-RNA, RNA-protein, and RNA-domain interactions detected by comparison of SHAPE reactivities in the presence and absence of transcription.
(A) The 2'-OH groups that show significant changes in SHAPE reactivity upon transcription inhibition and that are located at the surface of each ribosomal RNA domain are highlighted with spheres. Interacting groups, as inferred from the high-resolution structure, are emphasized by the coloring scheme shown at right. Domains are expanded in the same left-to-right orientation as in the standard view the intact 16S rRNA (see inset). Interactions with bridges to the 50S subunit are labeled. (B) Relationship between 16S rRNA helices that do not form stably in rapidly growing cells in the presence of transcription (magenta and cyan, as also shown in Fig. 2) juxtaposed with proteins in direct contact with these helices. (C) Positions of ribosomal proteins in direct contact with helices that change structure during assembly in a ribosome protein assembly map; yellow circles emphasize tertiary binding proteins.31 The four rows of proteins indicate early to late assembly intermediates, as measured by size of 30S-containing complexes in vivo.3
Almost half of the nucleotides protected upon rifampicin addition are in direct contact with ribosomal proteins and most of these primarily reflect RNA-protein interactions that occur in the 3' major domain (Fig. 5A, yellow spheres). Thus, protein association with the 3' domain appears to be a major rate-determining step for 30S subunit assembly in cells. We identified the small subunit proteins that are in direct contact with the 14 helices that undergo large conformational changes upon rifampicin addition (Figs. 2 and 3). Eight of the 20 small subunit proteins interact with the fourteen helices (Fig. 5C, circles). Strikingly, six of the eight proteins are those previously identified as tertiary-binding proteins31 (Fig. 5C, emphasized with yellow circles). In addition, S9, although not formally characterized as a tertiary-binding protein, participates in a network involving four of the tertiary-binding proteins (Fig. 5C, also emphasized with a yellow circle). The one additional protein-RNA interaction, involving S4 and the H16/17 RNA helix, undergoes a late conformational change distinct from the multiple changes involving the tertiary-binding proteins (Fig. 5C, green circle). Overall, the in-cell data imply that binding by the tertiary proteins is accompanied by widespread and previously unobserved structural changes in the 16S rRNA.
Discussion
SHAPE reveals intermediates in in-cell ribosome assembly
The 1M7 SHAPE reagent readily traverses the complex cell wall of the E. coli bacterium to modify RNA in a structure-selective manner. We used this reagent to obtain snapshots of the structure of 16S rRNA in bacteria at the mid-log phase of growth, when much of the rRNA has clearly not been fully assembled into ribosomes, and in the presence of a transcription inhibitor when most of the 16S rRNA has been incorporated into mature ribosomes. 1M7 elegantly balances the needs of experimental ease of use with high accuracy in probing nucleotide-resolution RNA dynamics.1,19,21
All experiments probing in-cell states likely report on complex ensembles of steady-state intermediates, including diverse RNA folding states and RNA-protein complexes comprised of heterogeneous complements of proteins. We found that 17% of 16S rRNA precursor transcripts have an incompletely processed 5' end and, thus, the “snapshot” taken using SHAPE in rapidly growing cells reflects 16S rRNA that is in various stages of assembly (Fig. 2). Our direct measurement of unprocessed RNA indicates a much larger fraction of 30S subunits are incompletely assembled in rapidly growing cells than generally assumed.13,32 Our results, although surprising, are consistent with recent studies showing high levels of ribosomal particles with incomplete complements of ribosomal proteins3 and with studies indicating that RNA is generally less structured in dividing cells.33
Critically, this work reveals a broad mechanism underlying the observation of increased RNA dynamics in cells. Upon inhibition of transcription using rifampicin, which results in conditions where the vast majority of 16S rRNA is incorporated into 30S subunits, SHAPE reactivities were ~99% consistent with the accepted secondary26 and tertiary25 structure (Fig. 3). Thus, many differences observed in growing cells reflect an on-going ribonucleoprotein assembly process.
