ABSTRACT
Temperate bacteriophages possess a genetic switch which regulates the lytic and lysogenic cycle. The genomes of the enterobacterial telomere phages N15, PY54 and ϕKO2 harbor a primary immunity region (immB) comprising genes for the prophage repressor, the lytic repressor and a putative antiterminator, similar to CI, Cro and Q of lambda, respectively. Moreover, N15 and ϕKO2 contain 3 related operator (OR) sites between cI and cro, while only one site (OR3) has been detected in PY54. Marine telomere phages possess a putative cI gene but not a cro-like gene. Instead, a gene is located at the position of cro, whose product shows some similarity to the PY54 ORF42 product, the function of which is unknown. We have determined the transcription start sites of the predicted repressor genes of N15, PY54, ϕKO2 and of the marine telomere phage VP58.5. The influence of the genes on phage propagation was analyzed in E. coli, Y. enterocolitica and V.parahaemolyticus. We show that the repressors and antiterminators of N15, ϕKO2 and PY54 exerted their predicted activities. However, while the proteins of both N15 and ϕKO2 affected lysis and lysogeny by N15, they did not affect PY54 propagation. On the other hand, the respective PY54 proteins exclusively influenced the propagation of this phage. The immB region of VP58.5 contains 2 genes that revealed prophage repressor activity, while a lytic repressor gene could not be identified. The results indicate an unexpected diversity of the growth regulation mechanisms in these temperate phages.
KEYWORDS: genetic switch, regulation, repressor, temperate, telomere phage
Introduction
Temperate phages can choose between a lytic and lysogenic pathway of development. During the lysogenic cycle most temperate phages are integrated into the bacterial chromosome. The temperate phages N15, PY54 and ϕKO2 isolated from E. coli, Yersinia enterocolitica and Klebsiella oxytoca, respectively, belong to a particular subgroup of lambdoid phages because at the lysogenic stage, their prophages replicate as linear plasmids with covalently closed hairpin ends (telomeres).1-5 The genomes of these phages can be divided into 2 arms separated by the telomere resolution site, which is essential for the conversion of the linear phage genome into the linear plasmid prophage. Sequence analyses disclosed that the left arm of the PY54 genome, which mainly contains virion structural genes is more closely related to ϕKO2 than to N15, whereas N15 and ϕKO2 show the strongest homologies in the right arm harboring genes involved in e.g., the generation and replication of the linear plasmid, phage immunity and host cell lysis. The plasmid prophages of N15 and ϕKO2 belong to the same incompatibility group while the PY54 prophage belongs to a separate group and thus is compatible with the 2 other plasmids.6
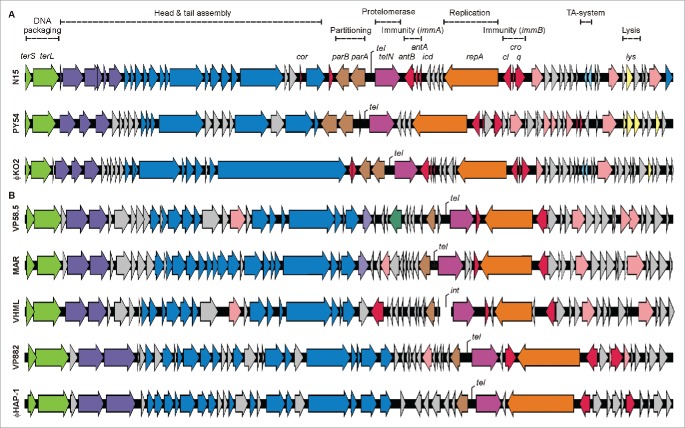
Telomere phages have also been found in marine bacteria. While the phages VP58.5, VP882 and vB_VpaM_MAR were isolated from V. parahaemolyticus strains,7-9 phage ϕHAP-1 was recovered from Halomonas aquamarina.10 VP58.5 and vB_VpaM_MAR are closely related phages,7,10 the same holds true for VP882 and ϕHAP-1.8,10 The marine telomere phages are also related to the V. harveyi (later V. campbellii) phage VHML but it is not clear whether this phage still exists.11-15 Particularly notable is the fact that unlike the siphoviridal phages N15, PY54 and ϕKO2, marine telomere phages are members of the family Myoviridae. However, even though there are only partial protein similarities between enterobacterial and marine telomere phages, all these phages share a similar genome organization (Fig. 1).1,3,4,8-10,13
Figure 1.
Genetic map of the telomere phages. Genome organization of the enterobacterial and marine telomere phages. ORFs depicted in gray encode hypothetical proteins whose functions are unknown.
