Abstract
Mutations of the retinoblastoma tumor suppressor gene (RB1) or components regulating the CDK-RB-E2F pathway have been identified in nearly every human malignancy. Re-establishing cell cycle control through CDK inhibition has therefore emerged as an attractive option in the development of targeted cancer therapy. The most successful example of this today is the use of the CDK4/6 inhibitor palbociclib combined with aromatase inhibitors for the treatment of estrogen receptor-positive breast cancers. Multiple studies have demonstrated that the CDK-RB-E2F pathway is critical for the control of cell proliferation. More recently, studies have highlighted additional roles of this pathway, especially E2F transcription factors themselves, in tumor progression, angiogenesis and metastasis. Specific E2Fs also have prognostic value in breast cancer, independent of clinical parameters. We discuss here recent advances in understanding of the RB-E2F pathway in breast cancer. We also discuss the application of genome-wide genetic screening efforts to gain insight into synthetic lethal interactions of CDK4/6 inhibitors in breast cancer for the development of more effective combination therapies.
Keywords: Retinoblastoma, E2F, metastasis, progression
Introduction
To maintain genome integrity, the mammalian cell cycle must be stringently regulated. In cancer, cell cycle regulation is perturbed through a variety of genetic mechanisms, including amplification, mutation and overexpression of the genes encoding the core components of the cell cycle machinery. These include the cyclins, cyclin dependent kinases (CDKs), CDK inhibitors (CKIs) and the retinoblastoma protein RB1, all of which contribute to activation of the downstream E2F transcription factors 3, 34. Activation of E2F in turn can cause unrestrained proliferation and ectopic cell divisions 14.
E2Fs are an evolutionarily conserved family of transcription factors that have been extensively studied in the context of development and cancer 14. E2F comprises a family of ten proteins encoded by eight distinct genes, and they are most well known for their function in cell cycle regulation. Most studies to date have shown E2Fs 1, 2, and 3a to be the transcriptional activators, E2F3b, 4, and 5 the passive repressors, and E2F6, 7a, 7b, and 8 the active repressors, however these classifications are oversimplified and context dependent 14. Indeed, in certain molecular settings, including in the mouse intestine, E2Fs are not required for proliferation 15. In this setting, recent reports have shown that E2f and Myc function together to control cell cycles in normal and Rb-deficient cells 30. In this study, Myc and E2f1-3 were critical for the transcriptional program required for normal cell divisions at the S/G2 boundary but had little effect at G1/S. Further, in Rb-deficient cells, Myc and E2f3 are taken from the S/G2 program essential for normal cell cycles and recycled for a G1-S program. Whether these observations are context dependent remains unknown; however, it demonstrates that our understanding about the unification of cell cycle networks through E2F is not yet mature and there is still much to discover.
Studies have further shown that RB1 cannot regulate all stages of the cell cycle alone. The p130 (RBL2) and p107 (RBL1) multi-subunit protein complex containing partner (DP), RB-like, E2F and MuvB (DREAM) represses most if not all cell cycle gene expression during quiescence 47. These studies revealed that the RB-like p130 and p107, together with BMYB (MYBL2) and Forkhead box M1 (FOXM1), coordinate cell cycle dependent gene expression through a common pathway. When RB is active, E2F transcription modules are in a repressed state, which is associated with the recruitment of repressive chromatin marks, histone modifiers, and chromatin remodeling proteins. Upon mitogenic stimulation, the serine/threonine-specific CDKs initiate a phosphorylation cascade that inactivates the RB1 protein and dissociates the entire repressive complex. This enables E2Fs to recruit transcriptional activators and alter transcription of genes involved in cell cycle progression, DNA synthesis and DNA replication. One of the more enigmatic findings in the field was the observation that mice lacking the E2f1 gene are prone to tumors in several organs including sarcomas, lung tumors, and lymphomas 62. This has highlighted E2F1 as both a tumor suppressor gene and an oncogene, depending on the context. Since E2F-RB1 complexes are repressive, loss of E2Fs could also lead to de-repression of genes involved in cell cycle progression, potentially explaining why loss of E2F can contribute to oncogenesis 45.
CDKs together with their cognate cyclins regulate appropriate cell cycle stages. There are four distinct stages in the mammalian cell cycle that operate to duplicate and divide the genetic material between two nascent daughter cells: the G1 (Gap phase 1), S phase (DNA synthesis), G2 (Gap phase 2), and M phase (mitosis). Entry into the cell cycle from quiescence requires the concerted action of both CDK4 and CDK6 together with D-type cyclins34 and cyclin E/CDK2 complexes. Together, these kinase complexes can phosphorylate RB1 to the extent that E2Fs are released to mediate transition into S phase. CDK2/cyclin A and CDK2/cyclin E complexes are active in S phase and beyond, while CDK1/cyclin B complexes are responsible for the final push into mitosis. There is some degree of redundancy in the system. Studies have suggested that mammalian cells require at least five CDKs to regulate interphase: CDK2, CDK3, CDK4, and CDK6, and finally CDK1 in mitosis. However evidence from mouse models has challenged that notion, since mice lacking individual CDKs survive in the absence of interphase CDKs 6, 7, 33, 40. Additional studies on mice lacking multiple CDKs also support the notion that CDK1 can execute all the events necessary to drive cell division, suggesting that for many cell types it is the only essential CDK 49. This begs the question which CDK inhibitor compounds would be most efficacious as anti-cancer therapeutics.
Genomic aberrations in the CDK-RB1-E2F pathway are common in breast cancer. An analysis of approximately 1,100 breast cancer samples from The Cancer Genome Atlas shows that CCND1 (encoding cyclin D1) and CCNE1 (encoding cyclin E) are frequently amplified, while RB1 and CDKN2A are recurrently lost due to gene deletion or mutation (Table 1). The cyclin gene amplifications show a strong correlation with breast cancer subtype: CCND1 amplification is frequent in ER-positive and HER2-amplified breast cancer, while CCNE1 amplification instead occurs mainly in triple negative breast cancer. In this dataset, the E2F transcription factors are not recurrently mutated or focally amplified or deleted. They are however frequently altered as part of large-scale chromosome aberrations, such as the common loss of 16q, which contains E2F4.