There is a strong 5’ to 3’ polarity in the extent to which nucleotide reactivities changed in the absence compared to the presence of rifampicin (Fig. 4). This is consistent with co-transcriptional folding and assembly into 30S subunits. In addition, domain junctions were punctuated by significant differences in nucleotide reactivity in the presence and absence of rifampicin, consistent with formation of local inter-domain interactions. In-cell SHAPE, coupled with chasing complex steady-state RNA and ribonucleoprotein ensembles into more structurally mature forms by transcription inhibition, is a broadly useful approach for defining RNA-protein assembly intermediates and inter-domain junctions in ribonucleoprotein complexes.
Domain-specific assembly features
For the 16S ribosomal RNA studied here, the differences in nucleotide reactivities in the presence and absence of rifampicin reflect distinct underlying physical processes in each ribosomal domain. In the 5' domain, all nucleotides whose reactivities were responsive to rifampicin showed decreases in reactivity. Moreover, over 80% (44/54) of these nucleotides are located in RNA elements that form intra-domain interactions (Figs 4B and 5A, 5’ domain panels). Thus, the 5' domain undergoes internal tightening and consolidation in structure in mature ribosomes compared to less mature forms. In contrast, for the central domain, roughly 65% (31/48) of the nucleotides that decreased in reactivity are involved in inter-domain or RNA-protein interactions, with the majority reflecting RNA-RNA interactions (compare Figs. 4B and 5A). One nucleotide reflects a direct interaction with the 50S subunit and forms part of the B2C inter-subunit bridge (Fig. 5A, purple sphere).34 The pattern is again distinctive in the 3' major and minor domains. Of the 47 nucleotides that decrease in reactivity in the presence of rifampicin and likely report RNA-protein interactions, 66% (31/47) are located in the 3′ major subunit (Fig. 5A, yellow spheres). In addition, six nucleotides which decreased in reactivity in the 3’ minor domain reflect direct interactions with the 50S subunit (Fig. 5A, purple spheres). Of these six nucleotides, five form the B5 and B6 inter-subunit bridges.34.
In sum, the numbers of rRNA nucleotides with differential reactivity by SHAPE in the absence and presence of rifampicin increase in the 5' to 3' direction. These changes show a striking structural polarity reflecting the assembly of the ribosome and begin predominantly with intra-domain RNA-RNA interactions (5' domain), followed by inter-domain RNA-RNA interactions (central domain), then intra-subunit (30S) RNA-protein interactions (3' major domain), and finally inter-subunit interactions (3' minor domain).
The predominant 30S state in growing bacteria is a late assembly intermediate
This and other recent work1 emphasizes that the cellular environment has large and poorly understood effects on RNA structure. For completely synthesized transcripts, the highly crowded environment in an E. coli cell appears to stabilize and favor more organized higher-order RNA structure.1 This effect undoubtedly influences bacterial ribosome structure, as well. Here we observed an additional effect of the cellular environment: the effect of on-going co-transcriptional assembly and structural biogenesis. During the snapshot taken during our in-cell probing experiment, the steady-state structure of 16S rRNA in healthy growing E. coli cells is heavily influenced by 16S rRNA that has not yet been fully assembled into mature 30S subunits.
Fourteen helical structures do not form stably in the intermediate state that predominates in rapidly growing cells (Fig. 2). Current evidence strongly suggests that folding of these helices coincides with a critical, late-stage, rate-determining step in ribosome assembly. First, eight of these helices (H2, H3, H12, H28, H34, H36, H38 and H44) are also unfolded when ribosomes are deproteinized.18 These unfolded structures thus comprise thermodynamically preferred RNA states. Second, chemical modification of seven of these same helices (H2, H12, H28, H31, H34, H36 and H44) interferes with ribosome assembly.35 Third, these helices are similar to those that are either improperly folded or interact with protein partners that are missing in immature 30S subunits created by deleting the ribosome late-assembly factors RbfA and RimM.4,36 Finally, six of the eight proteins that bind directly to the unfolded helices were defined as late-binding tertiary proteins in the original Nomura maps31 and have recently been shown to bind during late stages of ribosome assembly in vivo.3 (Fig. 5B and 5C, in yellow). Thus, although there are likely to be multiple species present, a significant fraction of the still-maturing interactions detected in the 2-min SHAPE snapshot correspond to events that occur at late stages in 30S subunit assembly.
