Abstract
Neutral red (NR) is a dye that must be actively imported into the cell, and, therefore, the dye has been used for decades to selectively stain living cells. In addition, NR can also be incorporated into virus particles, although the mechanism behind this is poorly understood. Once encapsulated into the virion, NR, a light sensitive dye, can be photoactivated to inactivate the virus. The proposed mechanism explaining this observation is that activation of NR allows the dye to cross-link viral genome to viral capsid and thus preventing viral uncoating and infection. To study the early events of murine norovirus (MNV)-host interaction, light-sensitive NR-containing MNV is used to distinguish between input virus (i.e., NR-containing virus) and replicated virus (i.e., NR-free virus). This protocol describes the incorporation of NR into MNV capsids and the use of these virions for detection of viral replication in a mouse and in tissue culture by standard plaque assay. The same technique is also used for study of poliovirus replication (1–3). Thus, there is the potential that this technique can be used for additional non-enveloped viruses. However, this has to be tested on a case-by-case basis as unpublished data on feline calicivirus suggests not all viruses may be able to stably incorporate NR into their capsid (J. Parker, personal communication).
Materials and Reagents
Neutral red 0.33% solution (Sigma-Aldrich, catalog number: N2889)
DMEM/High glucose (Hyclone, catalog number: SH30243.02)
2× MEM (Life Technologies, Gibco®, catalog number: 11935)
100× Penicillin and streptomycin (Hyclone, catalog number: SV30010)
100× Non-essential amino acids (Hyclone, catalog number: SH30238.01)
1 M HEPES (Hyclone, catalog number: SH30237.01)
200 mM (100x) L-glutamine (Hyclone, catalog number: SH30034.01)
Fetal bovine serum (FBS) (Life Technologies, Gibco®, catalog number: 10437 or Hyclone, catalog number: SH30070.02)
RAW 264.7 cell line (ATCC, catalog number: TIB-71)
Murine norovirus (MNV-1; GV/MNV1/2002/USA)
Sea Plaque Agarose (Lonza, catalog number: 50100)
Aluminum foil
1.0 mm Zirconia/Silica beads (BioSpec Products, catalog number: 11079110z)
Mice (6–8 weeks old Balb/cJ)
DMEM-10 (see Recipes)
DMEM-5 (see Recipes)
2× MEM media (500 ml) (see Recipes)
SeaPlaque agarose (see Recipes)
1:1 ratio of molten media (see Recipes)
Equipment
175 cm2 flask (Corning, catalog number: 3292)
Model 35 Speed Rocker (Labnet, catalog number: S2035)
Tissue culture incubator (Sanyo, catalog number: MCO-36M)
Magna Lyser Bead beater (F. Hoffmann-La Roche, catalog number: 03358968001)
Photographic safe red light
Procedure
-
Neutral red virus preparation
Seed RAW 264.7 cells in a total of 30 ml of DMEM-10 media at a density of 4 × 107 cells in a 175 cm2 flask and incubate in a tissue culture incubator at 37 °C and 5% CO2 overnight.
To obtain a virus stock, infect cells the next day with MNV by adding MNV at a multiplicity of infection (MOI) of 0.05 and incubate for 1 h in a tissue culture incubator at 37 °C and 5% CO2.
After the hour, add neutral red at 0.001% v/v from a 0.33% stock solution to the cells.
Wrap the entire flask with aluminum foil including the filter cap of the flask.
Incubate in a tissue culture incubator at 37 °C and 5% CO2 for 48 h.
-
After the 48 h, generate a freeze/thaw lysate. Freeze cells by moving the entire flask (including cells and media) covered with aluminum foil to −80 °C for at least 60 min. Then thaw flask contents at room temperature or in a tissue culture incubator at 37 °C until all ice has melted. Repeat freeze/thaw a second time. This will be your virus stock.
Note: After the second freeze/thaw cycle, virus stock should be aliquoted (in a darkened room) into single-use aliquots (e.g., 1 ml) and stored at −80 °C since light-sensitivity of virus stock decreases after repeated freeze/thaw cycles.
Block out all light in your tissue culture room to make a dark room and equip with a safe red light used in photography dark rooms.
Perform plaque assay to determine viral titer of your virus stock as previously described in detail (2) and as described briefly below. The day before the plaque assay seed 2 × 106 Raw 264.7 cells in DMEM-10/well in six well plates.
