INTRODUCTION
Pantothenate is vitamin B5 and is the key precursor for the biosynthesis of coenzyme A (CoA) and carrier proteins that have a phosphopantetheine prosthetic group. The phosphopantetheine moiety is donated to these proteins by CoA and is used to shuttle intermediates between the active sites of enzymes involved in fatty acid, non-ribosomal peptide and polyketide synthesis. CoA is an essential cofactor for cell growth and is involved in many metabolic reactions, including the synthesis of phospholipids, synthesis and degradation of fatty acids, and the operation of the tricarboxylic acid cycle. The prebiotic formation and stability of pantothenate precursors suggests that CoA function was important in the earliest metabolic pathways (87). From genetic footprinting experiments in E. coli, all five biosynthetic genes involved in CoA biosynthesis from pantothenate are essential (44), but the pantothenate biosynthetic genes can be dispensable in some organisms due to the expression of a pantothenate permease which transports the vitamin into the cell. The supply of pantothenate and the level of expression of pantothenate kinase (PanK) together determine the cellular CoA levels (19,118,138). The CoA biosynthetic pathway is a popular target for development of novel antibacterial agents, due to the distinctive differences between the bacterial and the mammalian proteins that catalyze the PanK reaction, and the phosphopantetheine adenylyltransferase (PPAT) reaction. The CoA biosynthetic pathway networks with other vitamin-associated pathways, for example, by altering the metabolic requirements for nutrients such as thiamine (32), or restriction of pantothenate production by the availability of methylene-tetrahydrofolate which is derived from folic acid (11).
BIOSYNTHESIS OF PANTOTHENATE and ITS PRECURSORS
Most bacteria, such as Escherichia coli, Salmonella typhimurium and Corynebacterium glutamicum (7,54,132), synthesize pantothenate from the amino acid aspartate and an intermediate in valine biosynthesis. In S. typhimurium, the acetohydroxy acid synthase isozyme I, followed by the dihydroxyacid dehydratase, provides most of the flux to valine and pantothenate (33), both of which are made from α-ketoisovalerate. Valine can revert back to α-ketoisovalerate, as mediated by the products of either the ilvE or avtA genes. The amino group of valine is replaced by a keto-moiety to yield α-ketoisovalerate, which, in turn, forms α-ketopantoate following transfer of a methyl group, then pantoate following reduction (Fig. 1). Aspartate is decarboxylated to yield β-alanine. The β-alanine and pantoate intermediates are then condensed to yield pantothenate (also termed pantothenic acid). Pantothenate is either used for CoA biosynthesis or released from the cell. Comparative analysis of the sequences and structures of the individual enzymes involved in pantothenate biosynthesis indicate that the pathway likely evolved via a patchwork mechanism, whereby the activities were recruited separately from diverse protein families (75). The level of pantothenate is low in all the archaebacteria, including methanogens (72), compared with that in eubacteria (95). Metabolic engineering of C. glutamicum to increase pantothenate production reveals several constraints on the biosynthetic pathway (49,107), most notably at the α-ketoisovalerate branch point (11).
Figure 1.
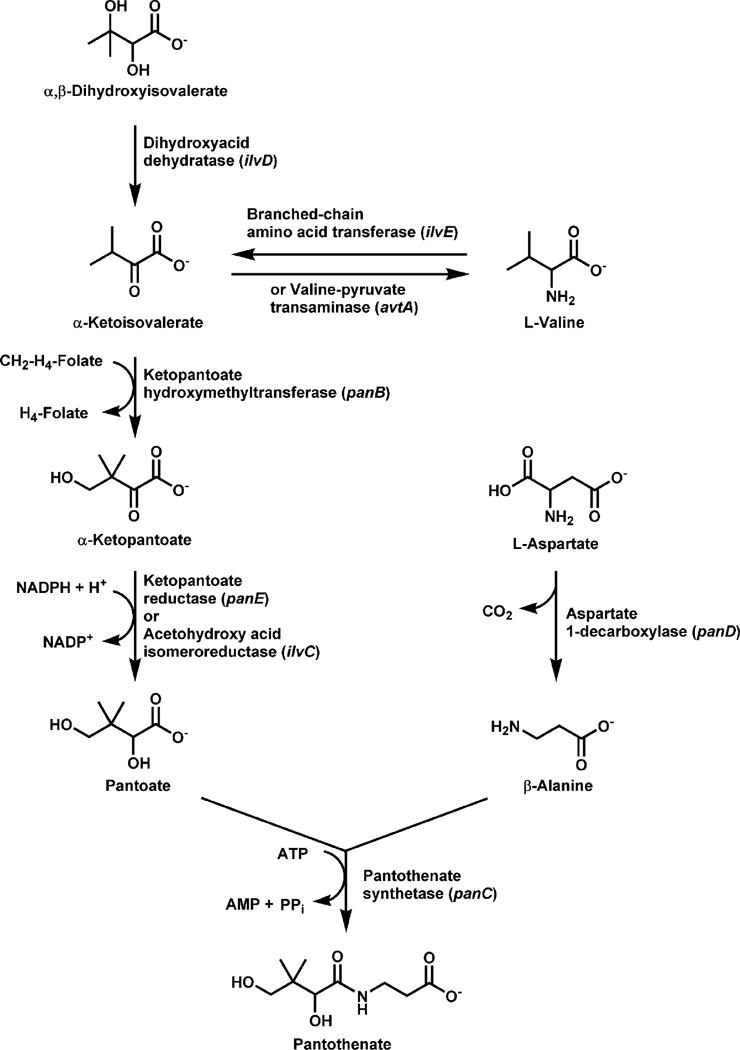
Pathway for the biosynthesis of pantothenic acid. Three enzymatic steps are required for the de novo formation of pantothenate. β-alanine is formed from aspartate by aspartate-1-decarboxylase, the product of the panD gene. Pantoate formation begins with the transfer of a methyl group to α-ketoisovalerate by ketopantoate hydroxymethyltransferase (the panB gene product) followed by reduction by ketopantoate reductase (the panE gene product). Pantothenate is then formed by the ATP-dependent condensation of β-alanine and pantoate by pantothenate synthetase (the panC gene product). Pantothenate is then either used for CoA biosynthesis or exported from the cell.
α-Ketopantoate Biosynthesis
The first step in the biosynthesis of D-pantoic acid is the formation of α-ketopantoate by the α-ketopantoate hydroxymethyltransferase (EC 2.1.2.11)(Fig. 1). The enzyme uses N5,N10-methylene tetrahydrofolate and catalyzes the conversion of α-ketoisovalerate to ketopantotate by transferring the carbon of the methylene moiety from the cofactor to the substrate (99). The idea that α-ketoisovalerate, an intermediate in the biosynthesis of valine, is also required for pantoate synthesis was first suggested by Maas and Vogel (81) and was later demonstrated in mutant strains with defects in ilvD, ilvE or avtA (153). The hydroxymethyltransferase is the product of the panB gene of E. coli which encodes a 28 kDa polypeptide estimated to be a large multimer by several techniques (60,99,100,129). The crystal structure of the native protein from E. coli (149) and M. tuberculosis (12) reveals that the enzyme forms a decameric complex, with subunits in opposing pentameric rings held together by a swapping of their C-terminal α-helices. A magnesium ion is coordinated in the active site and is required for catalysis (99). The affinity for ketoisovalerate is about 1 mM for the E. coli enzyme and 240 μM for the M. tuberculosis enzyme (152). The ketopantoate hydroxymethyltransferase exhibits end-product inhibition by ketopantoate, thus helping to regulate pantothenate production (99,134). The enzyme is also inhibited by pantothenate (>500 μM) and CoA (>1 mM), but by concentrations that are much higher than found in vivo (142), arguing against feedback regulation by these downstream components of the pathway.
The panB gene is in an operon together with the panC gene (60,106) and a promoter mutation leads to increased expression of both the hydroxymethyltransferase and the synthetase from S. typhimurium, which in turn increases pantothenate production (106). Overepxression of the panB gene alone, but not the panC (106), or supplementation with exogenous pantoate (19) increases CoA by 40–50%, suggesting that the supply of endogenous pantoate is limiting for pantothenate biosynthesis, and pantothenate utilization, in turn, is regulated at the PanK step.
Pantoate Biosynthesis
D-pantoate is synthesized from α-ketopantoate by α-ketopantoate reductase (EC 1.1.1.169), the product of the panE gene (Fig. 1), which transfers a hydrogen from NADPH to pantoate. The enzyme first purified from Stenotrophomonas (formerly Pseudomonas) maltophilia has a subunit molecular weight of 30.5 kDa and exists as a multimer in its native state (114). The monomer sizes of the reductase enzymes from E. coli and S. typhimurium are both about 33 kDa (40,84,167). The pH dependence of the reaction for the E. coli enzyme suggests the involvement of a general acid/base in the catalytic mechanism, where the conserved Lys176 and Glu256 residues are essential (166). The Lys moiety is located in a hinge region, or cleft, bending between the N- and C-terminal domains (15) and undergoes a conformational switch from a resting state to an active state which binds ketopantoate (15,74). The enzyme discriminates between ketopantoate and pantoate only with NADPH bound, which promotes cooperative interaction with the substrate.
The acetohydroxy acid isomeroreductase (EC 1.1.1.86) which is encoded by the ilvC gene and is involved in the biosynthesis of isoleucine and valine, is also capable of catalyzing the reduction of α-ketopantoate both in vitro and in vivo. In C. glutamicum, the ilvC gene product is the only enzyme capable of producing pantoate (85). Primerano and Burns isolated mutants in α-ketopantoate reductase and clarified the relationship between pantoate and branched-chain amino acid biosynthesis in S. typhimurium (101,102). Mutants in panE do not require pantoate when the ilvC gene is abundantly expressed, but when ilvC expression is low, panE mutants require pantoate for growth. The panE gene maps at 10 min and is allelic to apbA in S. typhimurium (40,41). A role for the apbA gene had been proposed an alternative pathway for thiamine biosynthesis, but in vivo labeling showed that pantoic acid, the product of the ApbA-catalyzed reaction, is not a direct precursor to thiamine. The conditional requirement of panE mutants for either thiamine or pantothenate is manifest only when flux through the purine biosynthetic pathway is reduced due to the lack of thiamine pyrophosphate, not decreased CoA (39). Enhanced expression of the panE gene by 3-fold results in about a 3-fold increase in pantothenate excretion from E. coli and even higher when the medium is supplemented with ketopantotate (30).
β-Alanine Biosynthesis
Aspartate was first suggested as the precursor to β-alanine on the basis of the conversion of aspartate to β-alanine by intact cells (21,148). Williamson and Brown (157) and Cronan (18) independently characterized an L-aspartate-1-decarboxylase activity from E. coli that converts aspartate to CO2 and β-alanine (EC 4.1.1.11) (Fig. 1). The decarboxylase has a molecular size of 58 kDa and has pyruvate moieties covalently bound to multiple identical subunits that associate as a tetramer. The pyruvate is involved in catalysis (156) and forms a Schiff base between enzyme and substrate (71) at the active sites which are located between adjacent subunits (3). The affinities of the enzymes from E. coli and M. tuberculosis for aspartate are similar, with Km values estimated at 151 μM (103) and 219 μM (13), respectively. The decarboxylase is translated as an inactive pro-protein (π-protein) of 13.8 kDa which subsequently undergoes an intramolecular rearrangement and is self-cleaved at the Gly24-Ser25 bond (103) to a mature form which has a β-chain (2.8 kDa) with a hydroxyl group at its C-terminus and an α-chain (11 kDa) which becomes activated by formation of the pyruvoyl group at its N-terminus. The α- and β-chains associate together and constitute a subunit of the tetramer (3,71). A crystal structure of the tetramer shows three cleaved subunits containing pyruvoyl moieties and one subunit with the ester intermediate. The molecular mechanism of self-processing relies on the conformational freedom of a loop preceding the cleavage site along with stabilization of an oxyoxazolidine intermediate by a Thr residue in the loop to form the ester intermediate (110).