Similar to other temperate phages, telomere phages possess a genetic switch between the lytic and the lysogenic cycle.16 Sequence analyses suggested that all telomere phages possess a primary immunity region (immB) which is comparable to the immunity region of lambda-like phages but exhibits a simpler arrangement. In the enterobacterial telomere phages, immB encodes products related to the prophage repressor CI, lytic repressor Cro and a putative antiterminator Q as well as operator sites located between cI and cro. Due to the close relationship between N15 and ϕKO2, the same repressor target specificity for these phages has been suggested.1 However, experimental data on this issue are still missing even though the function of the N15 and PY54 prophage repressors has already been demonstrated.3,6 Lytic repressor activity of the telomere phages has been demonstrated only for Cro of PY54.3 This protein revealed a high binding specificity for a single operator site (OR3) on the PY54 genome and did not bind to closely related N15 and ϕKO2 operator sites.17 Thus, PY54 Cro apparently suppresses cI transcription but not its own synthesis. Finally there are yet no data available about the repressor activities of the marine telomere phages.
In this work we studied the activity of the probable repressors and antiterminators of N15, PY54, ϕKO2 and VP58.5 by in vivo assays in E. coli, Yersinia, and Vibrio. For this purpose the correct start codons of the respective genes have been determined by RACE analyses. We show that the N15 and ϕKO2 regulatory proteins have the same specificities which diverge from those of PY54. The genetic switch of VP58.5 seems to be generally different as only genes mediating prophage repressor activity have been identified.
Results
In silico analysis of the immB regions
Sequence analyses of the hitherto described telomere phages disclosed a region (immB) on the genomes which probably harbors the genetic switch for the lysogenic and lytic cycle (Fig. 1).3,17 In the enterobacterial telomere phages N15, PY54 and ϕKO2, immB comprises genes potentially encoding the prophage repressor CI, Cro repressor and transcription antiterminator Q. In some other studies CI of N15 and ϕKO2 has been named CB but throughout this work it is termed CI. The CI and Cro repressors and the antiterminator Q of N15 and ϕKO2 are closely related, whereas the corresponding proteins of PY54 are more distantly related to the N15 and ϕKO2 repressors and Q, (Table 1).
Table 1.
Similarities of the immB gene products of N15, ϕKO2, PY54 and VP58.5.
| N15 | ϕKO2 | PY54 | VP58.5 | |
|---|---|---|---|---|
| N15 (CI) | 100% | 89% | 34% | – |
| N15 (Cro) | 100% | 88% | 31% | – |
| N15 (Q) | 100% | 71% | 31% | – |
| ϕKO2 (CI) | 89% | 100% | 34% | – |
| ϕKO2 (Cro) | 88% | 100% | 35% | – |
| ϕKO2 (Q) | 71% | 100% | 29% | – |
| PY54 (CI) | 34% | 34% | 100% | – |
| PY54 (Cro) | 31% | 35% | 100% | – |
| PY54 (ORF42) | – | – | 100% | 32% |
| PY54 (Q) | 31% | 29% | 100% | – |
| VP58.5 (CI) | – | – | – | 100% |
| VP58.5 (ORF44) | – | – | 32% | 100% |
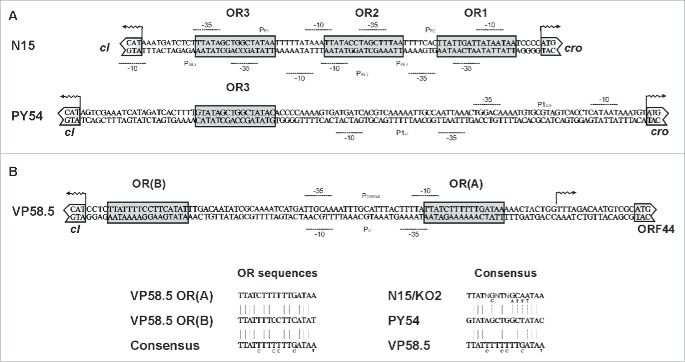
The genes cro and q of the enterobacterial telomere phages are arranged in one operon (Fig. 2A & B). In the PY54 operon cro and q are separated by an additional open reading frame (ORF42) whose function is unknown (Fig. 2B). Similar to the situation in lambda, the genes cI and cro are arranged in the opposite direction and separated by intergenic regions approximately 80 to 120bp in size. In N15 the intergenic region between cI and cro contains 3 operator sites (OR), which are related in sequence and share bases that are strictly conserved (Fig. 3A).16 The operators overlap with putative promoters of the repressor genes. In ϕKO2 3 very similar operator sites exist1 while PY54 exhibits only 1 site (OR3) that fits into this scheme.17
Figure 2.