Table 1. Recurrent genomic alterations in RB/E2F-related genes stratified by breast cancer subtype.
From The Cancer Genome Atlas, we gathered copy number (SNP6), and gene expression (RNA sequencing) data from 1,089 invasive breast carcinoma samples, and mutation data (DNA sequencing) from 993 samples. We selected the focally, recurrently amplified or deleted genes in the RB/E2F-pathway as identified by the RUBIC and GISTIC2 algorithms 35 (http://ccb.nki.nl/software/rubic/), and verified that they had a significant correlation between copy number with gene expression. For these recurrently altered genes, we tested whether the frequency in each subtype was significantly different using Fisher’s exact test. For RB1, we also compared the truncating mutation frequency, which includes nonsense mutations and frame shift insertions or deletions.
| Gene | Alteration | Frequency in subtype | P-value | ||
|---|---|---|---|---|---|
| ER-positive / HER2-normal | HER2-amplified | Triple-negative | |||
| CCND1 | Amplification | 22.1% | 25.9% | 2.9% | 6.1 · 10−13 |
| CCNE1 | Amplification | 2.1% | 5.2% | 16.3% | 4.0 · 10−12 |
| RB1 | Deletion | 2.7% | 2.2% | 4.4% | 0.39 |
| RB1 | Truncating mutation | 1.3% | 2.1% | 2.6% | 0.41 |
| CDKN2A | Deletion | 3.0% | 3.7% | 7.5% | 0.019 |
While this pathway has garnered attention for its unquestionable role in cell cycle regulation, recent advances in bioinformatics, mouse models, and DNA/RNA sequencing approaches have revealed a role for this pathway in other biological processes such as apoptosis, authophagy, angiogenesis, epithelial-to-mesenchymal transition (EMT) and metastasis. Since these biological processes are required in advanced, aggressive cancers, it suggests that the same proteins that initiate the tumor are in fact the same proteins that drive the progression of the disease, a notion that we first put forward over a decade ago 5. This article will review the studies of the RB-E2F pathway in breast cancer progression, and the implications for therapeutic targeting.
RB-E2F-pathway in breast cancer metastasis
The progression of cells to a more aggressive, metastatic state involves the upregulation of genes involved in angiogenesis, survival, tissue remodeling/invasion, and migration. Many studies have sought to identify specific transcription factors responsible for induction of these transcriptional changes and indeed, E2Fs have been identified in various cancer types as factors in tumor progression. In a study of HRAS-dependent invasion in breast cancer, E2Fs1-3 were shown to mediate invasion 64. E2Fs1-3 transcriptionally induce the expression of the β4 integrin subunit, which in turn enhances invasion mediated by the α6β4 integrin. In triple negative, basal-like breast tumors, there is often an epithelial to mesenchymal transition (EMT), associated with increased expression of mesenchymal genes and other EMT markers and hyperproliferation. This EMT utilizes a latent embryonic transcriptional program, thereby reprogramming an epithelial cell to a more motile, and aggressive state. The retinoblastoma protein controls the proliferation, differentiation, and survival of cells, and there is a central role for RB1 activity in the biology of stem and progenitor cells 48. Loss of RB1 function in stem or progenitor cells is a key event in the initiation of cancer. In a panel of breast cancer cell lines with inactive RB1, the mesenchymal phenotype and expression of genes involved in EMT has been observed 2. Further, this phenotype could be completely rescued following depletion of ZEB1, a transcriptional repressor of epithelial-cadherin. Studies in non small-cell lung cancer have shown that E2Fs can also transcriptionally upregulate the mesenchymal genes fibronectin, vimentin, and certain matrix metalloproteinase genes directly, though it is still unknown if this is a tissue-specific effect 26, 41. Finally, using an immortalized epithelial breast cancer line, MCF10A, Witkiewicz et al. found that loss of RB1 in the presence of ERBB2 overexpression altered key molecules needed for proper cellular organization and cell-to-cell adhesion60. Similar effects were observed in DCIS samples, where the loss of RB1 was associated with an increased risk of invasion.
Distant metastasis of breast cancer is one the leading causes of death for patients. Elegant studies from the Massague laboratory have revealed novel gene sets that mediate breast cancer metastasis to specific locations, albeit that we still do not fully understand which pathways govern this cascade 9, 27, 36. To study the role of the RB-E2F pathway in breast cancer, in vivo mouse models have recently been established. To determine which pathways are activated during Myc-induced mammary tumors, pathway activation predictions were generated focusing on activator E2f activity 22. Mice lacking various activator E2fs were crossed with mice expressing mammary-driven expression of the Myc oncogene (MMTV-Myc). E2f2 and E2f3 loss caused a significant delay in tumor onset. Further, gene expression analysis revealed that loss of E2f2 resulted in fewer tumors with EMT. This correlated with human breast cancer samples, where low probability of E2F2 activation was associated with increased relapse-free survival time. These data compliment other studies linking E2f2 to Myc-driven cancers 39.
This group later found that the MMTV-Myc transgenic mice crossed with E2f2 knockout mice had an increased percentage of lung metastasis 65. MDA-MB-231 cells with knockdown of E2F2 had increased migration and increased lung colonization in vivo. When tumors from MMTV-Myc and MMTV-Myc;E2f2-/- were compared with lung metastases samples, the authors identified PTPRD as a mediator of migration and lung colonization. Taken together, although the loss of E2f2 delays tumor onset, it results in increased metastasis in breast cancer, potentially functioning through a PTPRD dependent mechanism. This confounds the notion that inhibitors of the CDK-RB-E2F pathway will be useful for all breast cancers driven by different oncogenes and highlights the context dependency of E2F function.
Other studies have utilized the polyomavirus middle T oncoprotein (PyMT) model, which has been shown to activate multiple signaling pathways with relevance to human breast cancer 20. To identify pathways associated with the progression of breast cancer in this model, a number of gene expression data sets from genomic signaling signatures were analyzed 24. Although the tumors analyzed had a high degree of heterogeneity, nearly all samples had predicted E2f1 activity. When MMTV-PyMT mice were crossed with mice lacking E2f activators, E2f3 heterozygous mice had a significant delay in tumor onset. Gene expression studies revealed that E2f loss resulted in changes in genes critical to angiogenesis, ECM remodeling, tumor cell survival, and cell-cell interactions, suggesting that the major changes required for metastasis in the MMTV-PyMT model require E2f proteins.
