Summary
Microfluidic devices enable precise quantification of the interactions between anticancer bacteria and tumor tissue. Direct observation of bacterial movement and gene expression in tissue is not possible with either monolayers of cells or tumor-bearing mice. Quantification of these interactions is necessary to understand the inherent mechanisms of bacterial targeting and to develop modified organisms with enhanced therapeutic properties. Here we describe the procedures for designing, printing and assembling microfluidic tumor-on-a-chip devices. We also describe the procedures for inserting three- dimensional tumor-cell masses, exposing to bacteria, and analyzing the resultant images.
Keywords: microfluidic device, tumor-on-a-chip, tumor-targeting bacteria, PDMS, cancer, penetration, motility, accumulation, lithography
1. Introduction
Microfluidic devices are invaluable for understanding the mechanisms that control bacterial interaction with tumors [1]. Therapeutic bacteria have many advantages over standard chemotherapeutic drugs because of their unique properties [2]. In vivo, bacteria preferentially accumulate in tumors over other organs [3] and actively penetrate through tumor tissue [4]. Bacteria have also been engineered to produce anticancer agents inside tumors [5]. However, poor understanding of these mechanisms has slowed development of these therapies. It is not possible to study tumor accumulation and penetration in monolayers of cancer cells, because they do not contain microenvironment gradients or barriers to mass transfer [6]. Experiments with mice are costly, time consuming, and cannot be easily used to measure dynamic behavior [4]. Microfluidic devices are essential tools for quantifying bacteria behavior, because they are cheap, fast, and can be imaged in real time. Microfluidic devices are also essential components in the development of new therapeutic strategies, because they can rapidly evaluate genetic modifications that have been designed to improve bacterial performance.
Here we describe a procedure to create a microfluidic device that contains a tissue chamber with a flow channel on one side (Figure 1). This geometry exposes the contained cells to flowing medium, mimicking the interaction between cells and blood vessels [3]. The device is formed by imprinting a design in polydimethylsiloxane (PDMS), and adhering it to glass [7,8]. Cancer cells are inserted as spheroids and retained by a filter at the rear of the chamber [7]. The geometry of the device ensures that the cell mass is optically accessible through a glass slide (Figure 2). Because the design is vertically uniform (away from the glass), mass transfer creates linear microenvironment gradients that are identical in all z-planes. This uniformity enables analysis by standard epifluorescence microscopy [7].
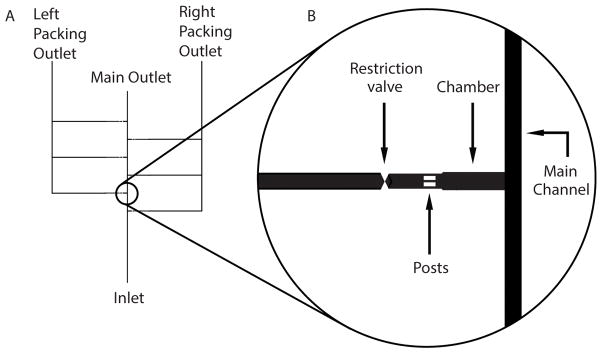
Figure 1. Schematic of Microfluidic Chamber and Channels.
A) Basic device containing a main channel with six alternating chambers connecting a main outlet and two packing outlets. B) Enlarged view highlighting the main channel, one chamber, posts, and restriction valve.
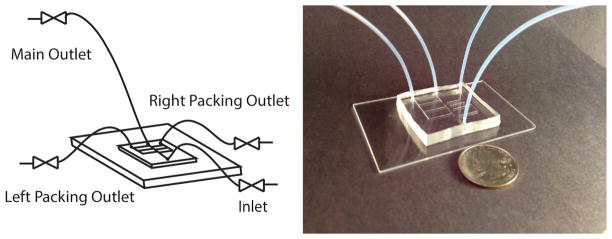
Figure 2. Schematic of Device Assembly and Tubing.
The device is composed of PDMS layer adhered to a glass slide with tubing attached to the main outlet, the packing outlets, and the inlet. Actual size, shown relative to a quarter.