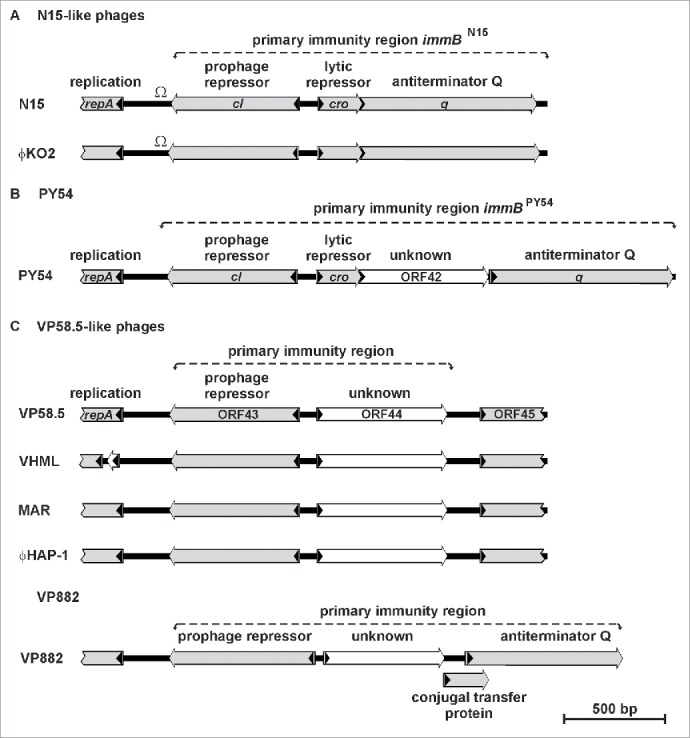
Organization of the primary immunity region (immB) of the telomere phages and of VHML. (A) immB of N15 and ϕKO2. (B) immB of PY54. (C) immB of the marine telomere phages VP58.5, vB_VpaM_MAR, VP882, ϕHAP-1 and of the related phage VHML.
Figure 3.
Sequence alignment of the intergenic regions of N15, PY54 and VP58.5 between the repressor genes cI and cro (ORF44). (A) Operator sequences of N15 determined by Lobocka et al.16 and similar sequences in the corresponding regions of PY54 and Vp58.5 are presented in light shaded boxes. Predicted promoters are indicated. (B) Consensus sequences of the operator sites. Nucleotides within the putative PY54 and VP58.5 operator sequences identical to those of the corresponding N15/ϕKO2 sites are marked by vertical lines.
The immB regions of the marine telomere phages also contain a putative prophage repressor gene but the product of this gene is not related to CI of the enterobacterial telomere phages (Table 1). Moreover, a cro-related gene could not be identified in marine telomere phages (Fig. 2C) while a possible q gene has only been detected in VP882.8 On the other hand, all marine telomere phages contain an ORF at the position of cro (e.g. ORF44 of VP58.5, Fig. 2C) whose product shows some relationship to the PY54 ORF42 product (Table 1). Differences between the immB regions of enterobacterial and marine telomere phages are also discernible in the intergenic region. The intergenic region of e.g., VP58.5 does not contain sequences with strong homologies to the operators of the enterobacterial telomere phages. However, 2 sites exhibiting some similarity to the aforementioned operators have been identified in VP58.5 (Fig. 3B). Since several possible start codons exist for the predicted repressor genes, the exact positions of the intergenic regions have not been defined in most telomere phages. Therefore, we first determined transcription start sites of the predicted repressor genes of N15, ϕKO2, PY54 and VP58.5 because these repressors should be studied under in vivo conditions.
Determination of the repressor genes' transcription start sites
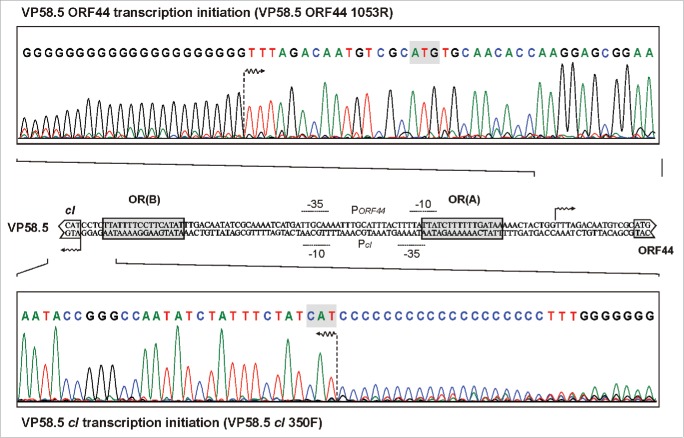
The transcription start sites of the mRNAs encoding CI and Cro (N15, ϕKO2, PY54)17 and CI and gp44 (VP58.5) were determined by sequencing of RACE products. The experiment was carried out using DNA fragments, which encompassed the intergenic region and partial sequences of the repressor genes. For the CI and Cro repressors of N15, ϕKO2 and PY5417 and for CI of VP58.5 only 1 species of a leaderless mRNA was identified beginning with the AUG start codon of cI or cro (Fig. 4). The data on the N15 repressors are in good agreement with the cI and cro start codons determined by in silico analysis.16 Unlike with the aforementioned repressor genes, the VP58.5 ORF44 mRNA revealed a short leader sequence (5´-TTTAGACA-3´) in front of the start codon. According to the obtained data, the intergenic region of N15, ϕKO2, PY54 and VP58.5 has a length of 81, 80, 104 and 106bp, respectively (Fig. 3).
Figure 4.
Determination of the transcriptional start sites of the VP58.5 immB genes. The upper and lower panel show sequences of the RACE-PCR products with poly(C) tail. In the middle the intergenic region between the cI and ORF44 start codon is shown. Putative operators and promoters and the transcriptional start sites of the genes are indicated.