Since ErbB2- and Ras-mediated mammary tumorigenesis in mice is dependent of cyclin D1, a known regulator of E2F activity, Wu et al. assessed whether E2f activators were also required in such mouse models 61. Using mice with epithelium-specific overexpression of ErbB2 or Myc, oncogenes that are overexpressed in up to 30% of human breast cancers, the authors created intercrosses with E2f1, E2f2, or E2f3 knockout mice that harbor the whey acidic protein (Wap) promoter to specifically delete E2f3 from the mammary epithelium (Wap-cre). Consistent with the results discussed above, loss of either E2f1 or E2f3 again significantly delayed tumor onset in both breast cancer models.
To further study the role of E2fs in in vivo mouse models, mice were created with ErbB2 expression driven by the MMTV promoter 1, which were subsequently crossed with mice lacking E2f activators. Contrary to the study from Wu et al., this study found that loss of any E2f delayed ErbB2-induced tumor onset. Tumors lacking E2f1 or E2f2 had a reduced metastatic capacity and E2f2 knockouts had fewer circulating tumor cells. Overall, these studies reveal the pivotal role of the cyclin D-RB-E2F regulatory pathway in proliferation, intravasation, survival in the bloodstream, and finally metastatic colonization of breast cancer cells in vivo. Moreover, these findings highlight the notion that the very same genetic lesions that drive the initial stages of tumor development can contribute to the later stages of tumor progression also.
Elegant work from Trikha et al highlights a role for E2f3 as a key transcription factor in tumor-associated macrophages (TAMs), which influences the tumor microenvironment and tumor cell metastasis 54. In this study the specific ablation of E2f3 in TAMs, but not in tumor epithelial cells, attenuated lung metastasis without affecting primary tumor growth. Though the loss of E2f3 had no impact on growth or survival of TAMs, this aberration significantly affected cytoskeleton rearrangements, cell migration and adhesion. Notably, the E2f3 TAM gene expression signature was sufficient to predict cancer recurrence and overall survival of estrogen receptor (ER)-positive breast cancer patients. This is the first study that explores the role of specific E2Fs in the tumor microenvironment of breast cancers.
RB-E2F in breast cancer differentiation, prognosis and therapy response
Breast cancer is a heterogeneous disease both in terms of intra- and intertumor heterogeneity. At least six distinct molecular subtypes have been identified on the basis of gene expression profiling, which include luminal A, luminal B, HER2-enriched, basal-like, claudin-low tumors, as well as a normal breast-like group 43. In recent years, a model has been proposed to explain intratumor heterogeneity, known as the cancer stem cell (CSC) model. In this model, a specific subpopulation of cells can drive progression of breast cancer. Moreover, cells have the capacity for self-renewal and phenotypic plasticity that is accompanied by the acquisition of mesenchymal characteristics at later stages 63. It should be noted that the finding that global breast cancer gene expression signatures (which measure the average patterns of gene expression in a tumor) can predict outcome, argues against this model 8, 23, 55, 56, 59. After all, if only a small subset of tumor cells would be capable of metastasis, such cells would not affect the global gene expression pattern of a breast tumor and it would consequently be impossible to predict outcome from the global gene expression pattern. A model that reconciles these seemingly conflicting data is that breast tumors with a “high risk” gene signature are less differentiated and that within such a tumor cell population de-differentiation to a breast cancer stem cell-like cell occurs more readily than in a more differentiated breast tumor having a “low risk” gene signature.
A bioinformatics approach found that histologically poorly differentiated tumors overexpressed genes normally enriched in embryonic stem (ES) cells, collectively called the ES signature, combined with preferential repression of PRC2 regulated genes 4. Among these transcriptional regulators were factors associated with proliferation, including E2F1, MYC, and FOXM1, and polycomb factors EZH2 and EED. An independent study found that cis-regulatory motifs bound by ELK1, E2F, NRF1 and NFY positively correlated with malignant progression of breast cancer 37. It was later shown that E2F activity and EZH2 could repress the polycomb repressive complex 2 (PRC2) in aggressive triple negative breast tumors 38.
The identification of signaling pathways that contribute to breast cancer progression has been greatly enhanced by advances in bioinformatics methods. Genomic signatures that identify poor prognosis breast cancer have been developed, and by applying these signatures to study activation of signaling pathways, pathways can be identified that contribute to metastasis 8, 23, 55, 56, 59. For instance, the MammaPrint 70 gene profile measures the mRNA of 70 genes and stratifies these patients into low-risk or high-risk groups. These and other subsequent prognostic methods have further highlighted the heterogeneity of human breast cancers. Network analysis of the 70 gene MammaPrint signature showed highly interconnected networks that center around known cancer-related genes including TP53, RB1, MYC, JUN, and CDKN2A 52. This finding is in agreement with the notion that components of the RB pathway are driving many facets of breast tumor progression by promoting a more stem cell-like phenotype within the tumor.
In patients with ER positive breast cancer, the ER antagonist tamoxifen is the primary therapeutic used in the clinic, despite the fact that nearly 50% of patients with metastatic disease do not respond. In addition, those patients who have an initial response will almost inevitably experience relapse 25. Microarray gene expression profiling of ER positive breast tumors has been used to identify gene signatures for prediction of clinical outcome of patients treated with tamoxifen. To better understand the mechanisms of tamoxifen resistance, these data sets were systematically analyzed 13, 25, 31, 32. In the case of tamoxifen resistant tumors, target genes for the transcription factors TFDP1, TFDP2, E2F1, and E2F4 were significantly enriched 25. Further, tamoxifen resistant tumors where enriched for genes involved in proliferation, DNA replication, and G1/S transition—all biological processes regulated by the RB-E2F pathway. In another study using modified DNA methylation-specific digital karyotyping and digital gene expression combined with massive parallel sequencing, four tamoxifen resistant cell lines and one parental cell line were used to study tamoxifen resistance. High expression of SOX2, and alterations of other SOX, E2F, and RB gene family members, were found in the tamoxifen resistant cells 29. Together, these studies highlight the potential role of the RB-E2F pathway in resistance to hormonal therapy in breast cancer.