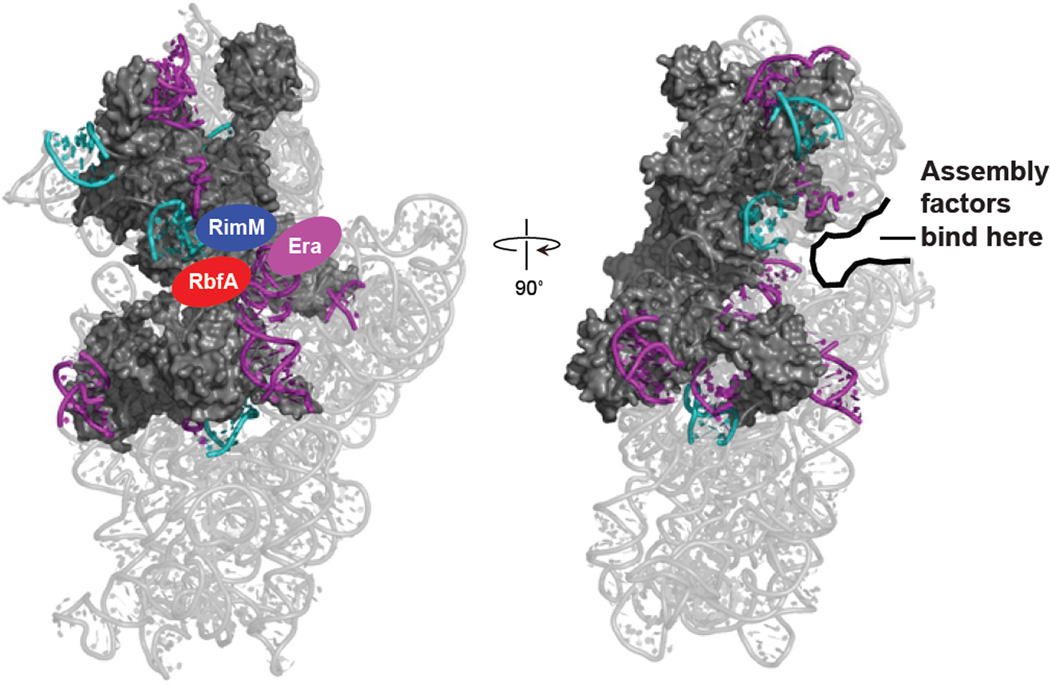
A molecular strut model for ribosomal assembly factors
The 14 incompletely folded helices, seven late-assembly proteins, and S4 form a C-shaped motif spanning the 5' and 3' domains of the 16S rRNA (Fig. 6). This region surrounds the mRNA binding channel, is important for tRNA binding and codon-anticodon decoding, and must be properly folded and arranged for ribosome function.37–39 Three factors, RbfA, RimM, and Era, have been shown to facilitate late-stage 30S subunit assembly. These three assembly factors bind at well-defined but distinct and largely non-overlapping sites, and can, in part, suppress deletion of or compensate for each other.6,40 These proteins all bind in the concave face of the C-shaped structure formed by the helices and proteins that we have shown rearrange in late stages of ribosome assembly (Fig. 6). We interpret the partially overlapping functions of these assembly factors in terms of a "molecular strut" model. Late stage assembly involves large-scale RNA structural rearrangements (Fig. 3) and protein assembly events (Fig. 5) that we hypothesize require, or are facilitated by, partial pre-organization of the C-shaped interaction network. In this model, assembly factors function in part to promote organization the C-shaped mRNA channel area by binding in the cleft. An individual factor could therefore compensate for contributions by another assembly factor, as long as this structural organization function were provided.
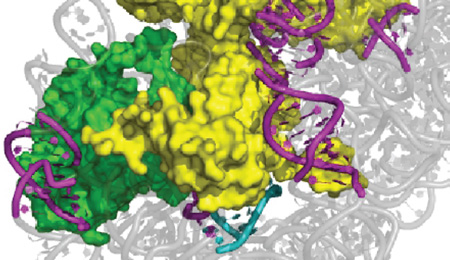
Figure 6. Positions of misfolded helices and late-binding proteins relative to late-stage ribosome assembly factors.