Specificity of the N15/ϕKO2 and PY54 repressors and antiterminators
To analyze the activity of the repressor proteins and antiterminators in E. coli and Yersinia, their promoterless genes were amplified by PCR and inserted into the expression vector pMS470Δ8Cat (Apr, Cmr) containing an inducible tac promoter.6 The resulting constructs and the vector without insert as control were introduced into E. coli C-1a and Y.enterocolitica 83/88/2 by electroporation. We could not perform experiments with K.oxytoca because for ϕKO2 a suitable indicator strain has not been identified.1,6 Escherichia coli transformants were infected by a N15 derivative in which a chloramphenicol resistance gene had been introduced while the corresponding Yersinia strains were infected by PY54 carrying a tetracycline resistance gene (see Materials and Methods).6 Escherichia coli strains containing the vector pMS470Δ8Cat were both lysed and lysogenized by N15. Strains which harboured the cI prophage repressor gene of N15 or ϕKO2 showed, compared to the control, an approximately 2 orders of magnitude enhanced lysogenization frequency while the lytic activity was slightly reduced (Table 2). On the other hand, cro and q of N15 and ϕKO2 mediated strong lysis whereas we could not detect any lysogenization of the bacteria. The q gene of both phages also complemented a cro mutation in N15 suggesting that it plays a similar role like in lambda.18 Unlike the immB genes of N15 and ϕKO2 none of the corresponding PY54 and VP58.5 genes influenced the lifestyle of N15. On the contrary, PY54 cI and cro/q boosted the lysogenic and lytic cycle, respectively, of PY54 while there was no visible effect with ORF42 and with the immB genes of the other phages (Table 3). We did not obtain divergent results under induced and non-induced conditions except for the finding that the N15, ϕKO2 and PY54 prophage repressors exerted a lytic activity after induction with IPTG. It can be surmised that at very high concentrations the CI proteins repress their own synthesis by binding to OR3.
Table 2.
Influence of the N15 and ϕKO2 immB genes on the propagation of N15.
| N15 |
||||
|---|---|---|---|---|
| Plasmid | Insert | Phage origin | Lysogenization | Lysis |
| pJH099-2 | None | None | 4.11 × 105(±2.02 × 105) | 3.39 × 108(±2.87 × 108) |
| pJH341-2 | cI | N15 | 2.44 × 107(±1.97 × 105) | 1.19 × 108(±1.11 × 108) |
| pJH342-2 | cro | N15 | – | 7.70 × 108(±2.89 × 108) |
| pJH344-2 | Q | N15 | – | 8.62 × 108(±3.11 × 108) |
| pJH141-2 | cI | ϕKO2 | 4.35 × 107(±2.03 × 103) | 1.98 × 108(±0.88 × 108) |
| pJH142-2 | cro | ϕKO2 | – | 4.76 × 108(±3.23 × 108) |
| pJH544-2 | Q | ϕKO2 | – | 5.37 × 108(±2.34 × 108) |
In vivo assays were performed with N15 Cmr phage mutant N15-D04.6 The given phage titers are mean values of 3 independent experiments. Standard deviations from the mean value are given in brackets.
Table 3.
Influence of the PY54 immB genes on the propagation of the phage.
| PY54 |
||||
|---|---|---|---|---|
| Plasmid | Insert | Phage origin | Lysogenization (SD) | Lysis (SD) |
| pJH099-2 | none | None | 6.21 × 105 (±2.55 × 105) | 4.85 × 108(±1.83 × 108) |
| pJH141-2 | cI | PY54 | 3.43 × 107(±0.96 × 107) | 1.12 × 108(±0.52 × 108) |
| pJH142-2 | cro | PY54 | – | 6.99 × 108(±1.12 × 108) |
| pJH149-2 | ORF42 | PY54 | 7.64 × 105(±2.04 × 105) | 5.73 × 108(±2.38 × 108) |
| pJH144-2 | q | PY54 | – | 8.81 × 108(±1.46 × 108) |
In vivo assays were performed with the PY54 Tcr phage mutant PY54-M1/283.
The VP58.5 immB region harbors 2 genes with prophage repressor activity
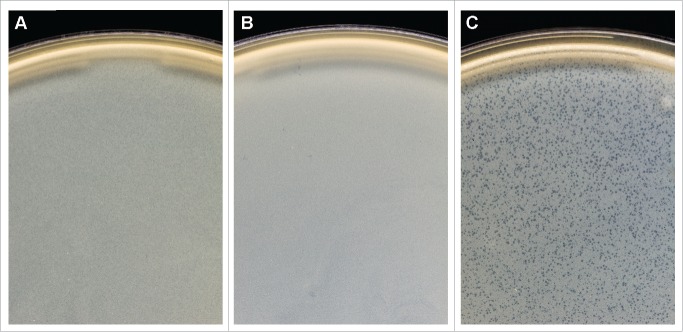
To study the activity of the regulatory proteins in Vibrio, the immB genes were inserted into the shuttle vector pVv3 containing the lac promoter19 and introduced into V. parahaemolyticus strain 37.5 by electroporation. Thereafter, transformants were infected with phage VP58.5. As expected the enterobacterial immB genes did not have any influence on VP58.5 propagation. However, surprisingly both cI and ORF44 of VP58.5 blocked the lytic activity of the phage. To verify that the ORF44 product has a prophage repressor-like activity, an ORF44-negative phage mutant was generated by inserting a chloramphenicol acetyltransferase gene (cat) into ORF44 (see Materials and Methods). This mutant produced significantly clearer plaques than the wild type phage (Fig. 5). By PCR and sequencing it was confirmed that the mutant contained the cat gene within ORF44, while cI of the mutant remained intact (data not shown). The mutant was also used to infect strains containing the intact ORF44 or cI. Both genes complemented the mutation of the phage. The data demonstrates that the immB region of Vp58.5 contains 2 genes exerting prophage repressor activity but no gene for a lytic repressor indicating that the genetic switch of this phage diverges fundamentally from those of the enterobacterial telomere phages.
