Abstract
The Liverpool Epidemic Strain (LES) is a polylysogenic, transmissible strain of Pseudomonas aeruginosa, capable of superinfecting existing P. aeruginosa respiratory infections in individuals with cystic fibrosis (CF). The LES phages are highly active in the CF lung and may have a role in the competitiveness of the LES in vivo. In this study, we tested this by competing isogenic PAO1 strains that differed only by the presence or absence of LES prophages in a rat model of chronic lung infection. Lysogens invaded phage-susceptible populations, both in head-to-head competition and when invading from rare, in the spatially structured, heterogeneous lung environment. Appreciable densities of free phages in lung tissue confirmed active phage lysis in vivo. Moreover, we observed lysogenic conversion of the phage-susceptible competitor. These results suggest that temperate phages may have an important role in the competitiveness of the LES in chronic lung infection by acting as anti-competitor weapons.
The LES is a transmissible strain of Pseudomonas aeruginosa that causes life-limiting chronic respiratory infections in individuals with CF (Fothergill et al., 2012). Unusually, the LES is capable of superinfection to displace other strains of P. aeruginosa, even after years of chronic colonisation (McCallum et al., 2001; Winstanley et al., 2009), but there is no evidence that it can be displaced by other strains (Fothergill et al., 2010; Mowat et al., 2011; Williams et al., 2015). The LES harbours five prophages, highly active in the CF lung (James et al., 2015), four of which have been shown by signature-tagged mutagenesis to be necessary for bacterial competitiveness in a rat model of chronic lung infection (Winstanley et al., 2009; Lemieux et al., 2015). Theoretical and empirical studies suggest that temperate phages could enhance the competitiveness of lysogens by killing phage-susceptible competitors by lysis (Bossi et al., 2003; Brown et al., 2006; Joo et al., 2006; Burns et al., 2014), a form of phage-mediated allelopathy (Stewart and Levin, 1984). However, it is unclear whether this ecological mechanism operates in the far more complex spatially structured and heterogeneous lung environment.
To test whether temperate phages increase competitive fitness during lung infection, we performed competition experiments between lysogenic and non-lysogenic strains of P. aeruginosa in a rat model of chronic lung infection (Winstanley et al., 2009). We constructed antibiotic-resistance-labelled PAO1 LES-Phage Lysogens (PLPLs) (James et al., 2012) using three of the LES phages (LESφ2, LESφ3 and LESφ4), both individually and in combination (for full methods, see Supplementary information). Initial in vitro experiments suggested that the triple lysogen (PAO1φtriple) was more invasive than any of the constituent single lysogens (Supplementary Figure S1), therefore PAO1φtriple was selected for use in the in vivo experiments. Competitors were embedded in agar beads in PLPL to PAO1φ− ratios of 1:5 or 1:1, to model invasion-from-rare and head-to-head competition, respectively. Rats were infected with inoculated agar beads by intubation and monitored for 7 days, after which they were killed. The densities of each competitor in the lungs were quantified (Supplementary Table S2) and the selection rate constant (rij) was calculated as described previously (Lenski et al., 1991).
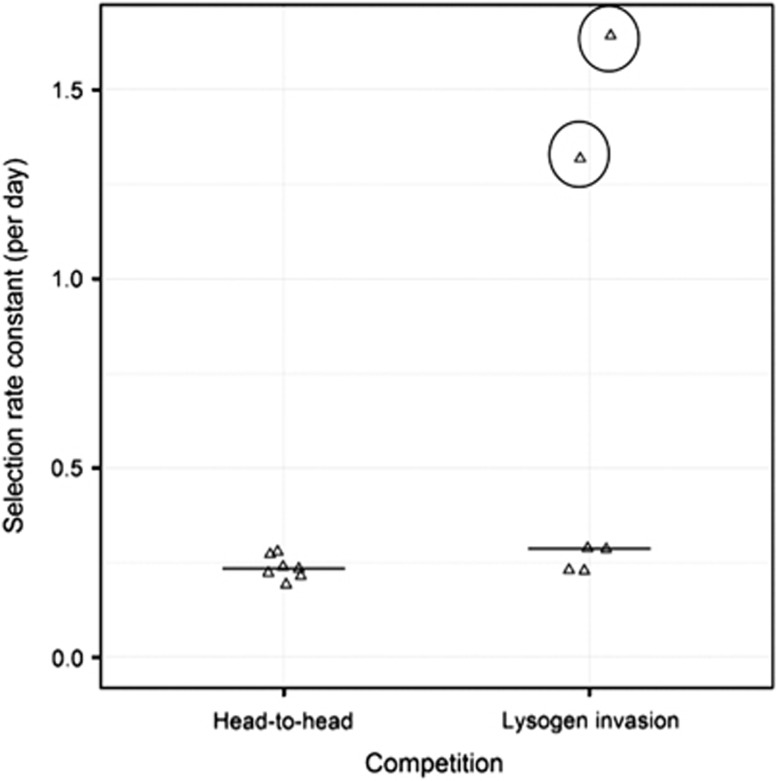
PAO1φtriple outcompeted PAO1φ− in vivo (Figure 1), at both initial starting frequencies (competition experiment, 1 sample t-test (alt=0), t6=20.3, P<0.001; invasion experiment, 1 sample t-test (alt=0), t3=15.8, P<0.01), but there was no effect of initial starting frequency on fitness (2-sample t-test; t6=1.08, P=0.32). Thus, temperate phages improved the competitiveness and invasiveness of lysogens against phage-susceptible populations in chronic lung infection. To confirm the role of phage lysis, we also measured levels of free infective phages in the lung homogenate. We observed appreciable densities of virions in the lungs in both treatments, with no significant difference in mean (±1 s.d.) phage-to-bacterium ratios between the competition (0.55±0.84) and invasion (0.47±0.44) treatments (2-sample t-test on log10+1 transformed data; t6=−0.11, P=0.91).