Positions of RbfA, RimM and Era are as defined by electron microscopy.36,41,42 Misfolded helices are in cyan and purple (see Fig. 2) and the proteins that directly contact these RNA elements are shown as gray surfaces.
Summary
In-cell SHAPE probing of growing E. coli cells showed that ribosomal assembly intermediates accumulate and may even predominate in rapidly growing cells. The SHAPE-detected transcriptional pulse-chase approach outlined in this work is both straightforward and information-rich and will likely find broad utility in understanding the steady-state RNA dynamics of other RNAs and RNA-protein complexes, and in other cell types.
Acknowledgments
This work was supported by the US National Science Foundation (MCB-1121024 to K.M.W.).
References
- 1.Tyrrell J, McGinnis JL, Weeks KM, Pielak GJ. Biochemistry. 2013;52:8777. doi: 10.1021/bi401207q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Swiatkowska A, Wlotzka W, Tuck A, Barrass JD, Beggs JD, Tollervey D. RNA. 2012;18:2187. doi: 10.1261/rna.034751.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen SS, Williamson JR. J Mol Biol. 2013;425:767. doi: 10.1016/j.jmb.2012.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clatterbuck Soper SF, Dator RP, Limbach PA, Woodson SA. Mol Cell. 2013;52:506. doi: 10.1016/j.molcel.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gourse RL, Gaal T, Bartlett MS, Appleman JA, Ross W. Annu Rev Microbiol. 1996;50:645. doi: 10.1146/annurev.micro.50.1.645. [DOI] [PubMed] [Google Scholar]
- 6.Shajani Z, Sykes MT, Williamson JR. Annu Rev Biochem. 2011;80:501. doi: 10.1146/annurev-biochem-062608-160432. [DOI] [PubMed] [Google Scholar]
- 7.Kaczanowska M, Ryden-Aulin M. Microbiol Mol Biol Rev. 2007;71:477. doi: 10.1128/MMBR.00013-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nierhaus KH. Biosystems. 1980;12:273. doi: 10.1016/0303-2647(80)90024-6. [DOI] [PubMed] [Google Scholar]
- 9.Granneman S, Baserga SJ. Exp Cell Res. 2004;296:43. doi: 10.1016/j.yexcr.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 10.Calidas D, Culver GM. RNA. 2011;17:263. doi: 10.1261/rna.2332511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Woodson SA. Acc Chem Res. 2011;44:1312. doi: 10.1021/ar2000474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mulder AM, Yoshioka C, Beck AH, Bunner AE, Milligan RA, Potter CS, Carragher B, Williamson JR. Science. 2010;330:673. doi: 10.1126/science.1193220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lindahl L. J Mol Biol. 1975;92:15. doi: 10.1016/0022-2836(75)90089-3. [DOI] [PubMed] [Google Scholar]
- 14.Blaha G, Stelzl U, Spahn CM, Agrawal RK, Frank J, Nierhaus KH. Methods Enzymol. 2000;317:292. doi: 10.1016/s0076-6879(00)17021-1. [DOI] [PubMed] [Google Scholar]
- 15.Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. J Am Chem Soc. 2005;127:4223. doi: 10.1021/ja043822v. [DOI] [PubMed] [Google Scholar]
- 16.McGinnis JL, Dunkle JA, Cate JH, Weeks KM. J Am Chem Soc. 2012;134:6617. doi: 10.1021/ja2104075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gherghe CM, Shajani Z, Wilkinson KA, Varani G, Weeks KM. J Am Chem Soc. 2008;130:12244. doi: 10.1021/ja804541s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deigan KE, Li TW, Mathews DH, Weeks KM. Proc Natl Acad Sci U S A. 2009;106:97. doi: 10.1073/pnas.0806929106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hajdin CE, Bellaousov S, Huggins W, Leonard CW, Mathews DH, Weeks KM. Proc Natl Acad Sci U S A. 2013;110:5498. doi: 10.1073/pnas.1219988110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilkinson KA, Gorelick RJ, Vasa SM, Guex N, Rein A, Mathews DH, Giddings MC, Weeks KM. PLoS Biol. 2008;6:e96. doi: 10.1371/journal.pbio.0060096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mortimer SA, Weeks KM. J Am Chem Soc. 2007;129:4144. doi: 10.1021/ja0704028. [DOI] [PubMed] [Google Scholar]