The activity of transcription factors can be altered, resulting in changes in expression of their target genes. Zhu et al developed a method, named REACTIN, which integrates transcription factor binding data with gene expression data to identify transcription factors with differential activity between disease and normal samples 66. When REACTIN was applied to normal and malignant breast epithelial samples, combined with ChIP-seq data from ENCODE, several transcription factors were identified as having higher activity in breast cancer samples, including E2F1 and E2F4. Furthermore, Cox proportional hazard models showed that although gene expression of E2F4 itself was not associated with patient survival in breast cancer, there was a significant correlation between the inferred regulatory activity of E2F4 and survival outcomes. Khaleel et al integrated additional ChIP-seq data for E2F4, as well as a larger set of gene expression and survival data from more than 1900 breast tumor samples 28. They showed that the inferred E2F4 regulatory activity remains prognostic independent of clinicopathological variables, clinical risk scores, Oncotype DX stratification and differences in treatment strategies. Intriguingly, E2F4 was also a prognostic factor across colon, glioblastoma, and bladder cancer. Together these studies and others highlight the importance of E2F4 activity in breast cancer and that this activity is robustly prognostic for patient survival over a variety of clinical contexts 44.
Publically available data sets have dramatically increased the richness of data for scientific research. Using eight publicly available gene expression data sets, Thomassen et al conducted a meta-analysis that used gene set enrichment with a subset of significantly differentially regulated genes, called GenMAPP, to rank pathway gene sets in metastasizing breast tumors compared to non-metastasizing tumors 51. They observed an up-regulation of genetic pathways involved in cell cycle regulation, glucose metabolism, cellular migration, proteasome, immune system, angiogenesis, and DNA repair in metastasized tumors. E2F, NFY, and YY1 were identified as the transcription factors responsible for upregulation of these pathways—again linking E2F to metastasis regulation in breast cancer.
Targeting the CDK-RB-E2F pathway in breast cancer
The frequent activation of the cell cycle through perturbations in the CDK-RB-E2F pathway in cancer has led to efforts to block this pathway pharmacologically. Kinase inhibitors are the furthest along in drug development, although several compounds targeting other components of the pathway are at various stages of development as well 50. Early CDK inhibitors, such as flavopiridol and roscovitine were effective at inhibiting the cell cycle and inducing cell death, yet have a wide range of biochemical targets. Flavopiridol targets CDKs 1, 2, 4, 6, 7 and 9. Despite this broad range of targets, it displays limited efficacy in a wide range of tumors 10. Further, the inhibition of CDK9 is likely the dominant effect, as studies have shown that inhibiting CDK9 can inhibit transcription and result in cellular toxicity 11, 12, 34, 57.
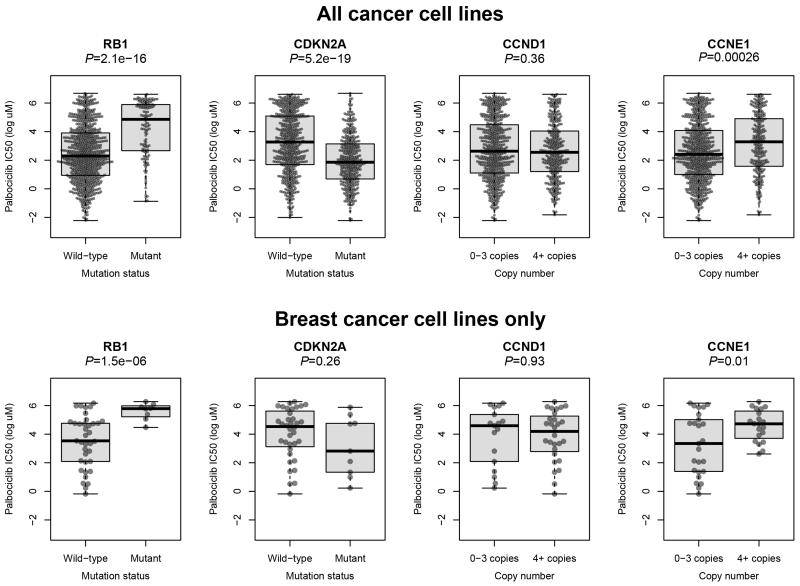
The most promising new drugs in the CDK inhibitor repertoire are undeniably the CDK4/6 inhibitors. These compounds are designed to target the ATP binding site of the CDK4-cyclinD and CDK6-cyclin D complexes. Over a decade after its initial synthesis by Pfizer in 2001, palbociclib is now the most advanced drug in its class 19, 21, 53. The success of palbociclib is attributed to the specificity of the compound; it inhibits CDK4 and CDK6 in the nanomolar range in a broad range of cell lines. Association analysis of genomic and drug profiles derived from the GDSC1000 (Sanger Cell line panel) highlights the importance of an intact RB1 gene for cells to be sensitive to palbociclib (Figure 1). Given that loss of CDKN2A (p16INK4A) could not stratify patient responses to palbociclib in the PALOMA-1 clinical trial or in breast cancer cell lines (Figure1), RB1 remains the only unequivocal biomarker of response.
Figure 1. Genomic biomarkers of sensitivity to palbociclib.
The Wellcome Trust Sanger Institute’s cell line drug sensitivity screen (GDSC1000) provided drug sensitivity estimates for palbociclib for 867 cell lines. Of these, 45 are breast cancer cell lines. We stratified these cell lines based on mutation or amplification status and tested whether the drug sensitivities differed between the groups using two-sided t-tests. In breast cancer, loss of RB1 shows the strongest correlation with palbociclib resistance.