Figure 5.
VP58.5 ORF44 confers prophage repressor activity. (A) Plaques of the VP58.5 wild type phage on V. parahaemolyticus strain 37.5. (B) Strain 37.5 containing VP58.5 ORF44 is not lysed by VP58.5. (C) A VP58.5 mutant possessing a defective ORF44 produces clear plaques on strain 37.5.
Discussion
Telomere phages are a particular group of temperate phages whose prophages are not integrated into the bacterial chromosome, but replicate as linear plasmids with terminal hairpins (telomeres). Thus far, 7 telomere phages isolated from Enterobacteriaceae and marine bacteria have been described.1,3,5,8-10,14 Vibrio phage VHML has not been reported to be a linear plasmid prophage13,14 but its close relationship to VP58.5 and vB_VpaM_MAR suggests that it also belongs to this group.7,9 In spite of the fact that the enterobacterial and marine telomere phages have different morphologies, their genome compositions reveal striking similarities. This particularly pertains to the content and order of specific genes (Fig. 1). The arrangement of genes for structural proteins, partitioning proteins, the protelomerase including its target site tel, the replication protein repA and lysis proteins is so similar that it appears likely that these phages share a common ancestor.3,20 At first glance this also applies to the primary immunity region immB of the telomere phages (Fig. 2), which resembles the genetic switch of many lambdoid phages. Binding of the N15 prophage repressor to 3 related operators located between cI and cro/q and overlapping with predicted promoters of the repressors genes indeed indicated the same principle of regulation.16 Moreover, due to the close relationship of N15 to ϕKO2 the same specificity has been proposed for the repressors of these phages.1 This assumption was confirmed in our study. The genes for the prophage repressor, lytic repressor and Q of ϕKO2 influenced lysis and lysogeny of N15 in the same way as their counterparts in N15. By contrast, none of the investigated PY54 genes affected the N15 life cycles but exerted specific activities in Yersinia strains infected with PY54. The only exemption was ORF42, which did not influence the lytic or lysogenic properties of the phage.
From the data it can be concluded that the repressors and antiterminators of N15 and ϕKO2 are too different from the corresponding PY54 proteins to share the same binding specificity. Moreover, the PY54 genome contains only one operator site (OR3) between cI and cro that is related to the N15/ϕKO2 operators. Previous studies demonstrated that in PY54 binding of the Cro repressor is restricted to this site upstream of cI.17 Other sites on the phage genome as well as N15/ϕKO2 operators were not bound by PY54 Cro. Thus, the PY54 Cro repressor is very specific in terms of recognition and binding to its targets. Preliminary data also revealed that the PY54 prophage repressor CI binds to 2 regions within the intergenic region of this phage that only share marginal sequence similarity.17 This finding confirms that the PY54 genetic switch diverges significantly from those of N15 and ϕKO2. Our studies on the predicted VP58.5 repressors showed that the switch of this phage is apparently even more diverse since it contains 2 genes exerting prophage repressor activity while a gene encoding a lytic repressor could not be detected. In addition, the intergenic region between cI and ORF44 of VP58.5 does not contain putative operator sites easily detectable from the DNA sequence. A 16bp imperfect palindrome (5´-TTATCTTTTTTGATAA-3´) immediately upstream of ORF44 exhibits some similarity to the N15 operator OR3 (5´-TTATA˜N6˜TATAA-3´) but is yet not clear whether this sequence is a binding site for CI or the ORF44 product. Nevertheless, due to the same mode of action of these proteins, the question arises how the lytic cycle of VP58.5 is induced. The same holds true for the closely related phages vB_VpaM_MAR and VHML and also for ϕHAP-1 that is more distantly related to VP58.5 but shows the same arrangement of genes within immB. An ORF whose product is similar to Q antiterminators has only been identified in the immB region of VP882 demonstrating that the genetic switch of marine telomere phages exhibits striking variations. We therefore searched for other VP58.5 genes that might exert lytic repressor activity by testing a gene library of the phage in V. parahaemolyticus but did not detect sequences, which enhanced VP58.5-induced cell lysis (data not shown). For V. campbellii phage VHML, a new hypothetical model of phage life cycle regulation has been proposed.11 According to this sequence-based model, a VHML-encoded adenine methyltransferase (DAM) methylates a rha antirepressor gene located on the phage genome. The VHML Rha antirepressor has homology with the Rha antirepressor of phage phi80. Once methylation is removed, homologous CI repressor protein becomes repressed and non-functional leading to the switching to the lytic cycle. Whether such an alternative regulation actually exists in VHML has still to be demonstrated by suitable experiments. VP58.5 contains a DAM gene almost identical to that of VHML, a rha homolog is, however, missing in this phage. Therefore, further studies are needed to elucidate how the lytic and lysogenic cycle are regulated in marine telomere phages.