Using this device, we have shown that bacterial motility is necessary for deep penetration into tumor tissue and that motile species are more effective at colonization [1]. Using a similar device, we have shown that chemotaxis and growth are necessary mechanisms for Salmonella accumulation in tumors [9,10]. Chemoreceptors attract bacteria to specific microenvironments created by dying cancer cells [10]. The ability to observe a live population in real time, permits modeling of the bacterial behavior and precise quantification of chemotaxis and growth [9]. Other groups have used similar devices to show that Salmonella have a preference for cancer hepatocytes compared to normal cells [11], and expression of invasin increases proliferation of E coli. in 3D tumor tissue [12].
A microfluidic device could be used to answer many unanswered questions about how bacteria interact with tumors. Devices could be used to study 1) penetration into tissue, 2) invasion into cancer cells, 3) production of drug molecules, and 4) control of gene expression. In addition, the response of cancer cells to invasion and bacterially produced molecules could be quantified in real time. A microfluidic device would be an essential component in the design of more effective bacteria by enabling visualization of engineered improvements in penetration, invasion, and drug production.
The procedure below outlines the steps necessary to design, fabricate and run a microfluidic device. The description is focused on a specific design with a single inlet, two outlets and a tissue-containing chamber. We have found this design to be simple to implement and stable for multiple days [7]. However, this soft-lithography technique is highly flexible and could be tuned for multiple applications by designing different architectures. The procedure is made up of four basic phases: 1) design and construction of the microfluidic device [steps 3.1–3.5]; 2) growth and insertion of cancer cells [steps 3.6–3.7]; 3) treatment with bacteria [step 3.8]; and 4) image acquisition and analysis [steps 3.9].
2. Materials
2.1 Mold and Device Components
4 inch diameter, 525 μm thick silicon wafers (University Wafer, South Boston, MA, USA)
SU-8 2050 permanent epoxy negative photoresist and developer (Microchem, Newton, MA, USA)
150 × 5 mm polystyrene petri dishes covered in aluminum foil, as light-block covers
100% silicone rubber, 732 multi-purpose sealant (Dow Corning, Midland, MI, USA)
Sylgard 184 silicone elastomer kit: silicone elastomer base and curing agent (Dow Corning, Midland, MI, USA)
1.5 mm biopsy punch drill bit
Microbore PTFE 0.032 inch ID, 0.056 inch OD tubing
Super Flangeless Fittings system, Tefzel, 1/16″ OD (Upchurch Scientific, Oak Harbor, WA, USA)
0.040 thru shut off valve (Upchurch Scientific, Oak Harbor, WA, USA)
2.2 Materials for Mammalian Cell and Bacterial Culture
Low-glucose medium: low-glucose Dulbecco’s Modified Eagle Medium (DMEM) with 10% fetal bovine serum (FBS), pH adjusted to 7.4, sterile filtered, and stored at 4°C.
HEPES-buffered medium: low-glucose DMEM with 10% FBS and 6g/L HEPES buffer.
PMMA/Ethanol solution: In a cell-culture hood, add 20 g/L Poly(methyl methacrylate) to pure ethanol. Store at 37°C and 5% carbon dioxide.
Phosophate buffered saline (PBS) without calcium or magnesium.
0.05% trypsin-EDTA 1x phenol red
LB medium: To 950 mL Nanopure water, add 10 g tryone, 5 g yeast extract, and 10 g sodium chloride. Use sodium hydroxide to adjust the pH to 7.0. Sterilize by autoclaving at 15 psi for 20 minutes. Store at room temperature.
3. Methods
When working with silicon wafers, use tweezers and avoid touching any area on the wafer. When the device is not in use for longer than an hour, cover it with plastic wrap.
3.1 Device Design
Draw design using a vector drawing program (see Note 1).
In the design, connect chambers to flow inlets and outlets at the front of the chamber and packing outlets on the rear of chamber. Chambers are 1000×300 μm, channels are 250 μm, posts are 195×65 μm, and restriction valves are a triangle and an inverted triangle spanning a 125×250 μm area (see Figure 1 and Note 2).
Print devices on high quality imagesetting film with 100 micrometer polyester base and 5080 dpi resolution (see Note 3).
3.2 Mold Fabrication
-
1
Using tweezers, wash each silicon wafer twice with toluene, isopropanol, and water. Spray solvents over entire surface and dry completely between solvents using air.