Upon treatment with palbociclib, breast cancer cell lines have a range of sensitivities, where sensitive cell lines have decreased levels of RB1 phosphorylation at serine 780 and 795 after treatment 18. In this study, cell lines that are luminal estrogen receptor-positive (ER+) subtype (including those that are HER2 amplified) were most sensitive, while non-luminal/basal subtypes were most resistant. RB1 and cyclin D1 were elevated and CDKN2A was decreased in the most sensitive lines, which correlated with G0/G1 arrest. Interestingly, palbociclib was synergistic with tamoxifen and trastuzumab in ER+ and HER2-amplified cell lines. Palbociclib enhanced sensitivity to tamoxifen in cell lines with conditioned resistance to ER blockade. Given that the cylinD1 promoter is a bona fide estrogen-regulated gene, there is a sound rationale for the combination of ER signaling blockade with CDK4/6 inhibitors 46. In this situation, cells undergo therapeutic modulation by lowering the expression of cyclin D1 through inhibition of ER function, followed by CDK4/6 inhibition by palbociclib, which cooperate to reduce CDK4/6 kinase activity to inhibit proliferation.
These promising preclinical data resulted in the first phase Ib study of palbociclib plus letrozole, an aromatase inhibitor used to treat hormone receptor-dependent breast cancers, versus letrozole alone. Following this small study, the cohort was expanded to a phase II study with 165 patients randomized to palbociclib plus letrozole or letrozole alone: the final analysis yielded an impressive 20.2 months versus 10.2 months: nearly doubling progression-free survival 19. The results of this trial led to the accelerated FDA approval of palbociclib; however, with the caveat that continued approval of palbociclib “may be contingent upon verification and description of clinical benefit in an ongoing confirmatory trial”. PALOMA-2, the Phase III trial, should report by the end of 2016. In addition, it is still unclear which molecular biomarkers can be used to further stratify patients who will benefit from CDK4/6 inhibitor treatment. In the PALOMA-1 study, patient selection based on cyclin D1 amplification or p16INK4A loss was not associated with improved outcome. Two other CDK4/6 inhibitors—one from Novartis (LEE 001) and one from Eli Lilly (LY 2835219)—are also in clinical testing for breast tumors and other cancers 16.
The need for combination therapies
Until recently, the development of highly selective cancer therapeutics has remained an elusive goal. Compared to cytotoxic chemotherapies, which are used for the treatment of most solid tumors, targeted therapies provide a new opportunity: to eliminate only the cancer cells by targeting cancer-specific biochemical defects. In this new era of precision medicine, patients can be treated based on the specific oncogenic addictions and pathways that are required for tumor growth, angiogenesis, and eventual metastasis. As discussed here, it appears that the CDK-RB-E2F pathway is a driver of multiple hallmarks of breast cancer and consequently could be a good target for therapy in this disease.
Although single agent targeted therapies have shown potent initial responses in the clinic, the emergence of acquired resistance to target therapy is inevitable. This has led to the realization that drug combinations will be required to control disease in a more effective manner. For example, activation of the phosphoinositide 3-kinase (PI3K) pathway occurs frequently in breast cancer; however, clinical effects of single-agent PI3K inhibitors are limited. Using a combinatorial drug screen on PIK3CA mutant cancers with resistance to PI3K inhibitors, CDK 4/6 inhibitors were shown to enhance sensitivity to PI3K inhibitors 58. Importantly, the combination of PI3K and CDK4/6 inhibitors overcomes intrinsic- and acquired resistance to these PI3K inhibitors.
To identify additional pathway dependencies that can be exploited to enhance the efficacy of CDK inhibitors for the treatment of breast cancer, the concept of synthetic lethality and genetic screening technologies can be applied. Synthetic lethality refers to a situation in which the inactivation of two genes (or pathways) individually is not lethal, but becomes lethal when combined. The first demonstration of synthetic lethality with potential clinical application was in breast cancer: BRCA1 mutant tumors become critically dependent on alternative DNA repair pathways that require the enzymes poly(ADP-ribose) polymerase (PARP) 1 and 2. Hence, such tumors are highly sensitive to PARP inhibitors 17. The recent approval of the PARP inhibitor olaparib in Europe for BRCA mutated ovarian cancer demonstrates the clinical utility of using this synthetic lethality approach. By analogy, one could also search for genes or pathways that are synthetic lethal with CDK4/6 inhibition in breast cancer. We have demonstrated recently the utility of functional genetic screens to show which drugs are most synergistic with BRAF inhibitors in BRAF mutant colon cancer 42. Using a similar approach, it should be possible to find the genes whose suppression is most synergistic with CDK4/6 inhibition in breast cancer. It will be particularly fruitful to carry out such synthetic lethality screens in triple negative breast cancer, as anti hormonal therapy will not show synergy with CDK4/6 inhibitors in this class of tumors.
Conclusions
Since the isolation of the RB1 gene in 1986 and the subsequent cloning of the first E2Fs in 1992, we have gained profound insights into the roles of the CDK-RB-E2F pathway in cancer. Indeed, we now know that in virtually all human malignancies this pathway is deregulated one way or the other, making this pathway an attractive target for cancer therapy. It has taken significant time before the first selective CDK inhibitors reached the clinic and the challenge will be how to best use these drugs. The logic to combine CDK4/6 inhibition with inhibitors of estrogen receptor signaling is obvious, as the gene encoding cyclin D1, which activates CDK4/6, is responsive to estrogen. In this sense, the combination therapy approach chosen for palbociclib is similar to other successful combination therapies in which two drugs are used to hit the same pathway at multiple levels, such as the use of BRAF and MEK inhibitors in BRAF mutant melanoma. However, synthetic lethal interactions of CDK4/6 inhibition may occur with hitherto unexplored signaling pathways and it will be a worthwhile effort to search in a systematic fashion for such synthetic lethal interactions in breast and other cancers. Finally, the notion discussed here that E2Fs contribute to later phases of the metastatic spread of breast cancer makes the case for using CDK4/6 inhibitors early on in the treatment of breast cancer to reduce not only proliferation, but also dissemination.