Aspartate decarboxylase is encoded by the panD gene located at 3.1 min on the E. coli chromosome and mutants require β-alanine or pantothenate for growth (18,157). The panD locus in S. typhimurium is located at 4.5 min and in both organisms the panD gene is adjacent to panB and panC. Insertional mutation of the panD gene in S. typhimurium confers sensitivity to suphometuron methyl, suggesting that CoA availability modulates the response to this herbicide (70). Inhibition of pantothenate production in intact cells by D-serine, β-hydroxyaspartic acid, or L-cysteic acid is consistent with inhibition of the decarboxylase enzyme (17,27,28,46,80,115). A screen for inhibitors of the aspartate decarboxylase revealed that the active site is highly stringent for the size of compounds and only the structural analogues of aspartate, glutamate and 2-aminomethylsuccinate will bind (152). A second pathway for the synthesis of β-alanine from dihydrouracil was proposed (116,117) but does not occur in E. coli, since Cronan showed that β-alanine auxotrophs (panD mutants) will not grow on dihydrouracil (18). The aspartate decarboxylase from C. glutamicum has more activity than the homolog from E. coli and overexpression of either enzyme increases de novo pantothenate production in E. coli (29). The C. glutamicum enzyme is expressed at a higher level due to more efficient translation of the protein. The amino acid carrier CycA mediates the transport of β-alanine into E. coli with a low affinity (Km 2.4 mM) (111) and supplementation of wild-type E. coli with high concentrations of β-alanine also increases pantothenate production (19,29).
Pantothenate Biosynthesis
Pantothenate synthetase (EC 6.3.2.1) catalyzes the ATP-dependent condensation of D-pantoate with β-alanine to form pantothenate (Fig. 1). Pantothenate synthetase is encoded by the panC gene located at min 3.1 on the chromosome in E. coli and at min 4.5 in S. typhimurium (19,79). Pantothenate synthetase from E. coli (89) and from Mycobacterium tuberculosis (165) has been purified and characterized. The reaction proceeds via formation of a pantoyl adenylate intermediate (155) following the binding of ATP first, then D-pantoate and the release of pyrophosphate. The binding of β-alanine, the second substrate, can only occur after formation of the pantoyl adenylate (150) and triggers the release of the products, pantothenate and AMP. The three-dimensional structure of the M. tuberculosis enzyme in complex with the pantoyl adenylate was determined (151) as part of the TB Structural Genomics consortium (45), and the active site residues required for the formation and stabilization of the intermediate were identified by mutagenesis (168).
The detailed reaction mechanism provided by this information led to the idea that nonreactive analogs of pantoyl adenylate would be highly specific inhibitors of the pantothenate synthetase. Inhibitors of pantothenate synthetase which have potential as antimicrobial agents have, in fact, been identified (137,154). Ten analogues of the reaction intermediate pantoyl adenylate, in which the phosphodiester is replace by either an ester or sulfamoyl group, are all modest inhibitors, with the the sulfamoyls being more potent (137). The clockwise gene order as determined by direct DNA sequencing is panB-panC-panD (19), where panB and panC lie adjacent to one another, but are separated from panD by an open-reading frame of unknown function, orf3, which is oriented in the opposite direction (86). Pantothenate synthetase activity is not tightly regulated in vivo since E. coli secretes into the medium most of the pantothenate that is synthesized (22,56,80), thus providing the vitamin to the mammalian host.
Pantothenate Transport
Pantothenate is taken up by virtually all bacteria and is essential for growth of those organisms lacking de novo pantothenate biosynthesis, such as Streptococcus pneumoniae, Lactobacillus lactis and Hemophilus influenzae (44). Bacillus subtilis is an exception, however, as efficient pantothenate uptake cannot be demonstrated (5). A pantothenate transport activity was first described in E. coli (92) and later identified as mediated by the pantothenate permease, also termed the PanF protein, encoded by the panF gene located at min 72 of the chromosome (Fig. 2). The panF gene is cotranscribed with the prmA gene which encodes a protein that methylates ribosomal proteins (146). The PanF protein is predicted to contain 12 transmembrane hydrophobic domains connected by short hydrophilic chains which is a topological motif characteristic of other cation-dependent permeases of the major facilitator superfamily of proteins (55,61,104). PanF uses a sodium-cotransport mechanism to concentrate pantothenate from the medium (55,145) which is highly specific for pantothenate, with a Kt of 0.4 μM and a maximum velocity of 1.6 pmol/min/108 cells (92,145). A similar transport system is present in S. typhimurium (S. D. Dunn and E. E. Snell, J. Supramol. Struct. 6:136, 1977). Overexpression of the PanF permease in E. coli produces a 10-fold increase in the rate of pantothenate uptake and concomitant elevation of the steady-state intracellular concentration of pantothenate (55). The CoA levels remain unaffected by overexpression, however, indicating that PanF activity does not regulate CoA biosynthesis. Conversely, CoA levels do not change the rate of pantothenate transport (55). Inactivation of the panF gene blocks uptake, but does not reduce the exit of pantothenate synthesized in the bacteria (144), indicating the existence of a distinct efflux system. The active transport of pantothenate into bacteria would suggest that the biosynthetic pathway leading to pantothenate is of limited interest for the development of novel antibacterial compounds. However, the development of an attenuated live vaccine against M. tuberculosis has been enabled by engineering the bacterium to rely solely on pantothenate transport for CoA biosynthesis (108). The pantothenate auxotroph of M. tuberculosis is severely limited in its growth (109) and its reduced replication in the host animal is sufficient to elicit an effective immune response but does not give rise to pathogenic infection. Thus, inhibition of pantothenate production in this bacterium would be an effective antibacterial strategy.
Figure 2.
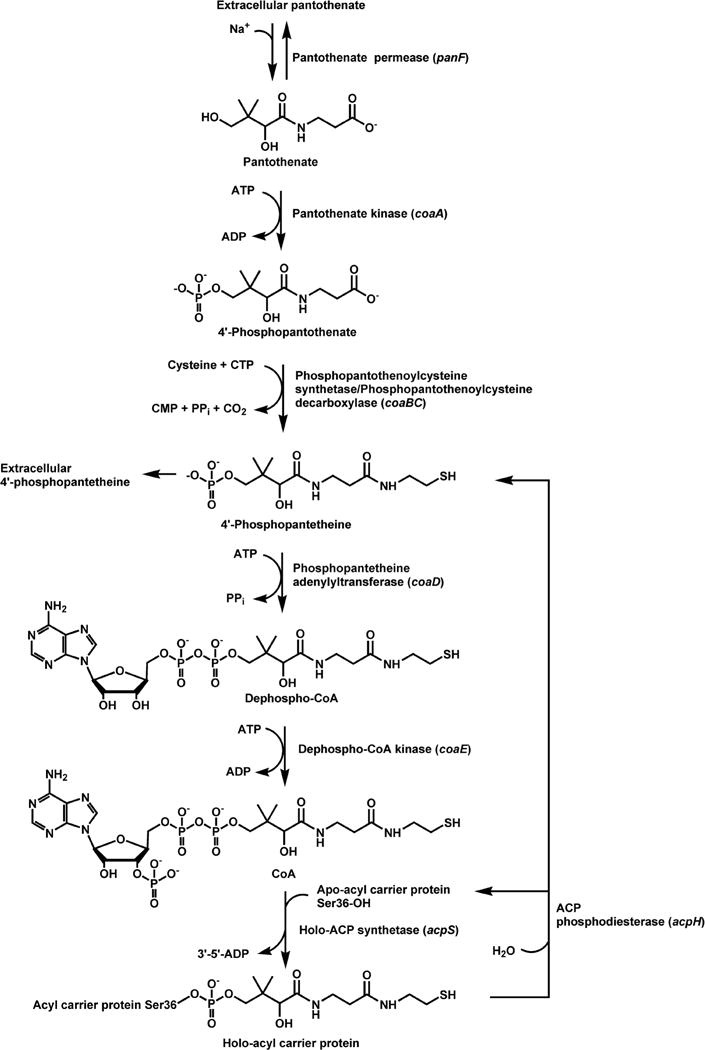
Pathway for CoA biosynthesis and the addition of the prosthetic group to ACP. Active uptake of pantothenate by a sodium-dependent permease (the panF gene product) is an alternate route to intracellular pantothenate, which is then either used for CoA biosynthesis or exported from the cells by a separate transport system. Pantothenate kinase (the coaA gene product) is the first, and most highly regulated, step in CoA biosynthesis and is regulated by feedback inhibition by CoA and its thioesters. Cysteine is then added to 4′-phosphopanthenate to generate 4′-phosphopantothenoylcysteine, which is then decarboxylated to yield 4′-phosphopantetheine. Both reactions are catalyzed by the bifunctional enzyme, phosphopantothenoylcysteine synthetase/phosphopantothenoylcyteine decarboxylase (the coaBC gene product). 4′-Phosphopantetheine adenylyltransferase (the coaD gene product) is a secondary regulatory point and is responsible for the formation of dephospho-CoA, which is then phosphorylated on the 3′-hydroxyl group to yield CoA by the dephospho-CoA kinase (the coaE gene product). CoA is then used as the 4′-phosphopantetheine donor in the synthesis of ACP from apo-ACP catalyzed by ACP synthase (the acpS gene product). The 4′-phosphopantetheine prosthetic group of ACP undergoes rapid metabolic turnover mediated by the ACP phosphodiesterase (the acpH gene product), generating apo-ACP and 4′-phosphopantetheine. The direct degradation of CoA to 4′-phosphopantetheine occurs during abrupt metabolic transitions. 4′-Phosphopantetheine is either reused for CoA synthesis or excreted from the cell. 4′-Phosphopantetheine accumulates in the growth medium since E. coli does not have an uptake system for this intermediate.
CoA BIOSYNTHESIS
Pantothenate Phosphorylation
Pantothenate kinase (PanK, also termed CoaA)(EC 2.7.1.33) catalyzes the ATP-dependent phosphorylation of pantothenate to 4′-phosphopantothenate, the first step in the biosynthesis of CoA (Fig. 2). Originally it was thought that the first phosphorylated intermediate in the pathway was 4′-phosphopantetheine; however this point was reinvestigated in 1958 by Brown (10), who convincingly showed that the formation of 4′-phosphopantothenate was required for the subsequent reactions in the CoA biosynthetic pathway. PanK is encoded by the coaA gene, and temperature–sensitive coaA mutants were isolated by Dunn and Snell in S. typhimurium (26) and subsequently by Vallari and Rock in E. coli (143). Actually, the first temperature-sensitive coaA mutants were isolated in 1966; however the biochemical defect was unknown and the mutated gene was termed rts (36). The rts and coaA mutations are alleles of the same gene (118), as indicated by the fact that the nucleotide sequences of rts and coaA are identical (37,120). The coaA mutants cannot be supplemented since E. coli does not incorporate extracellular phosphorylated intermediates such as 4′-phosphopantothenate, 4′-phosphopantetheine or dephosphoCoA. The coaA gene is located at min 90 of the E. coli chromosome between birA and thrU genes (120), it possesses its own promoter and produces a 1.1 kb transcript. The utilization of either of two translation initiation sites produces two PanK proteins that differ in eight amino acids at the amino terminus. The poor homology of the coaA promoter region to the consensus E. coli promoter sequences and the low frequency of optimal codon usage are consistent with a low level of PanK expression (120).
Jackowski and Rock (56) proposed PanK as a major rate-controlling step in CoA biosynthesis on the basis of the copious excretion of pantothenate from E. coli (22,56,80). Nonesterified CoA was five times more potent than CoA thioesters in inhibiting the kinase in extracts from E. coli (142). CoA is a competitive inhibitor with respect to ATP, thus providing a mechanism to coordinate CoA production with the energy state of the cell. The enzyme was purified to homogeneity from overexpressing strains and shown to be a homodimer of 36 kDa subunits (119). The purified enzyme exhibits cooperative binding of ATP and is competitively inhibited by CoA. Cooperative ATP binding made determination of the kinetic mechanism complicated, and, to overcome the problematic interpretation of the kinetic data, intragenic complementation was used to produce a chimeric heterodimer between a wild-type and an inactive subunit. The inactive subunit was expressed by a coaA plasmid construct in which the codon of the critical Lys101 in the predicted ATP binding site was mutated to a Met residue. Kinetic analysis of these chimeric molecules indicated the absence of cooperative ATP binding and revealed that the kinase reaction proceeds by a sequential mechanism, with ATP as the first substrate interacting with the protein. The Km affinity constants for ATP and pantothenate are 136 and 36 μM, respectively.