-
2
Center wafer on spin coater and apply vacuum. Pour approximately 4 mL of SU-8 2050 onto center of wafer. Spin at 500 rpm for 10 seconds with an acceleration of 100 rpm/second (Step 1) then at 1250 rpm for 30 seconds with an acceleration of 300 rpm/second (Step 2). These steps will achieve a thickness of 150 μm with SU-8 2050. Minimize light exposure during spin coating to avoid decomposition of SU-8.
-
4
Release vacuum and remove wafer using tweezers. Place wafer on a 65 °C slide heater, under a light-blocking plastic lid, and on top of a laboratory wipe for 5 minutes.
-
5
Remove wafer and allow to cool for 5 minutes. During this cooling step, increase heater setting to 95 °C. Place wafer back onto the heater for 30 minutes. Remove wafer again and allow to cool completely (see Note 4).
-
6
In a minimally lit area, securely attach design transparency on top of wafer using paper clips. Avoid sections of wafer with bubbles, dust, tweezers marks, and other imperfections. Cover any remaining exposed wafer surface with tin foil to ensure only desired features are exposed. An exposure of 260 mJ/cm2 is recommended for ideal crosslinking. For example, with a UV reading of 13.18 MW/cm2, a single exposure of 20 seconds is used (see Note 5).
-
7
Again, heat wafer to 65 °C for 5 minutes while covered (see Step 4), cool for 5 minutes, and heat to 95 °C for 12 minutes. Remove wafer and allow it to cool completely.
-
8
Using tweezers, place wafer on top of a magnetic stir bar in a high-walled glass dish. Add enough SU-8 2050 developer to just cover wafer. Turn magnetic stirrer to lowest setting for approximately 25 minutes. To verify that SU-8 2050 is completely developed, remove wafer using tweezers and spray with isopropanol. If a milky deposit forms, dry wafer with air and return to the developer. Once fully developed, clean wafer with isopropanol and dry with air.
-
9
Glue wafer to a plastic petri dish. Apply a circle of 100% silicone sealant approximately 1 cm from the features. Allow 24 hours for sealant to set before use.
3.3 Device Fabrication
Mix 10.8 g PDMS and 1.2 g silicone curing agent in a 50 mL centrifuge tube. Vortex on high for 1 minute. Pour mixture into mold, allowing as little as possible to spill over sealant edges (see Note 6).
To de-gas PDMS solution, cycle between applying and releasing vacuum suction within a vacuum chamber. When releasing vacuum, do so before bubbles spill over sealant edges. Continue until no bubbles are present at wafer surface or over any features. Some bubbles may remain near sealant edges.
Place mold with lid on a slide heater at 65 °C for at least 5 hours, preferably overnight.
Once cured, cut device away from mold using a scalpel and spatula, leaving approximately 0.5 cm edges from any desired features (see Figure 2).
Wrap the device in plastic wrap. Mark ends of each channel on plastic wrap with a dot. Punch these holes manually using a 1.5 mm biopsy punch (see Note 7).
Carefully remove debris using tweezers and a small needle. Clean with ethanol and air.
3.4 Plasma Treatment
Wash both device and a glass slide with acetone, isopropanol, and methanol in that order. Air dry completely between solvents.
Place device and glass slide inside an oxygen plasma cleaner with bonding surfaces face up.
Cycle between vacuum and oxygen to purge air from the chamber. Open vacuum valve and allow pressure to drop to 200 mtorr. Open oxygen valve and allow pressure to return to 1 atm. Close oxygen valve and allow pressure to drop to 1 torr. Open oxygen valve and allow pressure to return to 1 atm. Close oxygen valve and allow pressure to drop to 200 mtorr. Carefully open oxygen valve and balance oxygen and vacuum to maintain pressure at 200 mtorr.
Power on plasma cleaner and set to high for 2 minutes. A pink/purple glow should appear (see Note 8).
Power off the plasma cleaner, and close oxygen and vacuum valves. Once the door is able to open, gently but quickly place the two treated sides together. If necessary, gently tap device to induce bonding.
Wrap device with a laboratory wipe and place on a slide heater, glass slide down. Put a glass jar, with a few hundred milliliters of water, on top as a weight. Leave at 65 °C for an hour. Wrap in plastic wrap for storage.