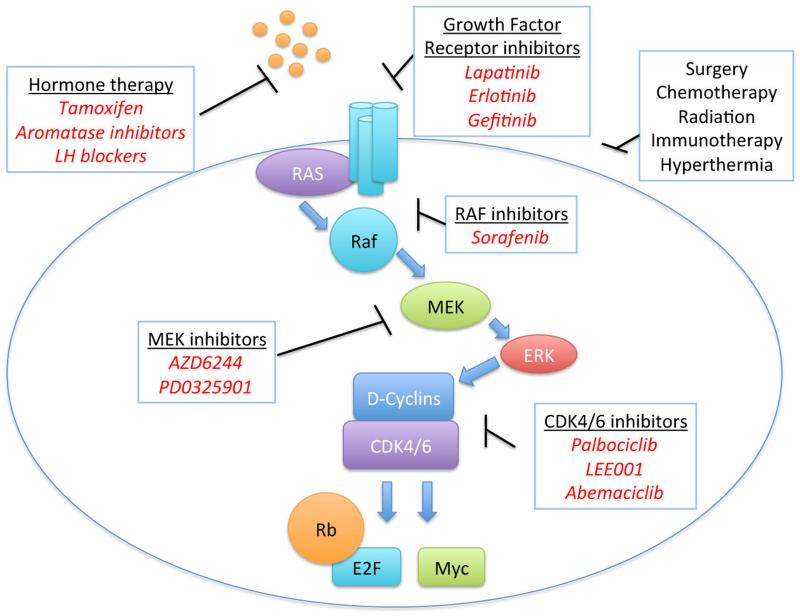
Figure 2. Therapeutic targeting of the CDK-RB-E2F pathway.
There are numerous strategies currently under investigation as anti-cancer therapies that target cell proliferation. Breast cancers are particularly dependent on this pathway. Current evidence suggests that targeting this pathway could inhibit initial tumor growth and also late-stage metastasis.
Acknowledgements
The work of R.B and L.W. is supported by grants of The Dutch Cancer Society and the and by the Cancer Genomics Netherlands consortium. M.J.G’s work is supported by the Wellcome Trust (102696), the Dutch Cancer Society (H1/2014-6919), Stand Up 2 Cancer, and a European Research Council Synergy grant.
Footnotes
The authors have no conflict of interest to disclose.
References
- 1.Andrechek ER. HER2/Neu tumorigenesis and metastasis is regulated by E2F activator transcription factors. Oncogene. 2015;34:217–225. doi: 10.1038/onc.2013.540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arima Y, Hayashi H, Sasaki M, Hosonaga M, Goto TM, Chiyoda T, et al. Induction of ZEB proteins by inactivation of RB protein is key determinant of mesenchymal phenotype of breast cancer. The Journal of biological chemistry. 2012;287:7896–7906. doi: 10.1074/jbc.M111.313759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker SJ, Reddy EP. CDK4: A Key Player in the Cell Cycle, Development, and Cancer. Genes & cancer. 2012;3:658–669. doi: 10.1177/1947601913478972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nature genetics. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernards R, Weinberg RA. A progression puzzle. Nature. 2002;418:823. doi: 10.1038/418823a. [DOI] [PubMed] [Google Scholar]
- 6.Berthet C, Aleem E, Coppola V, Tessarollo L, Kaldis P. Cdk2 knockout mice are viable. Current biology : CB. 2003;13:1775–1785. doi: 10.1016/j.cub.2003.09.024. [DOI] [PubMed] [Google Scholar]
- 7.Berthet C, Klarmann KD, Hilton MB, Suh HC, Keller JR, Kiyokawa H, et al. Combined loss of Cdk2 and Cdk4 results in embryonic lethality and Rb hypophosphorylation. Developmental cell. 2006;10:563–573. doi: 10.1016/j.devcel.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 8.Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 9.Bos PD, Zhang XH, Nadal C, Shu W, Gomis RR, Nguyen DX, et al. Genes that mediate breast cancer metastasis to the brain. Nature. 2009;459:1005–1009. doi: 10.1038/nature08021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bose P, Simmons GL, Grant S. Cyclin-dependent kinase inhibitor therapy for hematologic malignancies. Expert opinion on investigational drugs. 2013;22:723–738. doi: 10.1517/13543784.2013.789859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cai D, Latham VM, Jr., Zhang X, Shapiro GI. Combined depletion of cell cycle and transcriptional cyclin-dependent kinase activities induces apoptosis in cancer cells. Cancer research. 2006;66:9270–9280. doi: 10.1158/0008-5472.CAN-06-1758. [DOI] [PubMed] [Google Scholar]
- 12.Canduri F, Perez PC, Caceres RA, de Azevedo WF., Jr. CDK9 a potential target for drug development. Medicinal chemistry. 2008;4:210–218. doi: 10.2174/157340608784325205. [DOI] [PubMed] [Google Scholar]
- 13.Chanrion M, Negre V, Fontaine H, Salvetat N, Bibeau F, Mac Grogan G, et al. A gene expression signature that can predict the recurrence of tamoxifen-treated primary breast cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2008;14:1744–1752. doi: 10.1158/1078-0432.CCR-07-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen HZ, Tsai SY, Leone G. Emerging roles of E2Fs in cancer: an exit from cell cycle control. Nature reviews Cancer. 2009;9:785–797. doi: 10.1038/nrc2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chong JL, Wenzel PL, Saenz-Robles MT, Nair V, Ferrey A, Hagan JP, et al. E2f1-3 switch from activators in progenitor cells to repressors in differentiating cells. Nature. 2009;462:930–934. doi: 10.1038/nature08677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dickson MA. Molecular pathways: CDK4 inhibitors for cancer therapy. Clinical cancer research : an official journal of the American Association for Cancer Research. 2014;20:3379–3383. doi: 10.1158/1078-0432.CCR-13-1551. [DOI] [PubMed] [Google Scholar]
- 17.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 18.Finn RS, Dering J, Conklin D, Kalous O, Cohen DJ, Desai AJ, et al. PD 0332991, a selective cyclin D kinase 4/6 inhibitor, preferentially inhibits proliferation of luminal estrogen receptor-positive human breast cancer cell lines in vitro. Breast cancer research : BCR. 2009;11:R77. doi: 10.1186/bcr2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Finn RS, Crown JP, Lang I, Boer K, Bondarenko IM, Kulyk SO, et al. The cyclin-dependent kinase 4/6 inhibitor palbociclib in combination with letrozole versus letrozole alone as first-line treatment of oestrogen receptor-positive, HER2-negative, advanced breast cancer (PALOMA-1/TRIO-18): a randomised phase 2 study. The Lancet Oncology. 2015;16:25–35. doi: 10.1016/S1470-2045(14)71159-3. [DOI] [PubMed] [Google Scholar]
- 20.Fluck MM, Schaffhausen BS. Lessons in signaling and tumorigenesis from polyomavirus middle T antigen. Microbiology and molecular biology reviews : MMBR. 2009;73:542–563. doi: 10.1128/MMBR.00009-09. Table of Contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fry DW, Harvey PJ, Keller PR, Elliott WL, Meade M, Trachet E, et al. Specific inhibition of cyclin-dependent kinase 4/6 by PD 0332991 and associated antitumor activity in human tumor xenografts. Molecular cancer therapeutics. 2004;3:1427–1438. [PubMed] [Google Scholar]