- 22.Steen KA, Rice GM, Weeks KM. J Am Chem Soc. 2012;134:13160. doi: 10.1021/ja304027m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karabiber F, McGinnis JL, Favorov OV, Weeks KM. RNA. 2013;19:63. doi: 10.1261/rna.036327.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mathews DH, Disney MD, Childs JL, Schroeder SJ, Zuker M, Turner DH. Proc Natl Acad Sci U S A. 2004;101:7287. doi: 10.1073/pnas.0401799101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang W, Dunkle JA, Cate JH. Science. 2009;325:1014. doi: 10.1126/science.1175275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cannone JJ, Subramanian S, Schnare MN, Collett JR, D'Souza LM, Du Y, Feng B, Lin N, Madabusi LV, Muller KM, Pande N, Shang Z, Yu N, Gutell RR. BMC Bioinformatics. 2002;3:2. doi: 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gutell RR, Lee JC, Cannone JJ. Curr Opin Struct Biol. 2002;12:301. doi: 10.1016/s0959-440x(02)00339-1. [DOI] [PubMed] [Google Scholar]
- 28.Hartmann G, Honikel KO, Knusel F, Nuesch J. Biochim Biophys Acta. 1967;145:843. doi: 10.1016/0005-2787(67)90147-5. [DOI] [PubMed] [Google Scholar]
- 29.Gotta SL, Miller OL, Jr, French SL. J Bacteriol. 1991;173:6647. doi: 10.1128/jb.173.20.6647-6649.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roy-Chaudhuri B, Kirthi N, Culver GM. Proc Natl Acad Sci U S A. 2010;107:4567. doi: 10.1073/pnas.0912305107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Traub P, Nomura M. Proc Natl Acad Sci U S A. 1968;59:777. doi: 10.1073/pnas.59.3.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laughrea M, Tam J. Biochemistry. 1992;31:12035. doi: 10.1021/bi00163a011. [DOI] [PubMed] [Google Scholar]
- 33.Rouskin S, Zubradt M, Washietl S, Kellis M, Weissman JS. Nature. 2014;505:701. doi: 10.1038/nature12894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cate JH, Yusupov MM, Yusupova GZ, Earnest TN, Noller HF. Science. 1999;285:2095. doi: 10.1126/science.285.5436.2095. [DOI] [PubMed] [Google Scholar]
- 35.Xu Z, Culver GM. RNA. 2010;16:1990. doi: 10.1261/rna.2246710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guo Q, Goto S, Chen Y, Feng B, Xu Y, Muto A, Himeno H, Deng H, Lei J, Gao N. Nucleic Acids Res. 2013;41:2609. doi: 10.1093/nar/gks1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo Z, Noller HF. Proc Natl Acad Sci U S A. 2012;109:20391. doi: 10.1073/pnas.1218999109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Noeske J, Cate JH. Curr Opin Struct Biol. 2012;22:743. doi: 10.1016/j.sbi.2012.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jomaa A, Stewart G, Martin-Benito J, Zielke R, Campbell TL, Maddock JR, Brown ED, Ortega J. RNA. 2011;17:697. doi: 10.1261/rna.2509811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Donhofer A, Sharma MR, Datta PP, Nierhaus KH, Agrawal RK, Wilson DN. Encyclopedia of Life Sciences. Chichester: John Wiley & Sons, Ltd; 2009. [Google Scholar]
- 41.Sharma M, Barat C, Wilson D, Booth T, Kawazoe M, Hori-Takemoto C, Shirouzu M, Yokoyama S, Fucini P, Agrawal R. Mol. Cell. 2005;18:319. doi: 10.1016/j.molcel.2005.03.028. [DOI] [PubMed] [Google Scholar]
- 42.Datta P, Wilson D, Kawazoe M, Swami N, Kaminishi T, Sharma M, Booth T, Takemoto C, Fucini P, Yokoyama S, Agrawal R. Mol. Cell. 2007;28:434. doi: 10.1016/j.molcel.2007.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]