Conclusions
The genetic switch of most temperate phages characterized thus far is very similar to that of lambda, the paradigm of temperate phages, which has been thoroughly studied for decades. Though, some temperate phages do not follow the basic principles of the regulation of the lytic and lysogenic cycle found in lambda-like phages. We show that telomere phages, some of which are related to lambda, are diverse in terms of their regulatory genes and putative operator sequences residing in the primary immunity region immB. Among the telomere phages, the genetic switch of N15 and ϕKO2 is most similar to those of lambda-like phages. The N15 and ϕKO2 repressors and transcription terminators exhibited the same specificities which, however, diverged from those of the related phage PY54. Moreover, the arrangement and sequences of operator sites differ in N15/ϕKO2 and PY54. The genetic switches of marine telomere phages seem to be completely different because they apparently do not contain a cro-like gene. Instead, 2 genes mediating prophage repressor activity have been identified in the immB region of Vp58.5. The fact that immB of PY54 encodes a protein similar to the second prophage repressor (ORF44 product) of marine telomere phages indicates that the PY54 molecular switch is halfway between the molecular switches of N15/ϕKO2 and marine telomere phages. The data reveal that telomere phages show striking commonalities with respect to their genome organization but are obviously diverse regarding their gene regulation.
Materials and methods
Bacterial strains, bacteriophages and culture conditions
All strains, plasmids and bacteriophages used in this study are listed in Supplemental Material Table S1. E. coli strain Genehogs (Invitrogen-Thermo Fisher Scientific, Darmstadt, Germany) was used for cloning procedures. PY54 and N15 propagation was performed in Y.enterocolitica 83/88/2 and E. coli C1a, respectively.6 VP58.5 was propagated using V. parahaemolyticus strain 37.5 as host.9 Prophage induction was performed by treatment of lysogenic strains with 500ng/ml (N15, PY54) or 30ng/ml (Vp58.5) mitomycinC.6,9
If not stated otherwise, all strains were grown in lysogeny broth (LB).21 Solid media contained 1.8% (w/v) agar. For cultivation of V. parahaemolyticus strains, the medium was supplemented with 3% NaCl (w/v). When required, ampicillin and kanamycin were supplemented at 100 µgml−1 and chloramphenicol and tetracycline at 12.5 µgml−1.
Construction of phage mutants
The construction of the mutants N15-D04 (Cmr) and PY54-35Tc (Tcr) has been previously described.6,17 A VP58.5 mutant (VP58.5Km3) containing the kanamycin resistance gene (kan) of Tn5 was obtained using an in vitro transposon mutagenesis kit (EZ-Tn5™ <KAN-2> Insertion Kit, Biozym, Hessisch Oldendorf, Germany). One shot electrocompetent E. coli cells (Genehogs, Invitrogen) were transformed with the mutagenized Vp58.5 plasmid prophage and plated on agar containing kanamycin. To determine the position of the Km resistance gene within the plasmid prophage, EcoRV restriction fragments were ligated to the vector pLitmus38 (Apr; New England Biolabs, Frankfurt am Main, Germany). Upon transformation of E. coli strain Genehogs, transformants were selected on agar containing ampicillin and kanamycin. Recombinant plasmids were sequenced (MWG Eurofins, Ebersberg, Germany) applying primers deduced from the kanamycin resistance gene. Mutant VP58.5Km3 contains the kanamycin resistance gene in the intergenic region between ORF53 and ORF54 at nucleotide position 40,810. Introduction of the mutagenized Vp58.5 plasmid prophage into V. parahaemolyticus 37.5 was achieved by infection of the strain using a lysate obtained from the corresponding E. coli transformant. Mutant VP58.5Km3 was shown to have a phenotype similar to the wild type phage.