The crystal structures of E. coli PanK bound to a non-hydrolyzable ATP analog and to free CoA show that the phosphate groups of the two ligands occupy the same space at Lys101 (162). As a consequence, the binding of CoA prevents the binding of ATP, explaining the competitive nature of the CoA inhibition. The structure also reveals that the free thiol group of CoA fits tightly in the inhibitor binding site, hindering the space to accommodate acyl chains; this conclusion provides the structural basis for the more potent inhibition of PanK by nonesterified CoA than by CoA thioesters. In contrast to the CoA binding site, the pantothenate binding site is remarkably flexible, allowing for the binding and phosphorylation of a bulky pantothenate analog such as N-pentylpantothenamide (50). This molecule and similar N-alkylpantothenamides were first synthesized and found to inhibit E. coli growth in 1970 (16). However, the precise mechanism of action against the bacterium was elucidated more than 30 years later, when it was shown that N-pentylpantothenamide is a substrate for E. coli PanK and the downstream enzymes in the CoA biosynthetic pathway (125). N-pentylpantothenamide and, in general, the N-alkylpantothenamides block fatty acid biosynthesis in E. coli by being converted to CoA analogs that are incorporated into and inactivate acyl carrier protein (147,163).
The availability of genomic sequence information for a number of bacterial pathogens has uncovered new and different proteins that catalyze the first step in CoA biosynthesis. Three types of PanKs have been so far identified, and the well characterized E. coli enzyme is considered a type I, or prokaryotic, PanK. The PanK from Staphylococcus aureus is a type II, or eukaryotic-like enzyme. The S. aureus coaA gene was identified by comparative genomics and the encoded protein was found to be moderately related to eukaryotic PanKs and unrelated to the E. coli enzyme (44). S. aureus PanK has been expressed and purified to homogeneity (14,73). The enzyme is a dimer of 29 kDa subunits with Km affinity constants for ATP and pantothenate of 34 and 23 μM, respectively. The structure of the enzyme reveals two solvent exposed openings to the active site and suggests a non-ordered mechanism, with ATP entering from one opening and pantothenate binding through the other opening in a random sequence (48). The type II PanK accepts the N-alkylpantothenamides as substrates, and these compounds manifest more potent antimicrobial action against S. aureus than E. coli. (73,147). Unlike the type I PanK, the type II PanK from S. aureus is not subject to feedback inhibition by free CoA or CoA thioesters, and the organism accumulates high concentrations of intracellular CoA limited only by the pantothenate supply in the medium (73). S. aureus lacks glutathione (93), and instead this bacterium utilizes nonesterified CoA as the major intracellular thiol and a unique CoA disulphide reductase to maintain the redox potential of the cell (24,25). Therefore, the ability to accumulate CoA through lack of feedback inhibition of PanK represents a clear advantage in the physiological context of S. aureus. Bacterial type II PanK sequences are restricted to very few organisms including Bacillus anthracis (44); however, no 4′-phosphopantothenate was detected in reaction mixtures containing the predicted B. anthracis type II PanK and pantothenate (94). The existence of a third type of PanK was inferred by the fact that several organisms, including pathogens such as Pseudomonas aeruginosa and Helicobacter pylori, lack any recognizable PanK although they possess all the remaining enzymes of the CoA biosynthetic pathway (43,44,98). The first gene encoding a new type of PanK was discovered in B. subtilis and initially named coaX to distinguish it from the coaA gene encoding a type I PanK in the same organism (161). The observation that disruption of the coaA gene in B. subtilis gave a normal growth phonotype led to the conclusion that there could be another enzyme able to perform the same reaction. The coaX gene was identified by the screening of a B. subtilis gene library for the ability to complement a temperature-sensitive E. coli strain. B. subtilis coaX homologs were subsequently found in the vast majority of organisms classified as missing either a type I or a type II PanK. Type III PanKs from a variety of organisms have been expressed, purified and crystallized (9,48,94). The most remarkable difference between these enzymes and the types I and II PanKs is the low affinity for ATP. The Km values for ATP are in the millimolar range and the weak binding of the phosphoryl donor absolutely requires a monovalent cation such as potassium or ammonium. The type III PanK from P. aeruginosa is a dimer of 29 kDa subunits. The individual subunits of the S. aureus and P. aeruginosa PanKs fold in a very similar manner but assemble distinct dimers with unique enzymatic properties; in particular, the poor affinity and selectivity of the type III PanK for ATP can be explained by the wide and solvent-exposed binding site revealed by the crystal structure of the dimer (48). Conversely, pantothenate binds in a buried pocket at the dimer interface that would not accommodate bulkier analogs like the N-alkylpanothenamides. These compounds are not toxic for P. aeruginosa because they are not substrates for the type III PanK and cannot enter the CoA biosynthetic pathway. Hong and coworkers also showed that P. aeruginosa resistance to the N-alkylpanothenamides results from the lack of uptake of these compounds (48). Similarly to the type II S. aureus PanK, the type III enzymes are refractory to feedback inhibition by free CoA or CoA thioesters. Among the organisms that possess a type III PanK are those that rely on the CoA/CoA disulphide reductase system to maintain redox status of the cell, such as B. anthracis, but also organisms such as P. aeruginosa that utilize the more widespread glutathione/glutathione reductase system.
Formation of 4′-Phosphopantetheine
4′-phosphopantetheine is formed in two enzymatic steps (Fig. 2) (10). First, 4′-phosphopanothenate is condensed with cysteine by 4′-phosphopantothenoylcysteine synthetase (EC 6.3.2.5) to form 4′-phosphopantothenoylcysteine, which is then decarboxylated by 4′-phosphopantothenoylcysteine decarboxylase (EC 4.1.1.36) to generate 4′-phosphopatetheine. The 4′-phosphopantothenoylcysteine decarboxylase activity in E. coli was initially identified in fractions enriched in a 35 kDa protein containing a covalently bound pyruvoyl group (160); however, the enzyme was not purified to homogeneity and the conclusion turned out to be incorrect. More than a decade later the dfp gene (121,122), recently renamed coaBC, was found to encode a flavin mononucleotide-containing bifunctional enzyme responsible for both the 4′-phosphopantothenoylcysteine synthetase and the 4′-phosphopantothenoylcysteine decarboxylase activities in E. coli (66,127). Years before the discovery of the function of the dfp gene, a conditional-lethal dfp mutant, dfp-707, was isolated by Spitzer and coworkers who noticed that the mutation caused a slow cessation of DNA synthesis at 42 °C and that the dfp-707 strain required either β-alanine or pantothenate to grow at 30 °C in minimal medium (121,122). Another mutation in the same gene, named dfp-1, conferred auxotrophy but not the conditional lethality of dfp-707, allowing the dfp-1 mutant to grow in rich medium at 42 °C. The nutritional requirements of the both mutants were reminiscent of the panD mutants, but the aspartate-1-decarboxylase activity was similar to the wild-type strain and the authors did not investigate the connection to CoA biosynthesis any further. The 4′-phosphopantothenoylcysteine synthetase (CoaB) and the 4′-phosphopantothenoylcysteine decarboxylase (CoaC) activities encoded by the E. coli dfp gene were individually and independently discovered (66,127). These activities are fused together in almost all bacteria with the exception of enterococci and streptococci that possess separate genes (43,44). B. anthracis contains a gene predicted to encode a monofunctional 4′-phosphopantothenoylcysteine synthetase in addition to the bifunctional CoaBC enzyme. E. coli CoaBC is a homododecamer of 43 kDa subunits (66). Amino terminal sequencing of the endogenous protein identified ATG25 as the start codon of the coaBC gene and revealed the cleavage of the initiator methionine from the mature protein (127).
The individual 4′-phosphopantothenoylcysteine synthetase and 4′-phosphopantothenoylcysteine decarboxylase domains of the E. coli CoaBC have been expressed and purified (64,65). The flavin mononucleotide-binding amino-terminal domain of the enzyme spans residues 2 to 190, possesses 4′-phopshopantothenoylcysteine decarboxylase activity and it is responsible for the formation of dodecamers by the full-length bifunctional protein. Amplification and sequencing of the coaBC gene from the conditional lethal dfp-707 mutant identified a single point mutation in codon 11 that exchanges a glycine for aspartate, and the mutant protein was expressed and purified from cells grown at 30 and 37 °C (64). The 4′-phosphopantothenoylcysteine carboxylase activity of the mutant was not affected by the expression temperature but only very low amounts of protein were obtained at 37 °C, suggesting that the conditional lethality of E. coli dfp-707 results from decreased protein solubility or stability. From a mechanistic point of view, the decarboxylation of 4′-phosphopantothenylcysteine proceeds via a thioaldehyde intermediate formed by the flavin mononucleotide-dependent oxidation of the cysteine moiety of 4′-phosphopantothenoylcysteine. Decarboxylation of this intermediate followed by reduction through the flavin reduced form completes the catalytic cycle (124,126,128). The carboxy terminal domain of E. coli CoaBC encompasses residues 191–406, catalyzes the CTP-dependent condensation of 4′-phosphopantothenate and cysteine, and forms homodimers (65). The preference of CTP over ATP distinguishes the bacterial 4′-phosphopantothenoylcysteine synthetase from the human monofunctional homolog (10). Similarly to the dfp-707 mutant, the sequence analysis of the dfp-1 mutant revealed a single point mutation that exchanges Ala275 with a Thr (67). This Ala is part of the CTP binding site (123) but the molecular reason for the temperature sensitivity of the dfp-1 mutant needs to be elucidated in more detail.
The formation of 4′-phosphopantothenoylcysteine occurs via two half reactions: first an activated 4′-phosphopantothenoyl-cytidylate intermediate is formed which is then attacked by the amino group of cysteine to yield the final product (67). The 4′-phosphopantothenoyl-cytidylate intermediate copurifies with mutant forms of the E. coli enzyme containing different residues in place of Asn210 and has been unequivocally identified by mass spectrometry. The intermediate is also visible in the crystal structure of the Asn210Asp mutant obtained in the presence of CTP and 4′-phosphopanothenate (123). E. coli 4′-phosphopantothenoylcysteine synthetase discriminates between cysteine and serine or homocysteine, but can couple cysteamine and cysteine methyl ester to 4′-phophopanothenate. The available crystal structures of the Asn210Asp enzyme reveal a potential cysteine binding site where the thiol group would be tightly accommodated in a hydrophobic cavity with poor affinity for the more polar serine and with not enough space for larger side chains. Conversely, the carboxylate group of cysteine is predicted to be exposed to the solvent explaining why cysteamine and cysteine methyl esters can function as substrates. The crystal structure of the human 4′-phosphopantothenoylcysteine synthase is available (82); the major structural features are very similar between the human and the E. coli enzymes which likely share the same enzymatic mechanism. However, the two enzymes differ significantly in the nucleobase-binding parts of the respective nucleotide binding sites, explaining the preference of the human enzyme for ATP and of the bacterial enzyme for CTP.
Conversion of 4′-Phosphopantetheine to Dephospho-CoA
ATP:4′phosphopantetheine adenylyltransferase (EC 2.7.7.3), also known as CoaD or PPAT, catalyzes the Mg2+-dependent reversible transfer of the AMP moiety of ATP to 4′-phosphopantetheine to form dephospho-CoA (1,47). The enzyme was first isolated from Corynebacterium ammoniagenes (formely Brevibacterium ammoniagenes) as a trimer of 35 kDa subunits (83), however fractionation of E. coli cell-free extracts identified a smaller enzyme of 18 kDa with PPAT activity (42). N-terminal sequencing of the purified endogenous PPAT allowed the identification of kdtB as the coding gene, now renamed coaD. Recombinant E. coli PPAT forms homohexamers arranged as dimers of trimers, and has Km values of 220 and 7 μM for PPi and dephospho-CoA, respectively, in the reverse reaction.
Metabolic labeling experiments in E. coli detect accumulation of both intracellular and extracellular 4′-phosphopantetheine, suggesting that, in addition to the primary control exerted at the PanK level, PPAT is a secondary regulatory point in CoA biosynthesis (56,59) (see below). Similarly to PanK, regulation of the PPAT activity probably occurs through feedback inhibition by unesterified CoA (56,105,141), and consistent with this theory, the E. coli enzyme is purified with 0.5 moles of CoA/mole of enzyme (42). Crystal structures of the E. coli PPAT complexed with the substrates, product and the putative inhibitor CoA have been solved (51–53). These structures show that PPAT is an allosteric enzyme characterized by half-of-the-sites reactivity. In fact, only one of the trimers within the hexamer is in the substrate- or dephospho-CoA-bound conformation, whereas the other trimer is in its unbound, conformationally distinct state. This asymmetry in the binding to the two trimers is further confirmed by the CoA-bound structure of the enzyme. Specifically, the phosphopatetheine moiety of CoA binds to one trimer in a dephospho-CoA- or phosphopantetheine-like manner, with the adenylyl moiety disordered. This mode of binding, characterized by CoA occupying the substrate binding site, suggests a mechanism for the enzyme inhibition and produces a conformational change in the other trimer that forces CoA to bind in a ordered and unique conformation imposed by the presence of a phosphate group at the 3′ position of the ribose (52).