Figure 1.
Selection rate constant for the competition outcome between PAO1φ− and PAO1φtriple at different starting ratios of competitors. Each data point represents the outcome of competition in the lungs of an individual animal after 7 days, with the exception of the two circled data points. These represent two rats that were killed after 2 days, as they were showing symptoms of acute infection, with high bacterial loads (100-fold higher than other lungs after 7 days). These were excluded from statistical analyses.
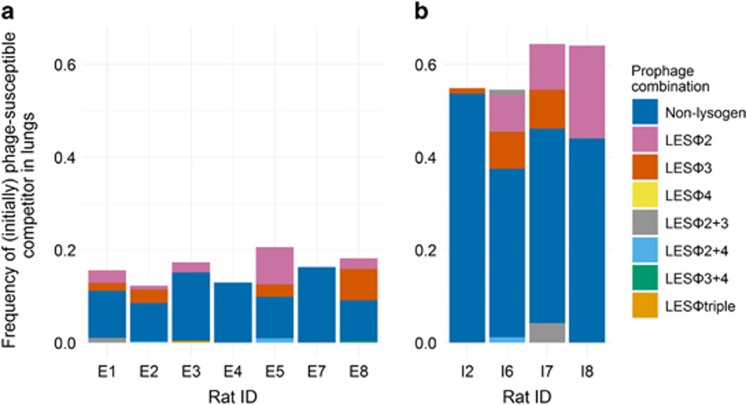
Phage-mediated invasion by lysogens can be limited by lysogenic conversion of the originally phage-susceptible competitor, creating phage-resistant lysogens (Gama et al., 2013). Having found that lysogenic conversion occurs at a very high frequency in vitro (Supplementary Figure S2), we investigated whether the phages also established lysogeny in vivo. We calculated for each animal the total lysogen frequency and proportion of each lysogen type for end point populations by screening 46 bacterial lung isolates of the initially phage-free competitor (PAO1φ−) using a multiplex PCR assay (Supplementary Methods and Supplementary Table S1). We observed appreciable rates of lysogenic conversion at both initial starting frequencies, but with substantial variation between animals (Figure 2). Although the mean (±1 s.d.) total frequency of lysogens was higher in head-to-head competition (0.89±0.03) compared with invasion-from-rare (0.56±0.07) treatment (2-sample t-test; t6=8.57, P<0.01) (due to a proportion of the PLPLφtriple competitor), the rate of lysogenic conversion of PAO1φ− (Figure 2) was similar in both treatments (Mann–Whitney U test; W=42.5, n1=7, n2=4, P=1.00). Lysogenic conversion of PAO1φ− was dominated by the formation of LESφ2 and LESφ3 lysogens, suggesting that these phages were most active in the lung, which is consistent with the high free-phage densities of these phages in human CF infections (James et al., 2015).
Figure 2.
Lysogenic conversion of PAO1φ− after 7 days in vivo. Prophage complement of streptomycin-labelled bacteria (initially PAO1φ−) isolated from rat lungs in (a) head-to-head competition and (b) invasion-from-rare treatments. Height of bars denote the frequency of the competitor out of total bacteria. Data are reported separately for each animal.
These data provide important experimental evidence supporting the role for phage-mediated allelopathy as a determinant of pathogen fitness in chronic lung infection. This extends previous studies using observational (James et al., 2015) and insect model approaches (Burns et al., 2014) to confirm, in a clinically relevant environment, that the LES-temperate phages are likely to have had a key role in the global spread of the LES. Crucially, we demonstrate that phage-mediated allelopathy allows lysogens to invade from rare, even in the complex, spatially structured, heterogeneous host lung environment, which has previously been theoretically predicted, but has never been demonstrated (Gama et al., 2013). In agreement with a recent observational clinical study of the ecological dynamics of the LES and its phages in CF patient sputa, we show the production of appreciable populations of free-phage virions by lysis in the lung (James et al., 2015). We observed lysogenic conversion in the lung, but at rates lower than those observed in liquid in vitro environments (Supplementary Figure S2), suggesting that lysogenic conversion may have been impeded in the lung environment. Consistent with this, recent evidence suggests that bacterial populations show strong regional structure within the CF lung, with low rates of mixing between regions of the lung (Jorth et al., 2015). Nevertheless, the transfer of genetic material among strains of P. aeruginosa within infections does raise concerns about the potential for the horizontal gene transfer of antibiotic resistance or virulence determinants (Penadés et al., 2015).