To obtain a VP58.5 mutant containing a defective ORF44, the immB region of the phage that had been amplified by PCR was inserted into the vector pVv3.19 After molecular cloning in E. coli K12, the construct was cleaved with BpiI, which cuts ORF44 twice. Following this, the chloramphenicol acetyltransferase gene (cat) of pBR329 was amplified by PCR and inserted into ORF44. Upon transformation of E. coli K12 the construct was isolated from a chloramphenicol resistant colony. Thereafter, it was introduced into V. parahaemolyticus 37.5 by electroporation applying the protocol of Klevanskaa et al.19 Transformants harboring the recombinant plasmid were infected with VP58.5. Lysates were purified and plated on V. parahaemolyticus 37.5. Some single clear plaques were isolated and used for the preparation of high titer lysates that were purified by standard procedures.21
In vivo assay for the immB genes
The influence of the putative regulatory proteins on PY54, N15 and VP58.5 propagation was investigated by infection of strains containing the respective gene inserted into the vector pMS470Δ8Cat (for studies in E. coli and Y. enterocolitica)22 and pVv3 (for studies in V. parahaemolyticus).19 To accomplish this, the regulatory genes were amplified by PCR using phage DNA as template. The forward and reverse primers contained embedded restriction sites for NdeI and HindIII (pMS470Δ8Cat) or BamHI and HindIII (pVv3), respectively. After digestion of the amplicons with the respective restriction endonucleases (New England Biolabs), each fragment was inserted into the corresponding sites of pMS470Δ8Cat and pVv3. Upon transformation of E. coli strain Genehogs, recombinant plasmids (Table S1) were isolated and verified by Sanger sequencing (MWG Eurofins). For in vivo experiments, the constructs were introduced into E. coli C-1a, Y.enterocolitica 83/88/2 and V. parahaemolyticus 37.5 by electroporation. Transformants were grown to an optical density of 1.0 (A588). Thereafter, a 100 µl aliquot of a diluted phage lysate and 100 µl of the test strain were incubated for 10min at room temperature. Phage titers (PFU) were determined by the standard soft agar overlay method.21 Lysogenization was studied by plating infected bacteria on agar containing the respective antibiotic.
Determination of transcription start sites
The transcription start sites of the immB genes were determined as previously described.17 Primers for the transcripts were deduced from the repressor genes and from vector sequences adjacent to the inserted fragments. Products were separated on an agarose gel and discrete bands were excised, purified using the QIAquick Gel Extraction kit (Qiagen, Hilden, Germany) and sequenced (MWG Eurofins, Ebersberg, Germany).
In silico analyses
Sequence analyses and alignments were carried out using the DSGene software (v2.5) of the Accelrys package. BLAST searches were performed at the NCBI database.23 For in silico promoter studies, the upstream sequence of the respective genes were analyzed for the existence of −35 and −10 consensus sequences and extended −10 sequences.24-26
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
The work of J.A. Hammerl was supported by a grant of the Deutsche Forschungsgemeinschaft (DFG) to Erich Lanka.
References
- [1].Casjens SR, Gilcrease EB, Huang WM, Bunny KL, Pedulla ML, Ford ME. The pKO2 linear plasmid prophage of Klebsiella oxytoca. J Bacteriol 2004; 186(6):1818-32; PMID:14996813; http://dx.doi.org/ 10.1128/JB.186.6.1818-1832.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Hertwig S, Klein I, Lurz R, Lanka E, Appel B. PY54, a linear plasmid prophage of Yersinia enterocolitica with covalently closed ends. Mol Microbiol 2003; 48(4):989-1003; PMID:12753191; http://dx.doi.org/ 10.1046/j.1365-2958.2003.03458.x [DOI] [PubMed] [Google Scholar]
- [3].Hertwig S, Klein I, Schmidt V, Beck S, Hammerl JA, Appel B. Sequence analysis of the genome of the temperate Yersinia enterocolitica phage PY54. J Mol Biol 2003; 331(3):605-22; PMID:12899832; http://dx.doi.org/ 10.1016/S0022-2836(03)00763-0 [DOI] [PubMed] [Google Scholar]
- [4].Ravin V, Ravin N, Casjens S, Ford ME, Hatfull GF, Hendrix RW. Genomic sequence and analysis of the atypical temperate bacteriophage N15. J Mol Biol 2000; 299(1):53-73; PMID:10860722; http://dx.doi.org/ 10.1006/jmbi.2000.3731 [DOI] [PubMed] [Google Scholar]
- [5].Rybchin VN, Svarchevsky AN. The plasmid prophage N15: a linear DNA with covalently closed ends. Mol Microbiol 1999; 33(5):895-903; PMID:10476025; http://dx.doi.org/ 10.1046/j.1365-2958.1999.01533.x [DOI] [PubMed] [Google Scholar]