Kinetic analysis of the reverse reaction catalyzed by the E. coli PPAT reveals the formation of a ternary complex between the enzyme and both pyrophosphate and dephospho-CoA before catalysis, and the structural analysis predicts a random ordered binding mechanism (42,53). Superposition of the ATP- and 4′-phosphopantetheine-bound PPAT structures supports a model in which the α-phosphate of ATP undergoes nucleophilic attack by the phosphate group of 4′-phosphopantetheine in an in-line displacement mechanism (51). No direct participation of active site residues in acid-base or covalent catalysis is necessary, and the sole role of the enzyme seems to be to properly orient the substrates and to stabilize the transition state in order to lower the reaction energy barrier. Histidine 18 is particularly important in the stabilization of the transition state. This residue corresponds to the last histidine of the highly conserved H/TxGH motif that characterizes the nucleotidyltransferase α/β phosphodiesterase superfamily (8) to which PPAT belongs (42). Crystal structures of the enzyme are also available from other organisms and they all reveal the same protein fold as the E. coli PPAT (91,133). In mammals, the PPAT is fused with dephospho-CoA kinase in a bifunctional enzyme designated CoA synthase (2,20,158,169). Similar to PanK, the PPAT domain of the human CoA synthase does not share any significant sequence similarity with its prokaryotic counterparts (20,43,44). This fact and the availability of crystal structures make the enzyme an attractive target for the design of selective antibacterial drugs (164).
Phosphorylation of Dephospho-CoA to CoA
The last step in CoA biosynthesis is catalyzed by dephospho-CoA kinase (DPCK, also termed CoaE)(EC 2.7.1.24) that adds an ATP-derived phosphate group to the 3′-hydroxyl of dephospho-CoA. The enzyme responsible for the dephospho-CoA activity in C. ammoniagenes was first isolated from cell-free extracts and used to determine the protein N-terminal sequence. A BLAST search using this information then allowed the identification of the yacE gene as encoding a highly homologous protein in E. coli. The yacE gene has been renamed coaE (88). E. coli dephospho-CoA kinase has been overexpressed and purified. The enzyme exhibits Km values of 0.74 and 0.14 mM for dephospho-CoA and ATP, respectively. DPCK is purified as a 22 kDa monomer, but in the presence of sulfate ions it forms trimers both in solution and in the crystal structure (96). The monomer fold identifies the enzyme as a member of the P-loop-containing nucleotide triphosphate hydrolase superfamily which includes several nucleotide and nucleoside kinases. The P-loop motif is involved in the binding of the ATP triphosphate group, as confirmed by the crystal structures of ATP-bound DPCK from H. influenzae and Thermus thermophilus HB8 (97,113). The three structures are very similar, but while E. coli DPCK crystallizes as a trimer, the structures of the H. influenzae and T. thermophilus HB8 enzymes contain one and three independent monomers in the asymmetric unit, respectively. The structures also reveal the three domain organization characteristic of nucleotide and nucleoside kinases: the nucleotide binding domain, the substrate binding domain and the lid domain. The latter two domains are expected to be very mobile and, upon binding of dephospho-CoA, to close over the catalytic site to direct the phosphate group transfer.
CoA METABOLISM
In proliferating bacteria with unlimited pantothenate availability, the CoA pool is large and more than enough to support rapid growth. Removal of the pantothenate supply does not result in a cessation of growth until the CoA pool is reduced by dilution through several generations (56). Regulation of the size of the CoA pool is largely accomplished by limitation of its synthesis through feedback inhibition rather than by degradation. Mechanisms to reduce CoA by degradation are in place, however, to respond to metabolic challenge, such as an acute change in carbon flux (58) or adaptation to pantothenate starvation (57).
Composition of the Intracellular CoA Pool
The composition of the E. coli intracellular CoA pool has been examined by using panD mutants to specifically label CoA and by estimating individual CoA thioesters by high pressure liquid chromatography (142). The size and composition of the CoA pool vary depending on the carbon source (Table 1). The CoA pool is highest in cells growing on glucose, and acetyl-CoA is the predominant species. In contrast, the amount of total CoA is much lower in cells growing on casein hydrolysate as the carbon source, indicating that amino acid biosynthesis requires an elevated CoA pool, particularly in the form of acetyl-CoA. Consistent with this interpretation, when pantothenate auxotrophs growing on glucose minimal medium are deprived of pantothenate, the succinyl-CoA pool becomes limiting for amino acid and protein synthesis, leading to growth arrest (58). Alternatively, growth on glucose may lead to acetate accumulation, which, in turn, may determine the size of the acetyl-CoA pool. The latter explanation is supported by the observation that the acetyl-CoA pool is rapidly reduced >30% upon withdrawal of supplement from an acetate auxotroph (141). The total amount of unesterified CoA drops correspondingly, with the excess CoA being hydrolyzed and 4′-phosphopantetheine effluxed out of the cell. On the other hand, elevation of intracellular CoA by overexpression of PanK in E. coli causes an increase in carbon flux to acetate production, provided that enough supplemental pantothenate precursor is available (118,138). Increased acetate production by the cell is not only reflected by increased acetyl-CoA levels, but also by an increase in excreted acetate (139).
TABLE 1.
Intracellular CoA pool composition in E. colia
| Carbon source | Concentration (μM)
|
||||
|---|---|---|---|---|---|
| Total CoA | CoASH | Acetyl-CoA | Succinyl-CoA | Malonyl-CoA | |
|
| |||||
| Glucose | 406 | 56 | 324 | 24 | 2 |
| Succinate | 368 | 119 | 137 | 94 | 18 |
| Acetate | 202 | 108 | 17 | 68 | 9 |
| Glycerol | 199 | 61 | 79 | 24 | 35 |
| Amino acids | 111 | 30 | 67 | 10 | 4 |
Data are derived from reference 142.
The concentration of nonesterified CoA in S. aureus reaches millimolar levels (24). In the absence of glutathione, this organism uses free CoA not only in metabolic cycles but also as the major low molecular weight thiol in the cell. The ratio of reduced to oxidized (disulphide) CoA is > 100 and maintained by a specialized CoA disulphide reductase. Accumulation of CoA to levels significantly higher than in E. coli is possible because S. aureus possesses a PanK refractory to feedback inhibiton by CoA (73). Glutathione is also missing in many Bacilli (34,35) that, instead, utilize a pantethine 4′,4′′-diphosphate- or CoA-disulfide reductase system (130,131). In the spore-forming bacterium B. megaterium, CoA is also used to form CoA-protein disulfides during spore formation. Accumulation of these mixed disulfides is proposed to maintain metabolic dormancy in the spores and/or to contribute to heat and radiation resistance by protecting labile protein thiol groups (112). Although there are reports of CoA-glutathione mixed disulfides in E. coli (76,77), these compounds are likely due to oxidation prior to analysis, and there is no evidence that they occur in vivo.
Regulation of CoA Levels by Feedback Inhibition
The phosphorylation of pantothenate catalyzed by PanK is the primary rate-limiting step in CoA biosynthesis in E. coli. This reaction is controlled through feedback inhibition of the enzyme by CoA and CoA thioesters, the end-products of the pathway. As previously mentioned, there are considerable differences in the size and composition of the CoA pool in E. coli cells grown on different carbon sources. A shift from glucose to acetate as the carbon source results in an increase in the nonesterified CoA/acetyl-CoA ratio from 0.7 to 4.3 (142) and in the reduction of ATP levels (78). This remodeling of the CoA pool composition is associated with the selective inhibition of pantothenate phosphorylation, consistent with nonesterified CoA being the most potent inhibitor of PanK in vivo. E. coli mutants which possess a PanK activity in crude extracts that is refractory to feedback inhibition by CoA have been isolated (141). Strains harboring this mutation [coaA16(Fr)] have CoA levels that are significantly (>2-fold) higher than in strains containing the wild-type kinase. A very similar result is obtained when a single mutation of Arg106 to Ala is introduced in E. coli PanK and the mutant is expressed at single copy levels in the coaA15(Ts) strain background at elevated temperature (105). Furthermore, corroboration of the conclusion that modulation of PanK activity by feedback regulation is the critical factor controlling the intracellular CoA concentration comes from studies of the effect of PanK overexpression on the size of the CoA pool (120). Strains expressing 76-fold more wild-type kinase exhibited only a 2.7-fold increase in the steady state CoA level. CoA regulates the PanK activity by competing for the ATP binding site (119,142,162), thus the activity of the enzyme can also be coordinated with the energy state of the cell, where an increase in ATP levels would displace the competitive inhibitor and resume the biosynthetic activity. Therefore, changes in the composition of the CoA pool and ATP levels function in concert to modulate the rate of CoA biosynthesis.
The secondary regulatory step in CoA biosynthesis is catalyzed by PPAT. Regulation at this step in E. coli is proven by the secretion of 4′-phosphopantetheine in the medium (56,59), and becomes more important when the primary regulatory step is disrupted (105) or when PanK is overexpressed (118). An increase in the amount of intracellular and extracellular 4′-phosphopantetheine under these circumstances reflects the restriction of the CoA precursor flux through PPAT. Excretion of 4′-phosphopantetheine is an irreversible event since E. coli is unable to uptake phosphorylated intermediates in CoA biosynthesis (59). The time- and concentration-dependent correlation between accumulation of intracellular CoA and exit of 4′-phosphopantetheine from the cells suggests that PPAT is regulated by free CoA (56,141). Consistent with this hypothesis, the enzyme is isolated and crystallized with bound unesterified CoA (42,52). Interestingly, S. aureus lacks both the PanK and the PPAT regulatory checkpoints. This conclusion is suggested by the fact that neither pantothenate nor 4′-phosphopantetheine accumulate inside or outside the cells in metabolic labeling experiments (73).
CoA Metabolism and Prosthetic Group Transfer
CoA is the source of the 4′-phosphopantetheinyl prosthetic group present in a number of proteins that function as acyl/aminoacyl/peptidyl group carriers. Examples are the carrier proteins of fatty acid synthases, nonribosomal peptide synthetases and polyketide synthases (63). The transfer of the 4′-phosphopantetheinyl moiety of CoA to a conserved serine residue of these carrier proteins releases 3′,5′-ADP, and is catalyzed by a class of enzymes called phosphopantetheinyl transferases (PPTases)(68,90). In E. coli, the acyl carrier protein (ACP) of fatty acid biosynthesis is specifically converted to holo-ACP by ACP synthase (EC 2.7.8.7) (31,69), also termed AcpS1 (38), which is a PPTase encoded by the acpS gene. Sequencing of the acpS gene from the E. coli strain MP4 reveals a Gly4Asp mutation which reduces the catalytic efficiency of the enzyme about 5-fold. Overexpression of the product of the gene yhhU can suppress the phenotype of acpS mutants, suggesting that the YhhU protein may also have a PPTase function. The EntD PPTase of E. coli activates the enterobactin synthetase (68). A third PPTase identified in E. coli K-12 and other E. coli strains by homology searches is encoded by the gene acpT (23). The AcpT protein modifies two carrier proteins encoded in O-island 128, a cluster of fatty acid biosynthesis-like genes located adjacent to acpT in the genome of the pathogenic E. coli strain O157:H7, but it cannot substitute fully for AcpS in its activity. Whereas in most cases the bacterial PPTases exhibit substrate specificity, in Pseudomonas aeruginosa the single PPTase is multifunctional (6).
The 4′-phosphopantetheine prosthetic group can be removed from E. coli ACP by AcpH, the ACP phosphodiesterase, also called ACP hydrolase (EC 3.1.4.14) (135,136,140). While AcpS is essential (44), AcpH is not and its distribution is limited to Gram-negative organisms (135). AcpH is the also the only PPTase identified thus far in bacteria that participates in the recycling of the 4′-phosphopantetheine moiety back to CoA (59). AcpH, a non-canonical member of the HD phosphatase phosphodiesterase family, cleaves both unacylated and acylated ACP species with chain lengths of 6–16 carbons (135) and its activity is dependent on Mn2+ ions (136). Metabolic radiolabeling following the starvation of pantothenate auxotrophs shows that the level of holo-ACP is maintained at the expense of CoA (4,56,59). The turnover of the ACP prosthetic group is four times faster than the rate of new ACP protein synthesis during recovery from CoA deprivation, and drops an order of magnitude during exponential growth when the CoA levels are high (59). The 4′-phosphopantetheine released by the AcpH can either re-enter the CoA biosynthetic pathway (Fig. 2) or irreversibly exit from the cell and thereby regulate the size of the CoA pool.