- 22.Fujiwara K, Yuwanita I, Hollern DP, Andrechek ER. Prediction and genetic demonstration of a role for activator E2Fs in Myc-induced tumors. Cancer research. 2011;71:1924–1932. doi: 10.1158/0008-5472.CAN-10-2386. [DOI] [PubMed] [Google Scholar]
- 23.Gatza ML, Lucas JE, Barry WT, Kim JW, Wang Q, Crawford MD, et al. A pathway-based classification of human breast cancer. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:6994–6999. doi: 10.1073/pnas.0912708107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hollern DP, Honeysett J, Cardiff RD, Andrechek ER. The E2F transcription factors regulate tumor development and metastasis in a mouse model of metastatic breast cancer. Molecular and cellular biology. 2014;34:3229–3243. doi: 10.1128/MCB.00737-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang L, Zhao S, Frasor JM, Dai Y. An integrated bioinformatics approach identifies elevated cyclin E2 expression and E2F activity as distinct features of tamoxifen resistant breast tumors. PloS one. 2011;6:e22274. doi: 10.1371/journal.pone.0022274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson JL, Pillai S, Pernazza D, Sebti SM, Lawrence NJ, Chellappan SP. Regulation of matrix metalloproteinase genes by E2F transcription factors: Rb-Raf-1 interaction as a novel target for metastatic disease. Cancer research. 2012;72:516–526. doi: 10.1158/0008-5472.CAN-11-2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kang Y, Siegel PM, Shu W, Drobnjak M, Kakonen SM, Cordon-Cardo C, et al. A multigenic program mediating breast cancer metastasis to bone. Cancer cell. 2003;3:537–549. doi: 10.1016/s1535-6108(03)00132-6. [DOI] [PubMed] [Google Scholar]
- 28.Khaleel SS, Andrews EH, Ung M, DiRenzo J, Cheng C. E2F4 regulatory program predicts patient survival prognosis in breast cancer. Breast cancer research : BCR. 2014;16:486. doi: 10.1186/s13058-014-0486-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin X, Li J, Yin G, Zhao Q, Elias D, Lykkesfeldt AE, et al. Integrative analyses of gene expression and DNA methylation profiles in breast cancer cell line models of tamoxifen-resistance indicate a potential role of cells with stem-like properties. Breast cancer research : BCR. 2013;15:R119. doi: 10.1186/bcr3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu H, Tang X, Srivastava A, Pecot T, Daniel P, Hemmelgarn B, et al. Redeployment of Myc and E2f1-3 drives Rb-deficient cell cycles. Nature cell biology. 2015;17:1036–1048. doi: 10.1038/ncb3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loi S, Piccart M, Sotiriou C. The use of gene-expression profiling to better understand the clinical heterogeneity of estrogen receptor positive breast cancers and tamoxifen response. Critical reviews in oncology/hematology. 2007;61:187–194. doi: 10.1016/j.critrevonc.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 32.Loi S, Haibe-Kains B, Desmedt C, Wirapati P, Lallemand F, Tutt AM, et al. Predicting prognosis using molecular profiling in estrogen receptor-positive breast cancer treated with tamoxifen. BMC genomics. 2008;9:239. doi: 10.1186/1471-2164-9-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malumbres M, Sotillo R, Santamaria D, Galan J, Cerezo A, Ortega S, et al. Mammalian cells cycle without the D-type cyclin-dependent kinases Cdk4 and Cdk6. Cell. 2004;118:493–504. doi: 10.1016/j.cell.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 34.Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: a changing paradigm. Nature reviews Cancer. 2009;9:153–166. doi: 10.1038/nrc2602. [DOI] [PubMed] [Google Scholar]
- 35.Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12:R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Niida A, Smith AD, Imoto S, Tsutsumi S, Aburatani H, Zhang MQ, et al. Integrative bioinformatics analysis of transcriptional regulatory programs in breast cancer cells. BMC bioinformatics. 2008;9:404. doi: 10.1186/1471-2105-9-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Niida A, Smith AD, Imoto S, Aburatani H, Zhang MQ, Akiyama T. Gene set-based module discovery in the breast cancer transcriptome. BMC bioinformatics. 2009;10:71. doi: 10.1186/1471-2105-10-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Opavsky R, Tsai SY, Guimond M, Arora A, Opavska J, Becknell B, et al. Specific tumor suppressor function for E2F2 in Myc-induced T cell lymphomagenesis. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:15400–15405. doi: 10.1073/pnas.0706307104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ortega S, Prieto I, Odajima J, Martin A, Dubus P, Sotillo R, et al. Cyclin-dependent kinase 2 is essential for meiosis but not for mitotic cell division in mice. Nature genetics. 2003;35:25–31. doi: 10.1038/ng1232. [DOI] [PubMed] [Google Scholar]
- 41.Pillai S, Trevino J, Rawal B, Singh S, Kovacs M, Li X, et al. beta-arrestin-1 mediates nicotine-induced metastasis through E2F1 target genes that modulate epithelial-mesenchymal transition. Cancer research. 2015;75:1009–1020. doi: 10.1158/0008-5472.CAN-14-0681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prahallad A, Sun C, Huang S, Di Nicolantonio F, Salazar R, Zecchin D, et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature. 2012;483:100–103. doi: 10.1038/nature10868. [DOI] [PubMed] [Google Scholar]