3.5 Device Assembly
Cut seven 18 inch pieces and one 24 inch piece of PTFE tubing. At one end of each piece, attach a ferrule fitting. Screw each fitted end into a 0.040 thru-hole valve such that three valves have both ends of 18 inch tubing and one valve has one 18 inch tubing and one 24 inch tubing (see Note 9).
Glue the device to the bottom of a well plate with super glue and place on microscope. Ensure that glue does not seep under the device around the features.
Arrange valves around the microscope such that the valve with 24 inch tubing is at least 11 inches above the device. Valves with all 18 inch tubing need not be arranged in any particular order (see Note 10).
Insert tubing from valves with 18 inch tubing into inlet and packing outlets of the device. Insert the 24 inch tubing to the main outlet (see Figure 2).
In a cell-culture hood, fill three 10 mL syringes attached to 20 gauge, 1½ inch needles with 5–7 mL 70% ethanol, 10% bleach, and HEPES-buffered medium. Remove air bubbles by tapping syringes (see Note 11).
Open inlet valve and all outlet valves. Insert the ethanol syringe into inlet tubing and gently push the syringe to flush the system. Close outlet valves and inlet valve in that order.
Repeat Step 6 with bleach and DMEM, in that order. Identify air bubbles and remove as many as possible during the medium flush. Do not inject entire medium syringe into the system as this will introduce air bubbles. Do not remove medium syringe attached to inlet tubing (see Note 12).
3.6 Cultivating Hanging-Drop Spheroids
Prepare a single-cell suspension of LS174T colon carcinoma cells by trypsinization. After centrifuging cells, resuspend pellet in 2–4 mL DMEM by pipetting up and down. Pipette vigorously with a micropipette to break up all cell clumps (see Note 13).
Count cell density using a hemocytometer, and create a 2 mL solution with a density of 300 cells/μL in DMEM.
Pipette 1 mL sterile water into each well of a 48 well plate. This water is critical for maintaining humidity and preventing evaporation of the small hanging drops.
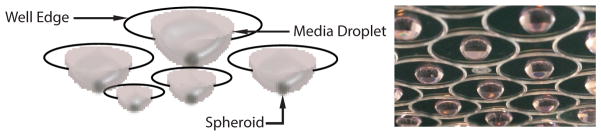
Pipette 20 μL drops of cell and medium solution onto the inside surface of a well plate lid. Gently touch the tip of a micropipette to just off center of each circle marked on the well plate lid. Inject cell-suspension medium toward the center of the marked circle (see Figure 3 and Note 14).
Carefully turn lid over and place on well plate such that no drops touch well edges.
Incubate at 37 ° C and 5% carbon dioxide. Final spheroid size depends on length of culture time. Five days produces 500 μm diameter spheroids (see Note 15).
Prepare PMMA flasks in a cell-culture hood by adding approximately 1 mL of PMMA/ethanol solution to each T25 flask. Leave caps loose and allow ethanol to evaporate overnight in a cell-culture hood.
In a cell-culture hood, remove lid from hanging drop well plate and turn upside down. Using a 20 μL micropipette, select individual spheroids from drops. Add spheroids to a PMMA- coated T25 flask containing 5 mL DMEM (see Note 16).
Incubate for 5 days, until spheroids are 500 μm in diameter, which is optimal for insertion into the microfluidic device.
Figure 3. Schematic and Image of Hanging Drop Spheroid Formation.
Spheroids are formed by suspending drops of medium from the lid of a well plate. Image shows LS174T tumor spheroids growing in media droplets, highlighting the well edge, media droplet, and spheroid.
3.7 Spheroid Insertion
In a cell-culture hood, add 1 mL of spheroid-containing medium, from a PMMA-coated flask, into a 60×15 mm petri dish. Add 5 mL room-temperature HEPES-buffered medium to a second 60×15 mm petri dish.
Carefully select spheroids using a 20 μL micropipette and add them to the second dish. Select spheroids based on size, uniformity, symmetry and optical density. (see Note 17)
Slowly draw spheroid-containing medium into a 10 mL syringe.
Attach a 20 gauge, 1½ inch needle to the syringe. With the needle facing up, allow spheroids to fall to the bottom of the syringe. Tap the syringe and gently push plunger to remove air bubbles.