CoA can also be hydrolyzed directly to yield 4′-phosphopantetheine and 3′,5′-ADP and this process does not involve ACP prosthetic group turnover (141). CoA degradation occurs when the level of acetyl-CoA falls, leading to a concomitant increase in nonesterified CoA. These results are corroborated by the finding that large amounts of 4′-phosphopantetheine are excreted also in an E. coli strain lacking AcpH (135). Recent data suggest that the pyrophosphatase responsible for CoA degradation could be a member of the nudix family (62,159).
Acknowledgments
Preparation of this chapter was supported by National Institutes of Health Grant GM062896 (S.J.), Cancer Center (CORE) Support Grant CA 21765, and the American Lebanese Syrian Associated Charities.
LITERATURE CITED
- 1.Abiko Y, Suzuki T, Shimizu M. Investigations on pantothenic acid and its related compounds XI. Biochemical studies 6. A final stage in the biosynthesis of CoA. J Biochem (Tokyo) 1967;61:309–312. doi: 10.1093/oxfordjournals.jbchem.a128549. [DOI] [PubMed] [Google Scholar]
- 2.Aghajanian S, Worrall DM. Identification and characterization of the gene encoding the human phosphopantetheine adenylyltransferase and dephospho-CoA kinase bifunctional enzyme (CoA synthase) Biochem J. 2002;365:13–18. doi: 10.1042/BJ20020569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Albert A, Dhanaraj V, Genschel U, Khan G, Ramjee MK, Pulido R, Sibanda BL, von Delft F, Witty M, Blundell TL, Smith AG, Abell C. Crystal structure of aspartate decarboxylase at 2.2 A resolution provides evidence for an ester in protein self-processing. Nat Struct Biol. 1998;5:289–293. doi: 10.1038/nsb0498-289. [DOI] [PubMed] [Google Scholar]
- 4.Alberts AW, Vagelos PR. Acyl Carrier Protein VIII. Studies of acyl carrier protein and coenzyme A in Escherichia coli pantothenate or β-alanine auxotrophs. J Biol Chem. 1966;241:5201–5204. [PubMed] [Google Scholar]
- 5.Baigori M, Grau R, Morbidoni HR, de MD. Isolation and characterization of Bacillus subtilis mutants blocked in the synthesis of pantothenic acid. J Bacteriol. 1991;173:4240–4242. doi: 10.1128/jb.173.13.4240-4242.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barekzi N, Joshi S, Irwin S, Ontl T, Schweizer HP. Genetic characterization of pcpS, encoding the multifunctional phosphopantetheinyl transferase of Pseudomonas aeruginosa. Microbiology. 2004;150:795–803. doi: 10.1099/mic.0.26823-0. [DOI] [PubMed] [Google Scholar]
- 7.Begley TP, Kinsland C, Strauss E. The biosynthesis of coenzyme A in bacteria. Vitam Horm. 2001;61:157–171. doi: 10.1016/s0083-6729(01)61005-7. [DOI] [PubMed] [Google Scholar]
- 8.Bork P, Holm L, Koonin EV, Sander C. The cytidylyltransferase superfamily: identification of the nucleotide-binding site and fold prediction. Prot Struct Funct Genet. 1995;22:259–266. doi: 10.1002/prot.340220306. [DOI] [PubMed] [Google Scholar]
- 9.Brand LA, Strauss E. Characterization of a new pantothenate kinase isoform from Helicobacter pylori. J Biol Chem. 2005;280:20185–20188. doi: 10.1074/jbc.C500044200. [DOI] [PubMed] [Google Scholar]
- 10.Brown GM. The metabolism of pantothenic acid. J Biol Chem. 1959;234:370–378. [PubMed] [Google Scholar]
- 11.Chassagnole C, Diano A, Letisse F, Lindley ND. Metabolic network analysis during fed-batch cultivation of Corynebacterium glutamicum for pantothenic acid production: first quantitative data and analysis of by-product formation. J Biotechnol. 2003;104:261–272. doi: 10.1016/s0168-1656(03)00146-9. [DOI] [PubMed] [Google Scholar]
- 12.Chaudhuri BN, Sawaya MR, Kim CY, Waldo GS, Park MS, Terwilliger TC, Yeates TO. The crystal structure of the first enzyme in the pantothenate biosynthetic pathway, ketopantoate hydroxymethyltransferase, from M tuberculosis. Structure (Camb) 2003;11:753–764. doi: 10.1016/s0969-2126(03)00106-0. [DOI] [PubMed] [Google Scholar]
- 13.Chopra S, Pai H, Ranganathan A. Expression, purification, and biochemical characterization of Mycobacterium tuberculosis aspartate decarboxylase. PanD Protein Expr Purif. 2002;25:533–540. doi: 10.1016/s1046-5928(02)00039-6. [DOI] [PubMed] [Google Scholar]
- 14.Choudhry AE, Mandichak TL, Broskey JP, Egolf RW, Kinsland C, Begley TP, Seefeld MA, Ku TW, Brown JR, Zalacain M, Ratnam K. Inhibitors of pantothenate kinase: novel antibiotics for Staphylococcal infections. Antimicrob Agents Chemother. 2003;47:2051–2055. doi: 10.1128/AAC.47.6.2051-2055.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ciulli A, Chirgadze DY, Smith AG, Blundell TL, Abell C. Crystal structure of Escherichia coli ketopantoate reductase in a ternary complex with NADP+ and pantoate bound: substrate recognition, conformational change, and cooperativity. J Biol Chem. 2007;282:8487–8497. doi: 10.1074/jbc.M611171200. [DOI] [PubMed] [Google Scholar]
- 16.Clifton G, Bryant SR, Skinner CG. N′-(substituted) pantothenamides, antimetabolites of pantothenic acid. Arch Biochem Biophys. 1970;137:523–528. doi: 10.1016/0003-9861(70)90470-4. [DOI] [PubMed] [Google Scholar]
- 17.Cosloy SD, McFall E. Metabolism of D-serine in Escherichia coli K-12: mechanism of growth inhibition. J Bacteriol. 1973;114:685–694. doi: 10.1128/jb.114.2.685-694.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cronan JE., Jr β-alanine synthesis in Escherichia coli. J Bacteriol. 1980;141:1291–1297. doi: 10.1128/jb.141.3.1291-1297.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cronan JE, Jr, Littel KJ, Jackowski S. Genetic and biochemical analyses of pantothenate biosynthesis in Escherichia coli and Salmonella typhimurium. J Bacteriol. 1982;149:916–922. doi: 10.1128/jb.149.3.916-922.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Daugherty M, Polanuyer B, Farrell M, Scholle M, Lykidis A, Crecy-Lagard V, Osterman A. Complete reconstitution of the human coenzyme A biosynthetic pathway via comparative genomics. J Biol Chem. 2002;277:21431–21439. doi: 10.1074/jbc.M201708200. [DOI] [PubMed] [Google Scholar]
- 21.David WE, Lichstein HC. Aspartic acid decarboxylase in bacteria. Proc Exp Biol Med. 1950;73:216–218. [Google Scholar]
- 22.Davis BD. Studies on nutritionally deficient bacterial mutants isolated by means of penicillin. Experimentia. 1950;6:41–50. [Google Scholar]
- 23.De Lay NR, Cronan JE. A genome rearrangement has orphaned the Escherichia coli K-12 AcpT phosphopantetheinyl transferase from its cognate Escherichia coli O157:H7 substrates. Mol Microbiol. 2006;61:232–242. doi: 10.1111/j.1365-2958.2006.05222.x. [DOI] [PubMed] [Google Scholar]
- 24.delCardayre SB, Stock KP, Newton GL, Fahey RC, Davies JE. Coenzyme A disulfide reductase, the primary low molecular weight disulfide reductase from Staphylococcus aureus. Purification and characterization of the native enzyme. J Biol Chem. 1998;273:5744–5751. doi: 10.1074/jbc.273.10.5744. [DOI] [PubMed] [Google Scholar]
- 25.delCardayre SB, Davies JE. Staphylococcus aureus coenzyme A disulfide reductase, a new subfamily of pyridine nucleotide-disulfide oxidoreductase. J Biol Chem. 1998;273:5752–5757. doi: 10.1074/jbc.273.10.5752. [DOI] [PubMed] [Google Scholar]
- 26.Dunn SD, Snell EE. Isolation of temperature-sensitive pantothenate kinase mutants of Salmonella typhimurium and mapping of the coaA gene. J Bacteriol. 1979;140:805–808. doi: 10.1128/jb.140.3.805-808.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Durham NN, Milligan R. Reversal of the D-serine inhibition of growth and division in a Flavobacterium. Biochem Biophys Res Commun. 1961;5:144–147. doi: 10.1016/0006-291x(61)90028-6. [DOI] [PubMed] [Google Scholar]
- 28.Durham NN, Milligan R. Mechanism of growth inhibition by D-serine in a Flavobacterium. Biochem Biophys Res Commun. 1962;7:342–345. doi: 10.1016/0006-291x(62)90311-x. [DOI] [PubMed] [Google Scholar]
- 29.Dusch N, Puhler A, Kalinowski J. Expression of the Corynebacterium glutamicum panD gene encoding L-aspartate-alpha-decarboxylase leads to pantothenate overproduction in Escherichia coli. Appl Environ Microbiol. 1999;65:1530–1539. doi: 10.1128/aem.65.4.1530-1539.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Elischewski F, Puhler A, Kalinowski J. Pantothenate production in Escherichia coli K12 by enhanced expression of the panE gene encoding ketopantoate reductase. J Biotechnol. 1999;75:135–146. doi: 10.1016/s0168-1656(99)00153-4. [DOI] [PubMed] [Google Scholar]
- 31.Elovson J, Vagelos PR. Acyl Carrier Protein X. Acyl carrier protein synthetase. J Biol Chem. 1968;243:3603–3611. [PubMed] [Google Scholar]
- 32.Enos-Berlage JL, Downs DM. Mutations in sdh (succinate dehydrogenase genes) alter the thiamine requirement of Salmonella typhimurium. J Bacteriol. 1997;179:3989–3996. doi: 10.1128/jb.179.12.3989-3996.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Epelbaum S, LaRossa RA, VanDyk TK, Elkayam T, Chipman DM, Barak Z. Branched-chain amino acid biosynthesis in Salmonella typhimurium: a quantitative analysis. J Bacteriol. 1998;180:4056–4067. doi: 10.1128/jb.180.16.4056-4067.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fahey RC. Novel thiols of prokaryotes. Annu Rev Microbiol. 2001;55:333–356. doi: 10.1146/annurev.micro.55.1.333. [DOI] [PubMed] [Google Scholar]
- 35.Fahey RC, Brown WC, Adams WB, Worsham MB. Occurrence of glutathione in bacteria. J Bacteriol. 1978;133:1126–1129. doi: 10.1128/jb.133.3.1126-1129.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Flaks JG, Leboy S, Birge EA, Kurland CG. Mutations and genetics concerned with the ribosome. Cold Spring Harbor Symp. Quant Biol. 1966;31:623–631. doi: 10.1101/sqb.1966.031.01.081. [DOI] [PubMed] [Google Scholar]
- 37.Flamm JA, Friesen JD, Otsuka JA. The nucleotide sequence of the Escherichia coli rts gene. Gene. 1988;74:555–558. doi: 10.1016/0378-1119(88)90189-8. [DOI] [PubMed] [Google Scholar]
- 38.Flugel RS, Hwangbo Y, Lambalot RH, Cronan JE, Jr, Walsh CT. Holo-(Acyl carrier protein) synthase and phosphopantetheinyl transfer in Escherichia coli. J Biol Chem. 2000;275:959–968. doi: 10.1074/jbc.275.2.959. [DOI] [PubMed] [Google Scholar]
- 39.Frodyma M, Rubio A, Downs DM. Reduced flux through the purine biosynthetic pathway results in an increased requirement for coenzyme A in thiamine synthesis in Salmonella enterica serovar typhimurium. J Bacteriol. 2000;182:236–240. doi: 10.1128/jb.182.1.236-240.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frodyma ME, Downs D. ApbA, the ketopantoate reductase enzyme of Salmonella typhimurium is required for the synthesis of thiamine via the alternative pyrimidine biosynthetic pathway. J Biol Chem. 1998;273:5572–5576. doi: 10.1074/jbc.273.10.5572. [DOI] [PubMed] [Google Scholar]