- 43.Prat A, Perou CM. Deconstructing the molecular portraits of breast cancer. Molecular oncology. 2011;5:5–23. doi: 10.1016/j.molonc.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rakha EA, Pinder SE, Paish EC, Robertson JF, Ellis IO. Expression of E2F-4 in invasive breast carcinomas is associated with poor prognosis. The Journal of pathology. 2004;203:754–761. doi: 10.1002/path.1573. [DOI] [PubMed] [Google Scholar]
- 45.Rowland BD, Denissov SG, Douma S, Stunnenberg HG, Bernards R, Peeper DS. E2F transcriptional repressor complexes are critical downstream targets of p19(ARF)/p53-induced proliferative arrest. Cancer cell. 2002;2:55–65. doi: 10.1016/s1535-6108(02)00085-5. [DOI] [PubMed] [Google Scholar]
- 46.Sabbah M, Courilleau D, Mester J, Redeuilh G. Estrogen induction of the cyclin D1 promoter: involvement of a cAMP response-like element. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:11217–11222. doi: 10.1073/pnas.96.20.11217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sadasivam S, DeCaprio JA. The DREAM complex: master coordinator of cell cycle-dependent gene expression. Nature reviews Cancer. 2013;13:585–595. doi: 10.1038/nrc3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sage J. The retinoblastoma tumor suppressor and stem cell biology. Genes & development. 2012;26:1409–1420. doi: 10.1101/gad.193730.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Santamaria D, Barriere C, Cerqueira A, Hunt S, Tardy C, Newton K, et al. Cdk1 is sufficient to drive the mammalian cell cycle. Nature. 2007;448:811–815. doi: 10.1038/nature06046. [DOI] [PubMed] [Google Scholar]
- 50.Singh S, Johnson J, Chellappan S. Small molecule regulators of Rb-E2F pathway as modulators of transcription. Biochimica et biophysica acta. 2010;1799:788–794. doi: 10.1016/j.bbagrm.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thomassen M, Tan Q, Kruse TA. Gene expression meta-analysis identifies metastatic pathways and transcription factors in breast cancer. BMC cancer. 2008;8:394. doi: 10.1186/1471-2407-8-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tian S, Roepman P, Van't Veer LJ, Bernards R, de Snoo F, Glas AM. Biological functions of the genes in the mammaprint breast cancer profile reflect the hallmarks of cancer. Biomarker insights. 2010;5:129–138. doi: 10.4137/BMI.S6184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Toogood PL, Harvey PJ, Repine JT, Sheehan DJ, VanderWel SN, Zhou H, et al. Discovery of a potent and selective inhibitor of cyclin-dependent kinase 4/6. Journal of medicinal chemistry. 2005;48:2388–2406. doi: 10.1021/jm049354h. [DOI] [PubMed] [Google Scholar]
- 54.Trikha P, Sharma N, Pena C, Reyes A, Pecot T, Khurshid S, et al. E2f3 in tumor macrophages promotes lung metastasis. Oncogene. 2015 doi: 10.1038/onc.2015.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van 't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 56.van de Vijver MJ, He YD, van't Veer LJ, Dai H, Hart AA, Voskuil DW, et al. A gene-expression signature as a predictor of survival in breast cancer. The New England journal of medicine. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 57.Vandenbosch R, Borgs L, Beukelaers P, Foidart A, Nguyen L, Moonen G, et al. CDK2 is dispensable for adult hippocampal neurogenesis. Cell cycle. 2007;6:3065–3069. doi: 10.4161/cc.6.24.5048. [DOI] [PubMed] [Google Scholar]
- 58.Vora SR, Juric D, Kim N, Mino-Kenudson M, Huynh T, Costa C, et al. CDK 4/6 inhibitors sensitize PIK3CA mutant breast cancer to PI3K inhibitors. Cancer cell. 2014;26:136–149. doi: 10.1016/j.ccr.2014.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.West M, Blanchette C, Dressman H, Huang E, Ishida S, Spang R, et al. Predicting the clinical status of human breast cancer by using gene expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:11462–11467. doi: 10.1073/pnas.201162998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Witkiewicz AK, Cox DW, Rivadeneira D, Ertel AE, Fortina P, Schwartz GF, et al. The retinoblastoma tumor suppressor pathway modulates the invasiveness of ErbB2-positive breast cancer. Oncogene. 2014;33:3980–3991. doi: 10.1038/onc.2013.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu L, de Bruin A, Wang H, Simmons T, Cleghorn W, Goldenberg LE, et al. Selective roles of E2Fs for ErbB2- and Myc-mediated mammary tumorigenesis. Oncogene. 2015;34:119–128. doi: 10.1038/onc.2013.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yamasaki L, Jacks T, Bronson R, Goillot E, Harlow E, Dyson NJ. Tumor induction and tissue atrophy in mice lacking E2F-1. Cell. 1996;85:537–548. doi: 10.1016/s0092-8674(00)81254-4. [DOI] [PubMed] [Google Scholar]
- 63.Ye X, Weinberg RA. Epithelial-Mesenchymal Plasticity: A Central Regulator of Cancer Progression. Trends Cell Biol. 2015;25:675–686. doi: 10.1016/j.tcb.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yoon SO, Shin S, Mercurio AM. Ras stimulation of E2F activity and a consequent E2F regulation of integrin alpha6beta4 promote the invasion of breast carcinoma cells. Cancer research. 2006;66:6288–6295. doi: 10.1158/0008-5472.CAN-06-0826. [DOI] [PubMed] [Google Scholar]
- 65.Yuwanita I, Barnes D, Monterey MD, O'Reilly S, Andrechek ER. Increased metastasis with loss of E2F2 in Myc-driven tumors. Oncotarget. 2015;6:38210–38224. doi: 10.18632/oncotarget.5690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhu M, Liu CC, Cheng C. REACTIN: regulatory activity inference of transcription factors underlying human diseases with application to breast cancer. BMC genomics. 2013;14:504. doi: 10.1186/1471-2164-14-504. [DOI] [PMC free article] [PubMed] [Google Scholar]