With the needle facing down, allow spheroids to fall to the middle of the syringe. Lay the syringe on its side. Gently shake and roll syringe to move spheroids such that they do not contact each other.
Before injecting spheroids, ensure there are no air bubbles in the syringe. If so, repeat Steps 3 and 4 (see Note 18).
Remove the medium-containing syringe and attach a spheroid-containing syringe to tubing, while keeping the spheroid-containing syringe lying sideways.
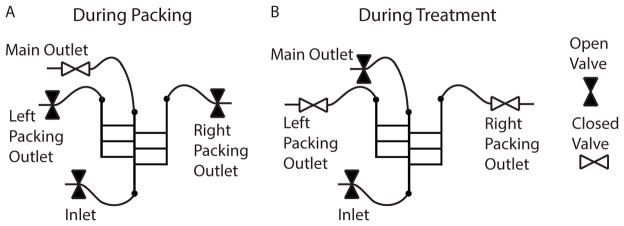
Open inlet and packing outlets. Do not open main outlet (see Figure 4-A).
Run approximately 1–2 mL medium from the spheroid syringe through the device. Observe air bubbles and remove as many as possible. Do not inject spheroids into the system.
Turn the syringe and needle face down. Allow spheroids to fall into the needle. Gently push spheroids through the needle. Watch spheroids flow through tubing and into the inlet of the device (see Note 19).
Slowly push spheroids through the device using steady hand pressure. Injecting medium too quickly will cause spheroids to be shredded by posts at the rear of chambers and exit through packing outlets (see Note 20).
Once spheroids have been packed, stop pushing medium through the tubing but leave a thumb on the syringe to maintain pressure in the device. Close packing outlet valves followed by the inlet valve. Leave the spheroid syringe in the inlet tube.
Fill a 10 mL syringe, with an attached 20 gauge, 1½ inch needle, with 10 mL medium. Fit syringe into a syringe pump.
Start an automated image-acquisition process (see Section 3.9, Steps 1–3).
Start the syringe pump with a flow rate of 3 μL/min. While watching spheroids through the microscope, slowly open the inlet valve, followed by the main outlet valve (see Note 21).
Inject medium into the device overnight to allow spheroids to grow into chambers.
Figure 4. Schematic of Spheroid Insertion.
Valves are variably opened and closed during (A) spheroid insertion into chambers, and (B) delivery of treatment to the tumor tissue. A) During insertion, the inlet and right and left packing outlet valves are open. B) During treatment, the inlet and main outlet valves are open.
3.8 Treatment with Bacteria
Grow bacteria in LB medium to mid logarithmic phase (0.2<OD600<0.5)
Centrifuge and resuspend bacteria in HEPES-buffered medium. Typical densities range from 105 to 107 CFU/mL.
Fill a 10 mL syringe attached to a 20 gauge, 1½ inch needle with bacteria solution.
Close main outlet valve followed by inlet valve. Stop the syringe pump.
Remove the medium syringe from the inlet valve and replace it with a bacteria syringe. Fit the bacteria syringe into the syringe pump.
Re-start the syringe pump. While watching spheroids, slowly open inlet valve followed by main outlet valve.
Administer bacteria for 1 hour (see Note 22).
Close main outlet valve followed by inlet valve. Stop the syringe pump program.
Remove bacteria syringe from inlet valve and replace it with a syringe containing only HEPES-buffered medium.
Fit medium syringe into syringe pump and re-start it. While watching spheroids, slowly open inlet valve followed by main outlet valve.
3.9 Image Acquisition and Analysis Time-lapse Images
Acquire images on an inverted microscope so that entire chamber, from rear to channel, is visible. If chamber is larger than a single image, tile multiple images together. Tiling requires alignment and calibration of an automated stage. Tiled images are create by acquiring an image, moving the stage one image width, and acquiring a second image (see Note 23 and Figure 5-A).
Program the image acquisition sequence to acquire images of all six chambers in series for every time interval (see Note 24).
After completion of the experiment, sort acquired images into individual stacks for each chamber. This creates a movie-like series of images.
For each chamber, rotate the images as a stack such that the rows of pixels line up with the edges of the chamber.
Identify the pixel columns that correspond to the front edge of the tissue and the rear of the chamber. Partition this area into columns of pixels (see Figure 5-A).