- 41.Frodyma ME, Downs D. The panE gene, encoding ketopantoate reductase, maps at 10 minutes and is allelic to apbA in Salmonella typhimurium. J Bacteriol. 1998;180:4757–4759. doi: 10.1128/jb.180.17.4757-4759.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Geerlof A, Lewendon A, Shaw WV. Purification and characterization of phosphopantetheine adenylyltransferase from Escherichia coli. J Biol Chem. 1999;274:27105–27111. doi: 10.1074/jbc.274.38.27105. [DOI] [PubMed] [Google Scholar]
- 43.Genschel U. Coenzyme a biosynthesis: reconstruction of the pathway in archaea and an evolutionary scenario based on comparative genomics. Mol Biol Evol. 2004;21:1242–1251. doi: 10.1093/molbev/msh119. [DOI] [PubMed] [Google Scholar]
- 44.Gerdes SY, Scholle MD, D’Souza M, Bernal A, Baev MV, Farrell M, Kurnasov OV, Daugherty MD, Mseeh F, Polanuyer BM, Campbell JW, Anantha S, Shatalin KY, Chowdhury SA, Fonstein MY, Osterman AL. From genetic footprinting to antimicrobial drug targets: examples in cofactor biosynthetic pathways. J Bacteriol. 2002;184:4555–4572. doi: 10.1128/JB.184.16.4555-4572.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goulding CW, Apostol M, Anderson DH, Gill HS, Smith CV, Kuo MR, Yang JK, Waldo GS, Suh SW, Chauhan R, Kale A, Bachhawat N, Mande SC, Johnston JM, Lott JS, Baker EN, Arcus VL, Leys D, McLean KJ, Munro AW, Berendzen J, Sharma V, Park MS, Eisenberg D, Sacchettini J, Alber T, Rupp B, Jacobs W, Jr, Terwilliger TC. The TB structural genomics consortium: providing a structural foundation for drug discovery. Curr Drug Targets Infect Disord. 2002;2:121–141. doi: 10.2174/1568005023342551. [DOI] [PubMed] [Google Scholar]
- 46.Grula EA, Grula MM. Inhibition in the synthesis of beta-alanine by D-serine. Biochim Biophys Acta. 1963;74:776–778. doi: 10.1016/0006-3002(63)91430-6. [DOI] [PubMed] [Google Scholar]
- 47.Hoagland MB, Novelli GD. Biosynthesis of coenzyme A from phosphopantetheine and of pantetheine from pantothenate. J Biol Chem. 1954;207:767–773. [PubMed] [Google Scholar]
- 48.Hong BS, Yun MK, Zhang YM, Chohnan S, Rock CO, White SW, Jackowski S, Park HW, Leonardi R. Prokaryotic type II and type III pantothenate kinases: The same monomer fold creates dimers with distinct catalytic properties. Structure. 2006;14:1251–1261. doi: 10.1016/j.str.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 49.Huser AT, Chassagnole C, Lindley ND, Merkamm M, Guyonvarch A, Elisakova V, Patek M, Kalinowski J, Brune I, Puhler A, Tauch A. Rational design of a Corynebacterium glutamicum pantothenate production strain and Its characterization by metabolic flux analysis and genome-wide transcriptional profiling. Appl Environ Microbiol. 2005;71:3255–3268. doi: 10.1128/AEM.71.6.3255-3268.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ivey RA, Zhang YM, Virga KG, Hevener K, Lee RE, Rock CO, Jackowski S, Park HW. The structure of the pantothenate kinase-ADP-pantothenate ternary complex reveals the relationship between the binding sites for substrate, allosteric regulator and antimetabolites. J Biol Chem. 2004;279:35622–35629. doi: 10.1074/jbc.M403152200. [DOI] [PubMed] [Google Scholar]
- 51.Izard T. The crystal structures of phosphopantetheine adenylyltransferase with bound substrates reveal the enzyme’s catalytic mechanism. J Mol Biol. 2002;315:487–495. doi: 10.1006/jmbi.2001.5272. [DOI] [PubMed] [Google Scholar]
- 52.Izard T. A novel adenylate binding site confers phosphopantetheine adenylyltransferase interactions with coenzyme A. J Bacteriol. 2003;185:4074–4080. doi: 10.1128/JB.185.14.4074-4080.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Izard T, Geerlof A. The crystal structure of a novel bacterial adenylyltransferase reveals half of sites reactivity. EMBO J. 1999;18:2021–2030. doi: 10.1093/emboj/18.8.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jackowski S. Biosynthesis of pantothenic acid and coenzyme A. In: Neidhardt FC, Curtiss R, Gross CA, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff W, Riley M, Schaechter M, Umbarger HE, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. American Society for Microbiology; Washington, D.C.: 1996. pp. 687–694. [Google Scholar]
- 55.Jackowski S, Alix JH. Cloning, sequence, and expression of the pantothenate permease (panF) gene of Escherichia coli. J Bacteriol. 1990;172:3842–3848. doi: 10.1128/jb.172.7.3842-3848.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jackowski S, Rock CO. Regulation of coenzyme A biosynthesis. J Bacteriol. 1981;148:926–932. doi: 10.1128/jb.148.3.926-932.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jackowski S, Rock CO. Turnover of the 4′-phosphopantetheine prosthetic group of acyl carrier protein. J Biol Chem. 1984;259:1891–1895. [PubMed] [Google Scholar]
- 58.Jackowski S, Rock CO. Consequences of reduced intracellular coenzyme A content in Escherichia coli. J Bacteriol. 1986;166:866–871. doi: 10.1128/jb.166.3.866-871.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jackowski S, Rock CO. Metabolism of 4′-phosphopantetheine in Escherichia coli. J Bacteriol. 1984;158:115–120. doi: 10.1128/jb.158.1.115-120.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jones CE, Brook JM, Buck D, Abell C, Smith AG. Cloning and sequencing of the Escherichia coli panB gene, which encodes ketopantoate hydroxymethyltransferase, and overexpression of the enzyme. J Bacteriol. 1993;175:2125–2130. doi: 10.1128/jb.175.7.2125-2130.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jung H. The sodium/substrate symporter family: structural and functional features. FEBS Lett. 2002;529:73–77. doi: 10.1016/s0014-5793(02)03184-8. [DOI] [PubMed] [Google Scholar]
- 62.Kang LW, Gabelli SB, Bianchet MA, Xu WL, Bessman MJ, Amzel LM. Structure of a coenzyme A pyrophosphatase from Deinococcus radiodurans: a member of the nudix family. J Bacteriol. 2003;185:4110. doi: 10.1128/JB.185.14.4110-4118.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kleinkauf H. The role of 4′-phosphopantetheine in the biosynthesis of fatty acids, polyketides and peptides. Biofactors. 2000;11:91–92. doi: 10.1002/biof.5520110126. [DOI] [PubMed] [Google Scholar]
- 64.Kupke T. Molecular characterization of the 4′-phosphopantothenoylcysteine decarboxylase domain of bacterial Dfp flavoproteins. J Biol Chem. 2001;276:27597–27604. doi: 10.1074/jbc.M103342200. [DOI] [PubMed] [Google Scholar]
- 65.Kupke T. Molecular characterization of the 4′-phosphopantothenoylcysteine synthetase domain of bacterial dfp flavoproteins. J Biol Chem. 2002;277:36137–36145. doi: 10.1074/jbc.M206188200. [DOI] [PubMed] [Google Scholar]
- 66.Kupke T, Uebele M, Schmid D, Jung G, Blaesse M, Steinbacher S. Molecular characterization of lantibiotic-synthesizing enzyme EpiD reveals a function for bacterial Dfp proteins in coenzyme A biosynthesis. J Biol Chem. 2000;275:31838–31846. doi: 10.1074/jbc.M004273200. [DOI] [PubMed] [Google Scholar]
- 67.Kupke T. Active-site residues and amino acid specificity of the bacterial 4′-phosphopantothenoylcysteine synthetase CoaB. Eur J Biochem. 2004;271:163–172. doi: 10.1046/j.1432-1033.2003.03916.x. [DOI] [PubMed] [Google Scholar]
- 68.Lambalot RH, Gehring AM, Flugel RS, Zuber P, LaCelle M, Marahiel MA, Reid R, Khosla C, Walsh CT. A new enzyme superfamily. Chem Biol. 1996;3:923–936. doi: 10.1016/s1074-5521(96)90181-7. [DOI] [PubMed] [Google Scholar]
- 69.Lambalot RH, Walsh CT. Cloning, overproduction, and characterization of the Escherichia coli holo-acyl carrier protein synthase. J Biol Chem. 1995;270:24658–24661. doi: 10.1074/jbc.270.42.24658. [DOI] [PubMed] [Google Scholar]
- 70.LaRossa RA, Van Dyk TK. Leaky pantothenate and thiamin mutations of Salmonella typhimurium conferring suphometuron methyl sensitivity. J Gen Microbiol. 1989;135(Pt 8):2209–2222. doi: 10.1099/00221287-135-8-2209. [DOI] [PubMed] [Google Scholar]
- 71.Lee BI, Suh SW. Crystal structure of the schiff base intermediate prior to decarboxylation in the catalytic cycle of aspartate alpha-decarboxylase. J Mol Biol. 2004;340:1–7. doi: 10.1016/j.jmb.2004.04.049. [DOI] [PubMed] [Google Scholar]
- 72.Leigh JA. Levels of water-soluble vitamins in methanogenic and non-methanogenic bacteria. Appl Environ Microbiol. 1983;45:800–803. doi: 10.1128/aem.45.3.800-803.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Leonardi R, Chohnan S, Zhang YM, Virga KG, Lee RE, Rock CO, Jackowski S. A pantothenate kinase from Staphylococcus aureus refractory to feedback regulation by coenzyme A. J Biol Chem. 2005;280:3314–3322. doi: 10.1074/jbc.M411608200. [DOI] [PubMed] [Google Scholar]
- 74.Lobley CM, Ciulli A, Whitney HM, Williams G, Smith AG, Abell C, Blundell TL. The crystal structure of Escherichia coli ketopantoate reductase with NADP+ bound. Biochemistry. 2005;44:8930–8939. doi: 10.1021/bi0502036. [DOI] [PubMed] [Google Scholar]
- 75.Lobley CM, Schmitzberger F, Kilkenny ML, Whitney H, Ottenhof HH, Chakauya E, Webb ME, Birch LM, Tuck KL, Abell C, Smith AG, Blundell TL. Structural insights into the evolution of the pantothenate-biosynthesis pathway. Biochem Soc Trans. 2003;31:563–571. doi: 10.1042/bst0310563. [DOI] [PubMed] [Google Scholar]
- 76.Loewen PC. Novel nucleotides from E. coli isolated and partially characterized. Biochem Biophys Res Commun. 1976;70:1210–1218. doi: 10.1016/0006-291x(76)91031-7. [DOI] [PubMed] [Google Scholar]
- 77.Loewen PC. Levels of coenzyme A-glutathione mixed disulfide in Escherichia coli. Can J Biochem. 1978;56:753–759. doi: 10.1139/o78-113. [DOI] [PubMed] [Google Scholar]
- 78.Lowry OH, Carter J, Ward JB, Glaser L. The effect of carbon and nitrogen sources on the level of metabolic intermediates in Escherichia coli. J Biol Chem. 1971;246:6511–6521. [PubMed] [Google Scholar]
- 79.Maas WK. Pantothenate studies III. Description of the extracted pantothenate-synthesizing enzyme of Escherichia coli. J Biol Chem. 1952;198:23–32. [PubMed] [Google Scholar]
- 80.Maas WK, Davis BD. Pantothenate studies. I. Interference by D-serine and L-aspartic acid with pantothenate bioysnthesis in Escherichia coli. J Bacteriol. 1950;60:733–745. doi: 10.1128/jb.60.6.733-745.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Maas WK, Vogel H. alpha-Oxoisovaleric acid, a precursor of pantothenic acid in Escherichia coli. J Bacteriol. 1953;65:388–393. doi: 10.1128/jb.65.4.388-393.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Manoj N, Strauss E, Begley TP, Ealick SE. Structure of human phosphopantothenoylcysteine synthetase at 2.3 Å resolution. Structure. 2003;11:927–936. doi: 10.1016/s0969-2126(03)00146-1. [DOI] [PubMed] [Google Scholar]
- 83.Martin DP, Drueckhammer DG. Separate enzymes catalyze the final two steps of coenzyme A biosynthesis in Brevibacterium ammoniagenes: purification of panthetheine phosphate adenylyltransferase. Biochem Biophys Res Commun. 1993;192:1155–1161. doi: 10.1006/bbrc.1993.1537. [DOI] [PubMed] [Google Scholar]
