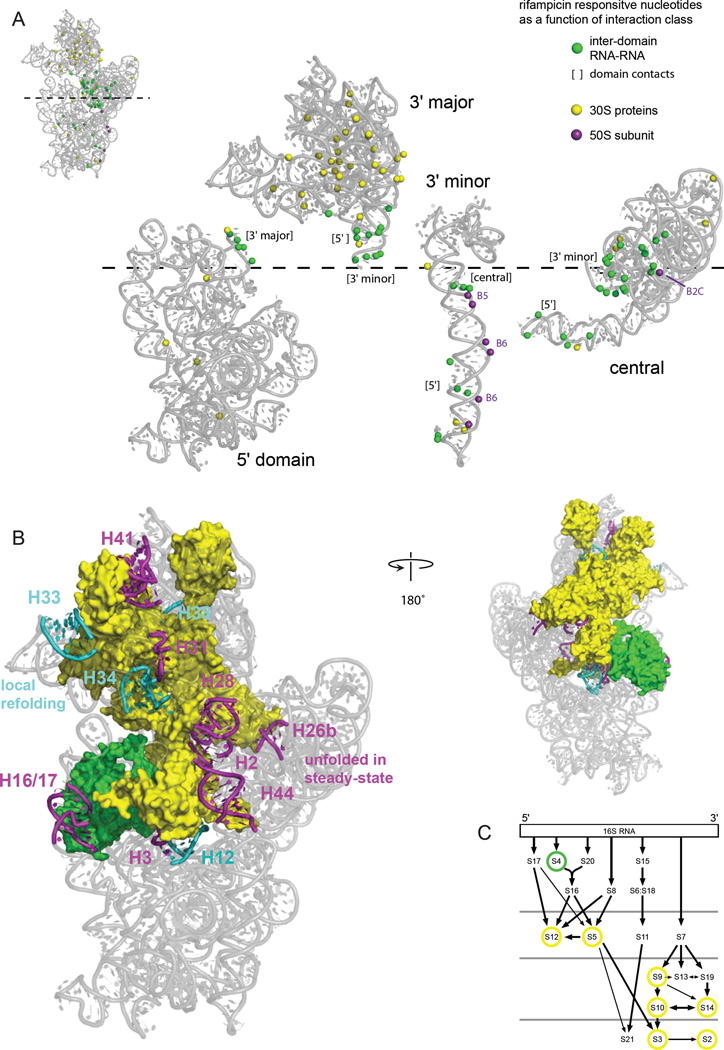
Figure 5. RNA-RNA, RNA-protein, and RNA-domain interactions detected by comparison of SHAPE reactivities in the presence and absence of transcription.
(A) The 2'-OH groups that show significant changes in SHAPE reactivity upon transcription inhibition and that are located at the surface of each ribosomal RNA domain are highlighted with spheres. Interacting groups, as inferred from the high-resolution structure, are emphasized by the coloring scheme shown at right. Domains are expanded in the same left-to-right orientation as in the standard view the intact 16S rRNA (see inset). Interactions with bridges to the 50S subunit are labeled. (B) Relationship between 16S rRNA helices that do not form stably in rapidly growing cells in the presence of transcription (magenta and cyan, as also shown in Fig. 2) juxtaposed with proteins in direct contact with these helices. (C) Positions of ribosomal proteins in direct contact with helices that change structure during assembly in a ribosome protein assembly map; yellow circles emphasize tertiary binding proteins.31 The four rows of proteins indicate early to late assembly intermediates, as measured by size of 30S-containing complexes in vivo.3