Average the fluorescence intensity of all pixels in each column individually from the front to the rear.
Convert pixel widths into absolute distances (i.e. micrometers).
This analysis on a stack of images creates an intensity profile as a function of distance for each image at each time point (see Figure 5-B).
Repeat for every chamber.
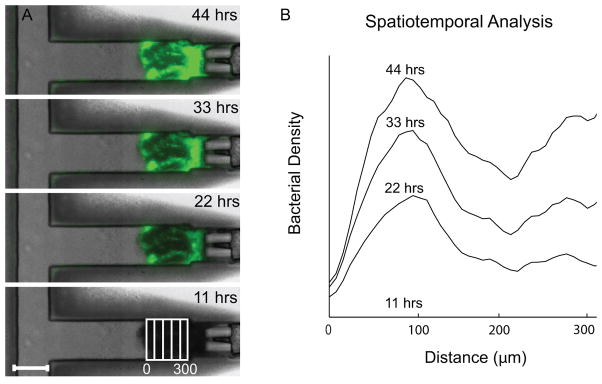
Figure 5. Salmonella Accumulated in Tissue Packed in a Device.
Results of a bacterial treatment to one chamber. A) Images of fluorescent bacteria (green) that have invaded into LS174T tumor tissue at four time points. Overlay on fourth image illustrates columns of pixels. The scale bar is 250 μm. B) Profiles of bacterial density as a function of distance from the front of the tissue and as a function of time.
Acknowledgments
We gratefully acknowledge financial support from the National Science Foundation (Grant No. 1159689) and the National Institutes of Health (Grant No. R01CA120825).
Footnotes
We use Adobe Illustrator to design our devices.
Our design has six chambers, three alternating either side of one flow channel. The number of channels and the arrangement of the flow channels can easily be changed in the drawing process to match desired experimental protocols. Our device also has channels approximately 20 mm long, spaced 5 mm apart, with connecting outlet channels approximately 50 mm long to allow for stable features and leave room for the 1.5 mm biopsy-punch holes.
No stroke should be used when designing devices to make measurements as accurate as possible. Colors should be inverted before printing so that features appear white and space appears black. We send our designs to PageWorks (Cambridge, MA).
If heating the slide heater to 95 °C is not achievable during the five minute cooling step, move on to the 30 minute heating step regardless of the temperature of the heater. Do not allow the heater to go above 95 °C.
An energy meter can be used to gauge the lamp strength and appropriate exposure times may be selected based upon the energy output. Using one wafer and only a small section of the desired features to test multiple exposure times can effectively determine an ideal time. Under or over exposure can cause malformed device features.
The recommended weight ratio for a microfluidic device made from PDMS and a silicone curing agent is 10:1. However, a 9:1 ratio ensures the device will be stiff enough to hold the tubing during experiments. Any amount of the PDMS and curing agent mixture (i.e. above or below a total of 12 g) can be used, as long as the appropriate ratio is maintained for the desired stiffness.
Holes can be punched in a device by hand using the 1.5 mm biopsy drill bit. They can also be done with a drill press that is powered off. When using a drill, hold the device down firmly as you release the drill.
Settings should be adjusted for individual oxygen plasma cleaners. Times varying from 2–8 minutes have shown successful results, as well as pressures ranging from 200–150 mtorr.
Attaching the valves to a plastic plate makes them easier to open and close. Attaching the plastic plate to a styrofoam block stabilizes the plate and valve to prevent unnecessary movement.
An insufficient pressure difference between the inlet and outlet of the device can cause spheroids to fall out of the chambers. The minimal height needed to maintain a sufficient pressure difference is 11 inches.
HEPES-buffered medium is used in device experiments primarily because it can maintain a pH of 7.5 under varying concentrations of carbon dioxide.
Opening and closing valves and repeatedly increasing and decreasing the medium flow rate can remove stubborn air bubbles.
LS174T cell are used because they readily form spheroids. Any cell line can be used that is sufficiently cohesive to aggregate into distinct cell masses.
Positioning the pipette slightly off center insures that the drops are formed at the center the circles.
The incubation time for hanging drops and spheroids in PMMA-coated flasks can vary from 3 to 5 days depending on cell growth rate. The time should be adjusted to produce final spheroids that are 500 μm in size.