- 84.Matak-Vinkovic D, Vinkovic M, Saldanha SA, Ashurst JL, von DF, Inoue T, Miguel RN, Smith AG, Blundell TL, Abell C. Crystal structure of Escherichia coli ketopantoate reductase at 1.7 A resolution and insight into the enzyme mechanism. Biochemistry. 2001;40:14493–14500. doi: 10.1021/bi011020w. [DOI] [PubMed] [Google Scholar]
- 85.Merkamm M, Chassagnole C, Lindley ND, Guyonvarch A. Ketopantoate reductase activity is only encoded by ilvC in Corynebacterium glutamicum. J Biotechnol. 2003;104:253–260. doi: 10.1016/s0168-1656(03)00145-7. [DOI] [PubMed] [Google Scholar]
- 86.Merkel WK, Nichols BP. Characterization and sequence of the Escherichia coli panBCD gene cluster. FEMS Microbiol Lett. 1996;143:247–252. doi: 10.1111/j.1574-6968.1996.tb08488.x. [DOI] [PubMed] [Google Scholar]
- 87.Miller SL, Schlesinger G. Prebiotic syntheses of vitamin coenzymes: II. Pantoic acid, pantothenic acid, and the composition of coenzyme A. J Mol Evol. 1993;36:308–314. doi: 10.1007/BF00182178. [DOI] [PubMed] [Google Scholar]
- 88.Mishra P, Park PK, Drueckhammer DG. Identification of yacE (coaE) as the structural gene for dephosphocoenzyme A kinase in Escherichia coli K-12. J Bacteriol. 2001;183:2774–2778. doi: 10.1128/JB.183.9.2774-2778.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Miyatake K, Nakano Y, Kitaoka S. Pantothenate synthetase from Escherichia coli [D-pantoate: β-alanine ligase (AMP-forming), EC 6.3.2.1] Methods Enzymol. 1979;62:215–219. doi: 10.1016/0076-6879(79)62221-8. [DOI] [PubMed] [Google Scholar]
- 90.Mootz HD, Finking R, Marahiel MA. 4′-phosphopantetheine transfer in primary and secondary metabolism of Bacillus subtilis. J Biol Chem. 2001;276:37289–37298. doi: 10.1074/jbc.M103556200. [DOI] [PubMed] [Google Scholar]
- 91.Morris VK, Izard T. Substrate-induced asymmetry and channel closure revealed by the apoenzyme structure of Mycobacterium tuberculosis phosphopantetheine adenylyltransferase. Protein Sci. 2004;13:2547–2552. doi: 10.1110/ps.04816904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nakamura H, Tamura Z. Pantothenate uptake in Escherichia coli K-12. J Nutr Sci Vitaminol. 1973;19:389–400. doi: 10.3177/jnsv.19.389. [DOI] [PubMed] [Google Scholar]
- 93.Newton GL, Arnold K, Price MS, Sherrill C, delCardayre SB, Aharonowitz Y, Cohen G, Davies J, Fahey RC, Davis C. Distribution of thiols in microorganisms: mycothiol is a major thiol in most actinomycetes. J Bacteriol. 1996;178:1990–1995. doi: 10.1128/jb.178.7.1990-1995.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nicely NI, Parsonage D, Paige C, Newton GL, Fahey RC, Leonardi R, Jackowski S, Mallett TC, Claiborne A. Structure of the type III pantothenate kinase from Bacillus anthracis at 2.0 Å resolution: Implications for coenzyme A-dependent redox biology. Biochemistry. 2007;46:3234–3245. doi: 10.1021/bi062299p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Noll KM, Barber TS. Vitamin contents of archaebacteria. J Bacteriol. 1988;170:4315–4321. doi: 10.1128/jb.170.9.4315-4321.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.O’Toole N, Barbosa JA, Li Y, Hung LW, Matte A, Cygler M. Crystal structure of a trimeric form of dephosphocoenzyme A kinase from Escherichia coli. Protein Sci. 2003;12:327–336. doi: 10.1110/ps.0227803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Obmolova G, Teplyakov A, Bonander N, Eisenstein E, Howard AJ, Gilliland GL. Crystal structure of dephospho-coenzyme A kinase from Haemophilus influenzae. J Struct Biol. 2001;136:119–125. doi: 10.1006/jsbi.2001.4428. [DOI] [PubMed] [Google Scholar]
- 98.Osterman A, Overbeek R. Missing genes in metabolic pathways: a comparative genomics approach. Curr Opin Chem Biol. 2003;7:238–251. doi: 10.1016/s1367-5931(03)00027-9. [DOI] [PubMed] [Google Scholar]
- 99.Powers SG, Snell EE. Ketopantoate hydroxymethyltransferase II. Physical, catalytic, and regulatory properties. J Biochem (Tokyo) 1976;251:3786–3793. [PubMed] [Google Scholar]
- 100.Powers SG, Snell EE. Purification and properties of ketopantoate hydroxymethyltransferase. Methods Enzymol. 1979;62:204–209. doi: 10.1016/0076-6879(79)62219-x. [DOI] [PubMed] [Google Scholar]
- 101.Primerano DA, Burns RO. Metabolic basis for the isoleucine, pantothenate or methionine requirement of ilvG strains of Salmonella typhimurium. J Bacteriol. 1982;150:1202–1211. doi: 10.1128/jb.150.3.1202-1211.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Primerano DA, Burns RO. Role of acetohydroxy acid isomeroreductase in biosynthesis of pantothenic acid in Salmonella typhimurium. J Bacteriol. 1983;153:259–269. doi: 10.1128/jb.153.1.259-269.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ramjee MK, Genschel U, Abell C, Smith AG. Escherichia coli L-aspartate-alpha-decarboxylase: preprotein processing and observation of reaction intermediates by electrospray mass spectrometry. Biochem J. 1997;323(Pt 3):661–669. doi: 10.1042/bj3230661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Reizer J, Reizer A, Saier MH., Jr The Na+/pantothenate symporter (PanF) of Escherichia coli is homologous to the Na+/proline symporter (PutP) of E. coli and the Na+/glucose symporters of mammals. Res Microbiol. 1990;141:1069–1072. doi: 10.1016/0923-2508(90)90080-a. [DOI] [PubMed] [Google Scholar]
- 105.Rock CO, Park HW, Jackowski S. Role of feedback regulation of pantothenate kinase (CoaA) in the control of coenzyme A levels in Escherichia coli. J Bacteriol. 2003;185:3410–3415. doi: 10.1128/JB.185.11.3410-3415.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rubio A, Downs DM. Elevated levels of ketopantoate hydroxymethyltransferase (PanB) lead to a physiologically significant coenzyme A elevation in Salmonella enterica serovar Typhimurium. J Bacteriol. 2002;184:2827–2832. doi: 10.1128/JB.184.10.2827-2832.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sahm H, Eggeling L. D-Pantothenate synthesis in Corynebacterium glutamicum and use of panBC and genes encoding L-valine synthesis for D-pantothenate overproduction. Appl Environ Microbiol. 1999;65:1973–1979. doi: 10.1128/aem.65.5.1973-1979.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sambandamurthy VK, Derrick SC, Jalapathy KV, Chen B, Russell RG, Morris SL, Jacobs WR., Jr Long-term protection against tuberculosis following vaccination with a severely attenuated double lysine and pantothenate auxotroph of Mycobacterium tuberculosis. Infect Immun. 2005;73:1196–1203. doi: 10.1128/IAI.73.2.1196-1203.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sambandamurthy VK, Wang X, Chen B, Russell RG, Derrick S, Collins FM, Morris SL, Jacobs WR. A pantothenate auxotroph of Mycobacterium tuberculosis is highly attenuated and protects mice against tuberculosis. Nat Med. 2002;8:1174. doi: 10.1038/nm765. [DOI] [PubMed] [Google Scholar]
- 110.Schmitzberger F, Kilkenny ML, Lobley CM, Webb ME, Vinkovic M, Matak-Vinkovic D, Witty M, Chirgadze DY, Smith AG, Abell C, Blundell TL. Structural constraints on protein self-processing in L-aspartate-alpha-decarboxylase. EMBO J. 2003;22:6193–6204. doi: 10.1093/emboj/cdg575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Schneider F, Kramer R, Burkovski A. Identification and characterization of the main beta-alanine uptake system in Escherichia coli. Appl Microbiol Biotechnol. 2004;65:576–582. doi: 10.1007/s00253-004-1636-0. [DOI] [PubMed] [Google Scholar]
- 112.Setlow B, Setlow P. Levels of acetyl coenzyme A, reduced and oxidized coenzyme A, and coenzyme A in disulfide linkage to protein in dormant and germinated spores and growing and sporulating cells of Bacillus megaterium. J Bacteriol. 1977;132:444–452. doi: 10.1128/jb.132.2.444-452.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Seto A, Murayama K, Toyama M, Ebihara A, Nakagawa N, Kuramitsu S, Shirouzu M, Yokoyama S. ATP-induced structural change of dephosphocoenzyme A kinase from Thermus thermophilus HB8. Proteins. 2005;58:235–242. doi: 10.1002/prot.20276. [DOI] [PubMed] [Google Scholar]
- 114.Shimizu S, Kataoka M, Chung MCM, Yamada H. Ketopantoic acid reductase of Pseudomonas maltophilia 845. J Biol Chem. 1988;263:12077–12084. [PubMed] [Google Scholar]
- 115.Shive W, Macow J. Biochemical transformations as determined by competitive analogue-metabolite growth inhibitions. J Biol Chem. 1946;162:451–462. [PubMed] [Google Scholar]
- 116.Slotnick IJ. Dihydrouracil as a growth factor for a mutant strain of Escherichia coli. J Bacteriol. 1956;72:276–277. doi: 10.1128/jb.72.2.276-277.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Slotnick IJ, Weinfeld H. Dihydrouracil as a growth factor for mutant strains of Escherichia coli. J Bacteriol. 1957;73:122–125. doi: 10.1128/jb.74.2.122-125.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Song WJ, Jackowski S. Cloning, sequencing, and expression of the pantothenate kinase (coaA) gene of Escherichia coli. J Bacteriol. 1992;174:6411–6417. doi: 10.1128/jb.174.20.6411-6417.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Song WJ, Jackowski S. Kinetics and regulation of pantothenate kinase from Escherichia coli. J Biol Chem. 1994;269:27051–27058. [PubMed] [Google Scholar]
- 120.Song WJ, Jackowski S. coaA and rts are allelic and located at kilobase 3532 on the Escherichia coli physical map. J Bacteriol. 1992;174:1705–1706. doi: 10.1128/jb.174.5.1705-1706.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Spitzer ED, Jimenez-Billini HE, Weiss B. beta-Alanine auxotrophy associated with dfp, a locus affecting DNA synthesis in Escherichia coli. J Bacteriol. 1988;170:872–876. doi: 10.1128/jb.170.2.872-876.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Spitzer ED, Weiss B. dfp Gene of Escherichia coli K-12, a locus affecting DNA synthesis, codes for a flavoprotein. J Bacteriol. 1985;164:994–1003. doi: 10.1128/jb.164.3.994-1003.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Stanitzek S, Augustin MA, Huber R, Kupke T, Steinbacher S. Structural basis of CTP-dependent peptide bond formation in coenzyme A biosynthesis catalyzed by Escherichia coli PPC synthetase. Structure (Camb) 2004;12:1977–1988. doi: 10.1016/j.str.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 124.Strauss E, Begley TP. Mechanistic studies on phosphopantothenoylcysteine decarboxylase. J Am Chem Soc. 2001;123:6449–6450. doi: 10.1021/ja016020y. [DOI] [PubMed] [Google Scholar]
- 125.Strauss E, Begley TP. The antibiotic activity of N-pentylpantothenamide results from its conversion to ethyldethia-coenzyme A, a coenzyme A antimetabolite. J Biol Chem. 2002;277:48205–48209. doi: 10.1074/jbc.M204560200. [DOI] [PubMed] [Google Scholar]