Acknowledgments
This work was funded by a project grant from The Wellcome Trust (089215/Z/09/Z to CW & MAB). EVD was funded by a studentship co-funded by the UK Medical Research Council and the University of Liverpool, and a research visit grant from the Microbiology Society. RCL was funded by Cystic Fibrosis Canada (grant number 2610) and by the Canadian Institute for Health Research (grant reference number 86644).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Bossi L, Fuentes JA, Mora G, Figueroa-Bossi N. (2003). Prophage contribution to bacterial population dynamics. J Bacteriol 185: 6467–6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SP, Le Chat L, De Paepe M, Taddei F. (2006). Ecology of microbial invasions: amplification allows virus carriers to invade more rapidly when rare. Curr Biol 16: 2048–2052. [DOI] [PubMed] [Google Scholar]
- Burns N, James CE, Harrison E. (2014). Polylysogeny magnifies competitiveness of a bacterial pathogen in vivo. Evol Appl 8: 346–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fothergill JL, Mowat E, Ledson MJ, Walshaw MJ, Winstanley C. (2010). Fluctuations in phenotypes and genotypes within populations of Pseudomonas aeruginosa in the cystic fibrosis lung during pulmonary exacerbations. J Med Microbiol 59: 472–481. [DOI] [PubMed] [Google Scholar]
- Fothergill JL, Walshaw MJ, Winstanley C. (2012). Transmissible strains of Pseudomonas aeruginosa in cystic fibrosis lung infections. Eur Respir J 40: 227–238. [DOI] [PubMed] [Google Scholar]
- Gama JA, Reis AM, Domingues I, Mendes-Soares H, Matos AM, Dionisio F. (2013). Temperate bacterial viruses as double-edged swords in bacterial warfare. PLoS One 8: e59043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James C, Fothergill J, Kalwij H, Hall A, Cottell J, Brockhurst M et al. (2012). Differential infection properties of three inducible prophages from an epidemic strain of Pseudomonas aeruginosa. BMC Microbiol 12: 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James CE, Davies EV, Fothergill JL, Walshaw MJ, Beale CM, Brockhurst MA et al. (2015). Lytic activity by temperate phages of Pseudomonas aeruginosa in long-term cystic fibrosis chronic lung infections. ISME J 9: 1391–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo J, Gunny M, Cases M, Hudson P, Albert R, Harvill E. (2006). Bacteriophage-mediated competition in Bordetella bacteria. Proc Biol Sci 273: 1843–1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorth P, Staudinger BJ, Wu X, Hisert KB, Hayden H, Garudathri J et al. (2015). Regional isolation drives bacterial diversification within cystic fibrosis lungs. Cell Host Microbe 18: 307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemieux A-A, Jeukens J, Kukavica-Ibrulj I, Fothergill JL, Boyle B, Laroche J et al. (2015). Genes required for free phage production are essential for Pseudomonas aeruginosa chronic lung infections. J Infect Dis 213: 395–402. [DOI] [PubMed] [Google Scholar]
- Lenski RE, Rose MR, Simpson SC, Tadler SC. (1991). Long-term experimental evolution in Escherichia coli. I. adaptation and divergence during 2, 000 generations. Am Nat 138: 1315–1341. [Google Scholar]
- McCallum SJ, Corkill J, Gallagher M, Ledson MJ, Hart CA, Walshaw MJ. (2001). Superinfection with a transmissible strain of Pseudomonas aeruginosa in adults with cystic fibrosis chronically colonised by P aeruginosa. Lancet 358: 558–560. [DOI] [PubMed] [Google Scholar]
- Mowat E, Paterson S, Fothergill JL, Wright EA, Ledson MJ, Walshaw MJ et al. (2011). Pseudomonas aeruginosa population diversity and turnover in cystic fibrosis chronic infections. Am J Respir Crit Care Med 183: 1674–1679. [DOI] [PubMed] [Google Scholar]
- Penadés JR, Chen J, Quiles-Puchalt N, Carpena N, Novick RP. (2015). Bacteriophage-mediated spread of bacterial virulence genes. Curr Opin Microbiol 23: 171–178. [DOI] [PubMed] [Google Scholar]
- Stewart FM, Levin BR. (1984). The population biology of bacterial viruses: why be temperate. Theor Popul Biol 26: 93–117. [DOI] [PubMed] [Google Scholar]
- Williams D, Evans B, Haldenby S, Walshaw MJ, Brockhurst MA, Winstanley C et al. (2015). Divergent, coexisting, Pseudomonas aeruginosa lineages in chronic cystic fibrosis lung infections. Am J Respir Crit Care Med 191: 775–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winstanley C, Langille MGI, Fothergill JL, Kukavica-Ibrulj I, Paradis-Bleau C, Sanschagrin F et al. (2009). Newly introduced genomic prophage islands are critical determinants of in vivo competitiveness in the liverpool epidemic strain of Pseudomonas aeruginosa. Genome Res 19: 12–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.