On the day of the plaque assay, perform 10-fold serial dilutions of your virus stock in DMEM-5. Make sure to adjust the volume for duplicate plaque assays, i.e., you will need a total volume of 2.5 ml for each dilution.
-
Next, take the six well plates with cells seeded on the day before, aspirate the media and infect cells in the dark in duplicate wells for each dilution with 0.5 ml/well, wrap in aluminum foil and rock for 1 h at room temperature with a speed rocker at ~18 oscillations per min.
Note: The 1 h incubation was determined experimentally to ensure efficient entry and uncoating of MNV. However, the length of time necessary for other viruses may vary.
Turn on the lights and leave the remaining 10-fold viral dilutions in the light for at least 10 min.
Repeat step 1-j. but keep plates exposed to light (i.e., do not wrap plates in aluminum foil).
After the hour, finish the plaque assay for both sets of plates with or without aluminum foil in the light by adding 2 ml/well of a 1:1 ratio of molten media (1:1 ratio of SeaPlaque agarose and 2× MEM). To allow agarose to solidify incubate plates at room temperature for 10 min. Then, move plates to a tissue culture incubator and incubate at 37 °C, 5% CO2 for 48 h.
-
After 48 h stain cells with a neutral red solution overlay (0.01% neutral red in 1× PBS), for 1–3 h prior to aspirating the solution and counting plaques as described previously in detail ( Gonzalez-Hernandez et al., 2012).
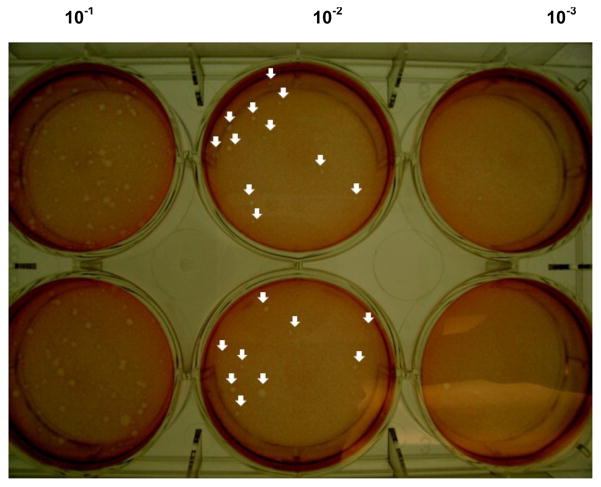
Note: To calculate viral titers and obtain plaque forming units per milliliter (PFU/ml), add the number of plaques in both wells at a single dilution and multiply by the dilution factor (i.e., 1 ml if 2 wells are infected with 0.5 ml). Figure 1 shows an example of a 6 well plate with plaques. To calculate the viral titer in the example: 11 + 9 = 20 plaques; 20 × 102 dilution factor = 2 × 103 PFU/ml. Two-to three-log reductions are typically observed in viral titers comparing total virus titers (i.e. obtained in dark) and light-insensitive (i.e. obtained in light) infections. Depending on the viral strain, log reductions may differ.
-
Tissue culture infection with neutral red virus
Seed 2 × 106 Raw 264.7 cells in six well plates the day before.
The next day, block out all light in your tissue culture room to make a dark room and equip with a safe red light used in photography dark rooms.
-
Infect with MNV containing NR at an MOI of 0.001 for 1 h.
Note: Pre-treatments with inhibitors of viral entry or uncoating can be performed before incubating with virus. Post-treatments with the same inhibitors are performed as a control after the 1 h incubation with virus as described (Perry, 2010).
-
Flash with light for various lengths of times between 0 to 60 min at the end of the infection time.
Note: For MNV, times between 0 and 60 min are typically used and at least 10 min has been sufficient to inactivate any leftover input virus (Perry, 2010).
Overlay with 2 ml of a 1:1 ratio of molten media (1:1 ratio of SeaPlaque agarose and 2× MEM), as you would do for plaque assays and as described (Gonzalez-Hernandez et al., 2012; Perry et al., 2012).
Incubate for 48–72 h in a tissue culture incubator at 37 °C and 5% CO2.
Count plaques by adding the number of plaques in both wells at a single dilution and multiplying by the dilution factor, as described above and previously (Gonzalez-Hernandez et al., 2012; Perry et al., 2012).
-
Mouse infection with neutral red virus
-
Inoculate mice perorally by pipetting virus directly into the mouth or by oral gavage with NR-MNV in a darkened room with a safe red light.