- 126.Strauss E, Begley TP. Stereochemical studies on phosphopantothenoylcysteine decarboxylase from Escherichia coli. Bioorg Med Chem Lett. 2003;13:339–342. doi: 10.1016/s0960-894x(02)01018-1. [DOI] [PubMed] [Google Scholar]
- 127.Strauss E, Kinsland C, Ge Y, McLafferty FW, Begley TP. Phosphopantothenoylcysteine synthetase from Escherichia coli. Identification and characterization of the last unidentified coenzyme A biosynthetic enzyme in bacteria. J Biol Chem. 2001;276:13513–13516. doi: 10.1074/jbc.C100033200. [DOI] [PubMed] [Google Scholar]
- 128.Strauss E, Zhai H, Brand LA, McLafferty FW, Begley TP. Mechanistic studies on phosphopantothenoylcysteine decarboxylase: trapping of an enethiolate intermediate with a mechanism-based inactivating agent. Biochemistry. 2004;43:15520–15533. doi: 10.1021/bi048340a. [DOI] [PubMed] [Google Scholar]
- 129.Sugantino M, Zheng R, Yu M, Blanchard JS. Mycobacterium tuberculosis ketopantoate hydroxymethyltransferase: tetrahydrofolate-independent hydroxymethyltransferase and enolization reactions with alpha-keto acids. Biochemistry. 2003;42:191–199. doi: 10.1021/bi020516q. [DOI] [PubMed] [Google Scholar]
- 130.Swerdlow RD, Green CL, Setlow B, Setlow P. Identification of an NADH-linked disulfide reductase from Bacillus megaterium specific for disulfides containing pantethine 4′,4″-diphosphate moieties. J Biol Chem. 1979;254:6835–6837. [PubMed] [Google Scholar]
- 131.Swerdlow RD, Setlow P. Purification and characterization of a Bacillus megaterium disulfide reductase specific for disulfides containing pantethine 4′,4″-diphosphate. J Bacteriol. 1983;153:475–484. doi: 10.1128/jb.153.1.475-484.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Tahiliani AG, Beinlich CJ. Pantothenic acid in health and disease. Vitam Horm. 1991;46:165–228. doi: 10.1016/s0083-6729(08)60684-6. [DOI] [PubMed] [Google Scholar]
- 133.Takahashi H, Inagaki E, Fujimoto Y, Kuroishi C, Nodake Y, Nakamura Y, Arisaka F, Yutani K, Kuramitsu S, Yokoyama S, Yamamoto M, Miyano M, Tahirov TH. Structure and implications for the thermal stability of phosphopantetheine adenylyltransferase from Thermus thermophilus. Acta Crystallogr D Biol Crystallogr. 2004;60:97–104. doi: 10.1107/s0907444903025319. [DOI] [PubMed] [Google Scholar]
- 134.Teller JH, Powers SG, Snell EE. Ketopantoate hydroxymethyltransferase I. Purification and role in pantothenate biosynthesis. J Biol Chem. 1976;251:3780–3785. [PubMed] [Google Scholar]
- 135.Thomas J, Cronan JE., Jr The enigmatic acyl carrier protein phosphodiesterase of Escherichia coli: Genetic and enzymological characterization. J Biol Chem. 2005;280:34675–34683. doi: 10.1074/jbc.M505736200. [DOI] [PubMed] [Google Scholar]
- 136.Thomas J, Rigden DJ, Cronan JE. Acyl carrier protein phosphodiesterase (AcpH) of Escherichia coli is a non-canonical member of the HD phosphatase/phosphodiesterase family. Biochemistry. 2007;46:129–136. doi: 10.1021/bi061789e. [DOI] [PubMed] [Google Scholar]
- 137.Tuck KL, Saldanha SA, Birch LM, Smith AG, Abell C. The design and synthesis of inhibitors of pantothenate synthetase. Org Biomol Chem. 2006;4:3598–3610. doi: 10.1039/b609482a. [DOI] [PubMed] [Google Scholar]
- 138.Vadali RV, Bennett GN, San KY. Cofactor engineering of intracellular CoA/acetyl-CoA and its effect on metabolic flux redistribution in Escherichia coli. Metab Eng. 2004;6:133–139. doi: 10.1016/j.ymben.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 139.Vadali RV, Bennett GN, San KY. Enhanced isoamyl acetate production upon manipulation of the acetyl-CoA node in Escherichia coli. Biotechnol Prog. 2004;20:692–697. doi: 10.1021/bp034326y. [DOI] [PubMed] [Google Scholar]
- 140.Vagelos PR, Larabee AR. Acyl carrier protein. IX. Acyl carrier protein hydrolase. J Biol Chem. 1967;242:1776–1781. [PubMed] [Google Scholar]
- 141.Vallari DS, Jackowski S. Biosynthesis and degradation both contribute to the regulation of coenzyme A content in Escherichia coli. J Bacteriol. 1988;170:3961–3966. doi: 10.1128/jb.170.9.3961-3966.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Vallari DS, Jackowski S, Rock CO. Regulation of pantothenate kinase by coenzyme A and its thioesters. J Biol Chem. 1987;262:2468–2471. [PubMed] [Google Scholar]
- 143.Vallari DS, Rock CO. Isolation and characterization of temperature-sensitive pantothenate kinase (coaA) mutants of Escherichia coli. J Bacteriol. 1987;169:5795–5800. doi: 10.1128/jb.169.12.5795-5800.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Vallari DS, Rock CO. Isolation and characterization of Escherichia coli pantothenate permease (panF) mutants. J Bacteriol. 1985;164:136–142. doi: 10.1128/jb.164.1.136-142.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Vallari DS, Rock CO. Pantothenate transport in Escherichia coli. J Bacteriol. 1985;162:1156–1161. doi: 10.1128/jb.162.3.1156-1161.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Vanet A, Plumbridge JA, Alix JH. Cotranscription of two genes necessary for ribosomal protein L11 methylation (prmA) and pantothenate transport (panF) in Escherichia coli K-12. J Bacteriol. 1993;175:7178–7188. doi: 10.1128/jb.175.22.7178-7188.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Virga KG, Zhang YM, Leonardi R, Ivey RA, Hevener K, Park HW, Jackowski S, Rock CO, Lee RE. Structure-activity relationships and enzyme inhibition of pantothenamide-type pantothenate kinase inhibitors. Bioorg Med Chem. 2006;14:1007–1020. doi: 10.1016/j.bmc.2005.09.021. [DOI] [PubMed] [Google Scholar]
- 148.Virtanen AI, Laine T. The decarboxylation of d-lysine and l-aspartic acid. Enzymologia. 1937;8:266–270. [Google Scholar]
- 149.von DF, Inoue T, Saldanha SA, Ottenhof HH, Schmitzberger F, Birch LM, Dhanaraj V, Witty M, Smith AG, Blundell TL, Abell C. Structure of E. coli ketopantoate hydroxymethyl transferase complexed with ketopantoate and Mg2+, solved by locating 160 selenomethionine sites. Structure (Camb) 2003;11:985–996. doi: 10.1016/s0969-2126(03)00158-8. [DOI] [PubMed] [Google Scholar]
- 150.Wang S, Eisenberg D. Crystal structure of the pantothenate synthetase from Mycobacterium tuberculosis, snapshots of the enzyme in action. Biochemistry. 2006;45:1554–1561. doi: 10.1021/bi051873e. [DOI] [PubMed] [Google Scholar]
- 151.Wang S, Eisenberg D. Crystal structures of a pantothenate synthetase from M. tuberculosis and its complexes with substrates and a reaction intermediate. Protein Sci. 2003;12:1097–1108. doi: 10.1110/ps.0241803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Webb ME, Smith AG, Abell C. Biosynthesis of pantothenate. Nat Prod Rep. 2004;21:695–721. doi: 10.1039/b316419p. [DOI] [PubMed] [Google Scholar]
- 153.Whalen WA, Berg CM. Analysis of an avtA::Mu d1(Ap lac) mutant: Metabolic role of transaminase C. J Bacteriol. 1982;150:739–746. doi: 10.1128/jb.150.2.739-746.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.White EL, Southworth K, Ross L, Cooley S, Gill RB, Sosa MI, Manouvakhova A, Rasmussen L, Goulding C, Eisenberg D, Fletcher TM., III A Novel Inhibitor of Mycobacterium tuberculosis Pantothenate Synthetase. J Biomol Screen. 2007;12:100–105. doi: 10.1177/1087057106296484. [DOI] [PubMed] [Google Scholar]
- 155.Williams L, Zheng R, Blanchard JS, Raushel FM. Positional isotope exchange analysis of the pantothenate synthetase reaction. Biochemistry. 2003;42:5108–5113. doi: 10.1021/bi0340853. [DOI] [PubMed] [Google Scholar]
- 156.Williamson JM. L-Aspartate α-decarboxylase. Methods Enzymol. 1985;113:589–595. doi: 10.1016/s0076-6879(85)13078-8. [DOI] [PubMed] [Google Scholar]
- 157.Williamson JM, Brown GM. Purification and properties of L-aspartate-α-decarboxylase, an enzyme that catalyzes the formation of β-alanine in Escherichia coli. J Biol Chem. 1979;254:8074–8082. [PubMed] [Google Scholar]
- 158.Worrall DM, Tubbs PK. A bifunctional enzyme complex in coenzyme A biosynthesis: purification of 4′ pantetheine phosphate adenylyltransferase and dephospho-CoA kinase. J Biochem (Tokyo) 1983;215:153–157. doi: 10.1042/bj2150153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Xu W, Shen J, Dunn CA, Desai S, Bessman MJ. The Nudix hydrolases of Deinococcus radiodurans. Mol Microbiol. 2001;39:286–290. doi: 10.1046/j.1365-2958.2001.02267.x. [DOI] [PubMed] [Google Scholar]
- 160.Yang H, Abeles RH. Purification and properties of Escherichia coli 4′-phosphopantothenoylcysteine decarboxylase: presence of covalently bound pyruvate. Biochemistry. 1987;26:4076–4081. doi: 10.1021/bi00387a050. [DOI] [PubMed] [Google Scholar]
- 161.Yocum RR, Patterson TA. 6,830,898 U S Patent. 2004
- 162.Yun M, Park CG, Kim JY, Rock CO, Jackowski S, Park HW. Structural basis for the feedback regulation of Eschericia coli pantothenate kinase by coenzyme A. J Biol Chem. 2000;275:28093–28099. doi: 10.1074/jbc.M003190200. [DOI] [PubMed] [Google Scholar]
- 163.Zhang YM, Frank MW, Virga KG, Lee RE, Rock CO, Jackowski S. Acyl carrier protein is a cellular target for the antibacterial action of the pantothenamide class of pantothenate antimetabolites. J Biol Chem. 2004;279:50969–50975. doi: 10.1074/jbc.M409607200. [DOI] [PubMed] [Google Scholar]
- 164.Zhao L, Allanson NM, Thomson SP, Maclean JK, Barker JJ, Primrose WU, Tyler PD, Lewendon A. Inhibitors of phosphopantetheine adenylyltransferase. Eur J Med Chem. 2003;38:345–349. doi: 10.1016/s0223-5234(03)00047-3. [DOI] [PubMed] [Google Scholar]
- 165.Zheng R, Blanchard JS. Steady-state and pre-steady-state kinetic analysis of Mycobacterium tuberculosis pantothenate synthetase. Biochemistry. 2001;40:12904–12912. doi: 10.1021/bi011522+. [DOI] [PubMed] [Google Scholar]
- 166.Zheng R, Blanchard JS. Identification of active site residues in E. coli ketopantoate reductase by mutagenesis and chemical rescue. Biochemistry. 2000;39:16244–16251. doi: 10.1021/bi002134v. [DOI] [PubMed] [Google Scholar]
- 167.Zheng R, Blanchard JS. Kinetic and mechanistic analysis of the E. coli panE-encoded ketopantoate reductase. Biochemistry. 2000;39:3708–3717. doi: 10.1021/bi992676g. [DOI] [PubMed] [Google Scholar]
- 168.Zheng R, Dam TK, Brewer CF, Blanchard JS. Active site residues in Mycobacterium tuberculosis pantothenate synthetase required in the formation and stabilization of the adenylate intermediate. Biochemistry. 2004;43:7171–7178. doi: 10.1021/bi049676n. [DOI] [PubMed] [Google Scholar]
- 169.Zhyvoloup A, Nemazanyy I, Babych O, Panasyuk G, Pobigailo N, Vudmaska M, Naidenov V, Kukharenko O, Palchevskii S, Savinska L, Ovcharenko G, Verdier F, Valovka T, Fenton T, Rebholz H, Wang ML, Shepherd P, Matsuka G, Filonenko V, Gout IT. Molecular cloning of CoA synthase: the missing link in CoA biosynthesis. J Biol Chem. 2002;277:22107–22110. doi: 10.1074/jbc.C200195200. [DOI] [PubMed] [Google Scholar]