We have found that spheroids toward the middle of the well plate are better formed.
Selecting spheroids by eye takes practice, but can be done reliably without use of a stereo microscope.
Using room-temperature medium decreases the formation of bubbles within the syringe.
If spheroids get stuck, 1) gently tap the base and tip of the needle (through the tubing); 2) gently tap the inlet valve while opening and closing it; 3) gently wiggle the inlet tube at the intersection with the device.
Multiple syringes of spheroids can be injected into the same device. If spheroids are lost or are unsatisfactory, they can be pushed through the post filters to clear these chambers. If some chambers have desirable spheroids, close the packing valves for these chambers. Injecting another syringe gently may allow the remaining chambers to be filled without damaging the already packed chambers.
Closing the main outlet valve and opening the packing outlet valves can force spheroids into chambers if they appear to be falling out while changing syringes or while opening the main outlet valve.
The ypical duration for bacterial administration is 1 hour. This is similar to the clearance time in mice. However, this time can be varied to suit the desired protocol.
Using 10x magnification we find that a 1000 μm chamber requires 2 images side-by-side.
Different time intervals are used for different applications. We have used less than 1 second intervals to capture fast events and as long as 1 hour for long, multiple-day studies.
References
- 1.Toley BJ, Forbes NS. Motility is critical for effective distribution and accumulation of bacteria in tumor tissue. Integr Biol. 2012;4:165–76. doi: 10.1039/c2ib00091a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Forbes NS. Engineering the perfect (bacterial) cancer therapy. Nature reviews Cancer. 2010;10:785–94. doi: 10.1038/nrc2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Forbes NS, Munn LL, Fukumura D, Jain RK. Sparse initial entrapment of systemically injected Salmonella typhimurium leads to heterogeneous accumulation within tumors. Cancer Res. 2003;63:5188–93. [PubMed] [Google Scholar]
- 4.Ganai S, Arenas RB, Sauer JP, Bentley B, Forbes NS. In tumors Salmonella migrate away from vasculature toward the transition zone and induce apoptosis. Cancer Gene Ther. 2011;18:457–66. doi: 10.1038/cgt.2011.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ganai S, Arenas RB, Forbes NS. Tumour-targeted delivery of TRAIL using Salmonella typhimurium enhances breast cancer survival in mice. Br J Cancer. 2009;101:1683–91. doi: 10.1038/sj.bjc.6605403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.St Jean AT, Zhang M, Forbes NS. Bacterial therapies: completing the cancer treatment toolbox. Curr Opin Biotechnol. 2008;19:511–7. doi: 10.1016/j.copbio.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walsh CL, Babin BM, Kasinskas RW, Foster Ja, McGarry MJ, Forbes NS. A multipurpose microfluidic device designed to mimic microenvironment gradients and develop targeted cancer therapeutics. Lab on a chip. 2009;9:545–54. doi: 10.1039/b810571e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Toley BJ, Ganz DE, Walsh CL, Forbes NS. Microfluidic Device for Recreating a Tumor Microenvironment. J Vis Exp. 2011 doi: 10.3791/2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kasinskas RW, Forbes NS. Salmonella typhimurium specifically chemotax and proliferate in heterogeneous tumor tissue in vitro. Biotechnology and Bioengineering. 2006;94:710–721. doi: 10.1002/bit.20883. [DOI] [PubMed] [Google Scholar]
- 10.Kasinskas RW, Forbes NS. Salmonella typhimurium lacking ribose chemoreceptors localize in tumor quiescence and induce apoptosis. Cancer research. 2007;67:3201–9. doi: 10.1158/0008-5472.CAN-06-2618. [DOI] [PubMed] [Google Scholar]
- 11.Hong JW, Song S, Shin JH. A novel microfluidic co-culture system for investigation of bacterial cancer targeting. Lab on a chip. 2013 doi: 10.1039/c3lc50163a. [DOI] [PubMed] [Google Scholar]
- 12.Elliott N, Lee T, You L, Yuan F. Proliferation behavior of E. coli in a three-dimensional in vitro tumor model. Integrative biology. 2011;3:696–705. doi: 10.1039/c0ib00137f. [DOI] [PMC free article] [PubMed] [Google Scholar]