- [6].Hammerl JA, Klein I, Appel B, Hertwig S. Interplay between the temperate phages PY54 and N15, linear plasmid prophages with covalently closed ends. J Bacteriol 2007; 189(22):8366-70; PMID:17827299; http://dx.doi.org/ 10.1128/JB.01066-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Alanis Villa A, Kropinski AM, Abbasifar R, Griffiths MW. Complete genome sequence of Vibrio parahaemolyticus bacteriophage vB_VpaM_MAR. J Virol 2012; 86(23):13138-9; PMID:23118463; http://dx.doi.org/ 10.1128/JVI.02518-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Lan SF, Huang CH, Chang CH, Liao WC, Lin IH, Jian WN. Characterization of a new plasmid-like prophage in a pandemic Vibrio parahaemolyticus O3:K6 strain. Appl Environ Microbiol 2009; 75(9):2659-67; PMID:19286788; http://dx.doi.org/ 10.1128/AEM.02483-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Zabala B, Hammerl JA, Espejo RT, Hertwig S. The linear plasmid prophage Vp58.5 of Vibrio parahaemolyticus is closely related to the integrating phage VHML and constitutes a new incompatibility group of telomere phages. J Virol 2009; 83(18):9313-20; PMID:19587034; http://dx.doi.org/ 10.1128/JVI.00672-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Mobberley JM, Authement RN, Segall AM, Paul JH. The temperate marine phage PhiHAP-1 of Halomonas aquamarina possesses a linear plasmid-like prophage genome. J Virol 2008; 82(13):6618-30; PMID:18448537; http://dx.doi.org/ 10.1128/JVI.00140-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Bochow S, Elliman J, Owens L. Bacteriophage adenine methyltransferase: a life cycle regulator? Modelled using Vibrio harveyi myovirus like. J Appl Microbiol 2012; 113(5):1001-13; PMID:22681538; http://dx.doi.org/ 10.1111/j.1365-2672.2012.05358.x [DOI] [PubMed] [Google Scholar]
- [12].Munro J, Oakey J, Bromage E, Owens L. Experimental bacteriophage-mediated virulence in strains of Vibrio harveyi. Dis Aquat Organ 2003; 54(3):187-94; PMID:12803382; http://dx.doi.org/ 10.3354/dao054187 [DOI] [PubMed] [Google Scholar]
- [13].Oakey HJ, Cullen BR, Owens L. The complete nucleotide sequence of the Vibrio harveyi bacteriophage VHML. J Appl Microbiol 2002; 93(6):1089-98; PMID:12452967; http://dx.doi.org/ 10.1046/j.1365-2672.2002.01776.x [DOI] [PubMed] [Google Scholar]
- [14].Oakey HJ, Owens L. A new bacteriophage, VHML, isolated from a toxin-producing strain of Vibrio harveyi in tropical Australia. J Appl Microbiol 2000; 89(4):702-9; PMID:11054176; http://dx.doi.org/ 10.1046/j.1365-2672.2000.01169.x [DOI] [PubMed] [Google Scholar]
- [15].Payne M, Oakey J, Owens L. The ability of two different Vibrio spp. bacteriophages to infect Vibrio harveyi, Vibrio cholerae and Vibrio mimicus. J Appl Microbiol 2004; 97(4):663-72; PMID:15357715; http://dx.doi.org/ 10.1111/j.1365-2672.2004.02362.x [DOI] [PubMed] [Google Scholar]
- [16].Lobocka MB, Svarchevsky AN, Rybchin VN, Yarmolinsky MB. Characterization of the primary immunity region of the Escherichia coli linear plasmid prophage N15. J Bacteriol 1996; 178(10):2902-10; PMID:8631680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Hammerl JA, Roschanski N, Lurz R, Johne R, Lanka E, Hertwig S. The molecular switch of telomere phages: High binding specificity of the PY54 Cro lytic repressor to a single operator site. Viruses 2015; 7(6):2771-93; PMID:26043380; http://dx.doi.org/ 10.3390/v7062746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Svenningsen SL, Semsey S. Commitment to lysogeny is preceded by a prolonged period of sensitivity to the late lytic regulator Q in bacteriophage lambda. J Bacteriol 2014; 196(20):3582-8; PMID:25092034; http://dx.doi.org/ 10.1128/JB.01705-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Klevanskaa K, Bier N, Stingl K, Strauch E, Hertwig S. PVv3, a new shuttle vector for gene expression in Vibrio vulnificus. Appl Environ Microbiol 2014; 80(4):1477-81; PMID:24362421; http://dx.doi.org/ 10.1128/AEM.03720-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Ziegelin G, Tegtmeyer N, Lurz R, Hertwig S, Hammerl JA, Appel B, Lanka E. The repA gene of the linear Yersinia enterocolitica prophage PY54 functions as a circular minimal replicon in Escherichia coli. J Bacteriol 2005; 187(10):3445-54; PMID:15866931; http://dx.doi.org/ 10.1128/JB.187.10.3445-3454.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Sambrook JR, D. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- [22].Strauch E, Voigt I, Broll H, Appel B. Use of a plasmid of a Yersinia enterocolitica biogroup 1A strain for the construction of cloning vectors. J Biotechnol 2000; 79(1):63-72; PMID:10817342; http://dx.doi.org/ 10.1016/S0168-1656(00)00216-9 [DOI] [PubMed] [Google Scholar]
- [23].Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 1997; 25(17):3389-402; PMID:9254694; http://dx.doi.org/ 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Harley CB, Reynolds RP. Analysis of E. coli promoter sequences. Nucleic Acids Res 1987; 15(5):2343-61; PMID:3550697; http://dx.doi.org/ 10.1093/nar/15.5.2343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Hawley DK, McClure WR. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res 1983; 11(8):2237-55; PMID:6344016; http://dx.doi.org/ 10.1093/nar/11.8.2237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Kumar A, Malloch RA, Fujita N, Smillie DA, Ishihama A, Hayward RS. The minus 35-recognition region of Escherichia coli sigma 70 is inessential for initiation of transcription at an “extended minus 10” promoter. J Mol Biol 1993; 232(2):406-18; PMID:8345519; http://dx.doi.org/ 10.1006/jmbi.1993.1400 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.