Note: We infected six-to eight-week old Balb/c mice perorally with NR-MNV at 105 pfu. Depending on the experiment, the length of infection will vary. We typically infect mice for 12–72 h. At the experimental endpoint (i.e. 12 h post-infection), sacrifice mice in a darkened room with a safe red light and harvest different regions of the gastrointestinal tract, each 1 cm in lengths. Place each tissue piece in an individual 2 ml centrifuge screw cap tube containing 0.4 – 0.5 g of silica beads and 1 ml of DMEM-10.
Homogenize tissues in the dark with a bead beater for 60 min at 6000 rpm, as described (Gonzalez-Hernandez et al., 2012).
Freeze (at −80 °C) and thaw tissues (at 37 °C) once before performing parallel plaque assays in the dark and following light exposure as described above.
-
Count plaques 48 h later.
Note: MNV-1 is known to undergo the first round of replication in ~12 h. Therefore, samples collected 3 h post-infection should only show input virus (i.e., virus containing NR and sensitive to light).
-
Figure 1. Representative image of a plaque assay plate after 48 h of MNV-1 infection.
Plaques are observed after staining with a neutral red solution overlay for 1–3 h. The image shows duplicate wells of three 10-fold viral dilutions: 10−1, 10−2 and 10−3 dilutions. Arrows indicate the formation of plaques and the viral titer of the sample is indicated below. Answer: 11+9= 20 × 102 = 2 × 103 PFU/ml.
Recipes
-
DMEM-10
DMEM/High glucose (1 L)
10% low-endotoxin FBS
1% HEPES
1% Penicillin and streptomycin
1% Non-essential amino acids
1% L-glutamine
-
DMEM-5
DMEM/High glucose (1 L)
5% low-endotoxin FBS
1% HEPES
1% Penicillin and streptomycin
1% Non-essential amino acids
1% L-glutamine
-
2× MEM media (500 ml)
10% low-endotoxin FBS
1% HEPES
1% Penicillin and streptomycin
2% L-glutamine
Equilibrate to 37 °C
-
SeaPlaque agarose
1.5 g SeaPlaque agarose
50 ml water
Autoclave solution
Equilibrate to 42 °C
-
1:1 ratio of molten media
Mix 50 ml of 2× MEM and 50 ml of SeaPlaque agarose a 1:1 before adding to the cells
Acknowledgments
This neutral red assay protocol for murine norovirus was adapted thanks to the original work for poliovirus that was previously described by Brandenburg et al. (2007) and Kuss et al. (2008). Additionally, this protocol was adapted from the work performed by Perry et al. (2010); Perry et al. (2012) and Gonzalez-Hernandez et al. (2012). Our work was funded by start-up funds from the University of Michigan and NIH grant AI080611 to C.E.W. M.B.G.-H. was funded by the University of Michigan Experimental Immunology Training Grant (NIH T32 A1007413-16), by the Molecular Mechanisms of Microbial Pathogenesis Training Grant (NIH T32 A1007528), and by the Herman and Dorothy Miller Fund for Innovative Immunology Research. J.W.P. was funded by the University of Michigan Human Genetics training grant (grant NIH T32 GM 07544) and the Molecular Mechanisms of Microbial Pathogenesis training grant (NIH T32 AI 007528).
References
- 1.Brandenburg B, Lee LY, Lakadamyali M, Rust MJ, Zhuang X, Hogle JM. Imaging Poliovirus entry in live cells. PLoS Biol. 2007;5(7):e183. doi: 10.1371/journal.pbio.0050183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gonzalez-Hernandez MB, Bragazzi Cunha J, Wobus CE. Plaque assay for murine norovirus. J Vis Exp. 2012;(66):e4297. doi: 10.3791/4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuss SK, Etheredge CA, Pfeiffer JK. Multiple host barriers restrict Poliovirus trafficking in mice. PLoS Pathog. 2008;4(6):e1000082. doi: 10.1371/journal.ppat.1000082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perry JW, Ahmed M, Chang KO, Donato NJ, Showalter HD, Wobus CE. Antiviral activity of a small molecule deubiquitinase inhibitor occurs via induction of the unfolded protein response. PLoS Pathog. 2012;8(7):e1002783. doi: 10.1371/journal.ppat.1002783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perry JW, Wobus CE. Endocytosis of murine norovirus 1 into murine macrophages is dependent on dynamin II and cholesterol. J Virol. 2010;84(12):6163–6176. doi: 10.1128/JVI.00331-10. [DOI] [PMC free article] [PubMed] [Google Scholar]